Abstract
Chordin is the prototype of a group of cysteine-rich domain-containing proteins that bind and modulate signaling of various TGFβ-like ligands. Chordin-like 1 and 2 (CHL1 and 2) are two members of this group that have been described in human, mouse and chick. However, in vivo roles for CHL1 and 2 in early development are unknown, due to lack of loss-of-function analysis. Here we identify and characterize zebrafish, Danio rerio, CHL (Chl). The chl gene is on a region of chromosome 21 syntenic with the area of murine chromosome 7 bearing the CHL2 gene. Inability to identify a separate zebrafish gene corresponding to the mammalian CHL1 gene suggests that Chl may serve roles in zebrafish distributed between CHL1 and CHL2 in other species. Chl is a maternal factor that is also zygotically expressed later in development, and has spatiotemporal expression patterns that differ from, but overlap those of zebrafish chordin (Chd), suggesting differences, but also possible overlap in developmental roles of the two proteins. Chl, like Chd, dorsalizes embryos upon overexpression, and is cleaved by BMP1, which antagonizes this activity. Loss-of-function experiments demonstrate that Chl serves as a BMP antagonist with functions that overlap and are redundant with those of Chd in forming the dorsoventral axis.
Keywords: Chordin-like, Zebrafish, Maternal factor, BMP1, Tolloid, Dorsoventral patterning, BMP antagonist, loss-of-function
Introduction
Modulation of signaling by ligands of the TGFβ superfamily, crucial to morphological events, is accomplished in part by various extracellular antagonists, which include chordin/sog, noggin, follistatin, and the DAN/cerberus family (Massague and Chen, 2000). Chordin (CHD), which is perhaps the best studied of these, acts by non-covalent binding of BMPs 2 and 4 in a latent complex, thus inhibiting BMP interaction with cell surface receptors (Piccolo et al., 1996). CHD has also been shown to bind BMP4-BMP7 heterodimers in the Xenopus system (Piccolo et al., 1997; Piccolo et al., 1996) and probably acts in zebrafish via binding Bmp2-Bmp7 heterodimers (Little and Mullins, 2009). CHD binds BMPs via four cysteine-rich domains (CRs 1–4), (Larrain et al., 2000), and cleavage of CHD downstream of CRs 1 and 3 by BMP1/Tolloid-like proteinases liberates BMPs from the latent complex (Piccolo et al., 1997; Scott et al., 1999). This conserved mechanism of binding and release underlies dorsoventral patterning in organisms ranging from Drosophila to mammals (Blader et al., 1997; Marques et al., 1997; Pappano et al., 2003; Piccolo et al., 1997; Scott et al., 1999). CHD is the prototype of a small group of extracellular proteins containing CRs that possess conserved CXXCXC and CCXXC motifs (Garcia Abreu et al., 2002). Most of the sequence homology existing between members of this protein group, a number of which have been shown to bind BMPs, is confined to the CRs (Garcia Abreu et al., 2002).
CHD-like 1 and 2 (CHL1 and CHL2), members of this group of proteins, each contain three CRs. Both long and short forms of CHL1 have been reported, differing in the lengths of sequences C-terminal to CR3 (Nakayama et al., 2001; Sakuta et al., 2001). The short and long forms have been designated by some as neuralin-1 (Coffinier et al., 2001) and ventroptin (Sakuta et al., 2001), respectively. Both forms bind BMP4 with high affinity, with somewhat lesser affinity shown by the short form for BMPs 5 and 6, and TGFβs 1 and 2, and no binding shown by the short form for activin (Nakayama et al., 2001; Sakuta et al., 2001). Lesser affinity is shown by the long form for BMP4/7 heterodimers, and no binding by the long form was detected for BMP7, TGFβ1, or activin A (Nakayama et al., 2001; Sakuta et al., 2001). Consistent with the binding data, both short and long forms induce secondary axes when overexpressed in Xenopus embryos (Nakayama et al., 2001; Sakuta et al., 2001). CHL1 RNA was not detected in 7-dpc mouse embryos (Nakayama et al., 2001), but was found in neural plate at late gastrulation (Coffinier et al., 2001), in somites at 9.5-dpc; and thereafter in presumptive and hypertrophic cartilage, neural crest cells and their derivatives, mesenchymal/stromal cells throughout the body, developing CNS, and in several adult organs (Coffinier et al., 2001; Kosinski et al., 2007; Nakayama et al., 2001). In chick, the long form was found in ventral retina, where it may help control topographic retinotectal projections by establishing dorsoventral and anteroposterior gradients of inhibition to signaling by BMP4 and related ligands (Sakuta et al., 2001).
CHL2 reportedly binds BMPs 2 and 4–7 and GDF5, but not activin A or TGFβs 1–3; blocks induction of osteogenic differentiation by BMPs 2, 4, 6 and 7; and induces secondary dorsal axes upon overexpression in Xenopus embryos (Nakayama et al., 2004). It has conversely been reported to bind activin A, but not BMP 2, 4 or 6 (Oren et al., 2004), although the latter study employed a slightly longer variant of CHL2, which may explain differences in reported CHL2 affinities. The slightly longer CHL2 variant has also been designated breast tumor novel factor 1 (BNF-1) and is over-expressed in tumors of human breast, lung and colon (Wu and Moses, 2003). CHL2 is also expressed in developing articular cartilage, suggesting a role in joint specification; it has been shown able to inhibit chondrocyte hypertrophic differentiation and formation of cartilaginous mineralized extracellular matrix; and it may play a role in regenerating osteoarthritic cartilage (Nakayama et al., 2004). CHL2 RNA has also been found in uterus and colon, connective tissues associated with reproductive organs, osteoblasts, epithelial cells of reproductive organs and bladder, and in other mouse and human soft tissues (Nakayama et al., 2004; Oren et al., 2004; Wu and Moses, 2003).
In the above studies, CHL 1 and 2 spatial and temporal expression profiles clearly differed from those of CHD, suggesting functional differences. Precise in vivo functions of the CHL proteins remain largely unknown, due in part to the lack of loss-of-function data for either protein. Here we identify zebrafish CHL (Chl), which has sequences and a protein domain structure similar to those of CHL1 and CHL2. Overexpression analysis shows that Chl, like zebrafish CHD (Chd), is capable of dorsalizing zebrafish embryos, although Chl-induced dorsalization is milder than that caused by Chd. In addition, it is shown that Chl and CHL1, like Chd, can be cleaved by BMP1, the expression of which leads to rescue of embryos dorsalized by Chl overexpression. Temporal and spatial expression patterns of Chl are shown to differ from, but to overlap those of Chd, consistent with differences and with the possibility of similarities in their developmental roles. Chl is shown to be both a maternal factor and zygotically expressed. Loss-of-function experiments demonstrate Chl function, including that of zygotically produced Chl, to overlap that of Chd in formation of the dorsoventral axis.
Materials and Methods
Identification of Chl Sequences
Mouse CHL2 amino acid sequences were used in a protein BLAST search of the Danio rerio genome database on the NCBI assembled genomes site, resulting in identification of a zebrafish “chordin-like 2” protein (GenBank accession no. XP_691539.2) predicted from automated computational analysis from sequences found on Chromosome 21 at location 19,863,200–19,876,300. A TBlastN protein vs translated DNA search of the zebrafish Zv8 whole genome assembly on the Sanger Institute site using mammalian CHL1 and CHL2 protein query sequences, identified the same gene on Chromosome 21, and no additional chordin-like genes. Although an Ensembl search of the Zv8 database with the term “chordin-like” gave two theoretical proteins, “chordin-like” and “chordin-like protein 2”, both of these are based on alternative predictions of intron/exon junctions of the same chl gene on chromosome 21, and no chordin-like proteins from additional genes were identified. Primers 5′-GAGAAGACGTACAGGCCAGGAGAT-3′ (forward) and 5′-GGGATGTTGGCACGGATACTGGTT-3′ (reverse), corresponding to predicted sequences, were used to PCR amplify a 750-bp fragment, using cDNA from total RNA of 48 hpf zebrafish as template. 5′ and 3′ RACE were then employed to obtain additional 5′ and 3′ cDNA sequences, using as template an adult zebrafish cDNA library (Open Biosystems). For 5′ RACE, a primary reaction was with vector primer (VP) 5′-GCGGATAACAATTTCACACAGGAAACA-3′ and gene specific primer (GSP) 5′-GGGATGTTGGCACGGATACTGGTT-3′; and 5μl of the product from this PCR was used as template for the second nested reaction with VP 5′-GCTATGACCATTAGGCCTATTTAGGT-3′ and GSP 5′-ACTGACGGGTTGGGAATAGCTCAT-3′. For 3′ RACE, a primary reaction was with GSP 5′-GAGAAGACGTACAGGCCAGGAGAT-3′ and VP 5′-CAGTTTTCCCAGTCACGACGTTGT-3′; and 5 μl of this product was used as template for the second nested reaction with GSP 5′-GATAGTTGGCATCCCTATTTGGAAC-3′ and VP 5′-AAACGACGGCCAGTGCCTAGCTTA-3′. 2,123 bp cDNA sequences obtained in the above manner, including full-length coding sequences, 61 bp of 5′ UTR and 697 bp of 3′ UTR; have been deposited with the EMBL/GenBank Data libraries under accession no. EU030433.
RT PCR
Embryos were collected from various stages (8 cell to 48 hpf) followed by total RNA extraction using Trizol Reagent (Invitrogen). SuperScriptIII (Invitrogen) reverse transcriptase was used to synthesize cDNA from 1μg of total RNA per stage. PCR was performed with primers 5′CGCATCTGTGCACGGATCGAGACTC 3′ (forward) and 5′CAGCCAAGAGTCTGTCACACCCGTC 3′ (reverse), for Chd; 5′GGTGATGACCTGGGAGCAAATGATG 3′ (forward) and 5′GACAAGTGTCCTGGTGTGGGAGATC 3′ (reverse), for Tcf3; and 5′GAGAAGACGTACAGGCCAGGAGAT 3′ (forward) and 5′GGGATGTTGGCACGGATACTGGTT 3′ (reverse), for Chl. Resulting amplicon sizes were 451-bp (Tcf3), 493-bp (Chd), and 806-bp (Chl). For the experiment in which specificity of the splice site MO SP-MO was tested, RNA was extracted from embryos treated with 0.5 mM SP-MO, and PCR was performed with primers 5′GAGAAGACGTACAGGCCAGGAGAT3′ (forward) and 5″CTTTGCACACTAGGCAGCAAGTATC3′ (reverse).
