Abstract
Streptavidin binds biotin-conjugates with exceptional stability, but dissociation does occur and can be limiting in imaging, DNA amplification, and nanotechnology. We identified a mutant streptavidin, which we call traptavidin, showing ~10-fold slower biotin off-rate, increased mechanical strength, and improved thermostability; this resilience should find diverse applications. We show that the motor protein FtsK could strip proteins from DNA, rapidly displacing streptavidin from biotinylated DNA; traptavidin resisted displacement and thus indicated the force generated by FtsK translocation.
The remarkably strong interaction between the small molecule biotin and the proteins streptavidin or (neutr)avidin is widely exploited in biological research1. Biotin-binding proteins have been isolated from a wide range of species but streptavidin shows the most stable binding to biotin-conjugates1. Streptavidin is used in imaging, protein purification and nano-assembly, while also showing success in cancer clinical trials. Streptavidin and biotin have low non-specific binding, while biotinylation generally does not disrupt biomolecule function. Alternative targeting methods, such as HaloTag or SNAP-tag, form irreversible covalent bonds to their ligand and are valuable for cellular labeling2. However, these domains do not have the resistance of streptavidin to temperature, pH, or denaturant, and so even though pre-bound ligand will remain attached under these harsh conditions, these covalent-binding proteins may unfold, aggregate and promote non-specific binding. Also, unlike SNAP-tag and HaloTag ligands, biotin can be precisely targeted to proteins in vitro, on cells and in living animals using biotin ligase1. An additional advantage is that a large variety of streptavidin- and biotin-conjugates are commercially available.
Despite its stable binding, the perception that the streptavidin-biotin interaction is essentially irreversible is far from correct. For example, in imaging, low endosomal pH led to dissociation of streptavidin being detected in 2 h, whereas the receptor of interest had a lifetime of ~4 days3. Also, nanoparticle attachment can cause a surprising decrease in streptavidin-biotin stability4; the Kd for a biotinylated peptide increased approximately a million-fold when streptavidin was attached to beads5. In the presence of shear-forces lower than a blood-capillary, streptavidin-coated beads do not attach to a biotinylated surface but instead roll across, with arrests of 20 ms to tens of seconds6. In addition, streptavidin cannot prevent the translocation of molecular motors such as helicases, RNA polymerase or DNA polymerase along DNA7. Streptavidin is used at high temperatures, such as for PCR, BEAMing and 454 DNA sequencing, but DNA has to be bis-biotinylated to reduce dissociation8.
A streptavidin mutant containing a cysteine formed a disulfide with a thiol-linked biotin-conjugate, giving controlled reversibility9, but this mutant only enhances binding to certain biotin-conjugates and in systems unaffected by changing redox, precluding use on cells. We therefore endeavored to engineer a streptavidin mutant that would bind more stably to any biotin-conjugate.
In a highly optimized system, almost any change reduces performance. Over two hundred mutants of streptavidin have been published but none have had improved biotin binding stability1. Streptavidin libraries have been screened for various properties by phage-display and in vitro compartmentalization, yielding, for example, a streptavidin variant with improved desthiobiotin binding, but no pair with as strong binding as wild-type streptavidin-biotin has been identified10, 11. Based upon this literature, we avoided mutations near the ureido or thiophene rings of biotin, which invariably impair binding1, and explored numerous mutations adjacent to the biotin carboxyl and in the L3/4 loop1, 10, 11. We randomized promising residues and evaluated purified proteins according to biotin-4-fluorescein off-rate, finding the lowest off-rate for the S52G R53D mutant of streptavidin (Fig. 1a), which we termed traptavidin. We hypothesize that the mutations in traptavidin reduce flexibility of the L3/4 loop (residues 45–50, Fig. 1a). Upon biotin binding, this loop becomes ordered and closes over the biotin-binding pocket12. A more ordered loop may reduce the entropic cost of biotin binding and inhibit dissociation, while decreasing the on-rate and enhancing thermostability.
