Abstract
Id proteins (Id1-4) are helix-loop-helix transcription factors that promote metastasis. It was found that Semaphorin 3F (SEMA3F), a potent inhibitor of metastasis, was repressed by Id2. High metastatic human tumor cell lines had relatively high amounts of Id2 and low SEMA3F levels compared to their low metastatic counterparts. No correlation between metastatic potential and expression of the other Id family members was observed. Furthermore, ectopic expression of Id2 in low metastatic tumor cells downregulated SEMA3F and, as a consequence, enhanced their ability to migrate and invade, two requisite steps of metastasis in vivo. Id2 overexpression was driven by the c-myc oncoprotein. SEMA3F was a direct target gene of the E47/Id2 pathway. Two E-box sites, which bind E protein transcription factors including E47, were identified in the promoter region of the SEMA3F gene. E47 directly activated SEMA3F promoter activity and expression and promoted SEMA3F biological activities, including F-actin depolymerization, inactivation of RhoA and inhibition of cell migration. Silencing of SEMA3F inhibited the E47-induced SEMA3F expression and biological activities, confirming that these E47-induced effects were SEMA3F-dependent. E47 did not induce expression of the other members of the SEMA3 family. Id2, a dominant-negative inhibitor of E proteins, abrogated the E47-induced SEMA3F expression and biological activities. Thus, high metastatic tumor cells overexpress c-myc, leading to upregulation of Id2 expression; the aberrantly elevated amount of Id2 represses SEMA3F expression and, as a consequence, enhances the ability of tumor cells to migrate and invade.
Keywords: Id2, Invasion, Migration, Semaphorin 3F, Transcriptional regulation
INTRODUCTION
Id (inhibitors of DNA binding/differentiation) proteins are helix-loop-helix (HLH) transcription factors that regulate cell proliferation and differentiation. Id proteins (Id1-4), which lack a DNA binding domain, exert this control by associating with the ubiquitously expressed E proteins (E12, E47, E2-2 and HEB) and preventing them from binding DNA either as homodimers or as heterodimers with tissue-restricted basic HLH (1).
Id proteins are mainly expressed during embryogenesis and are required for maintaining the timing of neuronal differentiation. However, expression of Id proteins is reactivated in many human cancers. For example, Id expression has been documented in prostate, breast, bladder, colon and pancreatic cancer, high-grade astrocytoma and T-cell lymphomas (2–7). Id proteins induce proliferation, anaplasia and invasiveness, and they serve as prognostic markers in several types of human cancers (1, 8, 9). More recent studies have established a direct connection between Ids and metastasis (10, 11). Furthermore, it was found that overexpression of Id1 and/or Id3 in breast cancer cells induced lung metastasis (12–14).
In promoting tumor cell proliferation, several direct target genes of the bHLH/Id pathway have been identified, including the cell cycle regulators p21, p15, p16 and p57 (15–17). However, less is known about the functional mediators responsible for the metastatic effects of Ids. In this regard, the pro-angiogenic and pro-metastatic genes downstream of Id proteins identified in tumor cells, such as VEGF, MMPs and EGFR, have not been shown to be directly regulated by the bHLH/Id pathway (18–20).
Semaphorin 3F (SEMA3F), first described as a negative mediator of axon guidance during neuronal development, is a functional inhibitor of tumor cell growth, tumor angiogenesis and tumor metastasis (21–25). A connection between SEMA3F and tumor formation became apparent when it was demonstrated that SEMA3F mapped to a region on human chromosome 3p21.3 that is commonly deleted in lung tumors (26, 27). A suppressive role in metastasis was proposed based on the observation that SEMA3F expression was strongly downregulated in high metastatic tumor cells and that SEMA3F overexpression inhibited metastasis in vivo (25). In tumor cells, SEMA3F promoter activity and SEMA3F expression are upregulated by p53 (28) and downregulated by ZEB-1 (29) transcription factors. However, the molecular pathways that silence SEMA3F expression in high metastatic tumor cells are still unknown.
SEMA3F acts directly on tumor cells expressing its receptor, neuropilin 2 (NRP2), to inhibit their tumor activities, such as adhesion, motility and invasion (25, 30). In addition to its effect on tumor cells, SEMA3F also acts directly on vascular endothelial cells (EC) expressing NRP2 to inhibit tumor angiogenesis (25). Thus, as a potential therapeutic, SEMA3F has the advantage as it can target both tumor and endothelial cells expressing NRP2.
In this report we have demonstrated that SEMA3F is a direct target gene of the E47/Id2 pathway. E47, a transcription factor that belongs to the E protein family of bHLH proteins, directly increased SEMA3F expression and biological activities. Conversely, Id2, a dominant-negative inhibitor of E proteins, abrogated these E47-induced effects. A novel finding was that high metastatic tumor cells overexpressed c-myc, leading to upregulation of Id2; upregulation of Id2 expression repressed SEMA3F expression and, as a functional consequence, enhanced the ability of tumor cells to migrate and invade, two requisite steps of metastasis in vivo. We conclude that the stimulation of tumor cell migration and invasion by Id2 occurs through transcriptional repression of SEMA3F.
MATERIALS AND METHODS
Cell culture
U87MG human glioma cells and human cell lines of low and high metastatic potential were cultured as in (25, 30).
Transfection
Human SEMA3F, E47, Id2, Id2-DBM and c-myc constructs have been previously described (16, 25, 31, 32). Cells were transfected using Fugene6 reagent (Roche Applied Science, Indianapolis, IN).
Adenovirus infection
E47 and Id2 adenovirus have been previously described (16). Cells were infected at a multiplicity of infection (MOI) of 100.
siRNA knockdown
siRNAs of Id2 (SMARTpool M-009864-00-005) and SEMA3F (SMARTpool M-017644-00-0005) were purchased from Thermo Fisher Scientific (Chicago, IL). siRNA control was purchased from Ambion Inc. (Austin, TX). Cells were transfected using SilentFect reagent (Bio-Rad Laboratories, Hercules, CA).