Morpholino (MO) Injections
Chl MOs were obtained from Gene Tools. A MO targeted against the Chl translation start site (TrSt-MO) was 5′ CAGGTCATTCTTTTCTTCATATTCC 3′, whereas a MO targeted against chl 5′-UTR sequences (5′1-MO) was 5′AAAAACTGGAGACGCAGGATTTGTC 3′. An RNA splice disrupting MO (SP-MO) 5′ GCTCATCTGATGATAGGAATATAC 3′, targets the exon 4 acceptor splice site. A standard control MO (StCon-MO) 5′ CCTCTTACCTCAGTTACAATTTATA 3′ has no target and no biological activity in zebrafish (Gene Tools). MOs were resuspended in sterile double distilled water at a concentration of 2 mM, diluted to the desired concentrations prior to injection, and injected with 0.1% phenol red tracer into one-cell stage embryos. Injection volume was 1 nl.
Chl - Chd Synergy and Rescue
A previously described Chd MO (Nasevicius and Ekker, 2000), diluted to a concentration (0.026 mM) that exhibited a mild ventralized phenotype when injected alone, was coinjected with 0.1 mM of the Chl 5′1-MO or with a subthreshold concentration of SP-MO (0.5 mM) into 1–2 cell stage embryos. Embryos were photographed and quantified at 24 hpf. Rescue experiments of the additive effects of Chd-MO combined with Chl 5′1 MO were performed using mouse CHL2, mouse CHL1-Fc, and Chl RNA. This was executed by coinjecting 0.026 mM of the Chd-MO combined with 0.1 mM of the 5′1-MO and 1 pg of mouse CHL2 RNA or with 80 pg or 1 ng of the mouse CHL1-Fc and Chl RNA constructs, respectively.
RNA Injections
Full-length Chl sequences or full-length mouse CHL2 sequences, both with C-terminal FLAG-epitopes and downstream of BM40 signal peptide sequences, were excised from protein expression vectors (see below) and then inserted between the EcoRI and XhoI sites of pCS2+, with partial filling of the EcoRI site with dATP and partial filling of an AflII site at the 5′ end of Chl or mouse CHL2 sequences with dTTP, to create compatible ends. The zebrafish BMP1 (Bmp1a) construct for in vitro expression of RNA has been described previously (Jasuja et al., 2006). To obtain full-length mouse CHL1 sequences with a C-terminal Fc domain (CHL1-Fc), a construct in which CHL1 CR3 domain sequences were cloned between the NheI and NotI sites of pcDNA4 was cut with NotI and EcoO109I, followed by insertion of a linker assembled using oligonucleotides 5′GCCCTAGGTCCCGTACGC3′ and 5′GGCCGCGTACGGGACCTAG3′. The resulting construct was cut with PpuMI and NotI and sequences encoding the murine Fc epitope, obtained from a previously described GDF11-Fc plasmid (Ge et al., 2005) by excision with PpuMI and NotI, were inserted. The resulting construct was then cut with NheI and Bsu15 and an NheI-Bsu15 restriction fragment encoding the remainder of CHL1 sequences was inserted. CHL1-Fc sequences were then cut from the resulting construct with NheI and XhoI and inserted between the NheI and XhoI sites in a pCS2+ vector containing BM40 signal peptide sequences (Scott et al., 2001). Full-length Chd sequences, missing only signal peptide sequences, were PCR amplified in two pieces, using primer sets 5′GCTAGCATCGAGACTCAAGACACCCGCGCT-3′ and 5′TGTGTGTGTCTGGCCTCCACGCGT-3′, for the 5′end, and 5′ACGCGTGGAGGCCAGACACACACA-3′ and 5′CCACTCTAGAGAGTGTCAGTGTCTCCA-3′ for the 3′end. Both PCR products were cloned into pGEM-T (Promega). The 3′end clone was cut with BstXI, treated with T4 DNA polymerase to remove an MluI site in the vector, and then cut with MluI and NotI. The 5′ end clone was cut with Bsp120I and MluI, and the insert band gel-purified and then ligated into the 3′end clone. The resulting construct was cut with NheI and XbaI to release the full-length Chd insert, which was then inserted between NheI and XbaI sites in a version of the CS2+ vector containing the BM40 signal peptide.
BssHII was used to linearize the Chl, mouse CHL1-Fc, mouse CHL2, and Bmp1a plasmids, NotI was used to linearize the Chd plasmid, and capped mRNA was transcribed using the mMessage mMachine SP6 kit (Ambion). Following transcription, capped mRNAs were injected into one to two cell stage embryos with 0.1% phenol red tracer.
Whole-Mount In Situ Hybridization
In situ hybridizations were performed as described previously (Pelegri and Maischein, 1998). The probe used for chd has been previously described (Schulte-Merker et al., 1994). Images were captured using a dissecting microscope (Leica, FLIII) and a color camera (Diagnostic Instruments Spot Insight). Chl probes, corresponding to sequences encoded by exons 1–5, were synthesized from a cDNA insert amplified with primers 5′-CAATGCGGTGCTTTGTTCAGGGTG-3′ (forward) and 5′-CTTTGCACACTAGGCAGCAAGTATC-3′ (reverse). The resulting insert was ligated into the pGEM-T vector (Promega), followed by linearization of the vector with NcoI or NotI and subsequent transcription with SP6 or T7 polymerases for antisense and sense RNA probes, respectively. Transcription employed the MAXIscript kit (Ambion) and digoxigenin-labeled nucleotides (Roche).
Protein Biochemistry
To produce recombinant Chl with a C-terminal FLAG tag, primers GCTATCTAGAAACTGAGGCATTACGACCTGGA (forward) and GATCGCGGCCGCTACTTGTCGTCATCGTCCTTGTAGTCAGATGTGCAAAGACT GTCCAGCTGT (reverse) were used to amplify a full-length Chl 1339-bp PCR product, minus native signal peptide sequences, that was then cut with XbaI and NotI and ligated between NheI and NotI sites of a modified pcDNA4/TO/myc-His vector (Steiglitz et al., 2004), such that CHL sequences were in frame and downstream of the signal peptide of the extracellular matrix protein BM40/SPARC, for optimization of secretion. To produce recombinant Bmp1a with a C-terminal FLAG tag, primers CCAGACCTTCGAAATCGAGGAGGA (forward) and GATGACCGGTCACTTGTCGTCATCGTCCTTGTAGTCCTTCTTACGGGAGTGGA GTGTGT (reverse) were used to PCR amplify a 280-bp fragment using a previously described vector for expression of His-tagged Bmp1a (Jasuja et al., 2006) as template. The 280-bp fragment was then cut with BstBI and AgeI and was ligated into the original Bmp1a-His construct, which had also been cut with BstBI and AgeI, to remove the corresponding fragment. To produce murine CHL2 (Nakayama et al., 2004) and the shorter, neuralin form of murine CHL1 (Coffinier et al., 2001; Nakayama et al., 2001) with C-terminal FLAG tags, primer sets 5′ AGTCAGCTAGCTGAACAAGTAAAACACTCAGACACA 3′ (forward CHL1), 5′GCATGCGGCCGCTACTTGTCGTCATCGTCCTTGTAGTCACCAAAGGCAGGGCCTCCAGCCAA 3′ (reverse CHL1); and 5′AGTCAGCTAGCTCAAGCACTATCCCGCTCGGGCAAA 3′ (forward CHL2), 5′GCATGCGGCCGCTACTTGTCGTCATCCGTCCTTGTAGTCTAATGTCTTGGTCACTTTGTCTGG 3′ (reverse CHL2) were used to amplify full-length CHL1 and CHL2 sequences, minus native signal peptide sequences, from mouse Marathon Ready cDNA (Clontech). These were inserted downstream of the BM40 signal peptide, between NheI and NotI sites of the modified pcDNA4/TO/myc-His vector (Steiglitz et al., 2004). Constructs were transfected into 293 T-Rex cells using LipofectAmine (Invitrogen) followed by selection with blasticidin and zeocin. Cells were treated with tetracycline, placed in serum-free media, and conditioned media was harvested at 24 and 48 h. FLAG-tagged proteins were purified from conditioned media via a FLAG affinity matrix column (Sigma) and concentrations of purified proteins were calculated by comparing intensities of Coomassie-stained bands from serial dilutions of each sample to those of a serially diluted BSA standard of known concentration. In vitro cleavage of Chl with Bmp1a was performed by incubating 200 ng Chl alone or in the presence of Bmp1a. For in vitro cleavage of CHL1 and CHL2, FLAG tagged human BMP1, TLD, TLL1 and TLL2 were prepared and purified as previously described (Scott et al., 1999). 150 ng of CHL1 or CHL2 was incubated overnight at 37 °C alone or in combination with each mammalian proteinase. All in vitro cleavage assays were at a 2:1 substrate to enzyme molar ratio at 37°C overnight in 50 mM CaCl2, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl. Reactions were stopped by adding 10× SDS sample buffer, 5% β-mercaptoethanol and boiling 5 min. Chl, Bmp1a samples were run on a 4–15% acrylamide SDS-PAGE gradient gel, while mammalian cleavage assay samples were run on a 12% SDS-PAGE gel. Electrophoresis was followed by transfer to PVDF membranes (Millipore) and immunoblotting with anti-FLAG antibody diluted 1:5000 for all proteins except for CHL1, which was detected with antibody to full-length CHL1 (R&D Systems) diluted 1:1000.
For amino acid sequence analysis, cleaved CHL1 was electrotransferred to Sequi-Blot membrane (Bio-Rad) and NH2-sequences were determined by automated Edman degradation at the Harvard University Microchemistry Facility using a PerkinElmer Life Sciences/Applied Biosystems Division Procise494 HT Protein Sequencing System.