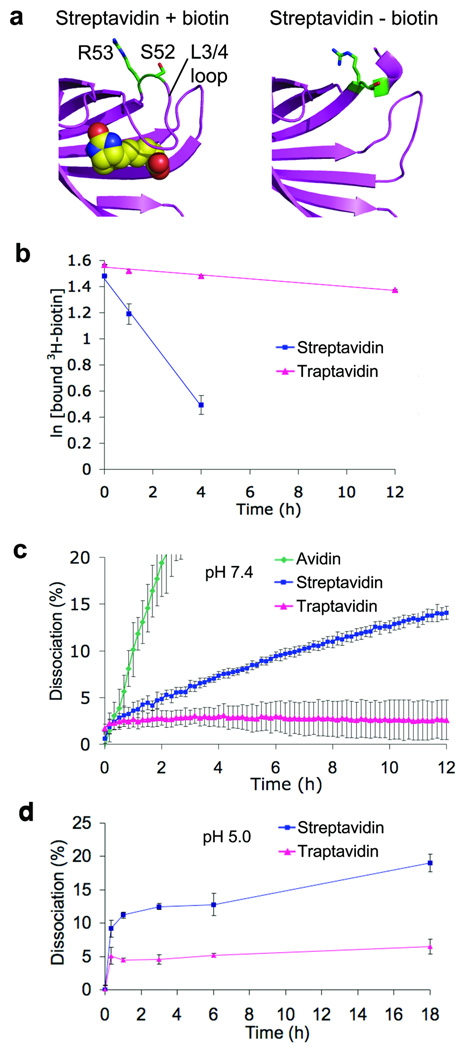
Figure 1. Traptavidin exhibits a slower off-rate from biotin-conjugates.
(a) PyMOL images of residues mutated to produce traptavidin (S52G R53D) in structures of streptavidin with biotin (1mk5) and without biotin (1swa). Biotin is shown as spheres of van der Waals radius. S52 and R53 are colored green. Without biotin the L3/4 loop is disordered and does not give clear electron density.
(b) Traptavidin exhibits a slower off-rate to biotin. Streptavidin and traptavidin were bound to 3H-biotin; excess non-radioactive biotin was then added and the fraction of 3H-biotin still bound was determined after varying times at 37 °C and pH 7.4. Mean of triplicate measurements ± 1 s.d. (Some error bars are too small to be visible.)
(c) Traptavidin exhibits a slower off-rate to a biotin-conjugate at neutral pH. Avidin, streptavidin and traptavidin bind to biotin-4-fluorescein, quenching its fluorescence. Upon addition of excess free biotin, biotin-4-fluorescein dissociates and the increase in fluorescence is observed at 37 °C and pH 7.4. Mean of triplicate measurements ± 1 s.d.
(d) Traptavidin exhibits a slower off-rate at weakly acidic pH. Streptavidin and traptavidin were analyzed as in (c) at 37 °C and pH 5.0. Mean of triplicate measurements ± 1 s.d.
The off-rate for free biotin at 37 °C and pH 7.4 was > 10-fold lower for traptavidin than streptavidin (4.2 ± 0.5 × 10−6 s−1 for traptavidin, 6.8 ± 0.3 × 10−5 s−1 for streptavidin, Fig. 1b). Since a substantial part of (strept)avidin’s binding energy comes from interaction with the carboxyl group of biotin1, it is important to establish how derivation at the carboxyl group changes binding strength. Traptavidin also showed a dramatically reduced off-rate to biotin-conjugates (Fig. 1c, P = 0.0008). After the ~2% dissociation at the initial time-point, there was little dissociation from traptavidin over the subsequent 12 h. In contrast, streptavidin dissociated steadily, while avidin dissociated even faster than streptavidin1. At pH 5, traptavidin dissociation was faster than at pH 7.4 but was still significantly slower than streptavidin (P = 0.001) (Fig. 1d). The on-rate of traptavidin for biotin-4-fluorescein was reduced two-fold, from 2.0 ± 0.1 × 107 M−1 s−1 for streptavidin to 1.0 ± 0.03 × 107 M−1 s−1 for traptavidin (P = 0.004), while the on-rate of traptavidin for 3H-biotin was also reduced (Supplementary Fig. 1). The slower on-rate of traptavidin means that longer incubations are required to reach equilibrium.