RNA isolation and analyses
Total RNA was isolated from the cells using the RNeasy kit (Qiagen, Valencia, CA). cDNA was prepared using Superscript II enzyme (Invitrogen Corp.). For real-time RT-PCR analysis, the DyNAmo Sybr-Green-based system from New England BioLabs, Inc. (Beverly, MA) was used. Reactions were run on a LightCycler (Roche Applied Science). Each experiment was done in duplicate and repeated three times. The P value is based on a Student’s t-test. Oligonucleotide primers for real-time and semi-quantitative RT-PCR reactions are listed in Tables S1, S2, respectively.
Immunoblot
Cell lysates
Cells were lysed and immunoblotted as described previously (30). Specific proteins were detected after incubation with anti-E47, anti-Id2, anti-c-myc (Santa Cruz Biotechnology, Inc.), anti-phospho-c-myc Thr58/Ser62 (Cell Signaling Technology, Inc. Danvers, MA) and anti-β-actin (Sigma-Aldrich Corp.) antibodies.
Conditioned media
Cells (2 × 105) were plated in 1.5 ml full media. After 3 days, 1.2 ml of conditioned media were collected. SEMA3F was immunoprecipitated overnight at 4°C with an anti-SEMA3F antibody (D-16, Santa Cruz Biotechnology, Inc.). Protein G slurry (50 μl) (GE Healthcare, Uppsala, Sweden) was added to the immunocomplexes, which were incubated for 1 h at 4°C. SEMA3F was detected after incubation with an anti-SEMA3F antibody generated for us by the GenScript Corp. This immunoprecipitation/immunoblot assay was highly specific for SEMA3F, since it did not detect other members of the SEMA3 family (Fig. S1). Each experiment was repeated three times.
Promoter luciferase constructs
A fragment of 1521 bp from the 5′-flanking region of the human SEMA3F gene was isolated from genomic DNA of U87MG glioma cells using the following primers: forward 5′-TCAAGTCAGCTCGAGAGTATCGAAGCTCTCTGAGG-3′; reverse 5′-CAGTGCTGCAAGCTTGTTCTTTGGCCTGCCTTTGT-3′. The SEMA3F promoter and the pGL3 vector (Promega Corp., Madison, WI) were digested with Xho I and Hind III and ligated together with the DNA ligation kit (Takara Bio Inc., Otsu, Japan). The SEMA3F promoter-luciferase deletion construct containing an internal deletion in the promoter was generated by PCR using the following forward primer: 5′-TCAAGTCAGCTCGAGCTGCCATTCAGTCAGCACTAGC-3′. Site-specific mutagenesis was carried out with the Quick-Change XL-kit (Stratagen, La Jolla, CA). The double mutated construct with mutations in both E-box sequences was generated by sequential mutation.
Luciferase reporter assay
U87MG cells were transfected with 10 ng of a Renilla luciferase vector (Promega Corp.), 0.5 μg of the SEMA3F promoter-luciferase constructs or pGL3 empty vector, and the indicated combination of plasmids expressing E47 and Id2. Lysates were prepared 36 h after transfection, and luciferase activities were measured with the dual-Luciferase reporter Assay System (Promega Corp.), with the reporter activities normalized to Renilla luciferase activity. Each transfection was done in duplicate and repeated at least three times.
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out using the Chromatin Immunoprecipitation Assay Kit from Active Motif (Carlsbad, CA). For immunoprecipitation, an anti-E47 monoclonal antibody (Santa Cruz Biotechnology, Inc.) and normal rabbit IgG as a negative control were used. Immunoprecipitated DNA was amplified by PCR using specific primers for the E-box sequence present in the human SEMA3F promoter: forward 5′-CCTGCAAGTGTGAACGTGTGC-3′; reverse 5′-CCTGTCATAGATGCTGCCTCC-3′ (224 bp PCR product).
Migration and invasion
Migration (2.5 × 104 for U87MG cells and 5 × 104 for A375P melanoma cells) and invasion (5 × 105 cells) assays were performed in Transwells (Corning Inc.) with an 8.0-μm pore size and coated with a 0.5% gelatin solution or with BD Matrigel™ Basement Membrane Matrix (0.1 mg/ml) (BD Bioscience, San Jose, CA), respectively. Cells in serum-free medium and increasing concentrations of SEMA3F and 20 μg/ml of either normal goat IgG or neutralizing anti-NRP2 antibody (R&D Systems, Minneapolis, MN) were added to the upper wells. Media containing 1% (migration) or 10% (invasion) fetal bovine serum were added to the lower wells. Cells that migrated through the filter after 16 h were stained and counted by phase microscopy as described earlier (30). SEMA3F was purified as described before (30). The experiment was repeated three times in duplicate. The results represent the average of the three experiments. The P value is based on a Student’s t-test.
RhoA activity
Rho activity assay was performed and quantified using the Rho activation assay kit as described previously (30).
Confocal microscopy
Cells were fixed and stained for F-actin as described before (30).
RESULTS
High metastatic tumor cells downregulate SEMA3F expression by upregulating Id2
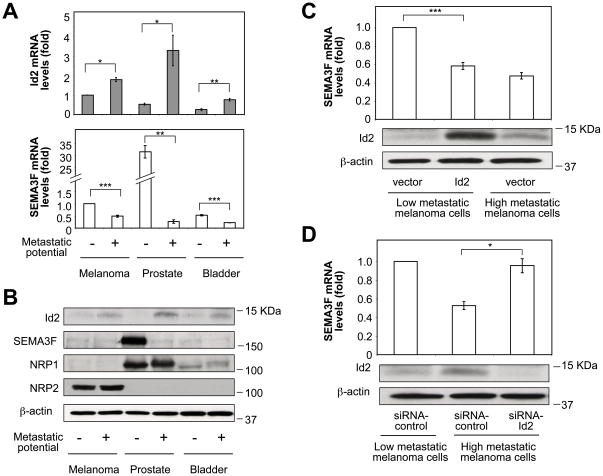
When matched pairs of human melanoma, prostate carcinoma and bladder carcinoma cell lines of low and high metastatic potential were compared for Id1-4 expression, it was found that Id2 was markedly upregulated in all three high metastatic tumor cell lines compared to their low metastatic counterparts, both at the mRNA (Fig. 1A, top panel) and protein (Fig. 1B) levels, suggesting a positive relationship between metastatic potential and Id2 expression. In contrast, no correlation between metastatic potential and expression of the other Id family members was observed (Fig. S2A).