Chl-GFP Fusion Construct
Sequences encoding green fluorescent protein (GFP) were fused to the C-terminus of Chl. This was done by using primers 5′-TCCAGAGCTCAAACCAGAGAGCAA-3′ (forward) and 5′-GTACCTCGAGTCCATGGATGTGCAAAGACTGTCCAGCTGT-3′ (reverse) to PCR amplify a 450-bp Chl fragment containing novel NcoI and XhoI sites, and then exchanging this SacI and XhoI fragment for the corresponding wild type fragment in the Chl pCS2+ vector (described above). GFP sequences were then inserted into the Chl CS2+ vector between the novel NcoI and XhoI sites. The Chl-GFP pCS2+ construct was linearized with BssHII and capped mRNA was transcribed using the mMessage mMachine SP6 kit (Ambion).
Results
Identification and cloning of zebrafish chordin-like (Chl)
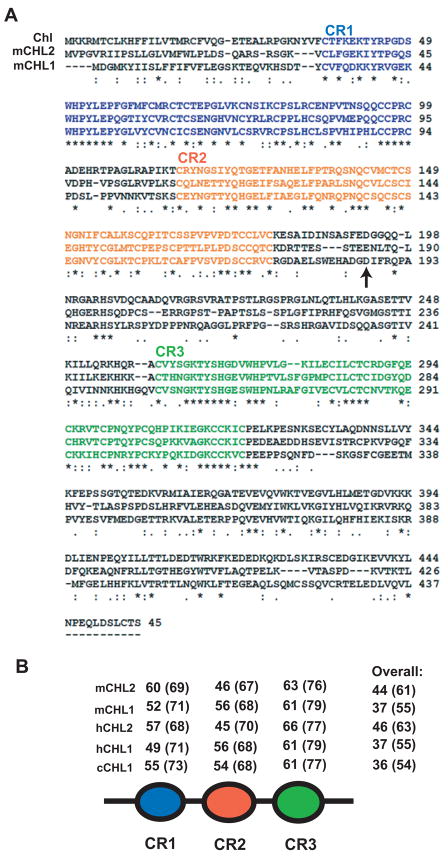
BLAST searches of zebrafish genome databases with mouse CHL2 amino acid sequences found only a single CHL sequence, which like CHL1 and CHL2 possesses three cysteine-rich domains (CRs) (Fig. 1). Searches with mouse CHL1 amino acid sequences, or mammalian CHL1 or CHL2 DNA sequences did not identify additional zebrafish CHL sequences. Alignment of the zebrafish Chl sequence with those of mouse CHL1 and CHL2 (Fig. 1A) revealed pronounced homology between the CRs of the three proteins, with highest levels of homology found between the CR3 domains of the three proteins (Fig. 1B). Homology between non-CR portions of the three proteins was markedly lower. Alignment of the Chl CRs with those of CHL proteins identified in various other species, showed a fair degree of conservation, with homologies ranging between 46–66% identity (Fig. 1B). Overall, Chl was found to be 37% identical/55 % similar to mouse CHL1 and 44% identical/61% similar to mouse CHL2.
Fig. 1.
Alignment of Chl and murine CHL deduced amino acid sequences. (A) Alignment of the Chl amino acid sequence with those of murine (m) CHL1 and CHL2. An arrow denotes the site at which CHL1 is cleaved by BMP1/Tolloid-like proteinases. (B) Schematic of shared CHL protein domain structure and comparison of percents identity and similarity (parentheses) between CRs of Chl and murine (m), human (h) and chick (c) CHL1 and CHL2. Alignment was with CLUSTAL W. Sequences of the longer/ventroptin-α form of CHL1 (Nakayama et al., 2001; Sakuta et al., 2001) and form of CHL2 of similar length (Nakayama et al., 2004), both of which are similar in length to Chl, were used for the alignment.
Chl exhibits a temporal expression profile distinct from, but overlapping that of Chd
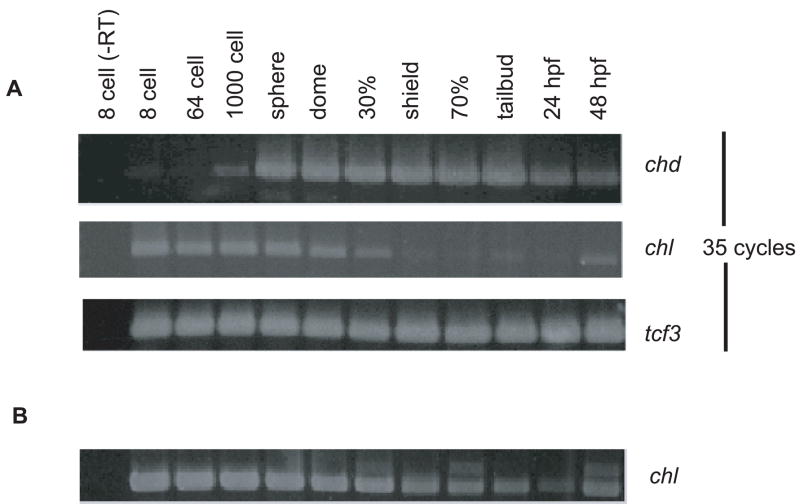
To determine the temporal pattern of Chl developmental expression and compare it to that of Chd, RT-PCR was performed on the RNA from a series of developmental stages, from the 8-cell stage to 48 hours post fertilization (hpf). As can be seen (Fig. 2A), temporal expression patterns for Chl and zChd differ, but overlap. An unexpected finding was the relatively high levels of Chl RNA at the 8- and 64-cell stages (1.25 and 2 hpf, respectively), suggesting Chl to be a maternally transmitted factor, as it appears before the midblastula transition (MBT, 2.75 hpf), at which point transcription of most zygotically expressed genes commences (Kane and Kimmel, 1993; Mathavan et al., 2005). Results also showed Chl RNA to be somewhat reduced by the dome stage and 30% epiboly (4.33 and 4.67 hpf, respectively), and at very much reduced levels by the shield stage (6 hpf) suggesting a fairly rapid turnover of maternal Chl transcripts. A lower level of Chl expression was also detectable from 30% epiboly through the tailbud stage (10 hpf), during the process of gastrulation, which occurs from 50% epiboly through the tailbud stage (Kimmel et al., 1995). Expression levels of Chl are at lowest levels at 24 hpf, but levels increase at 48 hpf, consistent with roles for Chl in later development. RT-PCR was performed for an increased number of cycles (40 cycles) to clearly demonstrate the presence of Chl RNA transcripts from shield stage through 24 hpf (Fig. 2B). Chd RNA transcripts are readily detectable at the 1000-cell stage, followed by maximum expression levels at the sphere stage, prior to the onset of gastrulation, and persisting through the tailbud stage, after which there is a somewhat of a decline in expression levels. These data support the possibility of both distinct and overlapping developmental roles for Chl and Chd.
Fig. 2.
Comparison of the temporal distributions of Chl and Chd expression during embryogenesis. (A) RT-PCR analysis of Chl and Chd RNA levels was performed for harvested embryos from 8-cell to 48 hpf stages. PCR was for 35 cycles. Expression levels of transcription factor 3 (tcf3) RNA are shown as a loading control. (B) RT-PCR was performed using 40 cycles to demonstrate presence of Chl RNA transcripts from shield stage through 24 hpf.
Spatial expression of chl compared to chd
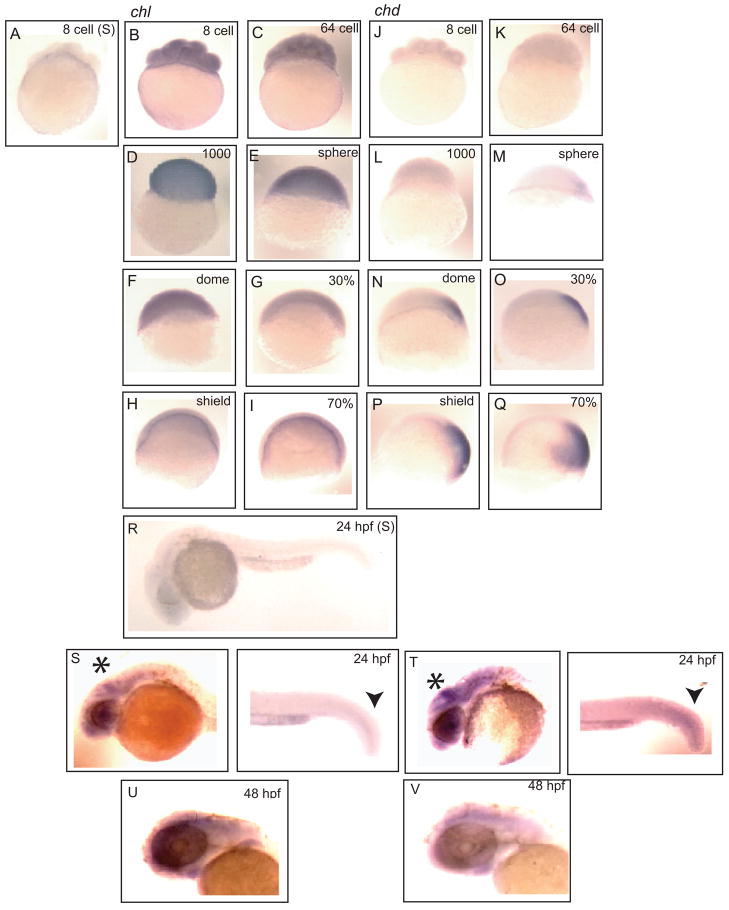
To explore the spatial distribution of Chl during embryogenesis, whole mount in situ hybridization was performed (Fig. 3) with a probe directed against the first five exons of the chl gene. Ubiquitous expression of Chl was observed at high levels throughout early embryogenesis (8-cell – dome stages), with expression levels tapering off at 30% epiboly, and at markedly reduced levels at the shield stage and midgastrulation (70% epiboly). In contrast, and consistent with RT-PCR results, strong Chd expression was readily detectable from sphere stage through 70% epiboly and, as expected (Miller-Bertoglio et al., 1997), was highly expressed throughout gastrulation on the dorsal aspect of the embryo (Fig. 3M – Q), in agreement with its role in dorsoventral patterning. Interestingly, at 24 hpf Chl and Chd showed similar expression patterns, with strongest signal for both localized to mid and hindbrain and tailbud (Fig. 3S and T). At 48 hpf, Chl and Chd signals are still seen to co-localize in brain (Fig. 3U and V).
Fig. 3.
