Summary
We describe a method for conditional regulation of gene expression based on the processing of an intron cassette by a splicing factor. The RNA processing factor MEC-8 is necessary for the function of the Caenorhabditis elegans touch receptor neurons; mec-8 mutants are touch insensitive. We show here that this insensitivity involves the loss of MEC-8-dependent splicing of mec-2, which encodes a component of the mechanosensory transduction complex. MEC-8 is needed to remove the ninth intron in mec-2 pre-mRNA to form the longest of three mRNAs, mec-2a. Without MEC-8, splicing causes the termination of the transcript. Inclusion of mec-2 intron 9 is sufficient to convey mec-8-dependent regulation on other genes and, in mec-8(u218ts) mutants, resulted in their temperature-dependent expression. Because mec-8 is expressed ubiquitously in embryos and extensively in larvae, this system should produce temperature-sensitive expression for most genes. We report a strain that exhibits temperature-dependent RNA interference.
Introduction
Conditional expression of gene activity provides a means to examine gene function that is restricted in time or cell type, and is particularly useful when complete elimination of gene activity leads to lethality. Several methods produce conditional gene expression. These methods include the Cre-lox system for inducible gene targeting in mice1, a heat-induced signal to target protein degradation in yeast2, a tetracycline-responsive promoter in mammalian cells3, and three heat-shock based systems for regulated gene expression in Caenorhabditis elegans4, 5, 6.
One of the first means of obtaining conditional gene expression was the use of temperature-sensitive mutations. Classic examples include studies of essential genes in Neurospora7 and of morphogenesis in the bacteriophage T4 (ref. 8). The usefulness of such mutations is limited, however, by their relative rarity; temperature-sensitive alleles do not exist for most genes. For example, in our own work identifying mutations affecting touch sensitivity in C. elegans, only seven of eighteen mapped genes (affected by 418 mutations) had temperature-sensitive alleles9.
In this paper we describe a method to generate splicing-dependent and temperature-sensitive expression of virtually any C. elegans gene for which a loss-of-function allele is available, using temperature sensitive MEC-8-dependent splicing. Unlike previous C. elegans methods4, 5, 6, which require heat shock to allow gene expression, our method employs a temperature shift at normal growth temperatures. The mec-8 gene is needed for touch sensitivity in C. elegans10. In addition, the MEC-8 protein, which has two RNA recognition motifs (RRMs), is required for the processing of the unc-52 transcript in the hypodermis11. We sought potential targets of mec-8 regulation among the genes needed for touch sensitivity. We found that MEC-8 is needed for the removal of the ninth intron of mec-2, which encodes a subunit of the channel complex needed for mechanosensory transduction12, 13. This discovery enabled us to generate a system that results in the mec-8-dependent expression of many C. elegans genes. Moreover, using the mec-8(u218ts) allele, which produces a temperature-sensitive phenotype9, we can express virtually any gene containing mec-2 intron 9 in a temperature-dependent manner.
Results
MEC-8 regulates alternative splicing of mec-2
The expression of mec-8 begins in the 50-cell embryo and is ubiquitous. As the animals develop, mec-8 expression persists in the hypodermis, intestine, the six touch receptor neurons (TRNs), chemosensory neurons of the head and tail, and in vulval nuclei (ref. 14; and our unpublished observations).
To identify mec-8 targets in the TRNs, where it is known to act cell-autonomously, we looked for differences in RT-PCR products of genes needed for touch sensitivity (listed in Methods) using mRNA from wild-type and mec-8 mutants. The only cDNAs that were differentially amplified were those of mec-2 (Fig. 1). The mec-2 gene produces three mRNAs, as identified by RT-PCR (www.wormbase.org). The largest (mec-2a: 1.4 kb) is formed from 13 exons12 (Fig. 2a). We could amplify mec-2a cDNA from wild type, but not from worms homozygous for the mec-8 null mutations u314 or e398 (Fig. 1a). The mec-2a mRNA was made, however, in mec-8 mutants rescued with mec-8(+) expressed in the TRNs (Fig. 1b). The mec-2b mRNA has exons 1 – 9 of mec-2a but lacks exons 10 – 13, containing instead a small exon (exon 9’) at its 3’ end. The inclusion of exon 9’ adds an in-frame stop codon and a consensus polyadenylation site to the mRNA. The mec-2c mRNA begins with an exon within intron 2 (exon 2’) and continues with exons 3 – 9’ as in mec-2b. Using cDNA-specific primers, we amplified mec-2b and mec-2c cDNAs in wild type and the two mec-8 null strains.
Figure 1. mec-2a mRNA is not produced in mec-8 mutants.
