Abstract
Loss of Hoxa1 function results in severe defects of the brainstem, inner ear and cranial ganglia in humans and mice as well as cardiovascular abnormalities in humans. Since Hoxa1 is expressed very transiently during an early embryonic stage, it has been difficult to determine whether Hoxa1 plays a direct role in the precursors of the affected organs or if all defects result from indirect effects due to mispatterning of the hindbrain. In this study we use a Hoxa1-IRES-Cre mouse to genetically label the early Hoxa1-expressing cells and determine their contribution to each of the affected organs, allowing us to conclude in which precursor tissue Hoxa1 is expressed. We found Hoxa1 lineage-labeled cells in all tissues expected to be derived from the Hoxa1 domain, such as the facial and abducens nuclei and nerves as well as r4 neural crest cells. Additionally, we detected the lineage in derivatives that were not thought to have expressed Hoxa1 during development. In the brainstem the anterior border of the lineage was found to be in r3, which is more anterior than previously reported. We also observed an interesting pattern of the lineage in the inner ear, namely a strong contribution to the otic epithelium with the exception of sensory patches. Moreover, lineage-labeled cells were detected in the atria and outflow tract of the developing heart. In conclusion, Hoxa1 lineage-tracing uncovered new domains of Hoxa1 expression in rhombomere 3, the otic epithelium and cardiac precursors, suggesting a more direct role for Hoxa1 in development of these tissues than previously believed.
Keywords: Hoxa1, hindbrain, inner ear, heart
INTRODUCTION
Homeobox (Hox) genes encode a family of transcription factors that regulate embryonic patterning and organogenesis (Alexander et al., 2009; Capecchi, 1997; Iimura and Pourquie, 2007). Hoxa1 is one of the earliest and most anteriorly expressed Hox genes (Murphy and Hill, 1991). Mice with a targeted disruption of Hoxa1 die shortly after birth from breathing defects, which are thought to result from mispatterning of the hindbrain (Chisaka et al., 1992; Lufkin et al., 1991). In addition, the inner ear fails to differentiate and cranial ganglia are smaller. Patients with homozygous truncating mutations in HOXA1 (Bosley-Salih-Alorainy syndrome (BSAS) or Athabascan brainstem dysgenesis syndrome (ABDS)) suffer from hypoventilation, deafness, facial weakness, vocal cord paralysis, swallowing dysfunction, carotid artery abnormalities and conotruncal heart defects (Bosley et al., 2008; Bosley et al., 2007; Holve et al., 2003; Tischfield et al., 2005).
Hoxa1 expression is first detected at embryonic day E7.5 in the primitive streak, in newly formed mesoderm and overlying neuroectoderm (Murphy and Hill, 1991). At E7.75, Hoxa1 expression reaches its anterior domain in the presumptive hindbrain, the embryonic precursor of the brainstem, and at E8.5, Hoxa1 has retreated from this region (Murphy and Hill, 1991). Thus, Hoxa1 is only expressed for about 12 hours in its most anterior domain.
The hindbrain or rhombencephalon is subdivided into eight transient swellings called rhombomeres, abbreviated r1-r8 (Lumsden and Keynes, 1989; Lumsden and Krumlauf, 1996). Loss of Hoxa1 function results in absence of r5 and size reduction of r4. This leads to loss of the abducens (6N) and strong reduction of the facial nuclei (7N), which is most likely the reason for horizontal gaze abnormalities and facial weakness in human patients. In addition to the defects in r4- and r5-derivatives, Hoxa1−/− mice also exhibit abnormal neurogenesis in r3, namely presence of cell patches with an r2 molecular identity, premature neuronal differentiation and abnormal navigation of motor axons (Helmbacher et al., 1998). Previous studies have described the anterior border of Hoxa1 expression to be in the hindbrain below the preotic sulcus at the future r3/r4 boundary (Barrow et al., 2000; Murphy and Hill, 1991). Therefore, the development of r3 was proposed to be non-autonomous and dependent on interactions with Hoxa1-expressing cells in r4 (Helmbacher et al., 1998).
Besides the hindbrain, another severely affected organ in Hoxa1−/− mice is the inner ear (Chisaka et al., 1992; Lufkin et al., 1991). The otic vesicle forms but fails to differentiate. Similarly, humans with mutations in HoxA1 have undifferentiated inner ears and are deaf. Development of the inner ear commences with an ectodermal thickening called the otic placode, which invaginates to form the otic cup and subsequently the otic vesicle. All components of the adult inner ear are derived from the otic ectoderm, including patches of sensory cells within the epithelium and sensory neurons in the spiral and vestibular ganglia, which innervate these patches. Since no Hoxa1 expression has been detected in the precursor of the inner ear, the abnormalities in Hoxa1−/− embryos were attributed to the disruption of hindbrain signals necessary for inner ear patterning (Mark et al., 1993). To date, little is known about how Hoxa1 performs its function during inner ear development and what signals are regulated by this gene.
Hoxa1−/− embryos also show defects in cranial ganglia and the stapes bone of the ear (Chisaka et al., 1992), structures that develop in part from neural crest cells (NCCs) which delaminate from rhombomere 4 after Hoxa1 expression has retracted from this region. This delamination takes place in two waves. The first wave migrates into BA2, where it forms cartilage, which differentiates into bone and connective tissue. The second wave of r4-NCC condenses lateral to the neural tube and gives rise to glia of the facio-acoustic ganglion complex (7/8G) (Baker et al., 1997; Kontges and Lumsden, 1996). All glia cells in this ganglion are derived from NCCs, whereas almost all neurons originate from ectodermal placodes (Barlow, 2002). The defects in NCC-derivatives in Hoxa1−/− mice lead to the hypothesis that Hoxa1 might specify the developmental program of cranial neural crest cells (Lufkin et al., 1991).
An even more dramatic phenotype is observed in embryos lacking both Hoxa1 and its paralogue Hoxb1, which in addition to neural crest defects almost completely lack the second branchial arch (BA2) and its mesodermal derivatives (Gavalas et al., 1998; Rossel and Capecchi, 1999). During development, cells from the cranial paraxial mesoderm surrounding r3-r6 migrate into the core of BA2 and give rise to the muscles of facial expression as well as the muscles of the jaw and upper neck. It has so far been controversial whether loss of mesodermal derivatives is secondary due to the absence of r4 NCCs in the double knockout, or if Hoxa1 (in redundancy with Hoxb1) plays a direct role in mesoderm development (Morrison, 1998).
A study in humans demonstrated that HOXA1 has a previously unrecognized role in development of the cardiovascular system (Tischfield et al., 2005). Humans with homozygous mutations in HOXA1 exhibit outflow tract (OFT) and internal carotid artery (ICA) abnormalities (Tischfield et al., 2005). The OFT develops from mesodermally-derived myocardial cells and is later infiltrated and remodeled by cardiac neural crest cells, originating in the hindbrain at the level of r6-r8 (Brown and Baldwin, 2006; Kirby and Waldo, 1995; Snider et al., 2007). Hoxa1 is expressed in the neural tube at the level from which cardiac NCCs arise (Murphy and Hill, 1991) in addition to the foregut and mesoderm adjacent to the cardiac field (Godwin et al., 1998; Ryckebusch et al., 2008). However, no Hoxa1 expression has been detected in myocardial precursors within the cardiac field (Godwin et al., 1998; Ryckebusch et al., 2008). Since the cardiovascular defects have not been analyzed in mice, it is unknown at which step of development and in what tissue Hoxa1 function is required.
In this study we present new insight into the role of Hoxa1 during embryogenesis by genetically labeling early Hoxa1 expressing cells, using the Cre/loxP system (Branda and Dymecki, 2004), and following their fate into later stages of development (Hoxa1-lineage tracing). Our analysis demonstrates that Hoxa1-lineage does not exhibit a sharp anterior border at the r3/4 boundary but extends into r3. We also find that Hoxa1-lineage gives rise to all neural crest cells which populate the second branchial arch and contribute to cranial ganglia. In contrast, no Hoxa1-lineage is detected in mesodermal derivatives of BA2. Interestingly, Hoxa1-lineage is seen in a restricted pattern in derivatives of the otic placode and myocardium, both structures that were not thought to express Hoxa1.