Figure 1. High metastatic tumor cells downregulate SEMA3F by upregulating Id2.
(A) Id2 and SEMA3F mRNA levels in paired low and high metastatic tumor cell lines. (B) Immunoblots showing Id2, SEMA3F, NRP1, NRP2 and β-actin protein. (C, D) SEMA3F mRNA levels in melanoma cells transfected with control or Id2 vectors (C) or with siRNA-control or siRNA-Id2 (D). Protein levels of Id2 and β-actin are shown in the panels below. *, P < 0.05; **, P < 0.01; ***. P < 0.001.
On the other hand, SEMA3F was markedly downregulated in high metastatic tumor cells, both at the mRNA (Fig. 1A, bottom panel) and protein (Fig. 1B) levels. This expression profile was highly evident for the prostate carcinoma cells, where SEMA3F mRNA and protein levels were high in low metastatic PC3M cells but almost undetectable in high metastatic PC3MLN4 cells (Fig. 1A, B, lanes 3, 4). A similar expression profile was observed at the SEMA3F mRNA level in melanoma (Fig. 1A, lanes 1, 2) and bladder carcinoma (Fig. 1A, lanes 5, 6) cells. SEMA3F protein expression in these cells was below the level of detection (Fig. 1B, lanes 1, 2, 5, 6). Nevertheless, melanoma cells were chosen for further studies because, unlike the other two cell types, they expressed relatively high levels of NRP2, the functional receptor of SEMA3F necessary for demonstrating SEMA3F biological activities (Fig. 1B, lanes 1, 2).
Ectopic expression of Id2 in low metastatic melanoma cells resulted in downregulation of SEMA3F expression to about the same level observed in high metastatic melanoma cells that express high endogenous Id2 levels (Fig. 1C). These results suggested that both cell types had sufficient amounts of Id2 to obtain maximal effects on SEMA3F repression. Conversely, silencing of Id2 in high metastatic melanoma cells induced SEMA3F expression (Fig. 1D). An upregulation of SEMA3F expression was also observed when Id2 was silenced in high metastatic prostate (Fig. S2B) and high metastatic bladder (Fig. S2C) carcinoma cells, suggesting that the regulation of SEMA3F expression by Id2 was not cell type specific. Taken together, these results show that, in tumor cells, there is an inverse relationship between Id2 and SEMA3F expression and that an aberrantly elevated amount of Id2 represses SEMA3F expression.
Id2 induces tumor cell migration and invasion by repressing expression of SEMA3F
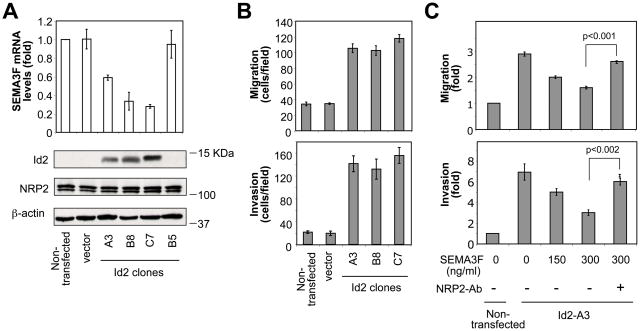
Next, we examined whether Id2 could promote a metastatic phenotype through transcriptional repression of SEMA3F. Low metastatic melanoma cells were stably transfected with Id2-DBM, a stable form of Id2. Stable clones A3, B8 and C7 expressed high levels of ectopic Id2 protein (Fig. 2A, bottom panel). SEMA3F mRNA levels were much diminished in these three Id2-clones (Fig. 2A, top panel), demonstrating a strong inverse correlation between Id2 and SEMA3F expression. In contrast, an Id2-DBM clone (B5) that failed to express detectable levels of ectopic Id2, expressed high levels of SEMA3F that were comparable to those of non-transfected and mock-transfected control cells. Importantly, Id2 did not affect the expression levels of NRP2 (Fig. 2A).
Figure 2. Id2 induces tumor cell migration and invasion through transcriptional repression of SEMA3F.
(A) SEMA3F mRNA and Id2, NRP2 and β-actin protein levels in various Id2-expressing clones. (B) Transwell cell migration and invasion assays of control cells and three Id2-expressing clones. (C) Transwell cell migration and invasion assays of control cells and a representative Id2-expressing clone (A3). Increasing concentrations of SEMA3F with anti-NRP2 antibody were added to the upper wells.
The three Id2-expressing clones displayed a three- to four-fold increased basal motility (Fig. 2B, top panel) and a seven-fold increase in invasiveness (Fig. 2B, bottom panel) compared to control cells. Although the three clones showed variable Id2 and SEMA3F levels, similar effects were observed at the migration and invasion levels, suggesting that the three clones had sufficient amounts of Id2 to obtain maximal migration and invasion.
To determine whether the Id2-induced cell migration and invasion were a direct result of SEMA3F inhibition, increasing concentrations of SEMA3F were added to the upper chamber of a Transwell. Exogenous SEMA3F inhibited the migration and invasion induced by Id2 in a dose-dependent manner (Fig. 2C). The SEMA3F inhibition of migration was NRP2-dependent, as shown by the loss of SEMA3F-induced inhibition of cell migration and invasion in the presence of an anti-NRP2 antibody (Fig. 2C). Thus, Id2 induces tumor cell migration and invasion through repression of SEMA3F expression.