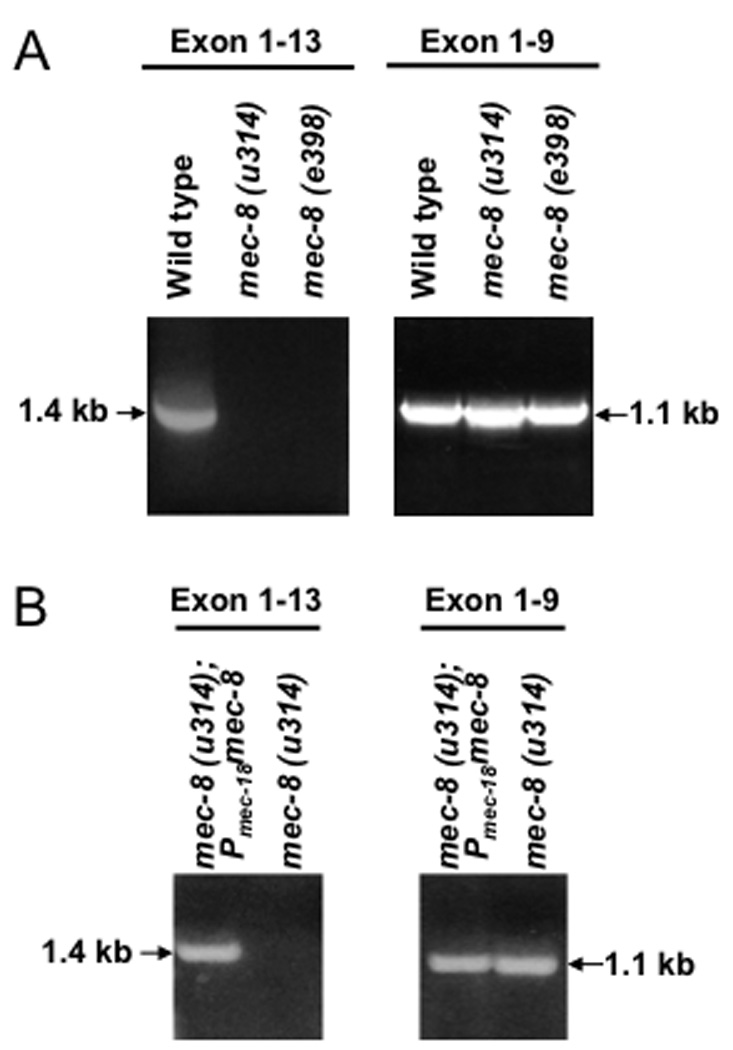
RT-PCR using primers that amplify exons 1–13 or 1–9 of mec-2 in wild type and the indicated mec-8 mutant strains (a) and in mec-8 mutant strains in which the expression of mec-8 is restored in TRNs (b). The 1.4 kb band corresponds to mec-2a and the 1.1 kb band to mec-2b.
Figure 2. MEC-8 regulates the alternative splicing of mec-2.
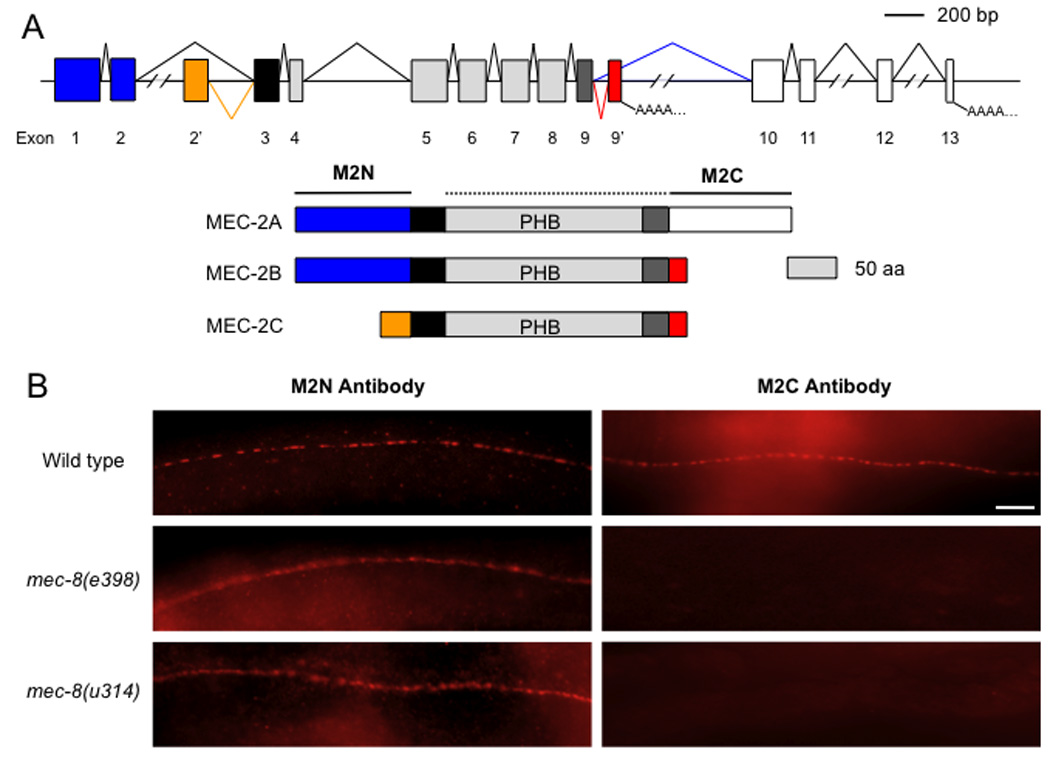
(a) mec-2 splicing pattern and MEC-2 proteins. Colors denote corresponding regions of the proteins and the transcripts (blue: MEC-2 N terminus; orange: exon 2’-encoded polypeptide; black: hydrophobic region; gray: PHB domain; dark gray: the portion of the PHB domain contributed by exon 9; red: exon 9’-encoded polypeptide; and white: MEC-2 C terminus). The mec-8-dependent splice is shown in green; the mec-2b- and mec-2c-specific splices are shown in red and orange, respectively. Distances between exons 2 and 2’, exons 9’ and 10, exons 11 and 12, and exons 12 and 13 are 6327, 1592, 1164 and 2678 bp, respectively. Regions of the proteins recognized by the M2N and M2C antibodies are indicated. (b) Immunofluorescent staining of worms of the indicated genotypes with M2N and M2C antibodies. Scale bar 10 µm
The three mec-2 mRNAs encode polypeptides with a prohibitin homology (PHB) domain preceded by a hydrophobic region that inserts the protein into the cytoplasmic leaflet of the plasma membrane. N-and C-termini surround the hydrophobic and PHB domains in MEC-2A (Fig. 2a). MEC-2B has the N-terminal but not the C-terminal domain, and MEC-2C lacks both domains (Fig. 2a).
