Abstract
The tropism of breast cancer cells for bone and their tendency to induce an osteolytic phenotype are a result of interactions between breast cancer cells and stromal cells, and are of paramount importance for bone metastasis. However, the underlying molecular mechanisms remain poorly understood. We hypothesize that tumor-stromal interaction alters gene expression in malignant tumor cells and stromal cells creating a unique expression signature that promotes osteolytic breast cancer bone metastasis and that inhibition of such interactions can be developed as targeted therapeutics. Microarray analysis was performed to investigate gene expression profiling at the tumor-bone (TB) interface versus the tumor alone area from syngenic mice injected with three different syngenic mammary tumor cell lines that differ in their metastatic potential. We identified matrix metalloproteinase (MMP)-13, receptor activator of NF-kB ligand (RANKL), and integrins binding sialoprotein (IBSP) to be genes upregulated at the tumor-bone interface and validated. To determine the functional role of MMP-13 in tumor-induced osteolysis, mice with Cl66 mammary tumors were treated with MMP-13 antisense oligonucleotides (MMP13-ASO) or controlled scrambled oligonucleotides (control-ASO). Knockdown of MMP-13 expression at the TB-interface leads to significant reduction in bone destruction, and the number of activated osteoclasts at the TB-interface. Further analysis to evaluate the mechanism of MMP-13 dependent osteolytic bone metastasis revealed that MMP13-ASO treatment decreased active MMP9, RANKL levels and TGF-β signaling at the TB-interface. Together, our data indicates that upregulation of MMP-13 at the TB-interface is important in tumor-induced osteolysis and suggests that MMP-13 is a potential therapeutic target for breast cancer bone metastasis.
Keywords: MMP13-ASO, Bone metastasis, soluble RANKL, Tumor-induced osteolysis
Introduction
Breast cancer is the most common cancer and the second leading cause of cancer-related death in women in the United States (1). Most complications of breast cancer are attributed to metastasis to distant organs including lymph nodes, liver, lung and bone (2, 3). In advanced stages of the disease nearly all breast cancer patients suffer with bone metastasis. Bone metastases in breast cancer are predominantly osteolytic and also cause skeletal lesions including pathological fracture, intractable bone pain, nerve compression and hypercalcemia (4, 5). These complications not only increase the risk of mortality but also cause a significant decrease in the quality of life (3).
Breast cancer cells show a strong predilection for bone (5). Arrival of tumor cells in the bone microenvironment initiates a “vicious cycle” of bi-directional interactions between tumor cells and stromal cells (3, 6). Tumor cells produce various factors such as parathyroid hormone-related peptide (PTHrP) (7, 8), interleukin (IL)-8, and IL-1 to stimulate osteoblasts to induce expression of receptor activator of NF-κB ligand (RANKL) to induce bone resorption (9). Increased bone resorption causes the release of sequestered factors that favor the growth of malignant tumor cells including bone derived growth factor (BDGF), fibroblast growth factor (FGF) and transforming growth factor–beta (TGF-β) (3). The underlying molecular mechanisms of tumor-bone interaction are poorly understood. In this report, we hypothesize that tumor-stromal interaction in the bone microenvironment alters gene expression in malignant tumor cells and stromal cells creating a unique expression signature that promotes osteolytic bone metastasis and that inhibition of such interactions can be targeted for development of novel therapeutics.
Extracellular Matrix (ECM) degradation, mediated by matrix metallo proteinases (MMPs), is an essential step in the growth, invasion and metastasis of malignant tumors. MMPs are a family of human zinc endopeptidases that can degrade virtually all ECM components (10). Apart from their ECM degradation functionality latest research in MMPs reveals their specific roles in cleaving several extracellular and membrane associated proteins and regulating cellular signaling pathways. MMP7 promotes osteolytic bone metastasis in prostate cancer through generation of sRANKL from membrane bound RANKL (11). MMP2 and MMP9 have been associated with tumor angiogenesis (12). Expression of these proteases is also associated with poor clinical outcome in various malignancies such as bladder, breast, lung cancer and head and neck squamous cell carcinomas (13, 14).
MMP13 was first identified from the overexpressing breast carcinomas (15). IL-1α and IL-1β are potential candidates for inducing expression of MMP13 in breast carcinomas (16). In case of Squamous Cell Carcinoma (SCC) MMP-13 is predominantly expressed by the tumor cells at the invading front and to some extent by stromal fibroblasts surrounding tumor cells (17, 18). Expression of MMP-13 in the head and neck SCCs correlates with the invasion and metastatic capacity (17, 18). In laryngeal and vulvar carcinomas the expression of MMP-13 colocalizes with the expression of MT1-MMP and MMP-2 suggesting that these three MMPs form a proteolytic cascade that leads to potent extracellular collagenolytic activity (19). In non-small cell lung carcinoma tumor cells expressing MMP13 have a potential to shed from the primary tumor and aggregate in the bone marrow and associated with poorer survival rates (20). But the specific role of MMP13 in malignant breast cancer remains unclear. Our study is focused on elucidating the role of MMP13 in the tumor-stromal interaction, with particular attention to the tumor-bone microenvironment.
We identified matrix metalloproteinase (MMP)-13, RANKL, and integrins binding sialoprotein (IBSP) were the genes upregulated at the tumor bone (TB)-interface. Moreover, we demonstrated that knockdown of MMP-13 expression at the TB-interface leads to a significant reduction in bone destruction and the number of activated osteoclasts at the TB-interface. MMP13-ASO treatment decreased the RANKL:OPG ratio, active MMP9 and TGF-β levels at the TB-interface. Together, our data demonstrated that upregulation of MMP-13 at the TB-interface is important for regulation of tumor-induced osteolysis and suggest that MMP-13 might be a potential novel therapeutic target for breast cancer bone metastasis.
Materials and Methods
Cells and Animals
Three murine mammary adenocarcinoma cell lines differing in their metastatic potential, 4T1 (highly metastatic), Cl66 (moderately metastatic) and Cl66M2 (poorly metastatic) were used in this study (21, 22). Cells were maintained in Dulbecco's Modified Eagle Media (DMEM) (Mediatech, Hendon, VA) with 5% serum supreme (Biowhitaker, Walkersville, MD), 1% vitamins, 1% L-glutamine and 0.08% gentamycin (Invitrogen, Carlsbad, CA).