Comparison of the spatial distributions of Chl and Chd expression during embryogenesis. Whole-mount in situ hybridization compares the spatial distribution patterns of Chl (B–I) and Chd expression (J–Q) in 8-cell stage through 70% epiboly embryos. (A) 8-cell embryo hybridized with a sense Chl control probe. Anti-sense probes were hybridized to (B and J) 8-cell, (C and K) 64-cell, (D and L) 1000-cell, (E and M) sphere stage, (F and N) dome stage, (G and O) 30% epiboly, (H and P) shield stage, and (I and Q) 70% epiboly embryos. (R) 24 hpf embryo hybridized with a sense Chl control probe. Signal resulting for hybridization to anti-sense probes for Chl (S and U) and Chd (T and V) is shown for 24 hpf (S and T) and the head regions of 48 hpf (U and V) embryos. Arrowheads mark overlapping Chl and Chd signals in 24 hpf tailbud. Asterisks mark midbrain and hindbrain structures enriched for both Chl (S) and Chd (T) signal at 24 hpf. Similar overlapping signals for Chl (U) and Chd (V) are evident in the brains of 48 hpf embryos.
Chl is a substrate of BMP1/Tolloid-like proteinases
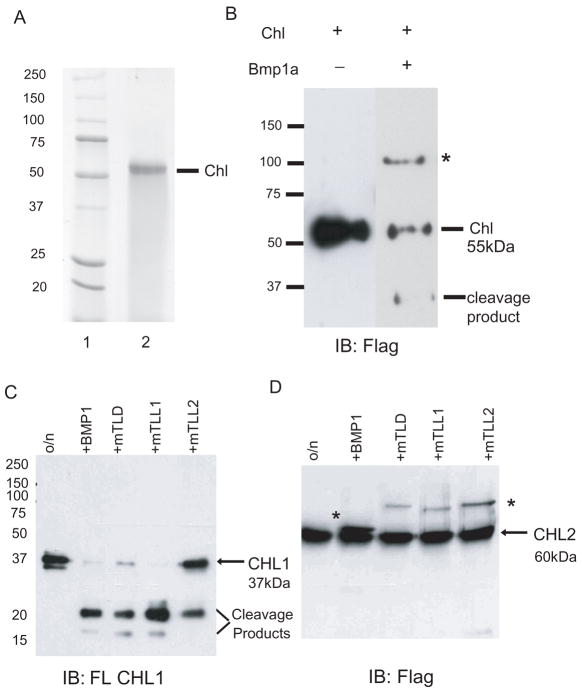
An important aspect of CHD function is its proteolytic cleavage by BMP1/Tolloid-like proteinases, for the activation of bound ligands (Hopkins et al., 2007). To determine whether Chl function might also be regulated by these proteinases, recombinant Chl was prepared and assayed for cleavage by zebrafish Bmp1a (Jasuja et al., 2006; Muraoka et al., 2006). As can be seen, the size of secreted recombinant Chl, purified from conditioned medium is ~55-kDa (Fig. 4A). Upon incubation of the recombinant Chl with Bmp1a in an in vitro cleavage assay (Fig. 4B), immunoblotting with antibody against the C-terminal FLAG epitope detected a single ~30-kDa cleavage fragment. Thus, Chl is indeed a substrate for BMP1/Tolloid-like proteinases, and the size of the cleavage product is consistent with a cleavage event that occurs between CRs 2 and 3. Cleavage sites for BMP1/Tolloid-like proteinases overwhelmingly have aspartate residues in the P1′ position, just N-terminal to the scissile bond (Hopkins et al., 2007). There are four aspartates between Chl CRs 2 and 3, all clustered within the first 28 amino acid residues just C-terminal to CR2 (Fig. 1), and it is probable that BMP1/Tolloid-like proteinases cleave at one or more of these sites.
Fig. 4.
Chl and CHL1 are substrates for BMP1/Tolloid-like proteinases. (A) A Coomassie stained SDS-PAGE gel is shown of protein molecular weight markers (lane 1) and of ~55-kDa secreted and purified Chl (lane 2). (B–D) Immunoblotting with anti-FLAG antibody (B and D) or anti-CHL1 antibody (C) is shown for in vitro assays in which recombinant Chl was incubated in the absence (−) or presence (+) of Bmp1a (B), or in which recombinant short, neuralin form of CHL1 (C) or CHL2 (D) was incubated in the absence (o/n) or presence of mammalian BMP1, mTLD, mTLL1, or mTLL2. Positions of the ~30-kDa Chl (B) and ~20-kDa and smaller CHL1 (C) cleavage products are shown. In B and D, asterisks mark the positions of FLAG-tagged BMP1 and related proteinases. Molecular masses (in kDa) are indicated for protein standards.
To determine whether mammalian CHL1 and CHL2 might be substrates for cleavage by BMP1/Tolloid-like proteinases, recombinant murine CHL1 and CHL2 were prepared and assayed for cleavage by each of the four different mammalian BMP1/Tolloid-like proteinases: BMP1, mammalian Tolloid (mTLD), mammalian Tolloid-like 1 (mTLL1) and mammalian Tolloid-like 2 (mTLL2) (Scott et al., 1999). The short form of CHL1 was used for these studies, as it was found to be markedly more stable, in the absence of added proteinase, than was the long form. As can be seen, three of the four mammalian proteinases readily cleaved CHL1 (Fig. 4C), but none of the four proteinases were observed to cleave CHL2 (Fig. 4D). Enough of the ~20 kDa CHL1 cleavage product was obtained to allow NH2-terminal amino acid sequencing, which gave the sequence DGDIFRQP. This shows cleavage to have occurred at Asp185, just C-terminal to CR2 (Fig. 1), one of several sequences noted by Nakayama et al as potential cleavage sites for BMP1/Tolloid-like proteinases (Nakayama et al., 2001). Thus, although Chl shows more sequence similarity to CHL2 than to CHL1, it is more similar to at least the short form of CHL1 in its ability to serve as a substrate for BMP1/Tolloid-like proteinases.
Chl overexpression results in embryos with a dorsalized phenotype that is counteracted by Bmp1a
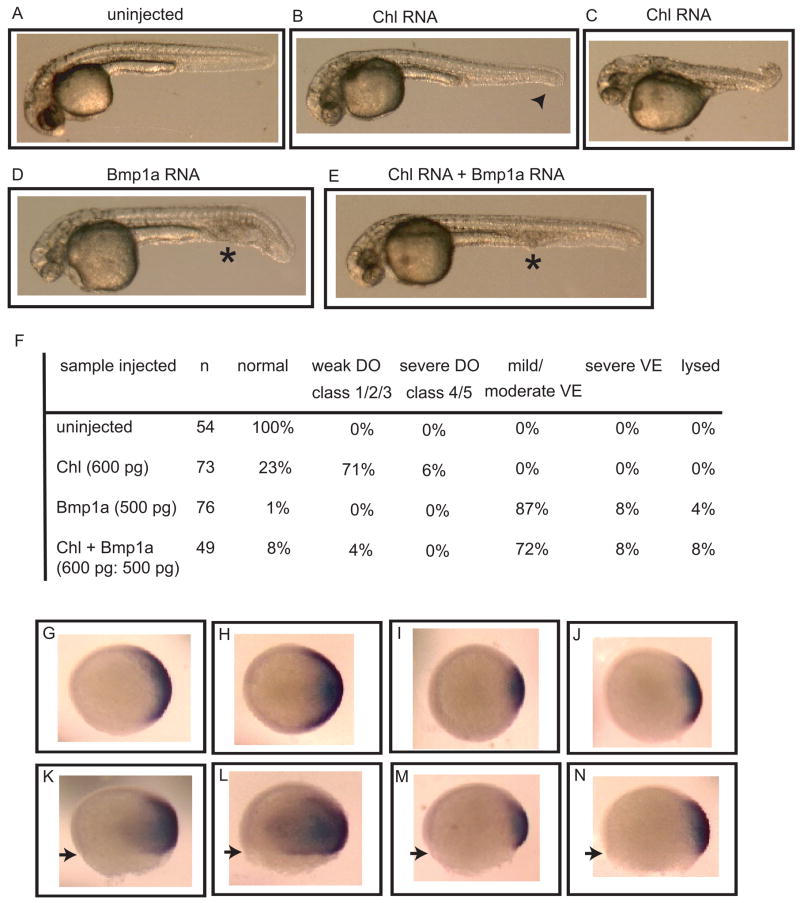
To gain further insights into Chl function, 500 pg Chl RNA was injected into one to two cell stage embryos for over-expression analysis, and phenotypes were analyzed 24 h later (Fig. 5). Embryos injected with Chl revealed a variety of dorsalized phenotypes, with most embryos showing mild class 1 to class 2 dorsalization (Fig. 5B), including partial to full loss of tail fin (Mullins et al., 1996); and a fraction showing moderate to severe (class 3 to class 5) dorsalization (Fig. 5C), characterized by complete tail loss to complete loss of ventral features and an upward twisting of the trunk. These data are consistent with the possibility that Chl, like CHL1 and CHL2 (Nakayama et al., 2001; Nakayama et al., 2004; Sakuta et al., 2001), is capable of binding and inhibiting the activity of TGFβ-like BMPs involved in ventralization during gastrulation.
Fig. 5.
Effects on dorsoventral patterning by overexpression of Chl in the presence or absence of Bmp1a. Phenotypes are shown of an uninjected 24 hpf control embryo (A), 24 hpf embryos with mild dorsalizaiton (B) or moderate dorsalization (C) after injection with 600 pg Chl RNA, and 24 hpf embryos injected with 500 pg Bmp1a RNA (D), or a combination of 600 pg Chl and 500 pg Bmp1a RNA (E). An arrowhead marks partial tail fin loss, asterisks mark blood pooling; signs of mild dorsalization or ventralization, respectively. (F) Quantification of phenotypes at 24 hpf. Severity of dorsalization (DO) is numerically classified as per Mullins et al. (Mullins et al., 1996). Degree of ventralization (VE) was not numerically classified. Whole-mount in situ hybridization is shown (animal view, panels G–J; lateral view, panels K–N) for expression of the dorsal marker Chd at 70% epiboly for uninjected embryos (G and K) or embryos injected, as above, with RNA for Chl alone (H and L), Bmp1a alone (I and M), or both Chl and Bmp1a (J and N). Arrowheads mark extent of epiboly. All embryos in panels G–N are from the same clutch of eggs and same developmental stage.