We had previously generated antibodies against the N terminus of MEC-2A (M2N) and against the PHB domain through the C-terminus of MEC-2A (M2C). We stained wild type and mec-8 mutants with the M2N and M2C antibodies and, as expected from the RT-PCR results, the M2N antibody labeled TRNs in wild-type and mec-8 worms; M2C antibody labeled only wild type TRNs (Fig. 2b). Since the antibodies were purified by preabsorption to extracts of mec-2 null mutants and since C. elegans has several PHB domain proteins15, the M2C antibody is likely to recognize only the C-terminus of MEC-2A, which is missing in mec-8 mutants.
mec-2 intron 9 is sufficient for mec-8-dependent splicing
To test whether mec-2 intron 9 contains all the necessary elements for conferring mec-8 dependency, we inserted mec-2 intron 9 before the coding sequence for Yellow Fluorescent Protein (YFP). We expressed intron 9::yfp in TRNs using the mec-18 promoter (Fig. 3a). No YFP fluorescence was observed in mec-8(e398) or mec-8(u314) worms transformed with this construct (Pmec-18intron 9::yfp). In contrast, fluorescence was seen in all six TRNs in the mec-8/+ progeny of transgenic worms when they were crossed with wild-type males (Fig. 3b). These results indicate that YFP expression was dependent on MEC-8.
Figure 3. mec-2 intron 9 conveys MEC-8 dependence.
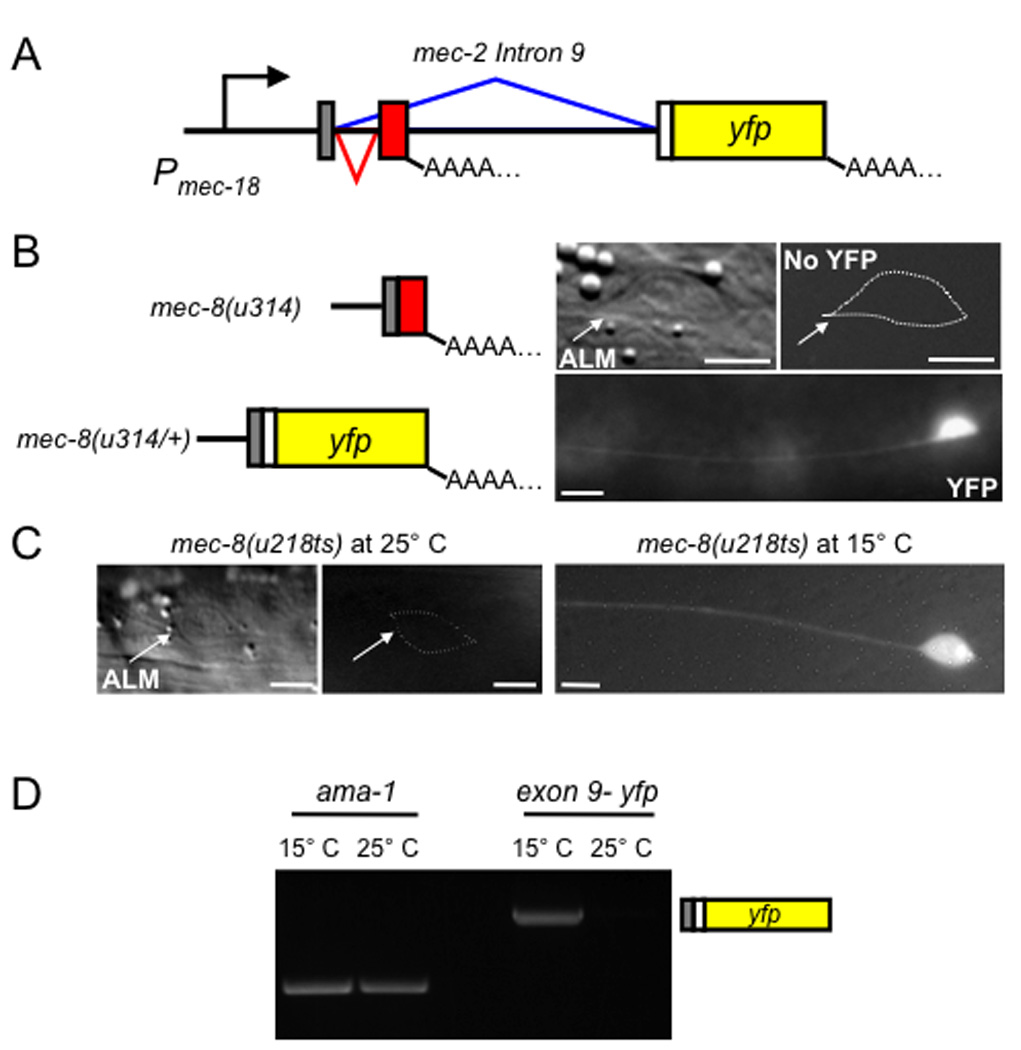
(a) Diagram of the intron 9::yfp construct under the control of the mec-18 promoter. Colors here and in (b) are the same as in Fig. 2; in addition, the YFP coding region and polypeptide are colored yellow. (b) YFP fluorescence in ALM TRNs in the indicated strains. The predicted mRNA is also depicted and for homozygous mec-8(u314) worms, a DIC image is shown. (c) Temperature-dependent expression of intron 9::yfp in a mec-8(218ts) mutant. Scale bars, 5 µm. (d) RT-PCR using primers for the last 18 nucleotides of mec-2 exon 9 and 863 nucleotides of yfp of worms grown at 15° C and 25° C. ama-1, encoding the large subunit of RNA polymerase II, is used as control.
The mec-8-dependent splicing of mec-2 intron 9 resulted in the temperature-dependent production of YFP fluorescence in worms with the temperature-sensitive mec-8 allele u218 (ref. 10; Fig. 3c). The u218 mutation produces an Ala278Thr change in the second RRM16. mec-8(u218ts) mutants are wild type at 15° C and touch insensitive (Mec) at 25° C. u218 mutants transformed with Pmec-18intron 9::yfp have fluorescent TRNs at 15° C, but not at 25° C (Fig. 3c). We also assessed the production of yfp-containing messages by RT-PCR of mRNA from u218 mutants expressing Pmec-18intron 9::yfp grown at different temperatures. Amplifiable material representing the splice from exon 9 to the yfp coding region was detected in worms grown at 15° C but not at 25° C (Fig. 3d). These results demonstrate that the control of MEC-8 over intron 9 splicing is very tight at the non-permissive temperature.