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nebraska Medical Center. Eight week old female BALB/c mice (NCI, Bethesda, MD) were used in this study. For in vivo experiments, tumor cells (5 × 104 cells/50ul) mixed in growth-factor reduced Matrigel (BD Biosciences, San Jose, CA) were injected directly onto the calvaria to mimic the close association of tumor cells and bone. Four-weeks post implantation of tumor cells, mice were sacrificed and tumor alone and TB-interface samples were collected. For immunohistochemical analysis, the samples were fixed in 4% paraformaldehyde at 4°C for 48 hours. The tissues were then transferred into a decalcification solution (15% EDTA with glycerol, pH 7.4) for four weeks and were subsequently paraffin embedded and processed for histology.
RNA and protein extraction were done by homogenization of tissue samples in liquid nitrogen. Total RNA was extracted using Trizol (Invitrogen, CA) following the manufacturer's instructions. The RNA concentration was quantified using a NANO drop ND-1000 Spectrophotometer (Nano Drop Technologies, Wilmington, DE).
Protein was extracted using T-PER tissue protein extractor solution (Pierce, Rockford, IL) following the manufacturer's provided protocol. Protein samples were quantified using a BCA protein assay kit (Pierce). Proteinase activity assays were performed with protein samples without protease inhibitors.
Microarray analysis and quantitative real time PCR (qRT-PCR)
Calcified frozen sections were serially sectioned in 10-μm thick slices and at least ten slides per mouse were micro-dissected with careful separation of the TB-interface and the tumor alone areas as described earlier (22, 23). Total RNA was extracted from each micro-dissected population, pooled and an equal amount of RNA was amplified using a probe amplification kit (Affymetrix, Santa Clara, CA). An Affymetrix Mouse Expression Array 430 was used for comparing gene expression profiles between the TB-interface and the tumor alone areas. A complete detection and analysis of signals for each chip was performed using Affymetrix GeneChip® Operating Software to generate raw expression data. A signal log ratio algorithm was used to estimate the magnitude of change of a transcript when two arrays were compared (experimental versus baseline). It was calculated by comparing each probe pair on the experimental array, here the tumor-bone interface, to the corresponding probe pair on the baseline array, here the tumor alone area, and considering the mean of the log ratios of probe pair intensities across the two arrays. The change is expressed as the log2 ratio. Thus a signal log ratio of 1.0 indicates an increase of transcript level by 2 fold and -1.0 indicates a decrease by 2 fold. For each set of tissue from 4T1, Cl66 and Cl66M2, the signal log ratio of the TB-interface versus the tumor alone area was calculated, and the genes were ordered from highest to lowest expression levels.
Gene expression analysis was confirmed using qRT-PCR for the TB-interface and tumor alone area samples. 5 μg of RNA from each sample was used to synthesize first strand cDNA. 2 μl of 1:100 diluted first strand cDNA were amplified in a 20 μl reaction with SYBR green master mix (Roche, Indianapolis IN) and 10 mM primer mix using a Bio-Rad iCycler (Bio-Rad, Hercules, CA). The following reaction conditions were used: initial denaturation at 95°C for 3 min, followed by amplification cycles with denaturation at 95°C for 60 s, annealing at 60°C for 60 s, and extension at 72°C for 60 s, and finally a long extension at 72°C for 2 min. Primers used for validation of gene expression are included in Supplementary Table 1. The fluorescence intensity of double-strand specific SYBR Green, reflecting the amount of formed PCR-product, was monitored at the end of each elongation step. The ct value for each gene was normalized with GAPDH expression for relative gene expression analysis.
Immunohistochemistry and TRAP staining
MMP-13 protein expression was evaluated by immunohistochemistry on tumor sections using a MMP-13-specific antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The sections were deparaffinized using EZ dewax solution (Biogenex, San Ramon, CA). For antigen retrieval, the sections were boiled in 10 mM citrate buffer (pH 6.0) for 10 min and endogenous peroxidase activity was blocked using 3% H2O2 for 5 min. The sections were then blocked in antibody diluent for one hour at room temperature. RANKL antibody was diluted 1:100 in blocking solution and sections were incubated overnight at 4°C. After washing, the slides were incubated with anti-goat biotinylated antibody for 30 min at room temperature. After washing, immunoreactivity was detected using Vectastain ABC and DAB substrate kits (Vector Laboratories, Burlingame, CA). Sections were counterstained with hematoxylin, dehydrated, and permanently mounted. The total number of TRAP-positive, multinucleated osteoclasts over the entire length of the tumor-bone interface were then counted for each section using a light microscope. The total number of osteoclasts was then divided by the length of the tumor-bone interface to get the number of osteoclasts per mm of tumor-bone interface.
For in vivo evaluation of TGF-β signaling, we performed immunohistochemistry for phosophorylated Smad-2 (P-Smad-2) (24, 25). Sections were blocked using goat serum diluted 1:500 for one hour at room temperature. Sections were then incubated overnight at 4°C with antibody directed against P-Smad-2 (Ser 465/467, Cell Signaling Technology, Danvers, MA) diluted 1:50 in blocking solution. After washing, sections were incubated for one hour at room temperature with biotinylated anti-rabbit IgG diluted 1:500.Tartrate resistant acid phosphatase (TRAP) staining was performed to detect activated osteoclasts in vivo according to the manufacturer's instructions (Sigma Chemicals, St. Louis, MO). Briefly, deparrifinized slides were rinsed with deionized water before incubating with TRAP containing buffer at 37°C for one hour, rinsed with deionized water and counter stained with Gill3 Hematoxylin solution for two minutes followed by aqueous mounting. Immunostained sections were examined under a Nikon light microscope and the number of TRAP positive multinucleated cells at the TB-interface was assessed at a magnification of 400× for each lesion.
Antisense oligonucleotide treatment for inhibition of tumor induced osteolysis
Antisense oligonucleotides (ASO) used in the therapeutic protocol were obtained from Isis Pharmaceuticals, Carlsbad, CA. Antisense oligonucleotides designed specifically to target MMP-13 were used throughout this study. 2′-methoxyethyl modified chimeric ASOs with a phosphorothioate backbone were synthesized as previously described (26). Briefly, these oligonucleotides are modified at the 2′ sugar position of the five bases at both the 3′ and 5′ ends with a methoxyethyl group. This modification greatly increases the stability of the oligonucleotides and the affinity for its target mRNA while reducing the amount of immune stimulatory and inflammatory effects that can be seen with oligonucleotides in mice (27). Active ASOs were identified by screening 46 ASOs designed to be specific for MMP-13. The efficacy of these ASOs was confirmed in concentration-response experiments and the most potent ASO was used for further experiments. Control-ASO did not match any known mRNA in the mouse genome.