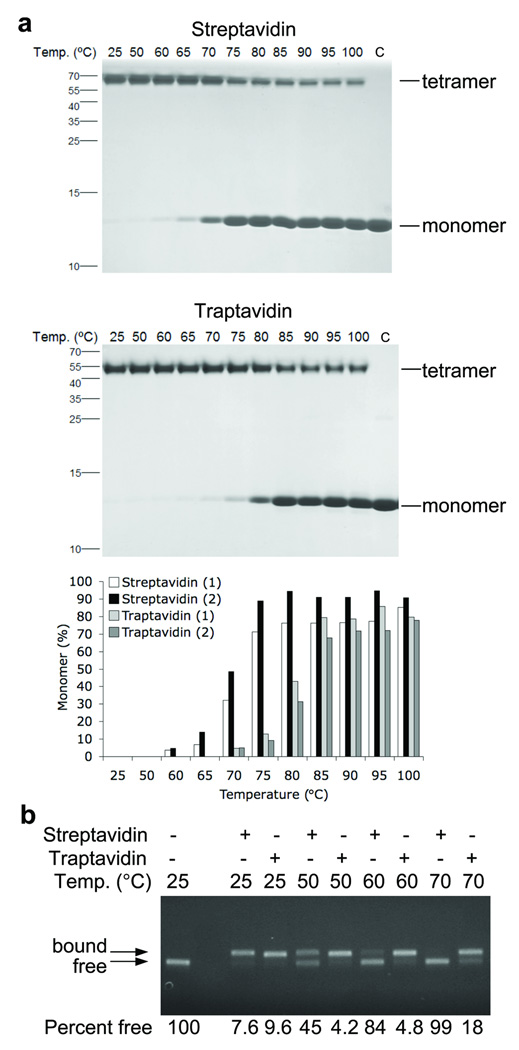
Streptavidin is often used at high temperature. Traptavidin had increased thermostability compared to streptavidin before splitting into monomers; the midpoint of transition was ~10 °C higher (Fig. 2a). We also assessed biotin-conjugate binding stability at elevated temperatures (Fig. 2b). At 70 °C there was complete dissociation from streptavidin but most ligand was still bound to traptavidin.
Figure 2. Traptavidin shows increased thermostability.
(a) Thermostability of tetramer structure. Streptavidin (upper panel) or traptavidin (middle panel) were incubated for 3 min at the indicated temperatures before SDS-PAGE and Coomassie staining. The positive control (C) was mixed with SDS before heating at 95 °C. Tetramer and monomer bands are indicated. The percentage monomer from duplicate gels is plotted in the lower panel.
(b) Thermostability of biotin-conjugate binding. Streptavidin or traptavidin were incubated with biotinylated DNA and heated for 3 min at the indicated temperature before agarose gel electrophoresis and fluorescent imaging of DNA. The left lane is a negative control with no streptavidin or traptavidin added. Bands corresponding to DNA that is free or bound to streptavidin or traptavidin are marked. The percentage of biotinylated DNA free from streptavidin or traptavidin is labeled under each lane.
Imaging of cell-surface proteins using biotin ligase and streptavidin is rapid and sensitive and the target-protein needs only to be modified with a 15 amino-acid tag. The altered charge of traptavidin may affect non-specific cellular binding, as seen for avidin compared to lower pI mutants1. We investigated whether traptavidin showed similar specificity to streptavidin on mammalian cells. We fused the type 1 insulin-like growth factor receptor (IGF1R) to the acceptor peptide (AP-IGF1R), biotinylated the AP with co-expressed biotin ligase (BirA-ER), and detected biotinylated AP-IGF1R with fluorescently-labeled traptavidin or streptavidin (Supplementary Fig. 2). Traptavidin showed high specificity for imaging, with a strong signal on cells expressing AP-IGF1R and BirA-ER and minimal binding when traptavidin was pre-blocked with biotin. Staining with traptavidin and streptavidin was comparable (Supplementary Fig. 2). However, with shorter staining times, the cell staining was more intense with streptavidin (data not shown), consistent with the slower on-rate of traptavidin.
The relationship between binding stability over time versus resistance to force is complex: force changes the height and landscape of the activation-energy for dissociation. We probed the mechanical strength of traptavidin at the single-molecule level by atomic force microscopy (AFM). Traptavidin had greater mechanical binding stability than streptavidin over a range of loading-rates (P < 0.0001) (Fig. 3a). We observed a distribution of binding strengths (Supplementary Fig. 3a) because of the significance of thermal-fluctuations to traversing the activation-barrier. From the relationship between loading-rate and rupture-force, we were able to estimate the difference in dissociation-rate between streptavidin and traptavidin at a given force (Supplementary Fig. 3b).