As Bmp1a is capable of in vitro cleavage of Chl (Fig. 4A), we sought to determine whether such cleavage might occur in vivo, thereby affecting Chl function. Towards this end, Chl was overexpressed in zebrafish embryos in the presence or absence of overexpressed Bmp1a. Injections of embryos with 500 pg Bmp1a RNA alone resulted in embryo ventralization, characterized by reduced head structures, thickening of the tail, and blood pooling at 24 hpf (Fig. 5D), consistent with the role of Bmp1a in dorsoventral patterning, and similar to previous data (Jasuja et al., 2006). Over-expression of Chl and Bmp1a together resulted in a majority of ventralized embryos, similar to those seen with Bmp1a alone (Figs. 5D and E), consistent with the possibility that in vivo cleavage by Bmp1a antagonizes the ability of Chl to dorsalize. Consistent with the above findings, whole-mount in situ hybridization for the dorsal marker Chd showed embryos in which Chl was overexpressed to display expansion of the Chd expression domain (Fig. 5, panels H and L), embryos in which Bmp1a alone was overexpressed to display reduction in the Chd expression domain (Fig. 5, panels I and M), and embryos in which Bmp1a and Chl were co-overexpressed (Fig. 5, panels J and N) to display reduction in the Chd expression domain, compared to embryos in which Chl alone was overexpressed (Fig. 5, panels H and L). Numbers of embryos and quantification of reductions and expansions of Chd domains for the experiments of figure 5G-N are displayed in Supplemental Table 1.
Chl knockdown
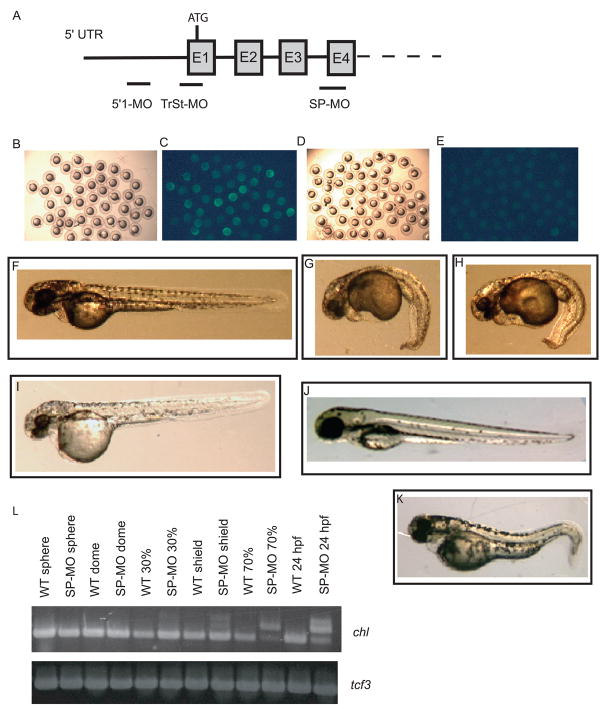
To determine the in vivo roles of Chl, loss-of-function experiments were performed in which endogenous Chl expression was knocked down, via independent use of two antisense MOs, designated 5′1-MO and TrSt-MO (Fig. 6A), designed to target 5′-UTR and translation start site sequences, respectively. To assess ability of these MOs for specific and efficient knockdown of Chl sequences, a construct was developed with GFP sequences fused 3′ of Chl 5′-UTR and coding sequences, for expression of RNA encoding a recombinant protein in which GFP is fused to the Chl C-terminus (Chl-GFP). Embryos were then injected at the one cell stage with 1 ng Chl-GFP RNA, plus 0.5 mM of 5′1-MO, or standard control (StCon) MO. Analysis at 70% epiboly showed that, whereas embryos injected with StCon-MO plus Chl-GFP RNA displayed easily detectable GFP signal (Fig. 6C), GFP signal was efficiently knocked down in embryos injected with 5′1-MO plus Chl-GFP RNA (Fig. 6E). Similar results were obtained using the TrSt-MO (Data not shown). Such experiments helped demonstrate the ability of the described MOs to recognize and effectively knock down Chl expression.
Fig. 6.
Treatment with Chl-specific anti-sense MOs. (A) A Schematic is shown representing MOs targeting the Chl translation start site (TrSt-MO), or a non-overlapping region of the chl 5′UTR (5′1-MO), or the acceptor splice site of chl exon 4 (E4). Brightfield (B and D) and darkfield (C and E) exposures are shown of 70% epiboly embryos injected at the one cell stage with 1 ng Chl-GFP RNA and 0.5 mM of either standard control (StCon) MO (B and C) or 5′1-MO (D and E). A comparison at 48 hpf is shown of embryos treated with 0.5 mM standard control (StCon) MO (F), 0.5 mM 5′1-MO (G), 0.5 mM 5′1-MO plus 0.25 mM p53-MO (H), 0.1 mM 5′1-MO (I), or 0.5 mM SP-MO (K), or with. (L) RT-PCR demonstrates the efficacy of SP-MO in inhibiting chl RNA splicing. Inhibition of splicing leading to accumulation of unspliced RNA, represented by one to two lower mobility bands, is clearly seen for shield stage, 70% epiboly, and 24 hpf embryos. Faint bands corresponding to unspliced RNA may also be discerned at 30% epiboly, especially upon comparison of lanes for samples from untreated (WT 30%) and splice site MO-treated (SP-MO 30%) embryos. Expression levels of transcription factor 3 (tcf3) RNA are shown as a loading control.
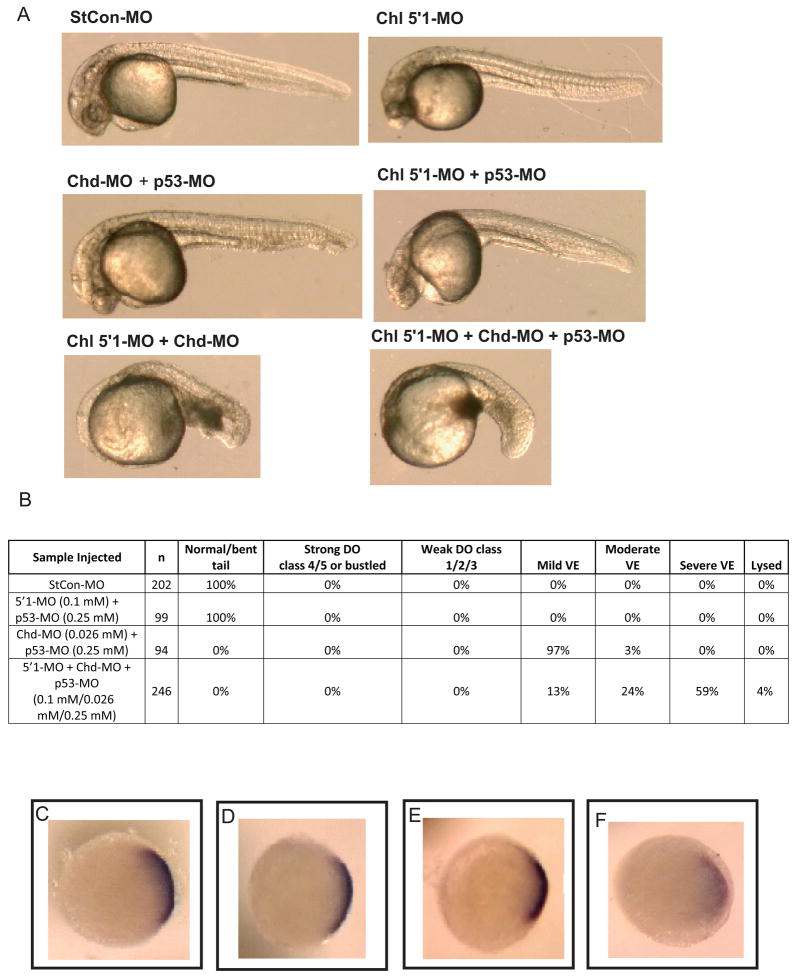
To examine the in vivo function of Chl, MOs were injected at a concentration of 0.5 mM (to a final concentration of 0.5 μM in the embryo) into one- to two-cell stage embryos for knockdown of endogenous Chl. Injection of 0.5 mM 5′1-MO (0.5 mM) resulted in arrest at about the shield stage and lysis of ~35% of embryos (n = 138), with remaining embryos surviving past 24 hpf. The latter embryos died at about 72 hpf and displayed a variety of defects that persisted from 24 to 72 hpf, and included reduced size in features such as head (microcephaly) and eyes, truncated tail (shortened body axis), and enlarged yolk sac (Fig. 6G). Similar results were obtained using TrSt-MO (data not shown). To address the possibility that non-specific, off-target effects of the Chl MOs might contribute to this phenotype, 0.5 mM 5′1-MO was co-injected with 0.25 mM of a p53 MO previously shown to suppress off-target effects (Robu et al., 2007). About 25% of the embryos of this group (n=146) underwent lysis, with the remainder surviving until ~72 hpf, and displaying a phenotype (Fig. 7H) identical to that seen in embryos injected with 0.5 mM 5′1-MO alone (Fig. 7G). Thus, evidence suggests the phenotype observed in 24 – 72 hpf embryos treated with 5′1-MO and TrSt-MO to be specific to Chl knockdown.
Fig. 7.