Animals bearing Cl66 mammary tumors were randomly divided into two treatment groups (Control-ASO and MMP13-ASO). The oligonucleotides were dissolved in physiological saline (0.9% NaCl) and were administered by intraperitoneal (i.p.) injection at a dose of 50 mg/kg/day starting at day 7 following tumor implantation for 5 days with two days off followed by another 4 days. Tumor growth was monitored and mice were sacrificed on day 28. Tumor alone and TB-interface samples were collected and processed for further analysis.
Gelatin Zymography
50 μg total protein isolated from either the TB-interface or tumor alone area from animals implanted with Cl66 tumors was subjected to electrophoresis on a 10% (w/v) polyacrylamide SDS gel containing 1 mg/mL porcine gelatin (Sigma-Aldrich, St. Louis, MO). At the completion of electrophoresis, the gel was washed with 2.5% Triton X-100 buffer for 30 minutes. After rinsing, the gel was incubated for 12 hours at 37°C in incubation buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 10 mM CaCl2, and 0.05% (w/v) NaN3. After rinsing, the gel was then stained using 0.025% Coomassie brilliant blue (Bio-Rad, Hercules, CA) and photographed using a Multi Image Light Cabinet (Alpha Innotech Corporation, San Leandro, CA). The volume of gelatinolytic activity was evaluated using ImageQuant® 5.1 (Molecular Dynamics, Sunnyvale, CA).
Pro-MMP9 Enzyme-linked Immunosorbant Assay (ELISA)
Tumor-bone lysates from control-ASO and MMP13-ASO treated mice (400 ng/μl) were used for analysis. The concentration of pro- MMP-9 in these samples was determined by Quantikine Mouse pro-MMP-9 ELISA according to the manufacturer's instructions (R&D Systems, MN). This assay is a quantitative ‘sandwich’ enzyme immunoassay. A curve of the absorbance of samples to the standard curve (plotted using recombinant pro-MMP9 protein), we determined the concentrations of pro-MMP9 in the tumor lysates.
Statistical analysis
For in vitro studies, the Student's t-test was used for statistical comparison. For in vivo studies, the Mann-Whitney U-test was used for statistical comparison. A p < 0.05 was considered significant.
Results
Gene expression profile at the tumor bone interface
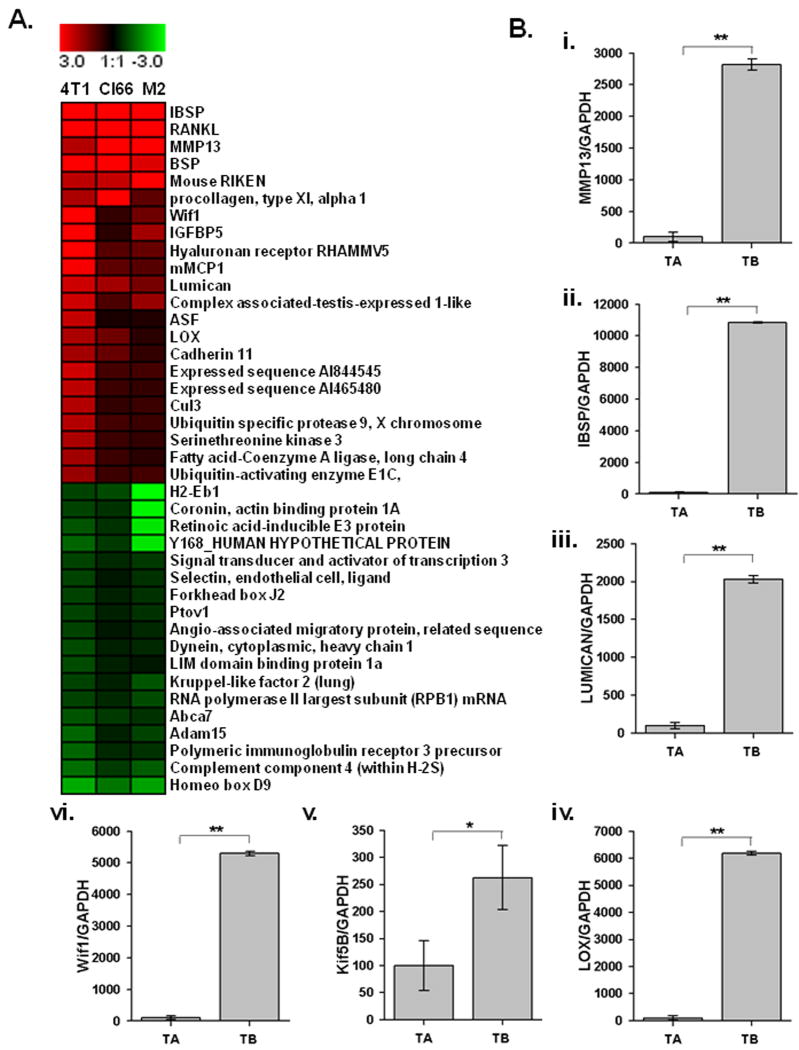
We analyzed gene expression patterns at the TB-interface compared to the tumor alone area using cDNA microarray. Mammary tumor cells with different metastatic potential, 4T1 (high), Cl66 (moderate), and Cl66M2 (low), were transplanted onto the calvaria of BALB/c mice. Histological analysis showed that all tumors exhibited tumor-induced osteolysis and osteoclast activation. Microarray analysis revealed the upregulation of 414 genes and the downregulation of 27 genes at the TB-interface compared to the tumor alone area. The highly upregulated genes were IBSP, RANKL, MMP-13, insulin growth factor binding protein (IGFBP)5, Lumican, Lysyl oxidase (Lox), Kinesin family 5B (Kif5b) and Wnt inhibitory factor 1(Wif1) (Figure 1A). The common upregulation of these genes in all three cell lines further suggested that they may play an important role in mammary tumor induced osteolysis.
Figure 1. Gene expression profile at the TB-interface.