MATERIALS AND METHODS
Gene targeting and genotyping
To generate the Hoxa1-IRES-Cre allele, a 7.9 kb ClaI fragment containing the Hoxa1 locus from 129/SV genomic DNA was subcloned. An AscI site placed 36 bp downstream of the stop codon was used to insert an IRES-Cre-frt-MC1-Neo-frt cassette (Arenkiel et al., 2003). The targeting vector was electroporated into R1 ES cells, which were cultured under positive selection using G418. Correctly targeted ES cell clones were identified by Southern hybridization (Fig. 1B) and used to generate chimeras, which were crossed to C57BL6 mice. The neomycin resistance gene was removed by crossing the mice to a FLPe deleter line (Rodriguez et al., 2000). Hoxa1-IRES-Cre homozygous mice are viable and fertile.
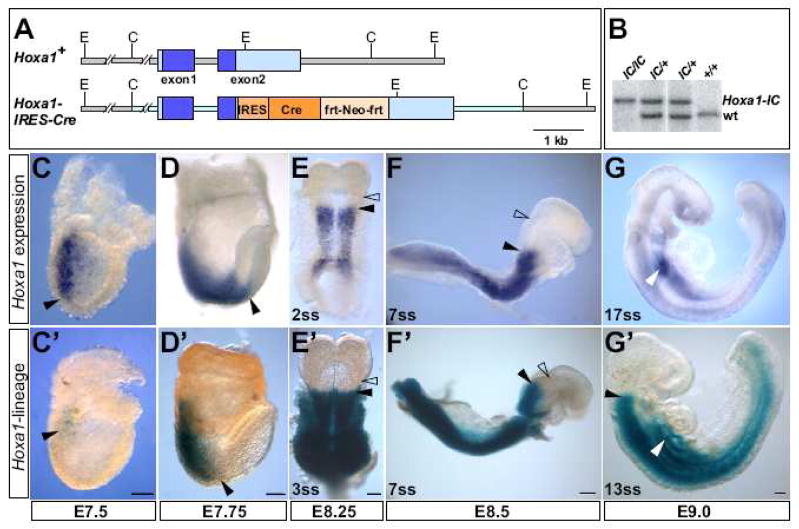
Fig. 1. Hoxa1 targeting and lineage analysis in the early embryo.
(A) Depiction of wild-type (Hoxa1+) and Hoxa1-IRES-Cre alleles. The IRES-Cre-fit-MC1-Neo-frt cassette (orange boxes) was inserted 36 bp downstream of the Hoxa1 stop codon. Grey lines. Hoxa1 genomic DNA: light blue lines, genomic region included in the targeting vector. C, ClaI; E, EcoRI. (B) Southern blot analysis to identify the Hoxa1-IRES-Cre (Hoxa1-IC) allele. DNA was digested with EcoRI and hybridized with a 5′ external probe to generate an 8.3 kb wt and a 9.8 kb Hoxa1-IRES-Cre band. (C–G) Hoxa1 expression analysis by RNA in situ hybridization. Hoxa1 expression starts at E7.5 in the posterior primitive streak and spreads anteriorly (C). By E7.75, Hoxa1 reaches its most anterior border (black arrowhead) in the future hindbrain (D) below the preotic sulcus (open arrowhead in E). At E8.5, expression has retreated from the hindbrain and remains in more posterior regions (F). At E9.0, expression is seen only in the posterior neural tube and in the foregut pocket (white arrowhead in G). (C′–G′) Hoxa1-lineage visualized by X-gal staining of Hoxa1-IRES-Cre; R26R-lacZ embryos. At E7.5, Hoxa1-lineage shows a slight delay compared to Hoxa1 expression (C′). Hoxa1-lineage displays the same anterior border as Hoxa1 expression between E7.75 and E8.25 (D′, E′). X-gal staining in later embryos highlights all regions of the embryo that are derived from the Hoxa1-expression domain (F′, G′). Black arrowheads in C–G′: anterior border of Hoxa1 expression or lineage; open arrowheads in E–F′: preotic sulcus; white arrowhead in G, G′: foregut pocket. In C, D and F anterior is to the right, in E to the top and in G to the left. Scale bars: 100 μm.
Genotyping was performed using multiplex PCR with the following primers: wt 5′ (AGCGATGAGAAAACGGAAG), wt 3′ (GGG ACG AGA AAG GAA GAG AG), Cre 5′ (CAA TAC CGG AGA TCA TGC AAG), generating a 220 bp wt and 382 bp engineered band. Lineage analysis was carried out using the previously described R26R-EYFP, R26R-lacZ and nLacZ lines (Haldar et al., 2008; Soriano, 1999; Srinivas et al., 2001). All mouse use complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee.
β-Galactosidase staining and RNA in situ hybridization
For β-gal staining, tissues were dissected in PBS, pH 7.4 with 2 mM MgCl2, fixed for 15 min to 2 hours depending on tissue size in 1% formaldehyde, 0.2% glutaraldehyde, 25 mM EGTA, 2 mM MgCl2, 0.02% NP40 in PBS, washed in PBS with 2 mM MgCl2, and moved into X-gal staining solution (0.8 mg/ml X-gal, 25 mM K3Fe(CN)6, 25 mM K4Fe(CN)6-3H2O, 2 mM MgCl2, 0.01% Na deoxycholate, 0.02% NP40 in PBS). Facial nerve staining was carried out after removal of the skin and surrounding tissues. Brains of adult mice were isolated after perfusion with 2% formaldehyde, cryosectioned, stained with X-gal and mounted in Celvol. All stainings were carried out over night at room temperature.
Whole-mount in situ hybridization with a digoxigenin-labeled antisense probe generated from a plasmid containing a 216 bp Hoxa1 exon1 fragment was carried out as described (Henrique et al., 1995).
Immunostaining and analysis
Tissues were fixed at 4°C for 1–2 hrs in 4% formaldehyde, rinsed in PBS, equilibrated to 30% sucrose and embedded in O.C.T. Cryosections were cut at 10 μm, washed in PBS and preincubated in blocking solution (2% BSA, 10% NGS, 0.1% Triton in PBS, pH 7.2). Primary antibodies were applied overnight at 4°C in a humid chamber, followed by secondary detection using Alexa Fluor conjugated (Molecular Probes) or DyLight conjugated (Jackson ImmunoResearch) secondary antibodies. Immunodetection was carried out using an SP5 confocal system (Leica) or an inverted microscope (Axiovert 200M, Zeiss) equipped with a SensiCam camera (The Cooke Coorperation). Data was acquired using the LAS AF or SlideBook™ software and processed using Adobe Photoshop. For hindbrain flatmounts embryonic hindbrains were isolated and the neural tube was cut along the roof plate. Brains were fixed, stained and visualized as above.
The following primary antibodies were used in this study: mouse anti-AP2 (DSHB; 1:75), chick anti-GFP (Aves Labs; 1:500), rabbit anti-GFP (Abcam; 1:4000 or Molecular probes; 1:2000), mouse anti-GFP (Molecular probes; 1:250), rabbit anti-Hox-B1 (Covance; 1:250), mouse anti-Islet1 (DSHB; 1:30), rat anti-MBP (Chemicon; 1:75), mouse anti-Myogenin (DSHB; 1:25), rabbit anti-p75 (Chemicon; 1:100), mouse anti-Pax7 (DSHB; 1:15), rabbit anti-Phox2b (kind gift of C. Goridis and J. P. Brunet; 1:1000), rabbit anti-Sox2 (Chemicon; 1:2000), rabbit anti-Sox9 (Chemicon; 1:1500), guinea pig anti-Sox10 (kind gift of M. Wegner; 1:150), mouse anti-Tuj1 (Covance; 1:1000).