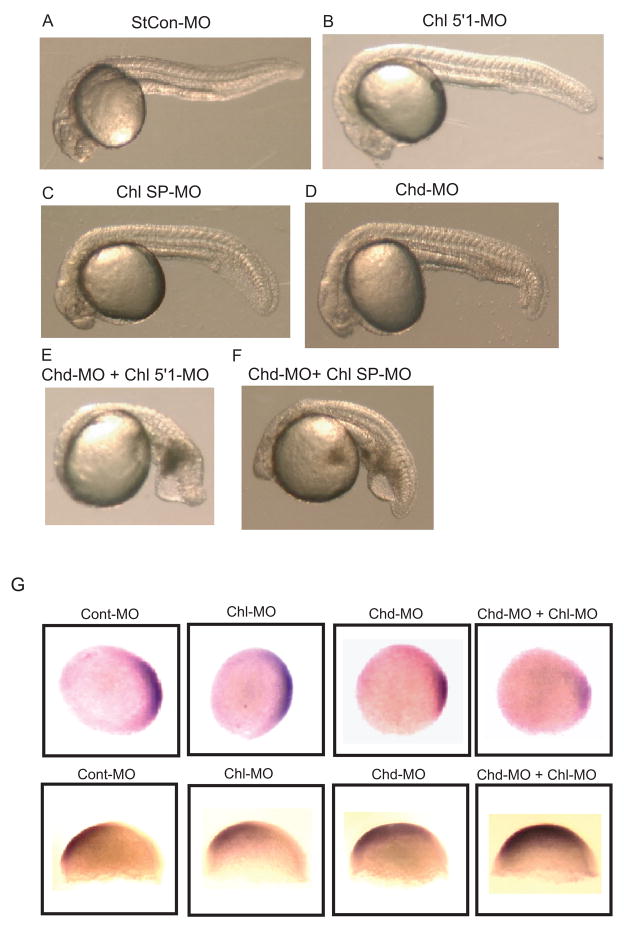
Effects in embryos co-injected with MOs against both Chl and Chd demonstrate overlapping roles for these genes in dorsoventral patterning. A comparison is shown at 24 hpf of embryos treated with 0.5013 mM StCon-MO (A), 0.1 mM Chl 5′1-MO plus 0.0013 mM StCon-MO (B), 0.5 mM Chl SP-MO plus 0.0013 mM StCon-MO (C), or 0.026 mM Chd-MO alone (D), or with combinations of 0.1 mM Chl 5′1-MO plus 0.026 mM Chd-MO (E), or 0.5 mM Chl SP-MO plus 0.026 mM Chd-MO (F). Single injections of 5′1-MO (B) and SP-MO (C) had added StCon-MO to match total amounts of MOs used in the co-injection experiments (E and F). (G) In situ hybridization shows altered expression patterns at 30% epiboly for the dorsal marker Chd (top panel) and at 70% epiboly for the ventral marker Gata2 (bottom panel) in response to treatment of embryos with the Chd MO (CHD-MO) and Chl 5′1′MO (Chl-MO), in comparison to embryos treated with StCon-MO (Cont-MO). Concentrations of MOs used were the same as those used in panel A. Note the increased reduction of the Chd expression domain and enlargement of the Gata2 expression domain in response to co-injection with the Chd and Chl MOs.
In contrast to results obtained with the above MOs, which would be expected to inhibit translation of both zygotic and maternal Chl mRNA, injection of embryos with 0.5 mM of an MO designed to interfere with correct RNA splicing (SP-MO), which would be expected to target zygotic but not maternal Chl transcripts (Draper et al., 2001), produced the relatively mild phenotype of curving of the tail, and sometimes of the body axis, at 48 hpf (Fig. 6I). Importantly, injection of embryos with lower concentrations of 5′1-MO or TrSt-MO (e.g. 0.25 mM) resulted in the same phenotype at 48 hpf (data not shown) as that achieved with 1 mM SP-MO, thus offering additional evidence that phenotypes achieved with the various MOs used in this study are specifically caused by knockdown of Chl expression. An RT-PCR control was used to demonstrate the efficacy of SP-MO for interfering in the splicing of Chl RNA, starting at 70% epiboly (Fig. 6J). As maternal RNA is already spliced, the latter results also demonstrate that zygotic Chl RNA expression is readily detectable by shield stage, and may be the predominant form of Chl RNA transcripts at 70% epiboly and at 24 hpf. Faint bands corresponding to unspliced and/or misspliced RNA, and thus zygotic transcription, may also be discernable at 30% epiboly, but were near the limits of detection (Fig. 6J).
Synergistic effects of Chl and Chd MOs demonstrate overlapping, redundant roles for these two gene products in dorsoventral patterning of the zebrafish embryo
Because RT-PCR and in situ hybridization results clearly showed coexpression of Chd and Chl from the 1000-cell stage onwards (Figs. 2 and 3), and because Chl overexpression dorsalizes embryos (Fig. 5), we tested the effect of simultaneously injecting Chl and Chd MOs, to determine whether these two genes might demonstrate functional overlap in formation of the dorsoventral axis. Injection of 0.1 mM Chl 5′1-MO (Fig. 7B), or of 0.5 mM Chl SP-MO (Fig. 7C) produced mild morphant phenotypes at 24 hpf; while injection of 0.026 mM Chd MO (Fig. 7D) produced a mild ventralized phenotype at 24 hpf. However, when 0.026 mM Chd was co-injected with either 0.1 mM 5′1-MO (Fig. 7E), or 0.5 mM SP-MO (Fig. 7F) severely ventralized phenotypes were obtained, in which reduced head structures, gross thickening of the tail, and blood pooling were observed at 24 hpf. Consistent with morphological results in figure 7A–F, in situ hybridization demonstrated synergistic effects from co-injection of Chl 5′1-MO and the Chd MO in reduction of the expression domain of the dorsal marker Chd and expansion of the expression domain of the ventral marker GATA2 (Fig. 7G, with quantification of data in supplemental Table S1). The fact that co-injecting 5′1-MO and the Chd MO results in significantly more severe effects than either MO injected alone supports the conclusion of overlapping, redundant Chl and Chd function in dorsoventral patterning.
As 5′1-MO targets 5′-UTR sequences, it would be expected to knock down expression of Chl from both maternally and zygotically transcribed RNA. In contrast, SP-MO, a splice site MO, would be expected to target zygotically, but not maternally transcribed Chl RNA, as the latter is already spliced. Thus, it is of interest that SP-MO, as well as 5′1-MO, is able to synergize with subthreshold amounts of Chd to produce severely ventralized phenotypes. These results are consistent with the interpretation that a portion of Chl dorsoventral patterning function, redundant with that of Chd, derives from zygotically transcribed RNA.
To address the possibility that non-specific, off-target effects of one or both MOs might contribute to the synergistic ventralizing effects caused by co-injection of Chl and Chd MOs, 5′1-MO and the Chd MO were co-injected with the p53 MO, described above. Co-injection of 5′1-MO and the Chd MO with the p53 MO had no effect on ventralization of embryos (Fig. 8), thus offering evidence that the ventralized phenotype achieved by co-injection of Chl and Chd MOs is specific.
Fig. 8.
Synergistic effects of Chl and Chd MOs in inducing ventralization does not involve p53-mediated off target effects. (A) A comparison is shown of 24 hpf embryos treated with 0.38 mM StCon-MO, 0.1 mM Chl 5′1-MO, 0.026 mM Chd-MO plus 0.25 mM p53-MO, 0.1 mM Chl 5′1-MO plus 0.026 mM Chd-MO, or with 0.1 mM Chl 5′1 plus 0.026 mM Chd-MO and 0.25 mM p53-MO. (B) Quantification of phenotypes at 24 hpf. Severity of dorsalization (D) is numerically classified as per Mullins et al. (Mullins et al., 1996). Degree of ventralization (VE) was not numerically classified. (C–D) Whole mount in situ hybridization is shown for expression of the dorsal marker Chl at 30% epiboly for embryos treated with 0.38 mM StCon-MO (C), 0.1 mM Chl 5′1-MO plus 0.25 mM p53-MO (D), 0.026 mM Chd-MO plus 0.25 mM p53-MO (E), or 0.1 mM Chl 5′1 plus 0.026 mM Chd-MO and 0.25 mM p53-MO.
Chl, murine CHL1 and CHL2, and Chd can rescue embryos ventralized by co-injection of Chl and Chd MOs
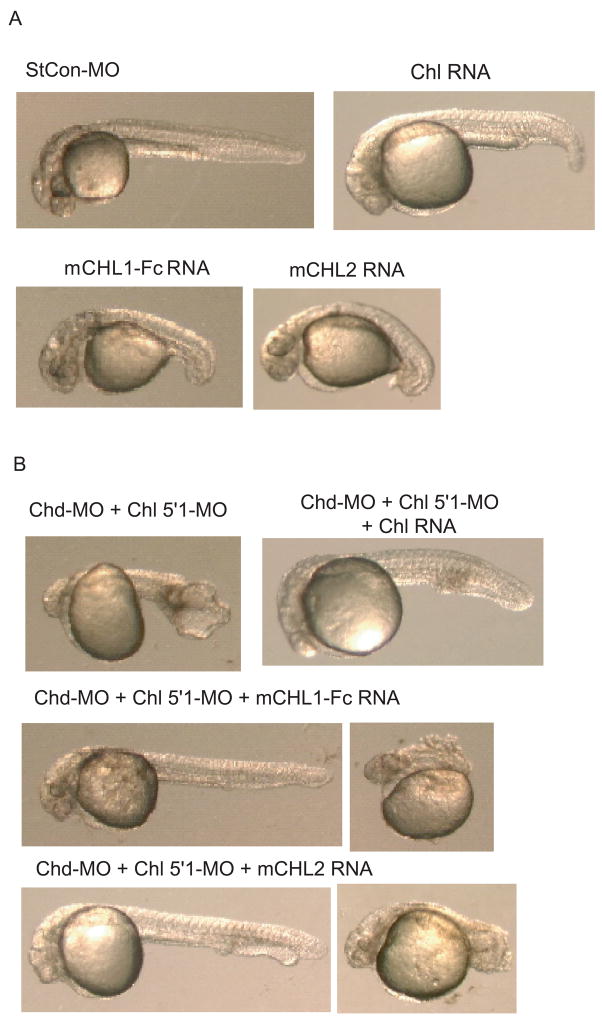
Consistent with the probability that Chl and murine CHL1 and CHL2 are all capable of antagonizing BMP signaling, overexpression of each resulted in dorsalization of zebrafish embryos (Fig. 9A). As a further test of the overlapping functions of Chl and Chd in dorsoventral patterning, and as a test for the specificity of knockdown with Chl MOs, we investigated whether embryos ventralized by co-injection of Chl and Chd MOs could be rescued by overexpression of Chl. As can be seen in figure 9B and Table 1, whereas the vast majority (98%) of embryos co-injected with Chl and Chd MOs are moderately to severely ventralized, co-overexpression of Chl in the same embryos achieves rescue to the extent that 11% of embryos have dorsalized phenotypes, and another 39% are only mildly ventralized. Interestingly, overexpression of murine CHL1 or CHL2 also rescued embryos (Fig. 9 and Table 1), implying some conservation of function between Chl and the two CHLs of higher vertebrates. It should be noted that while the murine CHL1 overexpressed in the experiments of figure 9 and Table 1 rescued more effectively than did Chl, this CHL1 was fused to an Fc domain to enhance stabilization of the protein, as overexpression of murine CHL1 not fused to an Fc domain was relatively ineffective at rescue (not shown). Thus, the relatively efficient rescue by CHL1-Fc was probably enhanced by the increased stability of the protein due to the added Fc domain. We speculate that the relatively strong ability of murine CHL2 to rescue embryos may be due to its imperviousness to cleavage by endogenous BMP1-like proteinases, as we have shown CHL2 to not be cleaved by such enzymes (Fig. 4). Chd RNA was also able to rescue the ventralization phenotype resulting from co-injection of Chl and Chd MOs (Supplemental Fig. 1), adding additional evidence that the ventralized phenotype is in all respects specific and not due to off target effects.