A. Changes in the gene expression profile at the TB-interface in comparison with the tumor alone area were determined using cDNA microanalysis using week 4 samples from 4T1, Cl66 and Cl66M2 tumors. B i to vi. mRNA expression of MMP-13, IBSP, Lumican, LOX, Kif5b and WIF1 was confirmed by real-time PCR analysis with gene specific primers. Relative expression of these genes from tumor alone and TB-interface samples were normalized to GAPDH expression. Real time gene expression data presented is representative of all three cell lines done in triplicate. Bars, SEM. **p <0.05.
We then used quantitative RT-PCR with gene specific primers to confirm gene expression at the TB-interface. Our data confirmed the upregulation of mRNA expression of MMP-13 (Figure 1B-i), IBSP (Figure 1B-ii), Lumican (Figure 1B-iii), Lox, (Figure 1B-iv) Kif5b (Figure 1B-v), and WIF1 (Figure 1B-vi) at the TB-interface.
Upregulation of MMP-13 at the TB-interface
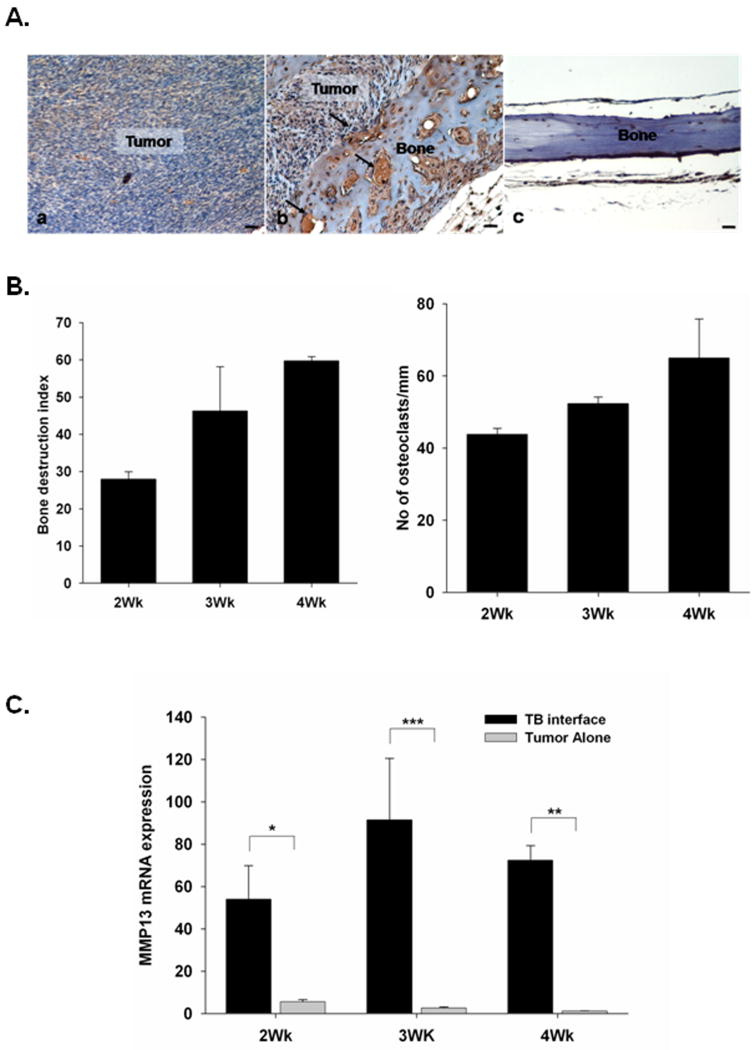
Recognizing the important role of proteases in tumor progression, we further evaluated the functional role of MMP-13 in mammary tumor induced osteolysis. We examined MMP-13 protein expression at the TB-interface by immunohistochemistry. MMP-13 specific antibody staining revealed a significant upregulation of MMP-13 in Cl66 tumors at the TB-interface (Figure 2A.a) but not at the tumor alone area (Figure 2A.b). We observed low levels of immunoreactivity in normal calvaria (Figure 2A.c). Antibody controls did not show any non-specific staining (data not shown). This protein level analysis further confirmed the earlier observation of increased MMP-13 transcript levels by cDNA microarray and qRT-PCR analyses. In addition, our immunohistochemical analysis reveals that MMP13 is expressed by tumor cells, osteoblasts and stromal cells at the tumor-bone interface, which are interacting with cells of bone microenvironment (Figure 2A.a).
Figure 2. MMP-13 expression at TB interface is associated with tumor-induced osteolysis.
A. Expression of MMP-13 at the tumor-bone interface. Immunohistochemistry for MMP-13 was done on sections from Cl66 tumor-bearing mice and non-tumor bearing mice. i. Tumor cells, osteoblasts and stromal cells at the tumor-bone interface were stained positive for the MMP13 (arrowheads). ii. Tumor cells in the tumor alone area did not stain for MMP-13. iii. immunostaining of normal bone showing very low MMP13 expression. Bar, 0.01 mm. B. Severity of tumor-induced osteolysis was computed by measuring the bone destruction index (BDI) on sections from Cl66 tumor-bearing mice at the 2nd, 3rd and 4th week after tumor implantation. We observed an increase in the BDI with time and an association with osteoclast homing at the TB-interface. Bars, SD. (n=5). C. Kinetics of MMP-13 expression at different time points post tumor implantation by RT-PCR. TB-interface and tumor alone area samples from Cl66-tumor bearing mice were analyzed for MMP-13 expression and an increase in expression was observed at the TB-interface at all time points. Relative MMP-13 expression was normalized to GAPDH expression. Bars, SD; *p< 0.05, **p < 0.01, ***p<0.005.
Kinetics of MMP-13 mRNA expression and tumor-induced osteolysis
In the next set of experiments, we examined the kinetics of MMP-13 expression and its association with tumor-induced osteolysis and the number of activated osteoclasts. Mice bearing Cl66 tumors were killed 2, 3 or 4-weeks after tumor implantation and tumors were examined for bone destruction, osteoclast number and MMP-13 expression at the TB-interface and tumor alone areas. We quantified the severity of lesions by measuring bone destruction index (BDI), which is a ratio of the length of the bone that is destroyed by the tumor to the total length of the bone at the TB-interface. Results shown in Figure 2B demonstrate an increase in bone destruction and osteoclast number over time. We observed higher levels of MMP-13 mRNA expression at the TB-interface as compared to tumor alone areas (Figure 2C).