RESULTS
Hoxa1-lineage overlaps with endogenous Hoxa1 gene expression at early embryonic stages
To analyze the contribution of Hoxa1 expressing cells in the early embryo to specific tissues and organs at later stages of development, we performed genetic lineage tracing using a novel Hoxa1-IRES-Cre allele. This allele was generated by targeting an IRES-Cre-Neo cassette (Arenkiel et al., 2003) to the 3′UTR of the Hoxa1 gene (Fig. 1A), allowing bicistronic expression of Hox-A1 and Cre recombinase from the Hoxa1 promoter. To study the fate of Hoxa1 expressing cells, Hoxa1-IRES-Cre mice were crossed to either R26R-EYFP (Mao et al., 2001), R26R-lacZ (Soriano, 1999) or nLacZ (Haldar et al., 2008) reporter lines, which express EYPF, β-gal or nuclear localized β-gal, respectively, upon recombination. The efficacy and specificity of the Hoxa1-IRES-Cre reporter was examined by comparing X-gal stained Hoxa1-IRES-Cre; R26R-lacZ embryos (Fig. 1C′–G′) to stage matched embryos in which Hoxa1 expression was visualized by RNA in situ hybridization (Fig. 1C–G). Hoxa1 mRNA expression is first seen at E7.5 in the posterior primitive streak (Fig. 1C), as described (Murphy and Hill, 1991). Hoxa1-lineage is detected at the same stage, with a brief temporal delay (Fig. 1C′), due to the cumulative delay of the Cre/loxP system (Danielian et al., 1998; Jukkola et al., 2005; Ohyama and Groves, 2004). Between E7.75 and E8.25, Hoxa1 mRNA expression is seen at the most anterior border in the future hindbrain (Fig. 1D, E). This border is below the preotic sulcus, a constriction which marks the future r2/r3 boundary (Fig. 1E). Hoxa1-lineage displays the same anterior border as Hoxa1 mRNA expression between E7.75 and E8.25 (Fig. D′, E′). Hoxa1 is expressed in its most anterior domain for only around 12 hours and by E8.5 mRNA expression has retreated from the hindbrain and remains in more posterior regions of the neural tube and mesoderm (Fig 1F). Although Hoxa1 becomes downregulated in the anterior region, cells that have expressed Hoxa1 are permanently marked by β-gal expression (Fig. 1F′). At E9.0, Hoxa1 mRNA expression remains only in the posterior neural tube and in the foregut pocket (Fig. 1G). X-gal staining can be detected in all regions of the embryo that are derived from the Hoxa1-expression domain (Fig. 1G′).
These results verify that the Hoxa1-IRES-Cre driver line recapitulates endogenous Hoxa1 activity and can be used to conduct in-depth lineage analysis. Due to the extremely transient expression of this gene, the Hoxa1-IRES-Cre line represents a valuable tool to permanently mark all Hoxa1-expressing cells and to follow their fate into later embryonic stages.
Cells derived from the Hoxa1-lineage are present in rhombomere 3
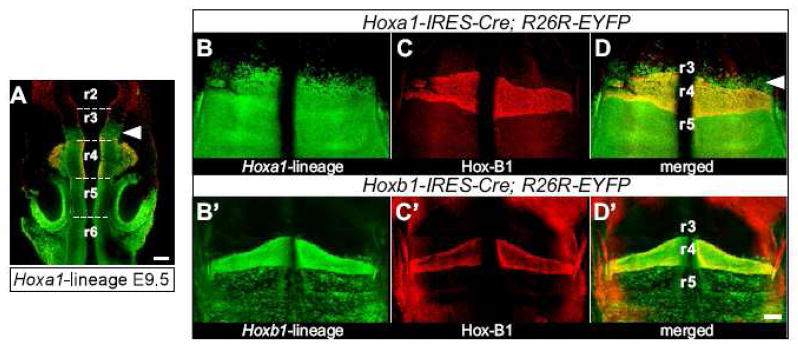
Since Hoxa1 is expressed very transiently in its most anterior domain (Fig. 1C–F) at a time before rhombomere boundaries are formed, we examined the anterior border of Hoxa1-lineage in older embryos. Lineage analysis in the hindbrain was carried out in Hoxa1-IRES-Cre; R26R-EYFP embryos at E9.5, after rhombomere boundaries have been established and can be seen as physical constrictions in the hindbrain (Fig. 2A). Interestingly, Hoxa1-lineage did not show a sharp anterior border at the r3/r4 boundary but was present in the caudal half of r3. When Hox-B1 immunostaining in r4 was used to mark the r3/r4 boundary in hindbrain flatmounts of E10.5 lineage-labeled embryos (Fig. 2B–D), Hoxa1-lineage was seen in r3 in all embryos examined (n=11).
Fig. 2. The anterior border of Hoxa1-lineage in the hindbrain is in r3.
(A) Hoxa1-lineage (green) is seen in the caudal half of rhombomere 3 in a dorsal view of an E9.5 embryo. At this stage, the rhombomeric constrictions are visible (dotted lines) and rhombomere 4 was labeled by immunostaining for Hox-B1 (red). (B–D) and (B′–D′) Hindbrain flatmounts of E10.5 Hoxa1-IRES-Cre; R26R-EYFP and Hoxb1-IRES-Cre; R26R-EYFP embryos, respectively. The lineage of each gene is shown in green (B, B′) and Hox-B1 expression in r4 in red (C, C′). The merged images demonstrate that while Hoxb1-lineage exhibits a sharp anterior border at the r3/r4 boundary (D′), Hoxa1-lineage is present in r3 (D). Scale bars: 100 μm.
We also analyzed the anterior border of Hoxb1-lineage (Arenkiel et al., 2003), the paralog of Hoxa1, which was reported to have the same anterior border as Hoxa1 at the r3/r4 boundary (Barrow et al., 2000). Very few if any GFP positive cells were detected in r3 of Hoxb1-IRES-Cre; R26R-EYFP embryos (Fig. 2D′) (n=10). These results demonstrate that in contrast to Hoxb1-lineage, which exhibits a sharp anterior border at the r3/r4 boundary, Hoxa1-lineage is present in rhombomere 3.
Hoxa1-lineage contributes extensively to the facial and abducens nuclei and gives rise to glia of the facial nerve
The most severe brainstem defects resulting from loss of Hoxa1 function are absence of the abducens and reduction of the facial nucleus and nerve (Chisaka et al., 1992; Lufkin et al., 1991). Here we examine the extent of Hoxa1-lineage contribution to neurons and glia of these nuclei.
Our analysis revealed that Hoxa1-lineage extensively labels the caudal brainstem of E12.5-E14.5 Hoxa1-IRES-Cre; R26R-lacZ embryos, including the region where the facial and abducens nuclei reside (Fig. 3A, B). On sections through the hindbrain of E11.5 embryos almost all neurons of the facial nucleus (7N), marked by Islet1 (Wang and Drucker, 1994) carry the lineage label (Fig. 3D–D″). Similarly, the majority of axonal projections of the facial nerve (7n), stained with the β-tubulin-specific antibody Tuj1 (Jepsen et al., 2000) are lineage-derived (Fig. 3E). Strong labeling of the abducens (6N) and facial nucleus (7N) was also seen on sections of the adult brain (Fig. 3F″, G).