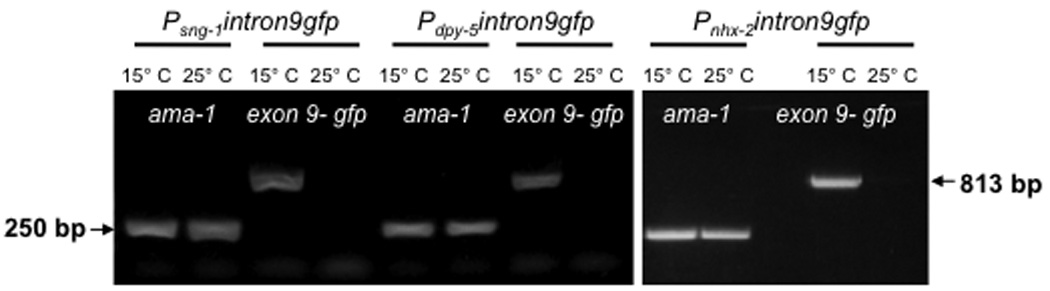
To show that mec-8 and intron 9 of mec-2 can be used to regulate gene expression in other tissues, we placed gfp in the intron 9 cassette driven by promoters specific for neurons (sng-1), hypodermis (dpy-5) and intestine (nhx-2) and injected the constructs into worms of the mec-8(u218ts) genotype. Worms expressing Psng-1intron9::gfp grown at 15° C had fluorescence in the neurons in the head and tail (including the TRNs) where mec-8 is expressed. Even though sng-1 is pan-neuronal, gfp was not seen in the ventral cord, where mec-8 is not expressed (ref. 14; and our unpublished observations). Worms grown at 25° C did not show fluorescent neurons (Fig. 4a). mec-8(u218) expressing Pdpy-5intron9::gfp and Pnhx-2intron9::gfp showed fluorescence in the hypodermis and intestine, respectively, at 15° C but not at 25° C (Fig. 4a). Consistent with these results, the expression of the mRNA for gfp under the sng-1, dpy-5 and nhx-2 was seen only at 15° C and not at 25° C (Fig. 4b). These experiments suggest that gene expression can be made conditional using intron 9 and mec-8(u218ts) for genes in many tissues.
Figure 4. MEC-8-dependent expression of mec-2 intron 9::gfp occurs in many tissues.
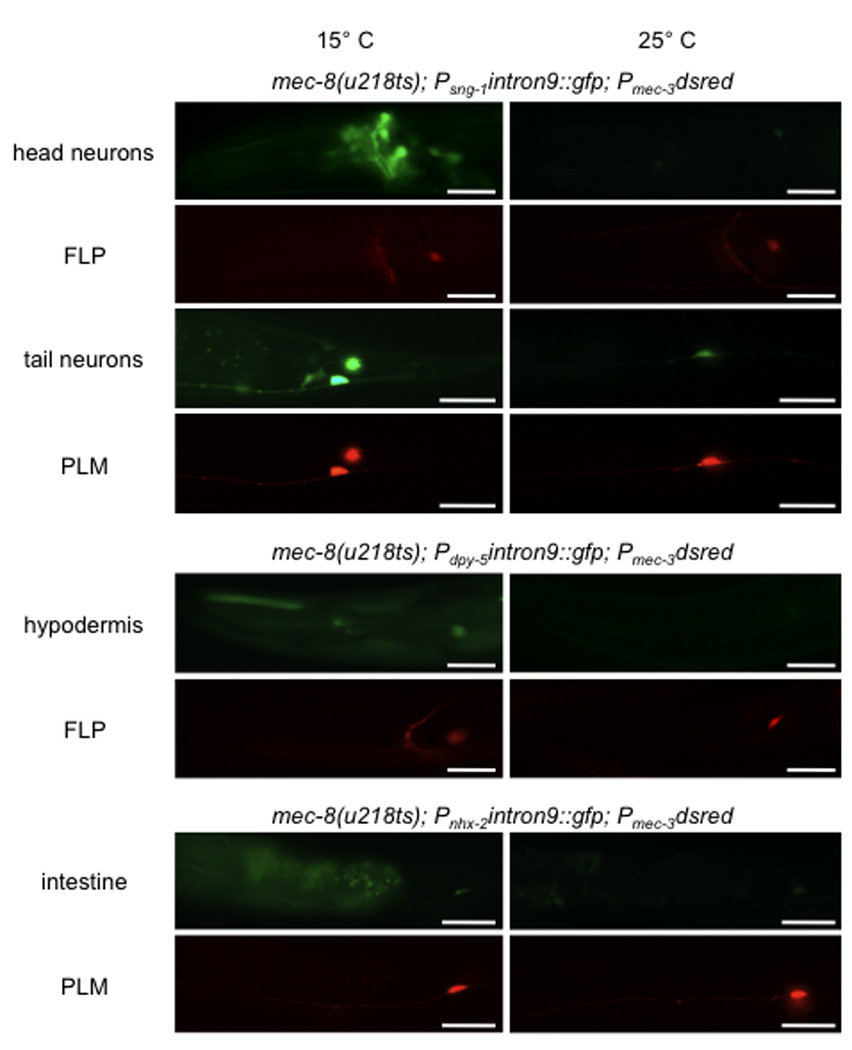
(a) Fluorescence micrographs of single worms showing temperature-dependent expression of intron 9::gfp (green) in neurons (using the sng-1 promoter), hypodermis (dpy-5 promoter), and the intestine (nhx-2 promoter). Expression of dsRed (red) in the FLP and PLM neurons (using the mec-3 promoter) is seen at both temperatures. Scale bars 25 µm. (b) RT-PCR on the indicated strains grown at 15° C or 25° C, using primers for the last 18 nucleotides of mec-2 exon 9 and for 863 nucleotides of gfp. The ubiquitously expressed ama-1 gene was used as a control.