Inhibiting MMP-13 expression abrogates mammary tumor-induced osteolysis and number of activated osteoclasts at the TB-interface
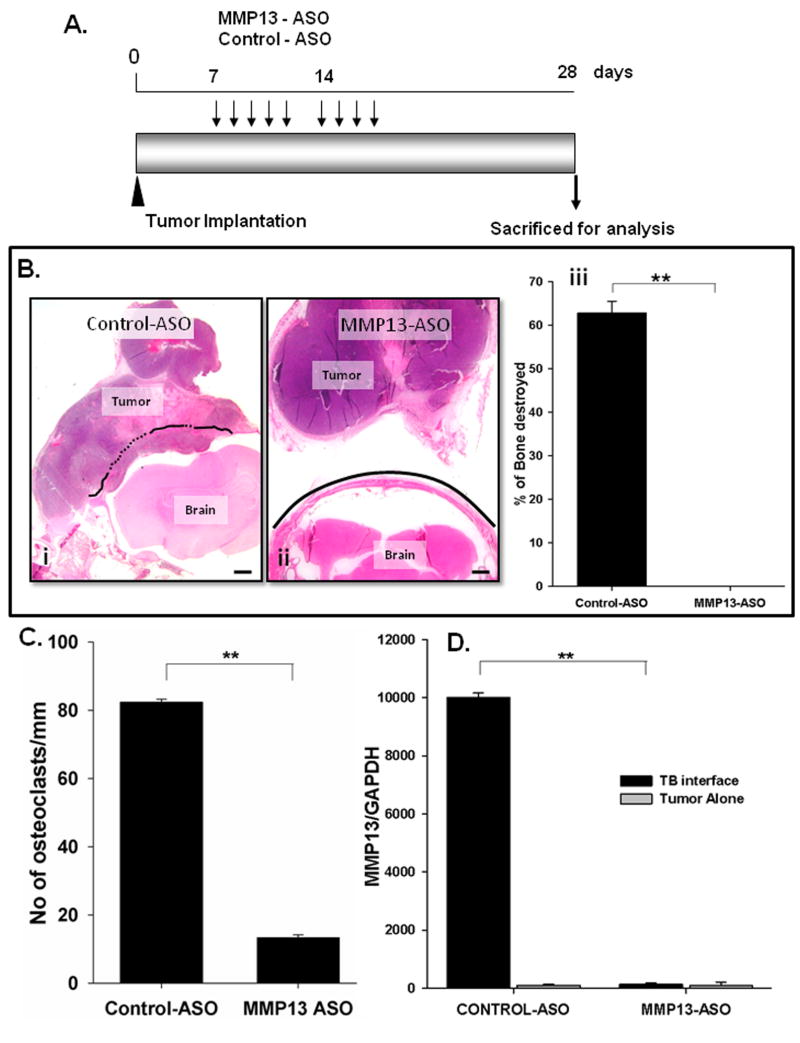
Next, we examined whether knocking down MMP-13 expression in Cl66 tumor bearing mice using ASOs inhibited mammary tumor-induced osteolysis. We did not observe any weight loss or toxicity in any of the treatment groups (data not shown). We observed smaller tumors in MMP13-ASO treated group as compared to Control-ASO treated group; however the decrease in tumor size was not significant (data not shown).
H&E staining of tumor sections demonstrated severe bone destruction in control-ASO treated tumors (Figure 3Bi) whereas MMP13-ASO treated tumors showed no osteolysis (Figure 3Bii). We observed a significant decrease in osteolysis in the MMP13-ASO treated group compared to the control-ASO treated group (Figure 3Biii). Similarly, we observed a significant decrease in the number of activated osteoclasts lining the TB-interface in the MMP13-ASO treated group (Supplementary Figure 1, Figure 3C).
Figure 3. Inhibition of MMP-13 in vivo reduces tumor induced osteolysis.
A. Experimental strategy for treatment of Cl66 tumor-bearing mice with MMP13-ASO and Control-ASO. B. H&E stained sections (20× magnification) of tumors from Control-ASO (i) and MMP13-ASO (ii) treated mice. Solid line represents the intact bone and dotted line represents the bone resorbed. (iii). Severity of the osteolytic lesion was measured by calculating the BDI, a significant reduction in the BDI was observed in MMP13-ASO treated mice compared to Control-ASO treated mice. Bars, SD; (n=5 per group and the study was repeated with same number of mice in each group). C. Number of TRAP positive osteoclasts in MMP13-ASO and Control-ASO treated mice. D. Expression of MMP-13 at the TB-interface of MMP13-ASO treated animal is significantly reduced compared to control-ASO treated mice. MMP-13 expression was determined by RT-PCR and normalized to GAPDH expression. The values are mean ± SEM. This is a representative of three experiments with similar results. Scale bar is 0.01 mm.
We examined the expression of MMP-13 in tumors from MMP13-ASO and control-ASO treated mice using qRT-PCR. The expression of MMP-13 at the TB-interface was significantly lower in the MMP13-ASO treated group as compared to the control-ASO treated group (Figure 3D). We observed very low levels of MMP-13 expression in the tumor alone area which was not altered by MMP13-ASO treatment (Figure 3D).