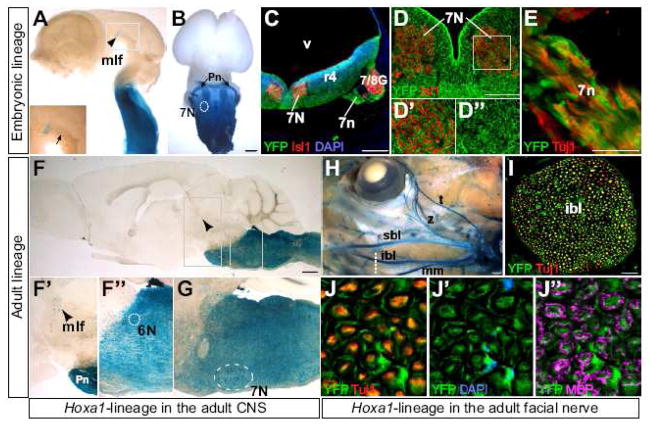
Fig. 3. Hoxa1-lineage contributes extensively to the facial, abducens and mlf nuclei and the facial nerve.
(A–E) Hoxa1-lineage in the embryonic nervous system. (A) Lateral view of a Hoxa1-IRES-Cre; R26R-lacZ E12.5 brain, with Hoxa1-lineage in the caudal brainstem and the medial longitudinal fasciculus (mlf) at the fore/midbrain boundary (arrowhead). Inset is a higher magnification of staining in the nucleus and tracts (arrow) of the mlf. (B) Ventral view of a brain from an E14.5 Hoxa1-IRES-Cre; nLacZ embryo, showing Hoxa1-lineage in the ventral brainstem including the facial nucleus (7N), as well as in migrating pontine nuclei (Pn). (C) Transverse section through the hindbrain of an E11.5 embryo at the level of rhombomere 4 (r4) showing the facial nucleus (7N) and nerve (7n) and the facio-acoustic ganglion complex (7/8G) (lineage in green, DAPI in blue, Isll in red). (D–D″) Higher magnifications of an adjacent section showing strong contribution of Hoxa1-lineage to neurons of the facial nucleus co-stained with Isll (red). (E) Higher magnification of Hoxa1-lineage in axons of the facial nerve co-stained with Tuj1 (red). (F–J″) Hoxa1-lineage in the adult nervous system. (F) Hoxa1-lineage in a sagittal section through the adult brain. (F′, F″) Higher magnification of X-gal positive cells in the mlf and the abducens nucleus (6N). (G) Hoxa1-lineage in the adult seventh nucleus. (H–J″) Hoxa1-lineage in the facial nerve. (H) X-gal staining of an adult Hoxa1-IRES-Cre; R26R head, showing the lineage in all branches of the facial nerve. (I) Cross-section through the inferior buccolabial branch (ibl) of the seventh nerve (dotted line in H) (Hoxa1-lineage in green Tuj1 in red). Higher magnification shows colocalization of Hoxa1-lineage with the neuronal marker Tuj1 (red) (J) and DAPI (blue), which marks Schwann cell nuclei (J′). Myelin basic protein (MBP) immunostaining (magenta) highlights the myelin sheet of Schwann cells (J″). Abbreviations: mm, marginal mandibular; sbl, superior buccolabial; t, temporal; v, ventricle; z, zygomatic. Scale bars in B, F, H: 500 μm; scale bars in C, I: 100 μm.
To analyze Hoxa1-lineage in the facial nerve in more detail, we stained heads of Hoxa1-IRES-Cre; R26R-lacZ adult mice with X-gal. All branches, namely the temporal, zygomatic, superior buccolabial, inferior buccolabial and the marginal mandibular show X-gal staining (Fig. 3H). We performed cross sections through different branches of the facial nerve (Fig. 3I–J and data not shown) and saw that the majority of axons express the lineage marker. In addition to axons of the facial nerve, we found that Hoxa1-lineage also gives rise to neural crest-derived Schwann cells, which ensheath these axons (Fig. 3I). This was demonstrated by performing co-staining using anti-MBP (myelin basic protein) (Fig. 3J″) and DAPI, to stain Schwann cell nuclei (Fig. 3J′).
Besides the hindbrain, Hoxa1 expression was reported in the medial longitudinal fasciculus (mlf) (McClintock et al., 2003), which is located at the fore/midbrain boundary and plays a role in the coordination of eye movement (Forlani et al., 2003). We see Hoxa1-lineage in the nucleus and axons of the mlf in dissected brains of E12.25 embryos (Fig. 3A) as well as on adult brain sections (Fig. 3F, F′). On sagittal sections of the entire adult Hoxa1-IRES-Cre; R26R-lacZ brain, the mlf is the most anterior structure in which Hoxa1-lineage is detected (Fig. 3F) demonstrating that Hoxa1 is not expressed in any regions of the brain besides the brainstem and the mlf.
Hoxa1-lineage gives rise to all r4-neural crest cells but not to mesodermal derivates of the second branchial arch
Hoxa1 is expressed in the neuroectoderm in presumptive r4 shortly prior to delamination of NCCs, which takes place at around E8.5 and is reduced in Hoxa1−/− embryos (Mark et al., 1993). In wholemount X-gal stained Hoxa1-IRES-Cre; R26R-lacZ embryos, labeled NCCs emerging from r4 can first be seen at E8.5 (9 somite stage; data not shown). Between E8.75-E9.5, a large stream of lineage labeled r4-NCCs migrates from the dorsal neural tube into BA2 (Fig. 4A, B and data not shown). In sections of E10.5 embryos, these cells are seen in BA2 as well as condensing next to the dorsal neural tube to contribute to the 7/8G complex (Fig. 4C). In order to examine if Hoxa1 is expressed in all or in a specific subpopulation of neural crest precursors, we performed co-staining of Hoxa1-lineage and the NCC markers AP2 alpha (Mitchell et al., 1991) and P75, the low-affinity neurotrophin receptor (Morrison et al., 1999; Mujtaba et al., 1998) (Fig. 4D and data not shown). Analysis of transverse sections of E9.0, E9.5 and E10.5 embryos through the entire length of r4 demonstrated that all NCCs are labeled by the Hoxa1-lineage.
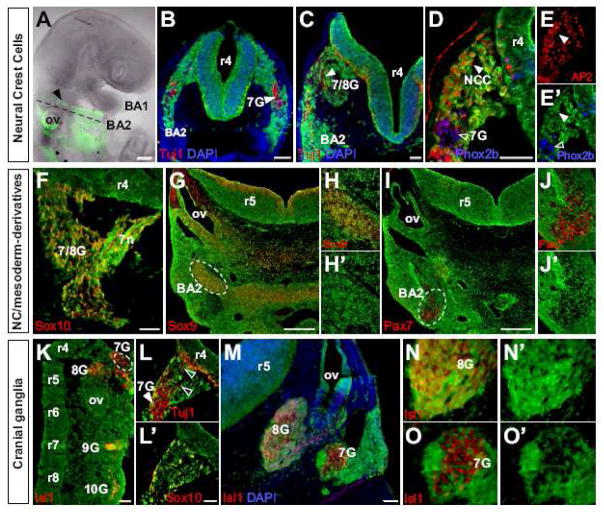
Fig. 4. Hoxa1-lineage gives rise to all r4 neural crest derivatives and neurons of specific cranial ganglia but not to myocytes in the second branchial arch.