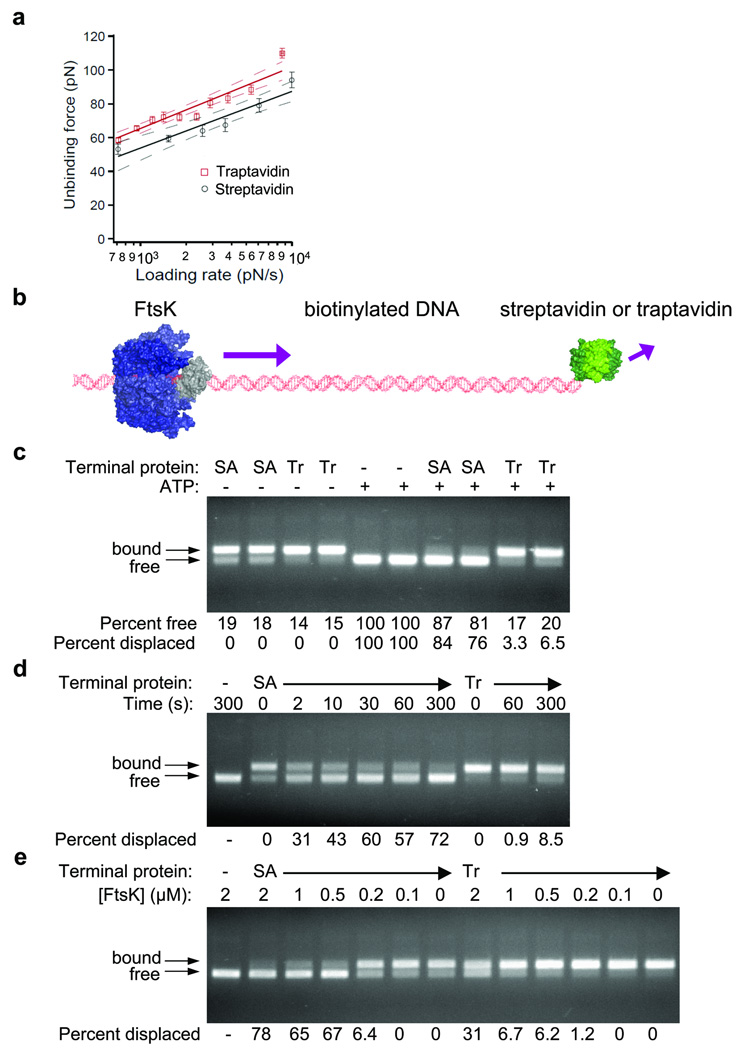
Figure 3. Traptavidin shows increased mechanical stability.
(a) Traptavidin resists AFM displacement. Single-molecule rupture forces between a biotinylated bead and an AFM tip coated with streptavidin (black circles) or traptavidin (red squares), acquired at a range of loading rates. Means are shown ± 1 s.e.m. (streptavidin n = 400, traptavidin n = 562), with a line of best fit (solid) and 95% confidence limits to this line (dashed) in black for streptavidin and in red for traptavidin.
(b) Cartoon of motor assay. DNA, containing a loading site for FtsK (αβ domains in blue, γ in gray) and a biotinylated thymidine near the terminus, was capped with traptavidin or streptavidin (green). In the presence of ATP, FtsK translocates along the DNA and collides with streptavidin or traptavidin.
(c) Traptavidin resists molecular motor displacement. Displacement of streptavidin (SA) or traptavidin (Tr) by FtsK after 180 s was determined by gel electrophoresis, with fluorescent visualization of DNA. FtsK does not remain bound to DNA upon electrophoresis, but bound streptavidin or traptavidin causes the gel shift indicated with arrows. Controls are shown without streptavidin or traptavidin or without ATP, preventing FtsK activity. The percentage of free DNA and the percentage of DNA displaced by FtsK for duplicate assays are indicated under each lane.
(d) Streptavidin is displaced by FtsK on the second time-scale. Streptavidin and traptavidin were incubated with FtsK for the indicated times and analyzed as in (c).
(e) Traptavidin is displaced by a high concentration of FtsK. Streptavidin and traptavidin were incubated for 180 s at the indicated concentration of FtsK and analyzed as in (c).
We applied traptavidin to study FtsK, one of the fastest molecular motor proteins, translocating along DNA at 5 kb/s13, 14. Before bacteria divide, FtsK runs along DNA until encountering XerC and XerD; then FtsK activates site-specific recombination by XerC/D, separating chromosome dimers and ensuring faithful partition of one chromosome to each daughter cell. DNA in vivo is bound to many proteins, including repressors, transcription factors, RNA polymerases and DNA-bending architectural proteins. To study how Pseudomonas aeruginosa FtsK copes with obstacles to its journey, we used a short DNA substrate containing KOPS, an 8 bp sequence that loads FtsK directionally13, 14, with a biotinylated nucleotide near the end, so that FtsK would load on to the DNA in a defined orientation and then translocate until encountering streptavidin or traptavidin (Fig. 3b).