Inhibition of MMP-13 decreases RANKL expression and regulates the RANKL:OPG ratio at the TB-interface
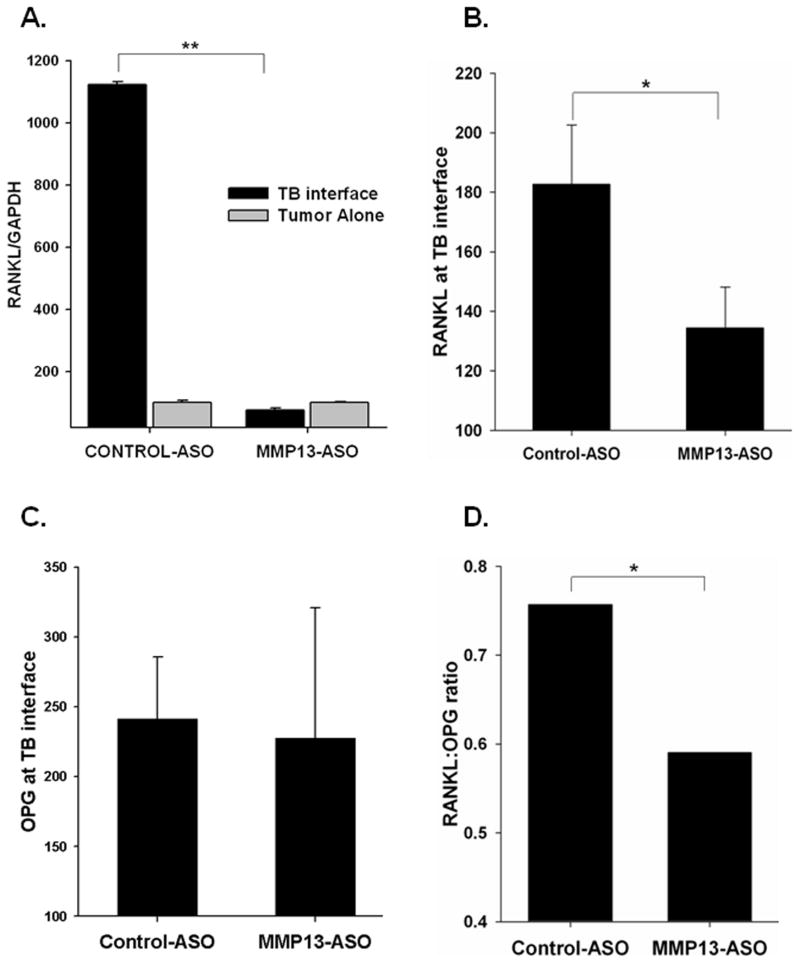
Previous observations from our laboratory have shown the upregulation of RANKL at the TB-interface in mammary tumor-induced osteolysis (28). RANKL interacts with its receptor RANK and activates osteoclasts. We evaluated RANKL mRNA levels in MMP13-ASO and Control-ASO treated mice and observed a significant decrease in RANKL expression at the TB-interface in the MMP13-ASO treatment group (Figure 4A).
Figure 4. Inhibition of MMP-13 in vivo regulates the RANKL:OPG ratio at the TB-interface.
RANKL mRNA (A) and protein (B) levels were significantly reduced in tumor-bone samples from MMP13-ASO treated mice compared to Control-ASO treated mice. C. OPG levels at the TB-interface did not differ between MMP-13 and Control-ASO groups as demonstrated by ELISA. D. The RANKL:OPG is significantly decreased at the TB-interface in MMP13-ASO treated mice. The values are mean ± SEM. This is a representative of three experiments with similar results.
The RANKL/OPG axis has been shown to play a pivotal role in osteolytic bone metastasis (29). We examined how the RANKL:OPG ratio at the TB-interface was altered due to MMP-13 ASO treatment. RANKL levels were significantly reduced due to MMP13-ASO treatment (Figure 4B), whereas we did not observe any significant difference in OPG levels between the two treatment groups (Figure 4C). With the decrease in the RANKL levels, we observed a significant decrease in the RANKL:OPG at the TB-interface in the MMP13-ASO treatment group (Figure 4D). These observations suggest a functional role for MMP-13 in tumor induced osteolysis mediated by the regulation of the RANKL/OPG axis.
Targeting MMP-13 inhibits MMP9 activation and TGFβ signaling
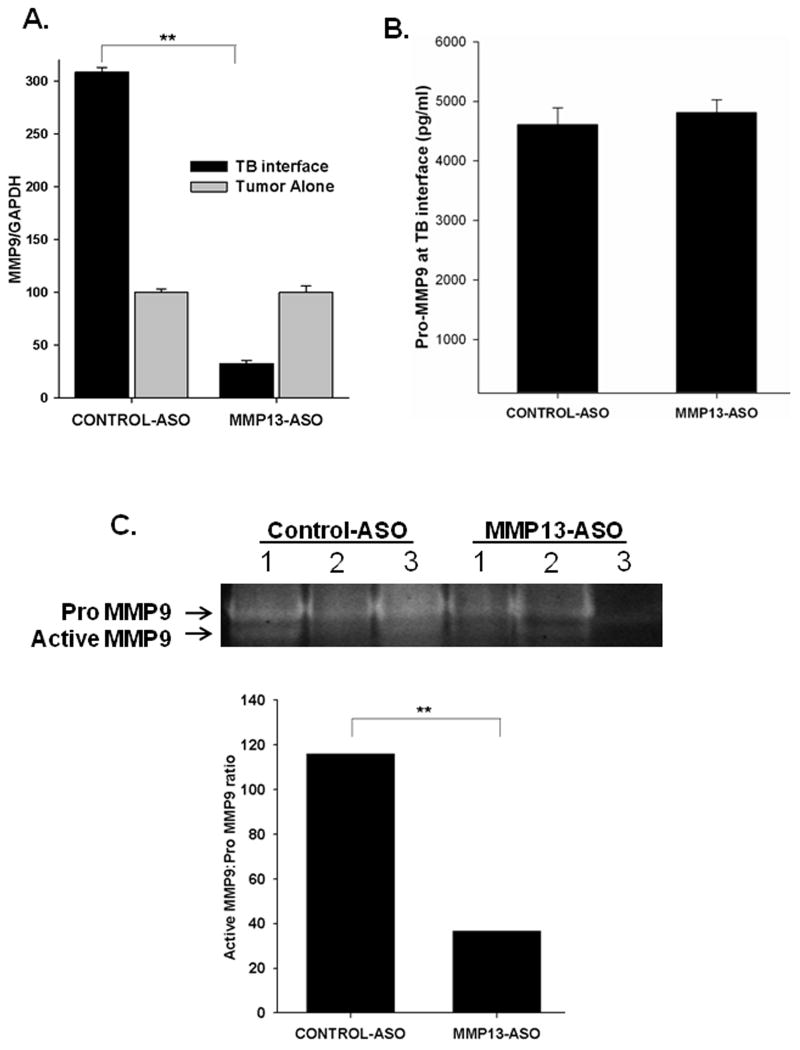
Previously, we have shown that MMP9 is also upregulated at the TB-interface in mammary tumor implanted mice (22). We observed a decrease in MMP9 mRNA expression at the TB-interface in the MMP13-ASO treated group as compared to the control-ASO treated group (Figure 5A). We evaluated the activity of MMP9 at the TB-interface of Cl66 tumor bearing mice treated with MMP13-ASO compared to Control-ASO. We observed a significant decrease in gelatinolytic activity (at 92kDa) in the MMP13-ASO treated mice. We observed a significant decrease in active:pro MMP9 enzyme levels in MMP13-ASO treated mice (Figure 5C). We did not observe significant difference in pro-MMP9 levels at tumor-bone interface between control-ASO and MMP13-ASO treated group (Figure 5B).