C-myc controls the upregulation of Id2 expression in high metastatic tumor cells
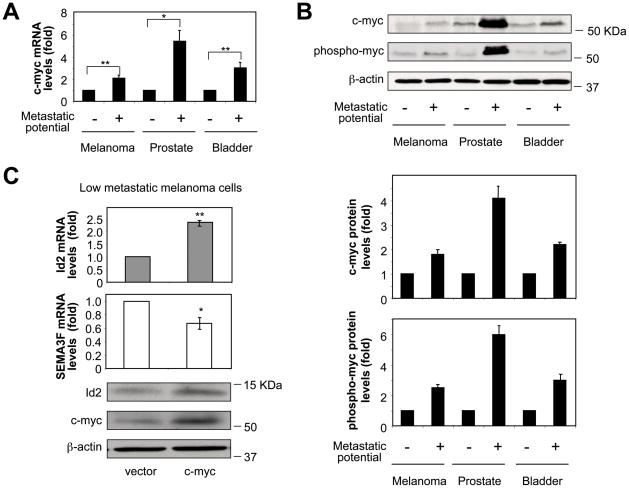
Id2 expression can be controlled by oncoproteins such as c-myc and n-myc (3, 32). Accordingly, we examined whether the high amounts of Id2 in metastatic tumor cells were a result of c-myc expression. When human tumor cell lines with low versus high metastatic potential were compared for c-myc expression, it was found that c-myc was upregulated in the three high metastatic tumor cell lines, both at the mRNA (Fig. 3A) and protein (Fig. 3B) levels. Furthermore, the amount of active phosphorylated c-myc was also increased in high metastatic tumor cells (Fig. 3B). The strongest effects were particularly evident in the prostate carcinoma cells (Fig. 3A, B, lanes 3, 4). Interestingly, whereas high metastatic prostate carcinoma cells carried an elevated copy number of the c-myc gene, high metastatic melanoma and bladder carcinoma cells did not (Fig. S3). When c-myc was overexpressed in low metastatic melanoma cells, it was found that Id2 expression was induced, both at the mRNA (Fig. 3C, top panel) and protein (Fig. 3C, bottom panel) levels, with consequent downregulation of SEMA3F expression (Fig. 3C, middle panel). It was concluded that Id2 overexpression and, as a result, SEMA3F downregulation, are driven by c-myc.
Figure 3. High metastatic tumor cells upregulate c-myc.
(A) C-myc mRNA levels in paired low and high metastatic tumor cell lines. (B) Immunoblots showing c-myc and phospho-c-myc protein levels. Quantitative results are shown in the panels below. (C) Id2 and SEMA3F mRNA levels in low metastatic melanoma cells transfected with control or c-myc vectors. Protein expression levels of Id2, c-myc and β-actin are shown in the panel below. *, P < 0.05; **. P < 0.02.
E47 induces SEMA3F expression and biological activities
Id proteins bind to E proteins such as E47 and inhibit their interaction with DNA, thus functioning as dominant negative inhibitors of bHLH-mediated transcription (1). To determine whether SEMA3F expression was regulated by E proteins, E47 was overexpressed in U87MG human glioma cells. U87MG cells expressed NRP2 and responded strongly to SEMA3F in a number of bioassays such as F-actin depolymerization, loss of stress fibers and inactivation of RhoA (30). Importantly, U87MG cells resembled the high metastatic tumor cell lines described above, in that they expressed high Id2 and undetectable SEMA3F levels (Fig. S4A). Furthermore, silencing of Id2 in U87MG cells induced SEMA3F expression (Fig. S4B).
Overexpression of E47 in U87MG cells increased SEMA3F mRNA levels significantly (Fig. S4C). However, expression of other members of the SEMA3 family (SEMA3A-E) was not induced by E47 (Fig. S4C), suggesting that the E47-mediated activation of SEMA3F expression was highly specific.
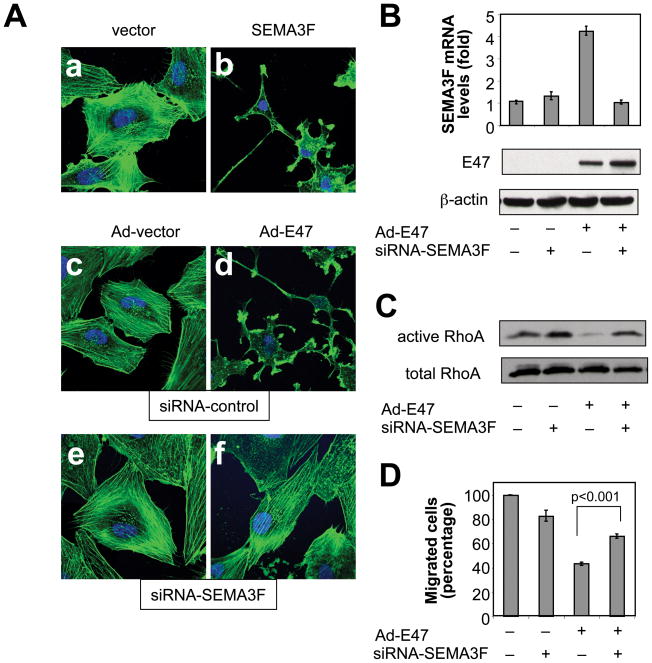
Previously, we showed that treatment of U87MG cells with SEMA3F protein induced morphological changes (30). Similarly, overexpression of SEMA3F in U87MG cells induced loss of F-actin stress fibers, reduced spreading and decreased cytoplasm (Fig. 4A, a vs. b). E47 adenovirus (Ad-E47) fully phenocopied the morphological changes induced by SEMA3F (Fig. 4A, b vs. d), suggesting that E47 could mimic the effects directly caused by SEMA3F. When E47-induced SEMA3F expression was inhibited by SEMA3F siRNA (Fig. 4B), it was found that the Ad-E47-induced F-actin depolymerization was inhibited (Fig. 4A, d vs. f), confirming that the effect of E47 on cell morphology was SEMA3F-dependent.