Despite the strength of the streptavidin-biotin interaction, FtsK displaced the majority of streptavidin from the DNA within 3 min, whereas traptavidin resisted displacement (P = 0.003) (Fig. 3c). Streptavidin displacement was detectable after only 2 s, but we observed little displacement of traptavidin even after 300 s (Fig. 3d). Increasing the FtsK concentration to 2 µM allowed substantial displacement of traptavidin (Fig. 3e), indicating that multiple FtsK motors could cooperate in exerting a stronger force. Traptavidin was equally a strong roadblock to Escherichia coli FtsK and with DNA biotinylated at the 5’-terminus rather than at an internal thymidine (Supplementary Fig. 4). Since FtsK rapidly broke the stable biotin-streptavidin interaction, FtsK in vivo should be sufficient to displace even strongly attached DNA-binding proteins.
The stall-force for FtsK measured by pulling on the DNA with optical tweezers was > 65 pN, at which point the DNA double-helix itself is deformed14. Traptavidin displacement should enable testing of higher forces without distorting the DNA. Streptavidin has previously been used as an obstacle to motors7, providing a simpler method to probe force-generation than single-molecule assays13, 14. However, only wild-type streptavidin and the weak nitroavidin have been used7, so traptavidin and the range of weaker streptavidin mutants1 could act as a calibration curve to dissect force-generation by proteins15.
Traptavidin binding was more stable for a range of biotin-conjugates (Supplementary Fig. 5), not just one particular ligand. Also, traptavidin can be recombinantly expressed in comparable yields to streptavidin and recombinant expression of streptavidin gives yields higher than purification from Streptomyces avidinii16. Therefore traptavidin has the potential to replace streptavidin in many applications where dissociation is a limitation, for example when used as a molecular anchor for arrays, surface-plasmon-resonance, or point-of-care diagnostics. Traptavidin-biotin recognition may also aid our understanding of the subtle intermolecular forces that govern interactions of extreme stability.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided by the Wellcome Trust (M.H., D.J.S., E.C.), the Biotechnology and Biological Sciences Research Council (C.E.C.), the National Institutes of Health (V.T.M., C.C.), the National Science Foundation (V.T.M.), the University of Miami (V.T.M.), and Worcester College Oxford (M.H.). We thank J.E. Graham and I. Grainge for reagents and helpful discussion.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
C.E.C., M.H., E.C., V.T.M. and C.C. performed the research; M.H., V.T.M. and D.J.S. designed research; all analyzed the data; M.H., D.J.S. and C.E.C. wrote the manuscript.
COMPETING INTERESTS STATEMENT
M.H. and C.E.C. are authors on a patent application regarding the streptavidin variant described in this study (United Kingdom Patent Application No. 0919102.4).
References
- 1.Laitinen OH, Hytonen VP, Nordlund HR, Kulomaa MS. Cell Mol. Life Sci. 2006;63:2992–3017. doi: 10.1007/s00018-006-6288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruneau E, Sutter D, Hume RI, Akaaboune M. J. Neurosci. 2005;25:9949–9959. doi: 10.1523/JNEUROSCI.3169-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swift JL, Heuff R, Cramb DT. Biophys. J. 2006;90:1396–1410. doi: 10.1529/biophysj.105.069526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buranda T, Lopez GP, Keij J, Harris R, Sklar LA. Cytometry. 1999;37:21–31. [PubMed] [Google Scholar]
- 6.Pierres A, Touchard D, Benoliel AM, Bongrand P. Biophys. J. 2002;82:3214–3223. doi: 10.1016/S0006-3495(02)75664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris PD, Tackett AJ, Raney KD. Methods. 2001;23:149–159. doi: 10.1006/meth.2000.1116. [DOI] [PubMed] [Google Scholar]
- 8.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu SC, Ng KK, Wong SL. Proteins. 2009;77:404–412. doi: 10.1002/prot.22446. [DOI] [PubMed] [Google Scholar]
- 10.Aslan FM, Yu Y, Mohr SC, Cantor CR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8507–8512. doi: 10.1073/pnas.0503112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M, Ellington AD. Chem. Biol. 2008;15:979–989. doi: 10.1016/j.chembiol.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag S, Le Trong I, Klumb L, Stayton PS, Stenkamp RE. Protein Sci. 1997;6:1157–1166. doi: 10.1002/pro.5560060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham JE, Sivanathan V, Sherratt DJ, Arciszewska LK. Nucleic Acids Res. 2010;38:72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pease PJ, et al. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 15.Crozat E, et al. EMBO J. 2010 in press. [Google Scholar]
- 16.Gallizia A, et al. Protein Expr. Purif. 1998;14:192–196. doi: 10.1006/prep.1998.0930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.