Hoxa1-lineage (green) in Hoxa1-IRES-Cre; R26R-EYFP embryos. (A–E′) Hoxa1-lineage gives rise to r4 neural crest cells but not to sensory neurons in the geniculate ganglion. (A) Lateral view of an E9.5 embryo, showing Hoxa1-lineage in r4-NCCs (arrowhead) migrating from the dorsal neural tube into the second branchial arch (BA2). (B) Section through r4 (dashed line in A) showing lineage-labeled r4 NCCs surrounding unlabeled early geniculate placode derived sensory neurons (7G; arrowhead; Tuj1 in red). (C) At E10.5, Hoxa1-lineage is found in BA2 and next to the dorsal neural tube, where the facio-acoustic ganglion complex (7/8G) forms. Tuj1 (red) marks neurons in 7/8G, which are not lineage-labeled (arrowhead). (D–E′) While all r4 NCCs (AP2 in red) are derived from Hoxa1-expressing cells (filled arrowheads), geniculate placode-derived neurons (Phox2b in blue) are not (open arrowheads). (F–H) Hoxa1-lineage in neural crest derivatives in the the 7/8G complex and BA2 in transverse sections of E11.5 embryos. Hoxa1-lineage gives rise to neural crest-derived glia cells (F; Sox10 in red) which ensheath the 7/8G complex as well as to chondrocytes (G; Sox9 in red) condensing in the second branchial arch. (H, H′) Magnified view of the circled area in G. (I) Mesoderm-derived myocytes in the core of BA2 (Pax7 in red) are not derived from Hoxa1-expressing cells (magnified view in J and J′). (K–O′) Hoxa1-expressing cells give rise to sensory neurons in the vestibular (8G), petrosal (9G) and nodose (10G) but not the geniculate (7G) ganglion. (K) Hoxa1-lineage in sensory neurons (Islet1 in red) of all cranial ganglia, except the geniculate ganglion (circled), in longitudinal section of an E10.5 embryo. (L, L′) Transverse section showing 7G neurons (L; Tuj1 in red) sending their projections to the neural tube (open arrowheads), along tracts of Hoxa1-lineage derived glia cells (L′; Sox10 in red). (M) Transverse section of an E11.5 embryo at the level of r5 and the otic vesicle (ov) (Isl1 in red). (N, N′) Higher magnification of the vestibular ganglion (8G), showing co-expression of Hoxa1-lineage and sensory neurons. (O, O′) Higher magnified image of the geniculate ganglion, in which sensory neurons are not derived from Hoxa1-expressing cells. All scale bars are 50 μm, except in A, G, I: 250 μm.
Next, we examined the contribution of Hoxa1-lineage to NCC-derivatives. Immunostaining for Sox10, a transcription factor expressed in glial progenitors (Britsch et al., 2001; Maka et al., 2005), on sections through the 7/8G complex, revealed that the majority of glia cells in this ganglion are derived from Hoxa1 expressing cells (Fig. 4F). We also performed immunostaining for NC-derived cartilage in BA2 using the chondrocyte marker Sox9 (Zhao et al., 1997) and saw that cartilage is labeled by the Hoxa1-lineage (Fig. 4G, H). This further confirms that Hoxa1 is expressed in all precursors of r4 NCCs, the ones that migrate into BA2 and form cartilage as well as those that give rise to glia in the 7/8G complex.
To address whether Hoxa1 plays a direct role in the development of BA2 mesoderm, we examined if Hoxa1-lineage can be detected in mesoderm-derived muscle cells. We performed co-immunostaining for YFP and the early muscle-markers Pax7 (Jostes et al., 1990) and myogenin (Wright et al., 1989) (Fig. 4I, J and data not shown). No co-staining of myocyte markers and the Hoxa1-lineage was detected, demonstrating that Hoxa1 is not expressed in BA2 muscle precursors.
Hoxa1-lineage is seen in sensory neurons of the vestibular, petrosal and nodose but not the geniculate ganglion
In Hoxa1 knockout mice, the geniculate ganglion (G7) is always present, while sensory ganglia G8-G10 are absent or reduced (Lufkin et al., 1991). G7 sensory neurons are derived from the geniculate epibranchial placode, neurons of the spiral and vestibular ganglion (G8) from the otic placode, while G9 and G10 neurons develop from the petrosal and nodose placodes, respectively. In order to understand the contribution of Hoxa1-lineage to these ganglia, we performed co-immunostaining of YFP and specific neuronal markers in sections of Hoxa1-IRES-Cre; R26R-EYFP embryos.
Tuj1 or Islet1 positive neuronal cells at the level of r4 are first detected at E9.0 (Fig. 4B and data not shown). These early sensory neurons are derived from the geniculate placode. Hoxa1-lineage is seen in NCCs surrounding these neurons (Fig. 4D, E) but not in the sensory neurons of the geniculate ganglion (7G), which were labeled using Islet1 (Fig. 4O), Phox2b (Pattyn et al., 1997) (Fig. 4D, E′) or Tuj1 (Fig. 4B, L). At E10.5, these non-lineage derived 7G neurons send their projections to the neural tube along Hoxa1-lineage labeled glia cells (Fig. 4L′).
In order to examine whether Hoxa1-lineage gives rise to neurons in other cranial ganglia, we immunostained longitudinal sections of E10.5 embryos with Islet1 (Fig. 4K) or Phox2b (data not shown). We detected Hoxa1-lineage in neurons of the petrosal, nodose and vestibular ganglia but not in neurons of the geniculate ganglion. The most surprising observation was that neurons of the otic placode-derived vestibular ganglion (8G) are labeled by the Hoxa1-lineage (Fig. 4K). Transverse sections of E11.5 embryos at the level of the developing otocyst (Fig. 4M) showed no co-expression of YFP and Islet1 in sensory neurons of the geniculate ganglion (Fig. 4O, O′). However, the majority of sensory neurons in the vestibular ganglion are derived from Hoxa1 expressing cells (Fig. 4N, N′).
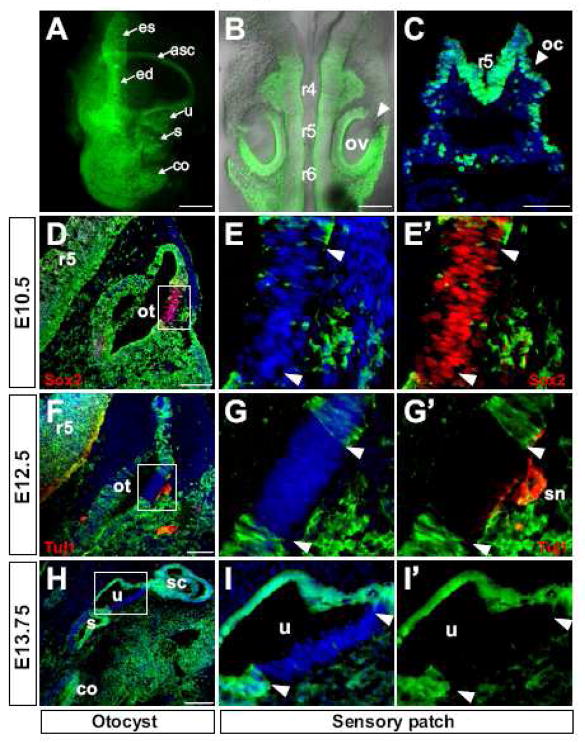
Hoxa1-lineage is found in a specific pattern in the otic epithelium
Although no Hoxa1 expression has been reported in the presumptive ear ectoderm, otic placode, otic vesicle or inner ear (Mark et al., 1993; Murphy and Hill, 1991), we found significant Hoxa1-lineage in the developing otic epithelium (Fig. 5). At E13.5, YFP expression was seen in the entire inner ear, namely the endolymphatic duct, the semicircular canals, the utricle, the saccule and the cochlea (Fig. 5A). In order to investigate the contribution of Hoxa1-lineage to the otic vesicle on a cellular level, cryosections of the developing inner ear of Hoxa1-IRES-CRE; ROSA-EYFP embryos at different stages of maturation were analyzed (Fig. 5C–I′). Hoxa1-lineage was detected in the epithelium of the otic cup at E8.75 (Fig. 5C). At E9.5 Hoxa1-lineage was seen in the otic vesicle, but was excluded from the anterodorsolateral region (Fig. 5B). Interestingly, the region from which Hoxa1-lineage was excluded, was identified as a sensory patch in sections of E10.5-E13.75 embryos (Fig. 5D–I′) by immunostaining for Sox2 (Fig. D–E′), a marker for sensory patches in the otic vesicle (Kiernan et al., 2005) and Tuj1, which marks sensory neurons and axons that innervate these patches (Fig. 5F–G′). In the more mature inner ear at E13.75, the lineage was absent from the utricular sensory patch (Fig. 5H–I′).