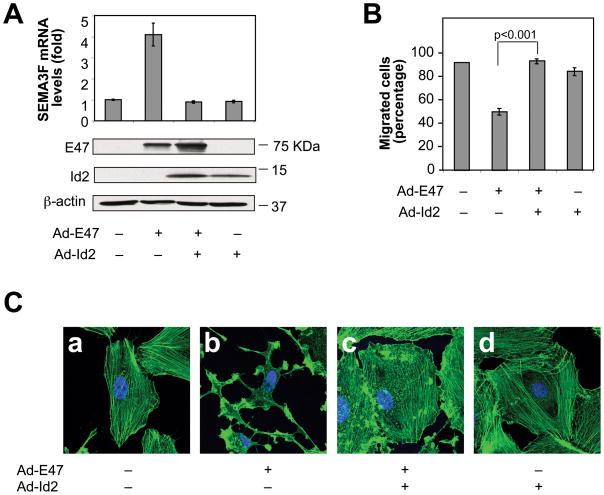
Figure 4. E47 induces SEMA3F expression and biological activities.
(A) Confocal microscopy of U87MG cells transfected with control (a) or pcSec-SEMA3F (b) vectors; transfected with siRNA-control and infected with Ad-vector (c) or Ad-E47 (d); transfected with siRNA-SEMA3F and infected with Ad-vector (e) or Ad-E47 (f). F-actin and nuclei were visualized using Alexa Fluor 488 phalloidin (green) and hoescht (blue), respectively. (B–D) U87MG cells were infected with Ad-vector (−) or Ad-E47 (+) and transfected with siRNA-control (−) or siRNA-SEMA3F (+). (B) SEMA3F mRNA and E47 and β-actin protein levels. (C) GTP-bound RhoA levels (active RhoA). Expression levels of total RhoA in lysates are shown in the panel below. (D) Transwell cell migration assay.
SEMA3F protein inactivated RhoA (30), a member of the Rho family of GTPases that stabilizes the F-actin cytoskeleton, and inactivated Rac1 (30), a regulator of lamellipodia formation (33). Similarly, overexpression of E47 in U87MG cells inactivated RhoA (Fig. 4C) and Rac1 (Fig. S4D). The E47-induced inactivation of RhoA and Rac1 was abrogated by SEMA3F siRNA, confirming that the E47-induced inactivation of RhoA and Rac1 was SEMA3F-dependent. On the other hand, Cdc42, which regulates filopodia, was not inactivated by SEMA3F (30) or E47 (Fig. S4D).
Exogenous SEMA3F protein inhibited U87MG cell migration (30). It was found that E47 overexpression inhibited U87MG cell migration by 57%. On the other hand, silencing of SEMA3F significantly rescued the inhibition in cell migration induced by E47, resulting in only a 34% inhibition (Fig. 4D).
These results indicate that E47 induces SEMA3F expression and biological activities and that E47-induced F-actin depolymerization, loss of stress fibers, inhibition of migration and inactivation of RhoA and Rac1 are mediated by SEMA3F.
Id2 inhibits the E47-mediated induction of SEMA3F expression and biological activities
Whether Id2 could inhibit the enhancing effect of E47 on SEMA3F expression and SEMA3F biological activities, was explored. SEMA3F mRNA levels were increased 4-fold in U87MG cells infected with Ad-E47, but this induction was fully inhibited when Ad-E7 was co-infected with Ad-Id2 (Fig. 5A).
Figure 5. Id2 inhibits E47-induced SEMA3F expression and biological activities.
U87MG cells were co-infected with Ad-vector (−) or Ad-Id2 (+) and with Ad-vector (−) or Ad-E47 (+). (A) SEMA3F mRNA and E47, Id2 and β-actin protein levels. (B) Transwell cell migration assay. (C) Confocal microscopy as in Fig. 4A.
E47-induced SEMA3F biological activities were also inhibited by Id2. Whereas E47 inhibited cell migration and induced F-actin depolymerization and loss of stress fibers, co-infection of Ad-E47 and Ad-Id2 completely abrogated the inhibition of cell migration (Fig. 5B) and the F-actin depolymerization (Fig. 5C, b vs. c) induced by E47. It was concluded that SEMA3F expression is regulated by the E47/Id2 pathway.
When using U87MG cells, Id2 by itself did not inhibit SEMA3F expression (Fig. 5A, lane 4 vs. lane 1), consistent with SEMA3F levels being very low, almost undetectable, in these cells. Consequently, Id2 by itself did not induce U87MG cell migration (Fig. 5B, lane 4 vs. lane 1).