Temperature-sensitive gene expression
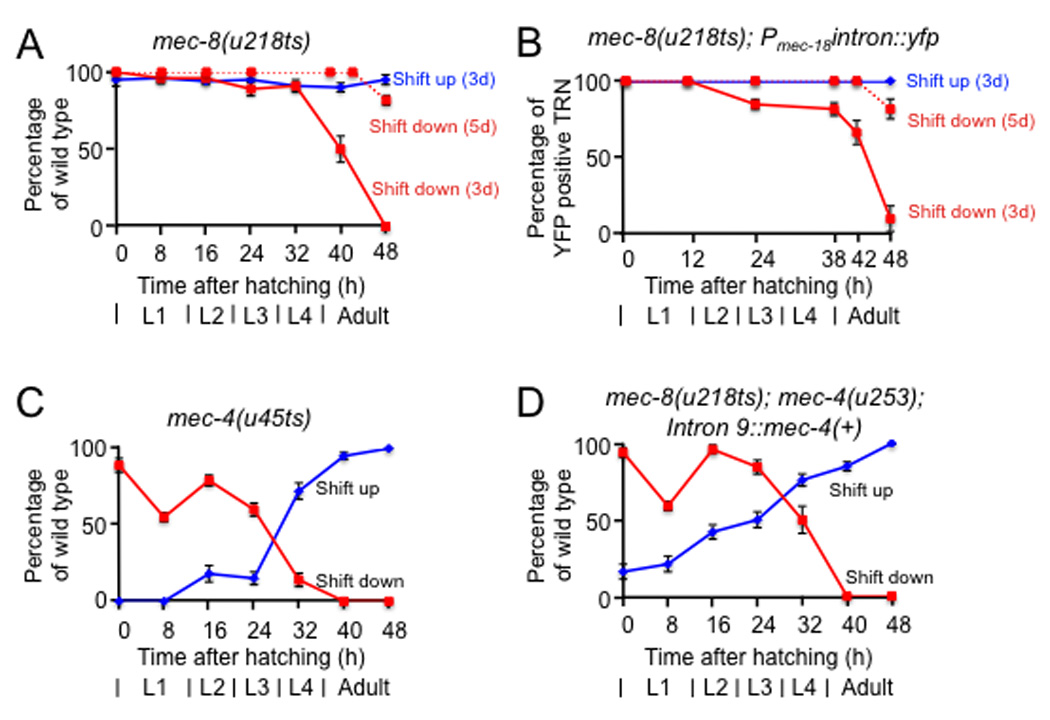
The finding that mec-2 intron 9 can convey mec-8 dependence suggests that intron 9 can be used to generate temperature-dependent constructs of potentially any C. elegans gene; these constructs can then be transformed into appropriate null backgrounds. Certain characteristics of the mec-8 gene and the mec-8(u218ts) allele suggest that intron 9 constructs of a gene of interest would mimic the expression of the endogenous gene product. First, suppression studies and temperature-shift experiments suggest that a relatively small amount of the wild-type MEC-8 may be needed for function, i.e., MEC-8 is not rate-limiting. Specifically, a single dose of a tRNA suppressor mutation can suppress an amber allele of mec-8, but not amber mutations in other mec genes9, 10. Second, shifting mec-8(u218ts) worms from the permissive to the restrictive temperature at hatching (so that only embryonic MEC-8 is available) results in touch sensitive adults (ref. 9; Fig. 5a) that detectably express YFP (Fig. 5b); thus, mec-8 displays considerable perdurance. Third, worms shifted from the restrictive to the permissive temperature as late larvae (Fig. 5a and 5b) produce sufficient mec-8 activity for touch sensitivity and YFP production in adulthood. Indeed, when mec-8(u218ts) worms were shifted to the permissive temperature as adults (Fig. 5a and 5b, dotted lines) and scored 48 hours later, they were both touch sensitive and YFP positive, an indication that MEC-8 is produced even after adulthood in the TRNs.
Figure 5. Intron 9-containing strains and strains with missense temperature-sensitive mutations show similar responses to temperature.
Panels a-d show the touch response of adults with the indicated genotypes that have been shifted up (blue lines and rhomboids) or down (red lines and squares) at the indicated times after hatching. Touch sensitivity and YFP fluorescence were scored at 72 hours (solid lines) and 96 hours (dotted lines) after hatching (n ≥ 3 sets of 30–50 worms). Error bars represent the standard error of the mean.
To test whether the use of intron 9 and mec-8(u218ts) could mimic the results of an endogenous temperature-sensitive mutation, we compared mec-8(u218ts); mec-4(u253) worms expressing Pmec-4intron9::mec-4, with mec-4(u45ts) worms. Although the temperature shift curves of touch sensitivity for mec-8(u218ts) and mec-4(u45ts) are quite different (Fig. 5a and 5c; ref. 9), worms with the intron 9::mec-4 construct showed temperature-shift touch-sensitivity curves that were essentially the same as those from mec-4(u45ts) worms (compare Fig. 5c and 5d). These results suggest that the timing of mec-4 expression, but not of mec-8, is critical for mec-4 function in touch sensitivity in adults.