Figure 5. MMP13-ASO treatment inhibited MMP9 mRNA expression and activity at the TB-interface.
A. qRT-PCR demonstrated that MMP13-ASO treatment significantly reduced mRNA expression of MMP9 in tumor-bone samples. B. Pro-MMP9 levels at TB interface determined by quantitative ELISA. The values mean ± SEM. This is representative of two experiments done in triplicate. C. A representative zymography showing pro- and active MMP9 levels. MMP9 gelatinolytic activity (92 kDa) is significantly reduced in tumor-bone samples from MMP13-ASO treated mice compared to control samples. The ratio of active MMP9 to pro-MMP9 is significantly reduced with MMP-13 inhibition. **p<0.01.
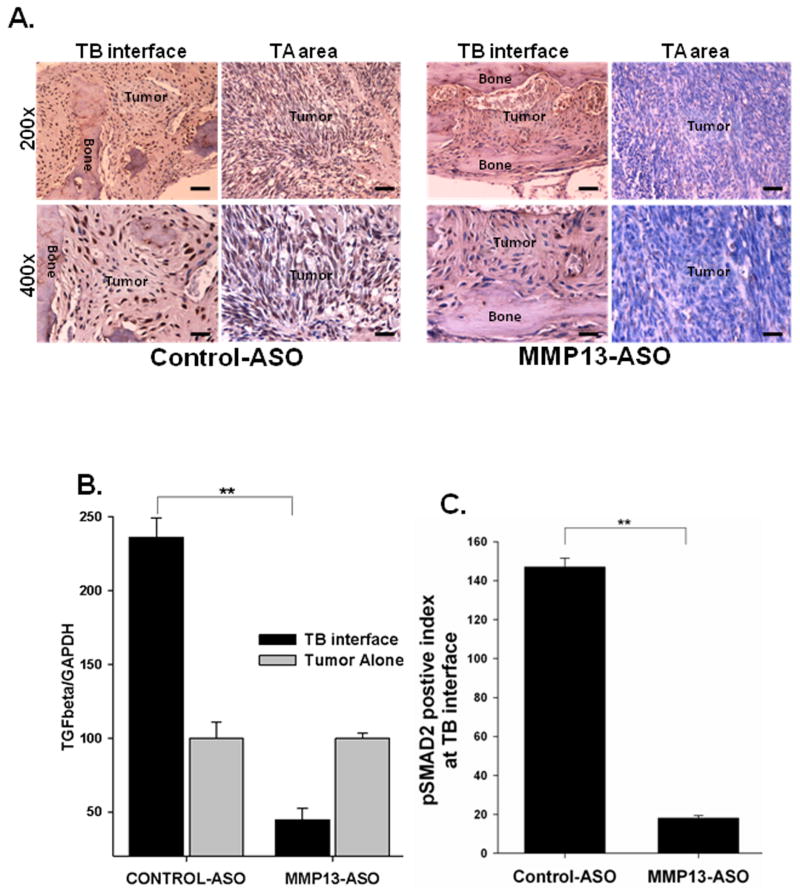
We have previously reported that, upregulation of TGFβ signaling during tumor-bone interaction promotes tumor induced osteolysis (23). We observed a significant decrease in tumor induced osteolysis with MMP-13 ASO treatment. To examine whether MMP-13 acts via TGFβ signaling in mammary induced osteolysis, we evaluated TGFβ expression and activity at the TB-interface of MMP13-ASO and control-ASO treated mice. We observed downregulation of TGFβ mRNA expression in MMP13-ASO treated mice (Figure 6B). We evaluated TGFβ signaling using immunohistochemistry for p-Smad2 (30). We observed a significant decrease in the pSmad-2 staining index and TGFβ signaling at the TB-interface of mice with MMP13-ASO treatment (Figure 6A&C).
Figure 6. Inhibition of MMP-13 blocks TGF-β signaling at the TB-interface.
A & C Sections from MMP13-ASO treated mice demonstrated a significantly reduced pSMAD2 staining index at the TB-interface. Scale bars represent 0.01mm. (n=5). B. qRT-PCR demonstrated a significant decrease in TGF-β mRNA expression at the TB-interface in MMP13-ASO treated mice compared to Control-ASO treated mice. The values are mean ± SEM. This is a representative of three experiments with similar results. **p<0.05.
Discussion
In this study, we evaluated the gene expression signature at the TB-interface and tumor alone areas in three different mammary adenocarcinoma cell lines differing in their metastatic potential. We identified MMP-13, RANKL, IBSP, Lumican, Kif5B, LOX and WIF1 as potentially important genes involved in tumor-induced osteolysis, which are commonly upregulated at the TB-interface in all three tumor types. Furthermore, overexpression of MMP-13 at the TB-interface promotes tumor induced osteolysis and knockdown of MMP-13 with ASO results in a significant inhibition of bone destruction. These data demonstrate that interaction of breast cancer cells with bone stromal cells results in an altered gene expression pattern that is critical for tumor-induced osteolysis.
We identified genes that are potentially involved in the modulation of tumor bone interaction during osteolytic bone metastasis using microarray analysis. We confirmed the microarray data by performing qRT-PCR analysis showing higher expression of MMP-13, RANKL, IBSP, Lox, Lumican, WIF1 and Kif5b mRNA expression at the TB-interface as compared to the tumor alone area. Our previous studies have established the importance of RANKL upregulation during TB-interaction in activating osteoclasts and promoting tumor induced osteolysis (22, 23). IBSP is a secreted, noncollagenous glycoprotein of bone matrix and is expressed in breast, lung, thyroid and prostate cancers that metastasize to bone (31). Expression of IBSP is associated with development of metastasis and poor prognosis (32, 33). Lumican is a small leucine rich proteoglycon abundantly present in breast tissue and it has been shown that lumican expression parallels tumor progression in breast carcinoma with higher lumican expression associated with higher tumor grade and lower estrogen receptor levels in the tumor (34). However, their role in bone metastasis remains unclear. LOX is an extracellular matrix remodeling enzyme and studies delineating the role of LOX in metastasis showed that it is essential for hypoxia-induced metastasis and inhibition of Lox proved to be promising in eliminating metastasis in mice with orthotopically grown tumors (35, 36) Silencing of WIF1 is associated with increased susceptibility to osteosarcoma (37). The role of the Kif5B gene in cancer progression is not well established. However, a recent report suggested a possible role for KIF5B as an oncofusion kinase (38). These studies suggest possible roles for genes upregulated at the TB-interface, however, little is known about their functional role in tumor-bone interaction during osteolytic metastasis.