SEMA3F is a direct transcriptional target of the E47/Id2 pathway
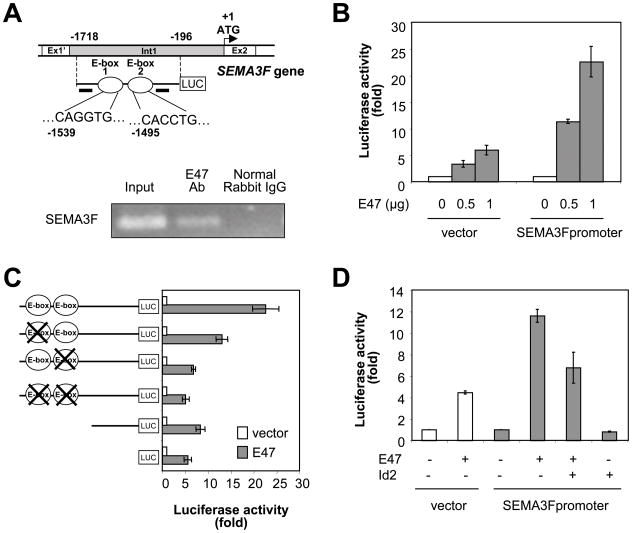
To test whether SEMA3F is an E47/Id2 direct target gene, we isolated a fragment extending 1521 bp in the 5′-flanking region of the human SEMA3F gene (−1718 to −196 relative to the start codon at position + 1). The SEMA3F promoter contained two E-boxes displaying the consensus CANNTG sequence that binds bHLH transcription factors. The two E-boxes, E-box 1 and E-box 2, were located at positions −1539 and −1495 upstream from the translation start site, respectively (Fig. 6A, top panel). Chromatin immunoprecipitation (ChIP) assays showed that E47 binds directly to the SEMA3F promoter in vivo (Fig. 6A, bottom panel). Since the two E-boxes were located in close proximity on the SEMA3F promoter, ChIP assays were not useful in determining whether only one or both E-boxes were capable of binding E47. Importantly, luciferase reporter assays using a SEMA3F promoter reporter construct showed that the two E-box sequences found in the SEMA3F promoter mediated a majority of the transcriptional activation of SEMA3F by E47. Ectopic expression of E47 increased SEMA3F promoter activity in a dose-dependent manner in U87MG cells (Fig. 6B). When compared to the wild type SEMA3F promoter, the constructs containing point mutations or deletions in both E-boxes displayed 77% and 75% reduction of its responsiveness to E47, respectively (Fig. 6C). Mutations of the individual E-boxes reduced activation by 42% (E-box 1) and 70% (E-box 2). On the other hand, co-transfection of E47 and Id2 prevented the E47-mediated induction of SEMA3F promoter activity (Fig. 6D), suggesting that the E47/Id2 pathway regulates SEMA3F expression at the transcriptional level.
Figure 6. Identification of SEMA3F as a direct target gene of the E47/Id2 pathway.
(A) Genomic organization of the SEMA3F gene. The two putative E-box sites, E-box 1 and E-box 2 are indicated by white circles. Horizontal black bars show the position of the primers used for the ChIP assay. ChIP assay is shown in the panel below. (B–D) Luciferase reporter assay. (B) U87MG cells were co-transfected with pGL3 luciferase empty or SEMA3F luciferase promoter vectors, and increasing amounts of E47. (C) U87MG cells were co-transfected with a luciferase construct (full-length wild-type SEMA3F promoter, SEMA3F promoter containing individual or combined point mutants for each of the two E-boxes or SEMA3F promoter lacking both E-boxes), and with control or pcDNA3-E47 vectors. E-boxes with point mutations are depicted as a circle with a large X. (D) U87MG cells were co-transfected with pGL3 luciferase empty or SEMA3F promoter-luciferase vectors, and the indicated combination of plasmids expressing E47 and Id2.
DISCUSSION
Id proteins (Id1-4) are helix-loop-helix transcription factors that promote metastasis (12–14, 34). However, little is known about the molecular targets and mechanisms that confer this metastatic capacity. Here we provide evidence that high metastatic human tumor cell lines overexpress Id2 compared to their low metastatic counterparts, leading to downregulation of SEMA3F expression, a potent metastasis inhibitor (25). Importantly, we have demonstrated that, in tumor cells, SEMA3F is a direct target gene of the E47/Id2 pathway. E47, a transcription factor that belongs to the E protein family of bHLH proteins, directly activated SEMA3F promoter activity and expression and, as a functional consequence, promoted SEMA3F biological activities, including depolymerization of the F-actin cytoskeleton, inactivation of RhoA and Rac1, and inhibition of cell migration. Id2, a dominant-negative inhibitor of E proteins (1), abrogated these E47-induced effects. In addition to binding E proteins, Id2 binds and opposes the function of the Rb tumor suppressor protein (35). Whether the Rb/Id2 pathway regulates SEMA3F expression is unknown.
C-myc oncoprotein promotes Id2 expression in high metastatic tumor cells. This is consistent with previous studies that demonstrated that Id2 expression can be controlled by c-myc and n-myc in neuroblastomas and T-cell lymphomas (3, 32). N-myc gene amplification drives Id2 overexpression in human neuroblastomas (32). We found that high metastatic prostate carcinoma cells carried an elevated copy number of the c-myc gene, which was consistent with the report that 92% of lymph node metastases from prostate cancer carried extra copies of c-myc oncogene (36). In contrast, high metastatic melanoma and bladder carcinoma cells did not amplify c-myc gene, suggesting that another mechanism may be involved in the upregulation of c-myc observed at the mRNA and protein levels in these high metastatic tumor cell lines. Together, these results demonstrate that in high metastatic tumor cells there is a pathway in which c-myc activates Id2 expression, leading to repression of SEMA3F expression.
The paired cell lines of low and high metastatic potential used in this study were derived from bladder carcinoma, prostate carcinoma and melanoma. Although the inverse correlation between c-myc/Id2 and SEMA3F expression was observed in the three cell types, this pattern was highly evident for the prostate carcinoma cells. High metastatic PC3MLN4 cells showed a 6-fold increase in Id2 mRNA levels and relatively high amounts of Id2 protein compared to low metastatic PC3M cells. SEMA3F mRNA levels were over 100-fold higher in low metastatic PC3M cells compared to high metastatic PC3MLN4 cells. Furthermore, whereas SEMA3F protein was readily detectable by immunoblot in low metastatic PC3M cells, it was not detectable in high metastatic PC3MLN4 cells.