Fig. 5. Hoxa1-lineage contributes extensively to the inner ear but is excluded from sensory regions.
Lineage analysis of Hoxa1-IRES-Cre; R26R-EYFP embryos. (A–C) Hoxa1-lineage (green) in the inner ear, otic vesicle and otic cup. (A) Dissected inner ear of an E13.5 embryo showing strong contribution of Hoxa1-lineage. (B) Dorsal view of an E9.5 embryo with Hoxa1-lineage in the otic vesicle (ov) but excluded from an anterodorsolateral patch (arrowhead). (C) The lineage-label can be detected as early as the otic cup (oc) stage at E8.75 (DAPI, blue). (D–I′) Strong Hoxa1-lineage contribution to the developing otocyst (ot) in transverse sections of E10.5, E12.5 and E13.75 embryos. Hoxa1-lineage is absent from an anterodorsolateral region that was identified as a sensory patch by Sox2 expression (red) which marks sensory cells in the otic epithelium (D) and Tuj1 (red) which marks sensory neurons (sn) and axons which innervate this patch (F). In more differentiated inner ears at E13.75 Hoxa1-lineage is absent from the utricular (u) sensory patch (H). (E, G, I) Higher magnification of the boxed areas in D, F, H, with DAPI counterstain (blue) (E, G, I) or YEP only (E′, G′, I′), showing absence of the lineage from the sensory patches. Arrowheads indicate the region that is devoid of Hoxa1-lineage. Abbreviations: asc, anterior semicircular canal; co, cochlea; ed, endolymphatic duct; es, endolymphatic sac; r4-r6, rhombomeres 4–6; s, saccule; sc, semicircular canals. All scale bars are 100 μm except A: 1mm and B: 200 μm.
Hoxa1-lineage is present in the outflow tract and atria of the heart
To examine the contribution of Hoxa1-lineage to the heart, we analyzed dissected hearts and cryosections of E10.5-E12.5 embryos (Fig. 6 and data not shown). Hoxa1-lineage was seen in the OFT and the atria but not the ventricles of the heart (Fig. 6A–C). The presence of Hoxa1-lineage in the atria is surprising because all cells in the atria are derived from the myocardium and no Hoxa1 expression has been detected in myocardial cells in previous studies (Godwin et al., 1998; Ryckebusch et al., 2008).
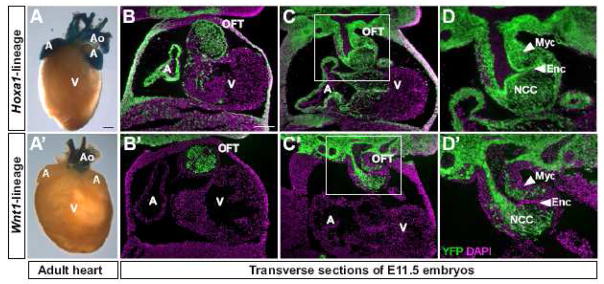
Fig. 6. Cardiac neural crest cells in the outflow tract are derived from Hoxa1-expressing cells.
(A, A′) X-gal staining of dissected adult hearts from Hoxa1-IRES-Cre; R26R-lacZ (A) and Wnt1-Cre; R26R-lacZ mice (A′). While Wnt1-lineage (A′), which represents the neural crest cell (NCC) lineage, is only seen in the aortic arch (Ao), Hoxa1-lineage (A) is found in the aortic arch and the atria (A) but not the ventricles (V) of the heart. (B–C′) Transverse sections of E11.5 embryos showing Hoxa1-lineage in the atrium and outflow tract (OFT) of the heart (B, C), compared to Wnt1-lineage, which is only present in cardiac NCCs in the OFT (B′, C′). Hoxa1- and Wnt1-lineage (green); DAPI (magenta). (D, D′) Enlarged view of the boxed areas in C and C′, demonstrating that the majority of cardiac neural crest cells in the OFT are derived from Hoxa1 expressing cells. Additionally, Hoxa1-lineage is detected in myocardial (Myc) and endocardial (Enc) cells. Scale bar in A: 1 mm, in B: 200 μm.
To assess whether cardiac neural crest cells in the outflow tract are derived from Hoxa1-expressing cells, we compared the distribution of the Hoxa1-lineage to that of the Wnt1-lineage (Danielian et al., 1998), which represents neural crest cells (Jiang et al., 2000) in coronal sections of E9.5-E11.5 embryos (Fig. 6 and data not shown). At E9.5, only a small number of Hoxa1- or Wnt1-lineage-labeled cells were seen in the truncus arteriosus region of the outflow tract (data not shown). At E10.5, Hoxa1- and Wnt1-lineage was seen in a similar pattern in the truncus arteriosus and conotruncus (data not shown). Comparison of Hoxa1-lineage (Fig. 6D) to Wnt1-lineage (Fig. 6D′) at E11.5 demonstrates that most if not all cardiac neural crest cells are labeled by the Hoxa1-lineage. In addition to cardiac neural crest cells, Hoxa1-lineage was also detected in the myocardium and endocardium of the outflow tract (Fig. 6D).
DISCUSSION
This study provides new insights into the role of Hoxa1 during development by uncovering the specific cell lineages in which Hoxa1 is expressed and the contribution of these cells to organs which are affected by loss of Hoxa1 function. After identification of a human syndrome caused by mutations in HOXA1 (Tischfield et al., 2005), several new questions arose which can only be answered in the mouse model. Our data prompt us to suggest a more direct role for Hoxa1 in development of the third rhombomere, cranial ganglia and the inner ear, all structures which were thought to only be indirectly affected by loss of Hoxa1. Additionally, we discovered the lineage of Hoxa1 in the heart, an organ which has not been studied in relation to Hoxa1, but was recently shown to be affected in humans with HOXA1 mutations (Bosley et al., 2008; Tischfield et al., 2005).
Hoxa1-lineage recapitulates endogenous gene activity
Hoxa1 is transiently expressed in the neuroectoderm, the lateral plate mesoderm as far anterior as the developing hindbrain and the presomitic mesoderm, in addition to the endoderm derived epithelium of the foregut pocket and the surface ectoderm adjacent to the gut-associated mesoderm (Murphy and Hill, 1991). As expected, Hoxa1-lineage in the early embryo showed the same pattern as Hoxa1 expression (Fig. 1). In older embryos, the lineage was seen in all tissues that are derived from domains which transiently express Hoxa1 in the early embryo. In the brain, Hoxa1-lineage showed an extensive contribution to the caudal brainstem with the most anterior structure labeled being the medial longitudinal fasciculus (Forlani et al., 2003) at the fore/midbrain boundary (Fig. 3). We observed a strong contribution of Hoxa1-lineage to the facial and abducens nuclei, both of which are severely affected in mice and humans with mutations in Hoxa1 (Mark et al., 1993; Tischfield et al., 2005). A more detailed analysis of the facial nerve revealed that Hoxa1-lineage contributes to all branches of the facial nerve and gives rise to both axons and NC-derived glia cells in this nerve (Fig. 3).
Our results show that the lineage marker is present in all structures expected to be derived from the Hoxa1-expression domain, making the Hoxa1-IRES-Cre line a useful tool to identify all tissues which transiently express Hoxa1. In addition to the expected regions, we detected Hoxa1-lineage in tissues not thought to be derived from Hoxa1-expressing cells, namely r3, the otic vesicle and cardiomyocytes. It seems likely that expression of Hoxa1 mRNA in the precursors of these tissues was missed in previous studies due to the very early and transient expression of this gene.