Temperature-sensitive RNAi
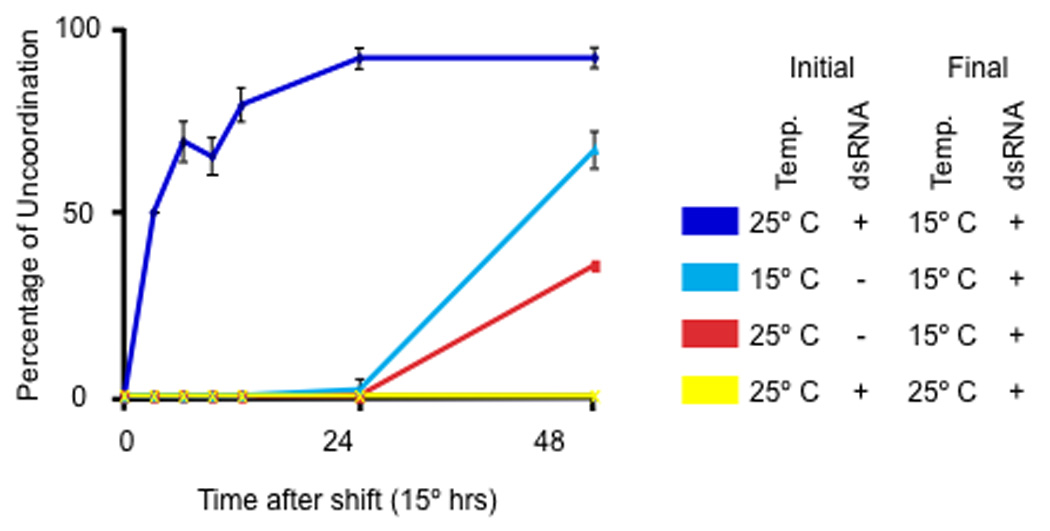
We have used the mec-2 intron 9 to produce temperature-dependent RNA interference (RNAi). We took advantage of the fact that RDE-1, a C. elegans Argonaut protein, is required for RNAi17. Transformation of mec-8(u218ts); rde-1(ne219) worms (ne219 is a null allele) with Prde-1intron 9::rde-1(+) resulted in worms that were responsive to RNAi at 15° C but not at 25° C (Table 1). We observed the relevant temperature-sensitive RNAi phenotype using bacteria making dsRNA for unc-22 (expressed in muscle), unc-52 (expressed in the hypodermis) and rpl-3 (needed for embryonic viability), indicating that the RNAi effects can be detected in many tissues. To test how quickly the RNAi phenotype could be detected, we fed newly hatched intron9::rde-1 worms with bacteria making dsRNA for unc-22 at 25° C for 24 hours and then shifted them to 15° C. The unc-22 twitcher phenotype was seen within 3 hours of the shift (Fig. 6). In comparison, worms placed on the bacteria at the time of the shift did not show a phenotype for at least 24 hours. Since RDE-1 is thought to act as part of the RISC complex18, worms presumably load with dsRNA at the restrictive temperature but cannot execute RNAi. Switching to the permissive temperature allows RNAi to proceed, thus making this method particularly useful for the study of late effects of genes whose loss is lethal.
Table 1.
RNAi responses in the presence or absence of rde-1 and mec-8.
| rde-1 allele | Intron9::rde-1 | mec-8 allele | RNAi (%)a | ||
|---|---|---|---|---|---|
| unc-22 | rpl-3 | unc-52 | |||
| + | − | + | 98 ± 5 | 97 ± 3 | 83 ± 9 |
| + | − | e398 | 99 ± 4 | 98 ± 3 | 86 ± 6 |
| ne219 | − | + | 0 | 0 | 0 |
| ne219 | + | + | 92 ± 7 b | 90 ± 7 b | 78 ± 8 b |
| ne219 | + | e398 | 0 | 0 | 0 |
| ne219 | + | u218 25°C | 0 | 0 | 0 |
| ne219 | + | u218 15°C | 90 ± 5 b | 87 ± 7 b | 79 ± 7 b |
Values indicate the percent of animals that show a phenotype. Values are the mean ± std. dev. (n = 3 sets of 30–50 worms). The phenotypes produced by the three dsRNAs were 1) unc-22 Twitcher by L4 and Paralyzed by adulthood, 2) unc-52 Paralyzed or Uncoordinated, rpl-3 Sterile or Growth defective (arrest as young larvae).
These worms responded in a less severe fashion than wild type and mec-8 mutants did at 15°C. The number of worms affected was not significantly different from wild type, but the phenotypes were less severe though still clear (for unc-22, the worms twitched throughout adulthood, for unc-52, worms were uncoordinated and paralyzed as later adults, and for rpl-3, worms were mostly sterile adults but some were arrested as late (L3 and L4) larvae.
Figure 6. Conditional RNAi is potent and rapidly triggered.
Conditional intron 9::rde-1(+) expression allows for rapid RNAi.
Newly hatched mec-8(u218ts); rde-1(ne219); Prde-1intron 9::rde-1(+) worms were grown for 24 hrs at 25°C (or for the equivalent developmental period – 48 hrs – at 15°C; Initial) and then placed at indicated temperature (Final). Worms were fed either bacteria with dsRNA for unc-22 (+) or control bacteria (OP50)(-). Worms were tested for the unc-22 twitcher phenotype (%Unc) at various times after the shift (times are given in 15°C equivalents). Error bars represent the standard error of the mean.
Discussion
We have developed a method to produce conditional mutants that depends on the correct processing of their transcripts by MEC-8. Taking advantage of the existence of a temperature-sensitive allele of mec-8, researchers can now produce tightly controlled and temperature-sensitive gene expression for virtually any gene in C. elegans. A further elaboration of this method was the generation of a strain that allows temperature-sensitive expression of RNAi.
Previous work by Lundquist et al.11 found that mec-8 facilitated exon skipping in unc-52 mRNAs, allowing splicing from exon 15 to exon 19 and exon 16 to exon 19. Similarly, our data show that mec-8 is required for the skipping of exon 9’ in mec-2, allowing the production of mec-2a mRNA. Since the C terminus of MEC-2A is needed for enhancing MEC-4 channel activity19, the loss of mec-2a mRNA underlies, at least in part, the touch-insensitive phenotype of mec-8 mutants. At this time we do not know whether MEC-8 regulates mec-2 processing by enhancing the exon 9 –10 splice or inhibiting the exon 9 –9’ splice. Based on examples of alternative splicing regulated by other RRM-containing proteins20, 21, however, we suggest that MEC-8 prevents the splice to exon 9’.