Fig. 9.
Chl, and murine CHL1 and CHL2 can rescue embryos ventralized by co-injection of Chl and Chd MOs. (A) Embryos were dorsalized by injection of RNA encoding Chl, murine CHL1 fused to an Fc domain (mCHL1-Fc), or CHL2 (mCHL2) (1 ng, 80 pg, or 1 pg, respectively). Also shown, for comparison, is a representative embryo injected with 0.1 mM of StCon-MO. (B) Embryos co-injected with Chd-MO and Chl 5′1-MO are for the most part moderately to severely ventralized, whereas overexpression of Chl RNA rescues embryos co-injected with Chd-MO and Chl 5′1-MO, such that many are only mildly ventralized (shown) or weakly or strongly dorsalized (not shown). Overexpression of murine CHL1-Fc (mCHL1-Fc) or murine CHL2 (mCHL2) also rescued embryos co-injected with Chd-MO and Chl 5′1-MO, resulting in a range of phenotypes that included mildly ventralized [left] and severely dorsalized [right].
Table 1.
| Sample Injected | n | normal/bent tail | strong dorsalized class 4/5 or bustled | weak dorsalized class 1/2/3 | mild ventralized | moderate ventralized | severe ventralized | lysed |
|---|---|---|---|---|---|---|---|---|
| StCon-MO | 96 | 100% | 0% | 0% | 0% | 0% | 0% | 0% |
| 5′1-MO (0.1mM) | 69 | 100% | 0% | 0% | 0% | 0% | 0% | 0% |
| Chd-MO (0.026 mM) | 88 | 0% | 0% | 0% | 98% | 2% | 0% | 0% |
| SP-MO (0.5mM) | 43 | 100% | 0% | 0% | 0% | 0% | 0% | 0% |
| 5′1-MO + Chd-MO (0.1 mM/0.026 mM) | 196 | 0% | 0% | 0% | 2% | 31% | 67% | 0% |
| SP-MO + Chd-MO (0.5 mM/0.026 mM) | 56 | 0% | 0% | 0% | 11% | 21% | 68% | 0% |
| Chl RNA (1 ng) | 68 | 0% | 7% | 93% | 0% | 0% | 0% | 0% |
| mCHL2 RNA (1 pg) | 41 | 0% | 19% | 49% | 0% | 0% | 0% | 32% |
| mCHL1-Fc RNA (80 pg) | 28 | 11% | 28% | 50% | 0% | 0% | 0% | 11% |
| 5′1-MO + Chd-MO + Chl RNA | 89 | 0% | 6% | 5% | 39% | 30% | 16% | 4% |
| 5′1-MO + Chd-MO + mCHL2 RNA | 52 | 0% | 14% | 42% | 17% | 4% | 0% | 23% |
| 5′1-MO + Chd-MO + mCHL1-Fc RNA | 26 | 0% | 12% | 27% | 27% | 19% | 4% | 11% |
Discussion
CHD/SOG is the prototype of a group of CR-containing proteins, members of which bind and modulate signaling by extracellular ligands of the TGFβ superfamily. Such modulation by CR-containing proteins is critical to formation of the dorsoventral axis in a broad range of species, as well as in wing vein patterning in Drosophila, and neural induction in vertebrates (De Robertis and Kuroda, 2004; Garcia Abreu et al., 2002). However, although experiments involving in vitro binding, overexpression in Xenopus embryos, and misexpression in chick retina have previously shown that vertebrate CR-bearing proteins CHL1 and CHL2 can bind BMP 2 and 4, and other TGFβ-like ligands (Nakayama et al., 2001; Nakayama et al., 2004; Sakuta et al., 2001), in vivo functions of CHL proteins, particularly in early development, are largely unknown due to lack of loss-of-function data.
The Danio rerio CR-protein Chl described here has slightly more homology to mammalian CHL2 than to mammalian CHL1. Moreover, comparison of zebrafish and murine genomic contigs found both the genes for Chl and murine CHL2 to be flanked by the genes for sialidase 3 and polymerase δ3, showing corresponding segments of R. danio chromosome 21 and M. musculus chromosome 7 to have shared synteny. However, although the gene for zCHL thus appears to correspond to the mammalian gene for CHL2, a separate zebrafish gene corresponding to mammalian CHL1 has not been identified, despite multiple Blast searches of the zebrafish genome databases with murine and human CHL1 and CHL2 amino acid and nucleotide sequences. Thus, it is not yet clear whether separate genes corresponding to CHL1 and CHL2 exist in zebrafish, or whether, instead, Chl may serve roles in zebrafish distributed between CHL1 and CHL2 in other vertebrate species. In support of the latter possibility, it is of interest that Chl, despite greater sequence homology to CHL2, resembles CHL1, rather than CHL2 in ability to be cleaved and thus regulated by BMP1/Tolloid-like proteinases.
Here we show temporal and spatial patterns of expression of Chl to differ from, but overlap those of Chd, suggestive of differences, but supporting the possibility of overlap, in developmental roles of these two proteins. Analysis of the temporal pattern of Chl expression, spanning the 8-cell stage to 48 hpf, found highest Chl RNA levels at the 8-cell stage; thus suggesting Chl to be maternally transmitted, as initiation of zygotic expression does not occur for most genes until the MBT at the 512- to 1000-cell stage (2.75 hpf) (Kane and Kimmel, 1993; Mathavan et al., 2005). Thus, some portion of Chl function is likely provided by maternally supplied Chl, although zygotically transcribed Chl RNA is also produced, and has a demonstrated role in dorsoventral patterning, as described in the current study. Although previous studies (Kosinski et al., 2007; Nakayama et al., 2001; Nakayama et al., 2004; Oren et al., 2004; Sakuta et al., 2001; Wu and Moses, 2003) have provided no indication that CHL1 or CHL2 gene products might be maternally provided, this may simply be because previous studies have not analyzed CHL1 or CHL2 expression as early in vertebrate development as was done here for Chl.
In normal vertebrate development, CHD, prototype of CR-containing proteins, is thought to dorsalize gastrula stage embryos by binding TGFβ-like BMPs, thus inhibiting their interactions with cognate cell surface receptors (Piccolo et al., 1996; Schulte-Merker et al., 1997). However, loss of CHD function in various model systems has shown surprisingly minor consequences to dorsoventral patterning (Bachiller et al., 2000; Hammerschmidt et al., 1996; Khokha et al., 2005). In zebrafish, for example, substantially more dorsal and dorsolateral structures persist even in strong Chd mutants (Hammerschmidt et al., 1996), than in embryos completely ventralized by overexpression of BMPs (Schmid et al., 2000). This has suggested that other gene products might functionally overlap with Chd in dorsalization of the early embryo. It has previously been shown that the extracellular BMP antagonists noggin and follistatin are together capable of providing some functional overlap with CHD in Xenopus (Khokha et al., 2005), although the morphant phenotypes in these triple knockdown experiments suggested the existence of additional BMP antagonists, needed to prevent formation of the most ventral fates (Khokha et al., 2005). In zebrafish, noggin1 (Nog1) and follistatin-like2 (Fstl2) appear to functionally overlap with Chd (Dal-Pra et al., 2006).
As a CR-containing protein, and in view of the fact that both CHL1 and CHL2 have been shown to be capable of interacting with various TGFβ-like ligands (Nakayama et al., 2001; Nakayama et al., 2004; Sakuta et al., 2001), it seems likely that Chl would exert its developmental effects, at least in part, by modulating the activity of such ligands. In fact, it has been proposed that in chick, CHL1 may be involved in modulating dorsoventral and anteroposterior patterning in the developing retina, via effects on signaling by BMP4 and related ligands (Sakuta et al., 2001). Here we demonstrate, via gain-of-function experiments, that Chl overexpression dorsalizes embryos, consistent with ability to bind and inhibit TGFβ-like BMPs. More importantly, loss-of-function experiments, employing coinjection of Chl and Chd MOs, show these two genes to have overlapping and cooperative functions crucial to correct dorsalization of the zebrafish embryo. Remarkably, the extent of ventralization achieved in embryos in which both Chl and Chd are simultaneously knocked down appears to exceed that previously achieved via triple knock down of Chd, Nog1 and Fstl2 (Dal-Pra et al., 2006), implying an important role for Chl in normal patterning of the embryonic dorsoventral axis, presumably, along with Chd, via antagonizing signaling by Bmp2-Bmp7 heterodimers, which have been shown to be responsible for eliciting the signaling response required for dorsoventral patterning in zebrafish (Little and Mullins, 2009).
Interestingly, simultaneous knock down of Chd and Chl via treatment with a Chd MO in conjunction with either a 5′-UTR Chl MO, expected to inhibit both zygotic and maternal Chl, or with a splice site Chl MO, expected to inhibit only zygotic Chl, both produced severe ventralization of embryos. These results suggest that an at least some component of Chl dorsalizing function, redundant with that of Chd, derives from zygotic Chl expression, which appears to commence at least as early as the shield stage (Fig. 6J). However, we note that in these experiments the dose of MO that could be used was lower for the 5′UTR Chl MO than for the splice site Chl MO (see below), and our results do not preclude potentially important roles for maternal Chl in either the initiation or maintenance of dorsoventral patterning.