Hoxa1-lineage is found in rhombomere 3
Our lineage analysis of E9.5 and E10.5 Hoxa1-IRES-Cre; ROSA-EYFP embryos and hindbrain flatmounts revealed that Hoxa1-lineage is present in the caudal half of rhombomere 3 (Fig. 2). Since Hoxa1 is only transiently expressed in its most anterior domain at a time before rhombomere boundaries have formed it has been difficult to determine its precise anterior expression border in the hindbrain. By examining the distribution of the Hoxa1-lineage marker at later embryonic stages when rhombomere boundaries are present, we were able to conclude that the anterior expression border of Hoxa1 is in the caudal half of the future r3 territory. Previous studies have described the anterior border of Hoxa1 expression to coincide with the future r3/r4 boundary (Barrow et al., 2000; Murphy and Hill, 1991) and put forward that the r3 defects seen in Hoxa1 mutants are due to an indirect effect from loss of Hoxa1 in prospective r4 (Helmbacher et al., 1998). However, our new findings suggest that Hoxa1 plays a direct role in the development of r3. This is supported by a recent study in zebrafish, which proposes a cell-autonomous involvement of Hox paralogous group 1 proteins in regulating r3 development (Wassef et al., 2008). Interestingly, the respiratory rhythm generator is induced by cells in r3 in the chick embryo (Coutinho et al., 2004). The same rhythm generator exists in mice (Champagnat and Fortin, 1997), and it is tempting to speculate that Hoxa1 might be necessary for proper specification of the rhythm inducing cells in r3.
Hoxa1-lineage gives rise to r4-NCCs and sensory neurons of specific cranial ganglia but not to BA2 myocytes
In Hoxa1 knockout mice, mispatterning of the hindbrain results in a size reduction of r4 and in the number of NCCs migrating from r4 into BA2 (Chisaka et al., 1992; Lufkin et al., 1991). Subsequently neural crest derivatives such as the stapes bone of the inner ear are missing and specific hindbrain ganglia are smaller. Hoxa1 is expressed in the neural tube in presumptive r4 shortly prior to the delamination of NCCs and it was suggested that Hoxa1 might specify gangliogenic neural crest cell precursors in the neural tube (Lufkin et al., 1991). In this study, we show that all r4-neural crest cells are derived from Hoxa1 expressing cells, suggesting that Hoxa1 is expressed in the entire pool of neural crest precursors and is not restricted to a specific subpopulation. This result does not rule out a role for Hoxa1 in neural crest development, but it demonstrates that its expression in r4-NC is not restricted and that it is therefore unlikely to specify a certain precursor population.
We also asked whether mesodermal-derived muscle cells in the second branchial arch are labeled by the Hoxa1-lineage. Hoxa1 is expressed in the presomitic mesoderm, but it was unclear if it is also expressed in cells of the cranial paraxial mesoderm, which migrate into the core of BA2 and give rise to muscles of the face and neck. Hoxa1/b1 double knockout mice lack BA2 and its mesodermal derivatives (Gavalas et al., 1998; Gavalas et al., 2001; Rossel and Capecchi, 1999), which raised the hypothesis that Hoxa1 might play a redundant role with Hoxb1 in pattering of BA2 mesoderm (Morrison, 1998). Our lineage analysis showed no contribution of Hoxa1-lineage to BA2 myocytes, demonstrating that Hoxa1 does not play a direct role in the development of BA2 mesoderm. Instead, the loss of BA2 mesodermal derivatives in the double knockout are likely secondary due to the absence of r4-NCCs that normally migrate into the branchial arch and that have been shown to play an instructive role in patterning of muscle tissue (Kontges and Lumsden, 1996).
Finally, we demonstrate that Hoxa1-lineage gives rise to sensory neurons of the petrosal (G9) and nodose (G10) ganglia, both of which are reduced in Hoxa1−/− embryos, but not to the geniculate (G7) ganglion, which is unaffected in the mutant (Mark et al., 1993). Our findings correlate with the reported phenotypes and suggest a selective expression of Hoxa1 in the petrosal and nodose but not the geniculate placode, even though all three placodes are derived from the epibranchial placode (Baker and Bronner-Fraser, 2001). Although Hoxa1-lineage does not give rise to neurons of the geniculate ganglion, we observed that these neurons send their projections to the neural tube along lineage-derived glia cells (Fig. 4L′). This fits with the finding that placodal derived neurons are guided to the hindbrain by tracks formed by NC-derived glia cells (Begbie and Graham, 2001). Surprisingly, we detect a strong contribution of Hoxa1-lineage to neurons of the otic placode-derived vestibular ganglion (G8), which suggests that Hoxa1 is expressed in the otic placode.
Hoxa1-lineage is found in the developing inner ear and is excluded from sensory regions
Our results show a strong contribution of Hoxa1-lineage to the developing inner ear. This was very surprising since no Hoxa1 expression has been reported in the precursor of the inner ear in previous studies (Mark et al., 1993; Murphy and Hill, 1991). Since all cellular components of the inner ear derive from the embryonic otic placode (except a minor contribution from melanocytes) (Torres and Giraldez, 1998), our results indicate that Hoxa1 is expressed in the otic epithelium.
Especially interesting was the absence of the lineage from an anterodorsolateral region corresponding to a sensory patch. Many genes which play a role in inner ear patterning (Pax2, Dlx3, Nkx5.1, Bmp4) are initially expressed ubiquitously at the placode and otic cup stage but then display more restricted expression domains in the otic vesicle (Bok et al., 2007; Torres and Giraldez, 1998). Therefore, it is intriguing that Hoxa1-lineage shows a restricted pattern, although it was seen as early as the otic cup stage. This suggests that Hoxa1 expression is already restricted prior to otic cup formation and therefore partitions the ear into lineage-restricted compartments at an early stage.
To date, the defects in the development of the inner ear in Hoxa1 mutants have been attributed solely to indirect effects. Two possibilities were proposed: (i) mispatterning of the hindbrain alters the positional specification of the otic placode and/or (ii) Hoxa1 is necessary to induce hindbrain signals important for inner ear development such as Fgf3 (Hatch et al., 2007; Hogan and Wright, 1992). Our findings now raise the possibility that Hoxa1 might instead or additionally play a direct role in early regional patterning of the otic epithelium. These findings will require further investigation to determine if Hoxa1’s role in inner ear development is restricted to the hindbrain or if it plays a direct role in the otic ectoderm.
To our knowledge this is the first Cre-driver that is active at the otic cup stage but is absent from future sensory regions. Therefore, the Hoxa1-IRES-Cre line provides a useful tool for the assembly of a temporal and spatial map of the developing sensory regions and for conditional gene inactivation in the inner ear.
Hoxa1-lineage in the outflow tract and atrium of the heart
Studies on patients with truncating mutations in HOXA1 identified a role for Hoxa1 in development of the cardiovascular system (Bosley et al., 2008; Tischfield et al., 2005). Human patients display defects of the outflow tract and internal carotid arteries (Tischfield et al., 2005), both of which are remodeled by cardiac neural crest cells, which delaminate from r6-r8 (Brown and Baldwin, 2006; Kirby and Waldo, 1995; Snider et al., 2007). Hoxa1 is expressed in the neural tube at this level and we find that indeed all cardiac neural crest cells in the outflow tract are derived from Hoxa1 expressing cells. This suggests that Hoxa1 might play a direct role in development of cardiac NCCs or their precursors, which could be the reason for the outflow tract defects in humans with mutations in HOXA1. However, Hoxa1-lineage is also present in the myocardium and endocardium of the outflow tract and might influence OF development in these tissues. The question in which tissue Hoxa1 is required for proper OF development can only be determined by inactivating the gene in specific precursor populations.
Additionally, we saw Hoxa1-lineage in the atria but not the ventricles of the heart. This demonstrates that Hoxa1 is not only expressed in cardiac neural crest but also in a subset of myocardial precursors, which was surprising, since no Hoxa1 expression had been detected in cardiac tissue in previous studies (Godwin et al., 1998; Ryckebusch et al., 2008). The regional restriction of Hoxa1-lineage to the atria is reminiscent of genes that play a role in patterning of the heart tube, such as Gata4, -5, -6 and Tbx5 (Bruneau et al., 1999; Jiang et al., 1998). Therefore, it is possible that Hoxa1 (redundantly with other genes) might play a role in craniocaudal patterning of the heart.