We have not tried to determine the minimal sequences needed for mec-8-dependent splicing of intron 9. The smallest number of exonic nucleotides in our constructs was 18 on each side (see Methods, TU#839). Before publication we distributed our constructs to Poon et al.22 who truncated the construct further (with 13 nucleotides at the 5' end and 15 at the 5' end). Their construct appeared to show a modest cold-sensitive expression (instead of heat-sensitive expression) in ventral cord neurons (which do not detectably express mec-8). Their modification appears to modify the splicing of intron 9.
Several characteristics of mec-8 contribute to the usefulness of this conditional system. First, many mec-8 mutants are viable at all growth temperatures. Second, a small amount of MEC-8 is sufficient for its function. Third, mec-8 is expressed in almost every cell and from the early embryo into adulthood. This last property allows conditional expression of the wild-type product from mec-8-dependent constructs at a wide variety of times using temperature shifts. Fourth, the use of mec-8 mutants and different promoters to drive expression of both mec-8(+) and intron 9-containing genes, provides a means of producing both spatial and temporal regulation of expression. For example, the use of different promoters with minimally overlapping expression patterns can permit cell-specific splicing. We have used such combinatorial expression to label specific cells with recGFP23 and to kill them with recCaspase24. Intron 9 and mec-8(+) would allow a similar recSplicing system to be used in C. elegans. For example, the use of a heat-shock promoter to drive the expression of an Intron 9-containing gene and a cell-specific promoter to drive the expression of mec-8(+) in a mec-8 mutant would result in temporal regulation in selective cells.
Since little MEC-8 is needed for touch sensitivity, a potential problem with using intron 9 and mec-8(u218ts) is that perdurance of MEC-8 may obscure the results of shifting from the permissive to the restrictive temperature. Nonetheless, the effect appears minimal, and could be potentially corrected by using a rapidly-degraded MEC-8, such as one with an attached RING domain from an E3 ubiquitin ligase25. Another potential problem is that comparisons are made between animals with and without mec-8 activity. For this reason, care must be taken to show that the mec-8 state does not affect the activity being studied.
Two other systems have used alternative splicing to control gene expression. The first relied on the temperature-dependent release from a block in splicing of an artificial intron to allow gene expression in yeast26. This method required transitions from 16° C to 36° C and resulted in 15–20% of the wild-type activity. The second method utilized sex-specific splicing of transformer in Drosophila to express genes only in females27. The method we describe here differs from these previous methods in that it does not require temperature extremes, appears to be highly efficient and tightly controlled, and is not restricted to one sex. In addition, the mec-8 system can be rapidly activated (within three hours at 15° C), whereas the artificial yeast intron system required growth for 3 days at 36° C.
We have not tested whether this system can be used in organisms other than C. elegans. Other splicing regulators that have identified targets, however, could be used to develop similar regulatory control in other animals and in cells in culture. Such components include Drosophila ELAV, for which temperature-sensitive alleles exist28, and either ewg intron 6 (ref. 29) or the nrg terminal neuronal intron20; and three sets of mammalian components, FOX-1/2 and the alternative neuronal intron of the Calcitonin/CGRP gene21, Nova-1 and clusters of the consensus binding sequence UCAY30, and ETR-3 and cardiac troponin T exon 5 (ref. 31).
Methods
C. elegans growth and strains
Wild-type C. elegans (N2) and strains with mutations in mec-8(u314, e398, or u218ts)I, rde-1(ne219)V, or mec-4(u45ts or u253)X9, 10, 17, were usually grown at 20°C as previously described32. For temperature sensitivity experiments, worms were tested after growth for several generations at either 15° C or 25° C.
Detection of mec-2 transcripts
RT-PCR was used to detect transcript differences between wild type and mec-8 mutants for the following genes: mec-1, mec-2, mec-3, mec-4, mec-5, mec-6, mec-7, mec-8, mec-9, mec-10, mec-12, mec-17, and mec-18. The presence of mec-2a, mec-2b, and mec-2c was tested using RT-PCR and RACE using the SMART RACE Amplification Kit (Clontech). Total RNA was isolated with TRIreagent (Sigma) and poly-A RNA with the Oligotex mRNA Maxi Kit (Qiagen). All the primers used in our experiments are listed in Supplementary Table 1. RACE products were TA cloned, sequenced and analyzed using Sequencher 4.5 (Gene Codes Corporation). We were not able to see mec-2c in our RACE experiments, probably because it is underrepresented compared with mec-2a and mec-2b.
Expression constructs and transformation
Three DNA fragments containing the 1.8 kb sequence of mec-2 intron 9 and different amounts of flanking exon codons were amplified by PCR from the plasmid DT#94 (ref. 12), which contains the full genomic sequence of mec-2, using primers that introduced 5’ and 3’ BamHI sites. The resulting PCR products were cut with BamHI and cloned between the mec-18 promoter and the yfp coding sequence in TU#739 (ref. 24) to generate TU#821 [Pmec-18intron 9::yfp(fragment I)], TU#838 [Pmec-18intron 9::yfp(fragment II)], and TU#839 [Pmec-18intron 9::yfp(fragment III)] or placed between the promoter and the genomic coding sequence of rde-1 [TU#820, Prde-1intron 9(fragmentII)::rde-1] or between promoter and the genomic coding sequence of mec-4 [TU#837, Pmec-4intron 9(fragment II)::mec-4,] in Fire vector pPD95.75 (www.ciwemb.edu/pages/firelab.html). rde-1 was amplified from genomic DNA with primers including PmlI/ BamHI sites for 2.1 kb of promoter region and BamHI/ ApaI sites for the coding sequence.