Diffusion of CHD from a dorsal organizer is thought to establish a gradient of antagonism to BMP signaling that, in large part, establishes patterning of the dorsoventral axis in vertebrates (Piccolo et al., 1996). Consistent with this model, in situ hybridizations in the present study showed Chd expression to be localized to the dorsal aspect of the embryo just prior to gastrulation, at the dome stage and 30% epiboly, and during gastrulation, at the shield stage and 70% epiboly (Fig. 3). In contrast, in situ hybridization did not detect dorsoventral asymmetry in Chl RNA distribution prior to gastrulation, as such RNA appeared to be ubiquitous in embryos from the 8-cell stage through 30% epiboly. However, Chl at these stages is likely predominantly maternal since, as noted above, zygotic Chl RNA thought essential to dorsoventral patterning is not expressed at readily detectable levels until during gastrulation, at the shield stage. It is not yet clear what role maternal Chl may contribute to dorsoventral patterning.
At the shield stage and 70% epiboly in situ hybridization signal for Chl was near the limits of detection. Thus, it is not yet certain whether or not dorsoventral asymmetry exists for zygotic Chl expression at these stages. However, even if there is an absence of dorsoventral asymmetry in Chl expression during gastrulation, Chl could still contribute to establishing a dorsoventral gradient of antagonism to BMP signaling, if functional Chl protein was asymmetrically distributed in the embryos. Such asymmetry could occur either via asymmetric dorsoventral binding of Chl protein to extracellular components, or via Chl cleavage by BMP1-like proteinases. In regard to the latter possibility, we show here that Chl is cleaved by such proteinases, and we have previously shown at least one of these proteinases to be preferentially expressed in ventral portions of the zebrafish embryo during gastrulation (Jasuja et al., 2006). Even in the absence of a dorsoventral gradient of Chl expression or protein, there is the possibility that Chl may aid in patterning the dorsoventral axis in a permissive way, by lowering baseline levels of BMP signaling throughout the embryo, thus aiding Chd in achieving a more complete inhibition of BMP signaling in dorsal regions.
In regard to possible roles for maternal Chl, RNA of which declines rapidly prior to gastrulation onset, we have found that higher concentrations (e.g. 1 mM) of the 5′-UTR MO used in the present study, or similarly high concentrations of two other non-overlapping MOs targeting either 5-UTR or translation start site sequences, results in developmental arrest in the blastula stage. This developmental arrest is due to a block in cell cycle progression, an effect not obtained using the SP-MO splice site MO specific for zygotic Chl (data not shown). It is not presently clear whether this effect is specific to knock down of maternal Chl, as efforts to rescue this particular phenotype via overexpression of Chl RNA were unsuccessful (our unpublished data). Nevertheless, given its initially high levels of expression in the pregastrula embryo, it seems likely that maternal Chl plays a role in these early stages. It is possible that this putative early role is functionally distinct from a latter role for Chl in dorsoventral patterning. Alternatively, maternal Chl may contribute, together with zygotic Chl, to the overall function of this gene in the regulation of dorsoventral patterning. The latter is a possibility that we cannot rule out, as such a role my be obscured by the cell cycle progression defect induced by high concentrations of 5′UTR or translation start site Chl MOs.
In summary, we describe the zebrafish CR-protein Chl and demonstrate it to be an important BMP antagonist with functions that overlap and synergize those of Chd in formation of the dorsoventral axis. Zygotically transcribed Chl, expression of which commences at least as early as the shield stage, seems important to this role in dorsoventral patterning of the axis, and this role in patterning, as for that of Chd, is likely affected via cleavage by BMP1-like proteinases. The roles of relatively high levels of maternally provided Chl remain to be defined. Beyond further exploring the roles of Chl in zebrafish development, it will be of great interest to determine whether CHL1 and/or CHL2 play similar roles in higher vertebrates.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01GM65303 (to F. P.), R01GM71679, R01AR53815, and R01AR53815-12S1 (to D. S. G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–61. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Blader P, Rastegar S, Fischer N, Strahle U. Cleavage of the BMP-4 antagonist chordin by zebrafish tolloid. Science. 1997;278:1937–40. doi: 10.1126/science.278.5345.1937. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Tran U, Larrain J, De Robertis EM. Neuralin-1 is a novel Chordin-related molecule expressed in the mouse neural plate. Mech Dev. 2001;100:119–22. doi: 10.1016/s0925-4773(00)00507-4. [DOI] [PubMed] [Google Scholar]
- Dal-Pra S, Furthauer M, Van-Celst J, Thisse B, Thisse C. Noggin1 and Follistatin-like2 function redundantly to Chordin to antagonize BMP activity. Dev Biol. 2006;298:514–26. doi: 10.1016/j.ydbio.2006.07.002. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–6. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Garcia Abreu J, Coffinier C, Larrain J, Oelgeschlager M, De Robertis EM. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene. 2002;287:39–47. doi: 10.1016/s0378-1119(01)00827-7. [DOI] [PubMed] [Google Scholar]
- Ge G, Hopkins DR, Ho WB, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol. 2005;25:5846–58. doi: 10.1128/MCB.25.14.5846-5858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development. 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–23. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasuja R, Voss N, Ge G, Hoffman GG, Lyman-Gingerich J, Pelegri F, Greenspan DS. bmp1 and mini fin are functionally redundant in regulating formation of the zebrafish dorsoventral axis. Mech Dev. 2006;123:548–58. doi: 10.1016/j.mod.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–11. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, Chen X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104:15418–23. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrain J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis EM. BMP-binding modules in chordin: a model for signalling regulation in the extracellular space. Development. 2000;127:821–30. doi: 10.1242/dev.127.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Mullins MC. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol. 2009;11:637–43. doi: 10.1038/ncb1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques G, Musacchio M, Shimell MJ, Wunnenberg-Stapleton K, Cho KW, O’Connor MB. Production of a DPP activity gradient in the early Drosophila embryo through the opposing actions of the SOG and TLD proteins. Cell. 1997;91:417–26. doi: 10.1016/s0092-8674(00)80425-0. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–44. [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–76. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Bertoglio VE, Fisher S, Sanchez A, Mullins MC, Halpern ME. Differential regulation of chordin expression domains in mutant zebrafish. Dev Biol. 1997;192:537–50. doi: 10.1006/dbio.1997.8788. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Nusslein-Volhard C. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- Muraoka O, Shimizu T, Yabe T, Nojima H, Bae YK, Hashimoto H, Hibi M. Sizzled controls dorso-ventral polarity by repressing cleavage of the Chordin protein. Nat Cell Biol. 2006;8:329–38. doi: 10.1038/ncb1379. [DOI] [PubMed] [Google Scholar]
- Nakayama N, Han CE, Scully S, Nishinakamura R, He C, Zeni L, Yamane H, Chang D, Yu D, Yokota T, Wen D. A novel chordin-like protein inhibitor for bone morphogenetic proteins expressed preferentially in mesenchymal cell lineages. Dev Biol. 2001;232:372–87. doi: 10.1006/dbio.2001.0200. [DOI] [PubMed] [Google Scholar]
- Nakayama N, Han CY, Cam L, Lee JI, Pretorius J, Fisher S, Rosenfeld R, Scully S, Nishinakamura R, Duryea D, Van G, Bolon B, Yokota T, Zhang K. A novel chordin-like BMP inhibitor, CHL2, expressed preferentially in chondrocytes of developing cartilage and osteoarthritic joint cartilage. Development. 2004;131:229–40. doi: 10.1242/dev.00901. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Oren A, Toporik A, Biton S, Almogy N, Eshel D, Bernstein J, Savitsky K, Rotman G. hCHL2, a novel chordin-related gene, displays differential expression and complex alternative splicing in human tissues and during myoblast and osteoblast maturation. Gene. 2004;331:17–31. doi: 10.1016/j.gene.2004.01.029. [DOI] [PubMed] [Google Scholar]
- Pappano WN, Steiglitz BM, Scott IC, Keene DR, Greenspan DS. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases. Mol Cell Biol. 2003;23:4428–38. doi: 10.1128/MCB.23.13.4428-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F, Maischein HM. Function of zebrafish beta-catenin and TCF-3 in dorsoventral patterning. Mech Dev. 1998;77:63–74. doi: 10.1016/s0925-4773(98)00132-4. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Lu B, Goodman S, Dale L, De Robertis EM. Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell. 1997;91:407–16. doi: 10.1016/s0092-8674(00)80424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–98. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuta H, Suzuki R, Takahashi H, Kato A, Shintani T, Iemura S, Yamamoto TS, Ueno N, Noda M. Ventroptin: a BMP-4 antagonist expressed in a double-gradient pattern in the retina. Science. 2001;293:111–5. doi: 10.1126/science.1058379. [DOI] [PubMed] [Google Scholar]
- Schmid B, Furthauer M, Connors SA, Trout J, Thisse B, Thisse C, Mullins MC. Equivalent genetic roles for bmp7/snailhouse and bmp2b/swirl in dorsoventral pattern formation. Development. 2000;127:957–67. doi: 10.1242/dev.127.5.957. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho KW, De Robertis EM, Nusslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–52. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Lee KJ, McMahon AP, Hammerschmidt M. The zebrafish organizer requires chordino. Nature. 1997;387:862–3. doi: 10.1038/43092. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Imamura Y, Clark TG, Steiglitz BM, Thomas CL, Maas SA, Takahara K, Cho KW, Greenspan DS. Mammalian BMP-1/Tolloid-related metalloproteinases, including novel family member mammalian Tolloid-like 2, have differential enzymatic activities and distributions of expression relevant to patterning and skeletogenesis. Dev Biol. 1999;213:283–300. doi: 10.1006/dbio.1999.9383. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Maas SA, Cho KW, Greenspan DS. Homologues of Twisted gastrulation are extracellular cofactors in antagonism of BMP signalling. Nature. 2001;410:475–8. doi: 10.1038/35068572. [DOI] [PubMed] [Google Scholar]
- Steiglitz BM, Ayala M, Narayanan K, George A, Greenspan DS. Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J Biol Chem. 2004;279:980–6. doi: 10.1074/jbc.M310179200. [DOI] [PubMed] [Google Scholar]
- Wu I, Moses MA. BNF-1, a novel gene encoding a putative extracellular matrix protein, is overexpressed in tumor tissues. Gene. 2003;311:105–10. doi: 10.1016/s0378-1119(03)00563-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.