Importantly, our results are specific for Id2 and SEMA3F. Among the four Id proteins (Id1-4), only Id2 was found to be overexpressed in high metastatic tumor cells, compared to their low metastatic counterparts. These results were in contrast with earlier studies showing that breast cancer cells metastatic to the lungs overexpressed Id1 and Id3 (12, 13). Thus, the effects of Id proteins on metastasis appear to be tumor cell and Id type dependent. SEMA3F belongs to the class 3 family of semaphorins, which consists of seven secreted proteins of approximately 100 KDa (SEMA3A to -G) (37). SEMA3 proteins contain a highly conserved amino-terminal 500 amino acid region, named the sema domain, that binds to NRPs. Nevertheless, the ability of E47 to induce SEMA3F expression was specific, since mRNA levels of the other members of the SEMA3 family were not induced by E47. Furthermore, the E47-induced SEMA3F activity was almost totally inhibited by SEMA3F siRNA, ruling out that other SEMA3s or other genes were involved. This is important since E47 is a ubiquitously expressed transcription factor that activates several target genes such as p21, p57, Notch1, Jagged2, Unc5A and adiponectin (16, 17, 31, 38). Interestingly, although transcription of SEMA3F and SEMA3B genes are regulated by methylation (39, 40) and p53 (28, 41), E47 increased the expression of SEMA3F but not SEMA3B, suggesting that SEMA3F and SEMA3B genes are regulated differently.
Given that SEMA3 proteins contain a highly conserved amino acid region, specific western blot analysis was technically challenging. We tested several commercial anti-SEMA3F antibodies and concluded that SEMA3F specificity was best determined with a combination of immunoprecipitation with one anti-SEMA3F antibody followed by immunoblot with a second anti-SEMA3F antibody. This combination did not detect other members of the SEMA3 family.
Whereas Id2 is mainly expressed during embryogenesis, expression of Id2 is reactivated in many human cancers, such as prostate and pancreatic cancer, high-grade astrocytoma and T-cell lymphomas (2, 3, 5, 7, 42). Here we have demonstrated that Id2 expression is upregulated in high metastatic subpopulations compared to their low metastatic matched counterparts, suggesting a positive correlation between metastatic potential and Id2 expression. This is an important finding since it is metastasis that is responsible for most cancer deaths. Cancer metastasis is a multi-step process through which tumor cells spread from the primary tumor site to distant organs (43–45). In order to metastasize, tumor cells must be able to migrate and survive in the circulation, invade the new organ, and reinitiate tumor growth to establish a metastatic colony. Whereas earlier studies have mainly implicated Id2 in tumor progression at the primary tumor site (6, 7, 42, 46, 47), we provide functional evidence for the involvement of Id2 in promoting cell migration and invasion in vitro, two requisite steps of metastasis in vivo, through transcriptional repression of SEMA3F. This is consistent with previous reports showing that knockdown of Id2 using small-hairpin RNAs (shRNA) in colon cancer cells decreased the number of detectable metastases to the liver (34) and that overexpression of SEMA3F in metastatic melanoma cells inhibited the ability of tumor cells to migrate and invade in vitro and to promote metastasis in vivo (25). Thus, the balance of Id2 versus SEMA3F in tumors may determine whether a tumor will metastasize or not.
In summary, our results identify SEMA3F, a potent inhibitor of metastasis, as a direct target of transcriptional repression by Id2. In high metastatic tumor cells, overexpression of c-myc upregulates Id2, which suppresses SEMA3F expression and activity. Furthermore, we demonstrate that SEMA3F is a major mediator of Id2 effects on cell migration and invasion, hallmarks of tumor metastasis in vivo. Thus, development of specific modifiers of this pathway that lower c-myc/Id2 expression and/or activity and, as a consequence, increase SEMA3F expression, could lead to novel therapeutic approaches to inhibit metastasis in the future.
Supplementary Material
Acknowledgments
We thank Dr. Diane Bielenberg, Dr. Andrew Dudley and Dr. Elena Geretti for valuable discussions. We thank Elke Pravda for technical assistance with the confocal microscope. We thank Kristin Johnson and Melissa Herman for preparation of the manuscript.
This work was supported by National Institutes of Health grants CA37392, CA45548 (M.K.) and CA085628, CA127643 (A.I.).
References
- 1.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–14. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 2.Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY. Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res. 2004;10:2044–51. doi: 10.1158/1078-0432.ccr-03-0933. [DOI] [PubMed] [Google Scholar]
- 3.Cotta CV, Leventaki V, Atsaves V, et al. The helix-loop-helix protein Id2 is expressed differentially and induced by myc in T-cell lymphomas. Cancer. 2008;112:552–61. doi: 10.1002/cncr.23196. [DOI] [PubMed] [Google Scholar]
- 4.Perk J, Gil-Bazo I, Chin Y, et al. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res. 2006;66:10870–7. doi: 10.1158/0008-5472.CAN-06-2643. [DOI] [PubMed] [Google Scholar]
- 5.Vandeputte DA, Troost D, Leenstra S, et al. Expression and distribution of id helix-loop-helix proteins in human astrocytic tumors. Glia. 2002;38:329–38. doi: 10.1002/glia.10076. [DOI] [PubMed] [Google Scholar]
- 6.Wilson JW, Deed RW, Inoue T, et al. Expression of Id helix-loop-helix proteins in colorectal adenocarcinoma correlates with p53 expression and mitotic index. Cancer Res. 2001;61:8803–10. [PubMed] [Google Scholar]
- 7.Kleeff J, Ishiwata T, Friess H, Buchler MW, Israel MA, Korc M. The helix-loop-helix protein Id2 is overexpressed in human pancreatic cancer. Cancer Res. 1998;58:3769–72. [PubMed] [Google Scholar]
- 8.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–30. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 9.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–33. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 10.Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Id proteins expression in prostate cancer: high-level expression of Id-4 in primary prostate cancer is associated with development of metastases. Mod Pathol. 2006;19:931–41. doi: 10.1038/modpathol.3800602. [DOI] [PubMed] [Google Scholar]