The mec-4 gene was amplified in two parts from genomic DNA TU#12 (ref. 33) using primers including HindIII/ XbaI sites for the 823 bp promoter region and BamHI/ EagI sites for the coding sequence. Tissue-specific constructs were made by inserting the intron 9 cassette (fragment III, cut BamHI from TU#839) into pPD95.75 creating TU#871. 2 kb of promoter sequences of sng-1, dpy-5 and nhx-2 were cloned into plasmid TU#871 using primers that created PstI sites for Psng-1, SphI/ XbaI sites for Pdpy-5; and SphI/ XbaI sites for Pnhx-2. The insertion of the mec-2 sequences before yfp (fragments I and III) or gfp (fragment III) introduced several amino acids to YFP and GFP (52 for fragment I and 6 for fragment III). Fragment II, which was inserted before the genomic sequences for rde-1 and mec-4, did not introduce a new start codon, so their translation start was unaltered.
We generated transgenic worms by microinjection34. The injection mix contained 10– 40 ng/µl of one of the intron 9 constructs; combined with either 40 ng/µl of the dominant Roller marker plasmid34 (for the YFP vector), 20 ng/µl of pCW2.1 (a ceh-22::gfp plasmid; ref. 35) for the rde-1 and yfp plasmid, 20 ng/µl of Pmyo-3gfp for the mec-4 plasmid or 10 ng/µl of Pmec-3dsred (a gift from I. Topalidou) for sng-1, dpy-5 and nhx-2 plasmids. pBSK (Stratagene) plasmid was used as filling DNA to a final concentration of 100 ng/µl for all injection mixes. At least 5 stable lines were generated for each construct and all behaved similarly.
Detection of MEC-8-dependent splicing
Production of mec-2 exon-9::yfp or mec-2 exon-9::gfp mRNA at 15° C and 25° C was assessed by RT-PCR using primers for the last 18 nucleotides of mec-2 exon 9 and for 863 nucleotides of yfp or gfp. The reverse primer amplifies both yfp and gfp messages, since it does not target the regions of divergence.
Immunochemistry
Whole-mount immunochemistry was carried out as in Ruvkun et al.36. We used acetone powder-purified MEC-2 antibody (ref. 37; 1:1000) and rhodamine-conjugated goat anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA; 1:1000). Both MEC-2 antibodies were used in parallel and gave the same results in wild type. MEC-2 antibody-staining pattern was scored in blind tests.
Phenotype characterization
Temperature shifts and YFP expression: Worms synchronized at hatching were grown at the appropriate temperatures for at least two generations. They were shifted from 15° C to 25° C or 25° C to 15° C at the indicated times, and tested for touch sensitivity and/or YFP expression as egg-laying adults. YFP fluorescence was observed using a Zeiss Axiophot II or a Leica stereodissecting microscope equipped for fluorescence. DIC optics were used to observe the TRNs when YFP was absent.
RNAi sensitivity: RNAi responses were tested by growth on bacteria making dsRNA for unc-22, unc-52, or rpl-3 according to Timmons and Fire38. All RNAi experiments were performed in triplicates in at least 3 independent experiments on different days. Thirty to fifty worms were used and scored in each plate. Temperature-sensitive mutants were grown at 25° C or 15° C for more than two generations prior to the experiments. The scoring was performed blindly.
The time course of RNAi induction was determined using worms grown at 25° C in the presence of unc-22 dsRNA expressing bacteria or OP50 (for three generations) and were synchronized and re-fed as L1 larvae in the desired condition. After 24 hrs, worms were moved to 15° C and scored for the unc-22 twitcher phenotype at intervals of 3 hrs for the first day, 12 hrs for the second day, and 24 hrs for subsequent days.
Supplementary Material
Figure 7.
Acknowledgments
We thank Mingxia Huang for providing valuable insight in the work with mec-2, and Larry Chasin, Jim Manley, and members of the Chalfie lab for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health grant GM30997 to M.C.
Footnotes
Author contributions
A.C., C.M. and M.C. designed experiments; A.C. performed the experiments; A.C. and M.C. wrote the manuscript.
References
- 1.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 2.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 3.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis MW, Morton JJ, Carrol D, Jorgensen EM. Gene Activation Using FLP Recombinase in C. elegans. PLoS Genetics. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voutev R, Hubbard AJ. A “FLP-Out” System for Controlled Gene Expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horowitz NH, Leupold U. Some recent studies bearing on the one gene one enzyme hypothesis. Cold Spring Harb. Symp. Quant. Biol. 1951;16:65–74. doi: 10.1101/sqb.1951.016.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Epstein RH, et al. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp. Quant. Biol. 1963;28:375–394. [Google Scholar]
- 9.Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- 10.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 11.Lundquist EA, et al. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122:1601–1610. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- 12.Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 13.O'Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat. Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 14.Spike CA, Davies AG, Shaw JE, Herman RK. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development. 2002;129:4999–5008. doi: 10.1242/dev.129.21.4999. [DOI] [PubMed] [Google Scholar]
- 15.Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 16.Davies AG, Spike CA, Shaw JE, Herman RK. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics. 1999;153:117–134. doi: 10.1093/genetics/153.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 18.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 19.Goodman MB, et al. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 20.Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 2001;15:2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon VY, Klassen MP, Shen K. UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature. 2008;455:669–673. doi: 10.1038/nature07291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. U S A. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyurovsky MV, et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol. Cell. 2003;12:875–887. doi: 10.1016/s1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimatsu T, Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989;244:1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- 27.Fu G, et al. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol. 2007;25:353–357. doi: 10.1038/nbt1283. [DOI] [PubMed] [Google Scholar]
- 28.Samson ML, Lisbin MJ, White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141:1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soller M, White K. ELAV inhibits 3'-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen KB, et al. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. U S A. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 32.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 34.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 36.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, et al. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr. Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