In conclusion, we demonstrate that Hoxa1-lineage is present in r3, the inner ear and the heart, all tissues which were not thought to be derived from Hoxa1-expressing cells. Therefore, our study opens up new avenues for further investigations on the role of Hoxa1 in these tissues.
Acknowledgments
We thank B. R. Arenkiel for generation of the Hoxa1-ICN targeting vector and members of our tissue culture and mouse facility, in particular S. Barnett, C. Lenz and K. Lustig for ES cell culture, injection and mouse care. We thank P. Tvrdik for the Hoxa1 cDNA clone and C. Goridis, J. P. Brunet and M. Wegner for antibodies. This manuscript was improved by helpful comments from A. M. Boulet, L. C. Murtaugh and D. Kopinke, whom we additionally thank for help and support throughout this study. N. Makki was supported by the Boehringer Ingelheim Fonds PhD fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–56. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–86. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M, Le Douarin NM, Teillet MA. Early- and late-migrating cranial neural crest cell populations have equivalent developmental potential in vivo. Development. 1997;124:3077–87. doi: 10.1242/dev.124.16.3077. [DOI] [PubMed] [Google Scholar]
- Barlow LA. Cranial nerve development: placodal neurons ride the crest. Curr Biol. 2002;12:R171–3. doi: 10.1016/s0960-9822(02)00734-0. [DOI] [PubMed] [Google Scholar]
- Barrow JR, Stadler HS, Capecchi MR. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000;127:933–44. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- Begbie J, Graham A. Integration between the epibranchial placodes and the hindbrain. Science. 2001;294:595–8. doi: 10.1126/science.1062028. [DOI] [PubMed] [Google Scholar]
- Bok J, Chang W, Wu DK. Patterning and morphogenesis of the vertebrate inner ear. Int J Dev Biol. 2007;51:521–33. doi: 10.1387/ijdb.072381jb. [DOI] [PubMed] [Google Scholar]
- Bosley TM, Alorainy IA, Salih MA, Aldhalaan HM, Abu-Amero KK, Oystreck DT, Tischfield MA, Engle EC, Erickson RP. The clinical spectrum of homozygous HOXA1 mutations. Am J Med Genet A. 2008;146A:1235–40. doi: 10.1002/ajmg.a.32262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosley TM, Salih MA, Alorainy IA, Oystreck DT, Nester M, Abu-Amero KK, Tischfield MA, Engle EC. Clinical characterization of the HOXA1 syndrome BSAS variant. Neurology. 2007;69:1245–53. doi: 10.1212/01.wnl.0000276947.59704.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CB, Baldwin HS. Neural crest contribution to the cardiovascular system. Adv Exp Med Biol. 2006;589:134–54. doi: 10.1007/978-0-387-46954-6_8. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–8. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Hox genes and mammalian development. Cold Spring Harb Symp Quant Biol. 1997;62:273–81. [PubMed] [Google Scholar]
- Champagnat J, Fortin G. Primordial respiratory-like rhythm generation in the vertebrate embryo. Trends Neurosci. 1997;20:119–24. doi: 10.1016/s0166-2236(96)10078-3. [DOI] [PubMed] [Google Scholar]
- Chisaka O, Musci TS, Capecchi MR. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–20. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- Coutinho AP, Borday C, Gilthorpe J, Jungbluth S, Champagnat J, Lumsden A, Fortin G. Induction of a parafacial rhythm generator by rhombomere 3 in the chick embryo. J Neurosci. 2004;24:9383–90. doi: 10.1523/JNEUROSCI.2408-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–6. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development. 2003;130:3807–19. doi: 10.1242/dev.00573. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–36. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128:3017–27. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- Godwin AR, Stadler HS, Nakamura K, Capecchi MR. Detection of targeted GFP-Hox gene fusions during mouse embryogenesis. Proc Natl Acad Sci U S A. 1998;95:13042–7. doi: 10.1073/pnas.95.22.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–45. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development. 2007;134:3615–25. doi: 10.1242/dev.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbacher F, Pujades C, Desmarquet C, Frain M, Rijli FM, Chambon P, Charnay P. Hoxa1 and Krox-20 synergize to control the development of rhombomere 3. Development. 1998;125:4739–48. doi: 10.1242/dev.125.23.4739. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–90. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hogan B, Wright C. Developmental biology. The making of the ear. Nature. 1992;355:494–5. doi: 10.1038/355494a0. [DOI] [PubMed] [Google Scholar]
- Holve S, Friedman B, Hoyme HE, Tarby TJ, Johnstone SJ, Erickson RP, Clericuzio CL, Cunniff C. Athabascan brainstem dysgenesis syndrome. Am J Med Genet A. 2003;120:169–73. doi: 10.1002/ajmg.a.20087. [DOI] [PubMed] [Google Scholar]
- Iimura T, Pourquie O. Hox genes in time and space during vertebrate body formation. Dev Growth Differ. 2007;49:265–75. doi: 10.1111/j.1440-169X.2007.00928.x. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rose DW, Rosenfeld MG. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–63. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–16. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Tarzami S, Burch JB, Evans T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev Genet. 1998;22:263–77. doi: 10.1002/(SICI)1520-6408(1998)22:3<263::AID-DVG8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Jostes B, Walther C, Gruss P. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech Dev. 1990;33:27–37. doi: 10.1016/0925-4773(90)90132-6. [DOI] [PubMed] [Google Scholar]
- Jukkola T, Trokovic R, Maj P, Lamberg A, Mankoo B, Pachnis V, Savilahti H, Partanen J. Meox1Cre: a mouse line expressing Cre recombinase in somitic mesoderm. Genesis. 2005;43:148–53. doi: 10.1002/gene.20163. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–5. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–5. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–42. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–19. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–8. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–15. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev Biol. 2005;277:155–69. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–6. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, Gorry P, Lumsden A, Chambon P. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–38. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Jozefowicz C, Assimacopoulos S, Grove EA, Louvi A, Prince VE. Conserved expression of Hoxa1 in neurons at the ventral forebrain/midbrain boundary of vertebrates. Dev Genes Evol. 2003;213:399–406. doi: 10.1007/s00427-003-0335-7. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–19. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- Morrison AD. 1 + 1 = r4 and much much more. Bioessays. 1998;20:794–7. doi: 10.1002/(SICI)1521-1878(199810)20:10<794::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96:737–49. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- Mujtaba T, Mayer-Proschel M, Rao MS. A common neural progenitor for the CNS and PNS. Dev Biol. 1998;200:1–15. doi: 10.1006/dbio.1998.8913. [DOI] [PubMed] [Google Scholar]
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–9. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–75. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–40. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–40. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008;105:2913–8. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider P, Olaopa M, Firulli AB, Conway SJ. Cardiovascular development and the colonizing cardiac neural crest lineage. Scientific World Journal. 2007;7:1090–113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2005;37:1035–7. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Torres M, Giraldez F. The development of the vertebrate inner ear. Mech Dev. 1998;71:5–21. doi: 10.1016/s0925-4773(97)00155-x. [DOI] [PubMed] [Google Scholar]
- Wang M, Drucker DJ. The LIM domain homeobox gene isl-1: conservation of human, hamster, and rat complementary deoxyribonucleic acid sequences and expression in cell types of nonneuroendocrine lineage. Endocrinology. 1994;134:1416–22. doi: 10.1210/endo.134.3.7907017. [DOI] [PubMed] [Google Scholar]
- Wassef MA, Chomette D, Pouilhe M, Stedman A, Havis E, Desmarquet-Trin Dinh C, Schneider-Maunoury S, Gilardi-Hebenstreit P, Charnay P, Ghislain J. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development. 2008;135:3369–78. doi: 10.1242/dev.023614. [DOI] [PubMed] [Google Scholar]
- Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–17. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–86. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]