- 11.Yuen HF, Chan YP, Chan KK, et al. Id-1 and Id-2 are markers for metastasis and prognosis in oesophageal squamous cell carcinoma. Br J Cancer. 2007;97:1409–15. doi: 10.1038/sj.bjc.6604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–11. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong S, Itahana Y, Sumida T, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100:13543–8. doi: 10.1073/pnas.2230238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagliuca A, Gallo P, De Luca P, Lania L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors’ promoter activity and negatively affect cell growth. Cancer Res. 2000;60:1376–82. [PubMed] [Google Scholar]
- 16.Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26:4351–61. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–96. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desprez PY, Lin CQ, Thomasset N, Sympson CJ, Bissell MJ, Campisi J. A novel pathway for mammary epithelial cell invasion induced by the helix-loop-helix protein Id-1. Mol Cell Biol. 1998;18:4577–88. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y, Wang G, Ling MT, et al. Significance of Id-1 up-regulation and its association with EGFR in bladder cancer cell invasion. Int J Oncol. 2006;28:847–54. [PubMed] [Google Scholar]
- 20.Ling MT, Lau TC, Zhou C, et al. Overexpression of Id-1 in prostate cancer cells promotes angiogenesis through the activation of vascular endothelial growth factor (VEGF) Carcinogenesis. 2005;26:1668–76. doi: 10.1093/carcin/bgi128. [DOI] [PubMed] [Google Scholar]
- 21.Capparuccia L, Tamagnone L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment--two sides of a coin. J Cell Sci. 2009;122:1723–36. doi: 10.1242/jcs.030197. [DOI] [PubMed] [Google Scholar]
- 22.Roth L, Koncina E, Satkauskas S, Cremel G, Aunis D, Bagnard D. The many faces of semaphorins: from development to pathology. Cell Mol Life Sci. 2009;66:649–66. doi: 10.1007/s00018-008-8518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang R, Davalos AR, Hensel CH, Zhou XJ, Tse C, Naylor SL. Semaphorin 3F gene from human 3p21.3 suppresses tumor formation in nude mice. Cancer Res. 2002;62:2637–43. [PubMed] [Google Scholar]
- 24.Kessler O, Shraga-Heled N, Lange T, et al. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res. 2004;64:1008–15. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 25.Bielenberg DR, Hida Y, Shimizu A, et al. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–71. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekido Y, Bader S, Latif F, et al. Human semaphorins A(V) and IV reside in the 3p21.3 small cell lung cancer deletion region and demonstrate distinct expression patterns. Proc Natl Acad Sci U S A. 1996;93:4120–5. doi: 10.1073/pnas.93.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche J, Boldog F, Robinson M, et al. Distinct 3p21.3 deletions in lung cancer and identification of a new human semaphorin. Oncogene. 1996;12:1289–97. [PubMed] [Google Scholar]
- 28.Futamura M, Kamino H, Miyamoto Y, et al. Possible role of semaphorin 3F, a candidate tumor suppressor gene at 3p21.3, in p53-regulated tumor angiogenesis suppression. Cancer Res. 2007;67:1451–60. doi: 10.1158/0008-5472.CAN-06-2485. [DOI] [PubMed] [Google Scholar]
- 29.Clarhaut J, Gemmill RM, Potiron VA, et al. ZEB-1, a Repressor of the Semaphorin 3F Tumor Suppressor Gene in Lung Cancer Cells. Neoplasia. 2009;11:157–66. doi: 10.1593/neo.81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu A, Mammoto A, Italiano JE, Jr, et al. ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. J Biol Chem. 2008;283:27230–8. doi: 10.1074/jbc.M804520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasorella A, Stegmuller J, Guardavaccaro D, et al. Degradation of Id2 by the anaphase- promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–4. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 32.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–8. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 33.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–79. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 34.Gray MJ, Dallas NA, Van Buren G, et al. Therapeutic targeting of Id2 reduces growth of human colorectal carcinoma in the murine liver. Oncogene. 2008;27:7192–200. doi: 10.1038/onc.2008.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–84. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–31. [PubMed] [Google Scholar]
- 37.Bielenberg DR, Klagsbrun M. Targeting endothelial and tumor cells with semaphorins. Cancer Metastasis Rev. 2007;26:421–31. doi: 10.1007/s10555-007-9097-4. [DOI] [PubMed] [Google Scholar]
- 38.Doran AC, Meller N, Cutchins A, et al. The helix-loop-helix factors Id3 and E47 are novel regulators of adiponectin. Circ Res. 2008;103:624–34. doi: 10.1161/CIRCRESAHA.108.175893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusy S, Potiron V, Zeng C, et al. Promoter characterization of Semaphorin SEMA3F, a tumor suppressor gene. Biochim Biophys Acta. 2005;1730:66–76. doi: 10.1016/j.bbaexp.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Nair PN, McArdle L, Cornell J, Cohn SL, Stallings RL. High-resolution analysis of 3p deletion in neuroblastoma and differential methylation of the SEMA3B tumor suppressor gene. Cancer Genet Cytogenet. 2007;174:100–10. doi: 10.1016/j.cancergencyto.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Ochi K, Mori T, Toyama Y, Nakamura Y, Arakawa H. Identification of semaphorin3B as a direct target of p53. Neoplasia. 2002;4:82–7. doi: 10.1038/sj.neo.7900211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruyama H, Kleeff J, Wildi S, et al. Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am J Pathol. 1999;155:815–22. doi: 10.1016/S0002-9440(10)65180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 44.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 46.Langlands K, Down GA, Kealey T. Id proteins are dynamically expressed in normal epidermis and dysregulated in squamous cell carcinoma. Cancer Res. 2000;60:5929–33. [PubMed] [Google Scholar]
- 47.Nishimori H, Sasaki Y, Yoshida K, et al. The Id2 gene is a novel target of transcriptional activation by EWS-ETS fusion proteins in Ewing family tumors. Oncogene. 2002;21:8302–9. doi: 10.1038/sj.onc.1206025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.