Matrix metalloproteinases have been implicated in tumor progression and metastasis in several tumor types (11, 39-41). Our present data demonstrated the upregulation of MMP-13 at the TB-interface in three different mammary tumors. MMP-13 expression was higher in both tumor cells and osteoblasts at the TB-interface. Previous studies have shown expression of MMP-13 in several cancers including breast (15), head and neck squamous carcinoma (18), melanoma and chondrosarcoma (42). A previous report suggests involvement of MMP-13 in the degradation of basement membrane during bone metastasis of breast cancer (41). We have observed an association between MMP-13 expression, the number of osteoclasts and tumor induced osteolysis. Recent study has shown the upregulation of MMP13 in clinical bone metastatic samples from breast cancer patients (43). Our present data and the previous reports suggest an important role for the expression of MMP13 at the TB-interface (11, 39-41, 44), hence in this report we focused on delineating the functional analysis of MMP13 in tumor induced osteolysis.
To further strengthen our findings, we used MMP13-ASOs to target MMP-13 expression in vivo. Treatment of mice with MMP13-ASOs significantly inhibited tumor-induced bone resorption as well as number of activated osteoclasts at the TB-interface. The mechanism underlying MMP-13 dependent osteoclast activation remains unclear. Previous reports have shown that RANKL/RANK/OPG signaling is important in osteoclast activation and subsequent bone destruction (45). It is possible that, MMP-13 inhibition might alter the expression of RANKL and OPG at the TB-interface. We observed that targeting MMP-13 decreased RANKL expression at TB-interface without affecting OPG levels. As explained previously, the relative RANKL:OPG ratio is an important factor in driving osteolysis, and we observed a significant decrease in the RANKL:OPG ratio at the TB-interface in MMP13-ASO treated mice. These observations suggest the potential role of MMP-13 in RANKL-dependent osteoclast activation and tumor-induced osteolysis. The actual mechanism through which MMP-13 is involved in the regulation of the RANKL/OPG axis at the TB-interface merits further investigation.
MMP-13 has been shown to play a central position in the MMP activation cascade (46). MMP-13 activates MMP9 by cleaving pro MMP9 (47). In our previous studies, we have shown the upregulation of pro- and active-MMP9 during tumor-bone interaction (22). In this report, we observed a significant decrease in active MMP9 levels at the TB-interface in MMP13-ASO treated mice. MMP9 has been shown to recruit osteoclasts during development of long bones (48). These studies suggest that MMP-13 might indirectly potentiate osteoclast recruitment and activation by regulating activation of MMP9 at the TB-interface during tumor-bone interaction.
In a recent report, we demonstrated the role of TGFβ signaling during tumor-bone interaction in promoting mammary tumor growth and osteoclast activation (23). In this report, we analyzed the effect of MMP-13 knock-down on TGFβ signaling by pSmad-2 staining and observed a significant decrease in the pSmad2 activity in the MMP13-ASO treated group as compared to control treated group. The decrease in TGFβ signaling could be due to lower levels of active MMP9 at the TB-interface, which has been shown to be critical in TGFβ activation (49, 50). In our previous report, we did not observe increase in TGFβ mRNA expression at the TB-interface (23). However, TGFβR1 expression and signaling was increased at the TB-interface as compared to the tumor alone area (23). Our data suggest that increased MMP-13 levels at tumor bone interface potentiate TGFβ signaling via activation of MMP9. In our previous study, we evaluated the functional role of Cathepsin G in osteolytic bone metastasis of breast cancer and observed that Cathepsin G was involved in activating MMP9 by cleaving pro-MMP9. Inhibition of Cathepsin G resulted in increased latent TGFβ levels. There by Cathepsin G mediated activation of MMP9 led to activation of TGFβ and ultimately promotes tumor-induced osteolysis (50). Findings from our previous and present study allow us to speculate that, MMP9 might be playing as a central molecule in tumor-induced osteolytic cascade, where it is activated from several factors including Cathepsin G and MMP13 and leading to enhancement of TGFβ signaling at the tumor bone interface ultimately contributing to the osteolytic bone metastasis.
In conclusion, our present study demonstrated that tumor-bone interaction during osteolytic bone metastasis alters the gene expression signature at the TB-interface which further promotes bone resorption and establishment of osteolytic bone metastasis. These findings delineate the potential role of MMP-13 as one of the key regulators, commonly upregulated at the TB-interface, during tumor bone interaction in osteolytic bone metastasis. MMP-13 expression contributes to the osteolytic process by regulating RANKL:OPG levels, activating MMP9 and increasing TGF-β signaling. Our study provides data for understanding the mechanistic role of MMP-13 in osteolysis observed during bone metastasis. Additional studies are required to understand the regulation of MMP-13 expression during osteolytic bone metastasis and the development of targeted therapeutics.
Supplementary Material
Acknowledgments
This work was supported in part by Susan G. Komen for the Cure grant KG090860, Cancer Glycobiology Program from Nebraska Research Initiative and by grant CA72781 (R.K.S.), and Cancer Center Support Grant (P30CA036727) from National Cancer Institute, National Institutes of Health, and the Department of Defense (DOD) [Breast Cancer Research Program (BCRP) Predoctoral Traineeship Award (BC083293)] (K.C.N.).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009 doi: 10.3322/caac.20006. caac. [DOI] [PubMed] [Google Scholar]
- 2.Boyce BF, Yoneda T, Guise TA. Factors regulating the growth of metastatic cancer in bone. Endocr Relat Cancer. 1999;6:333–47. doi: 10.1677/erc.0.0060333. [DOI] [PubMed] [Google Scholar]
- 3.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2:584–93. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 4.Mundy GR. Mechanisms of bone metastasis. Cancer. 1997;80:1546–56. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1546::aid-cncr4>3.3.co;2-r. [DOI] [PubMed] [Google Scholar]
- 5.Coleman RE. Skeletal complications of malignancy. Cancer. 1997;80:1588–94. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1588::aid-cncr9>3.3.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Yoneda T, Hiraga T. Crosstalk between cancer cells and bone microenvironment in bone metastasis. Biochemical and Biophysical Research Communications. 2005;328:679–87. doi: 10.1016/j.bbrc.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 7.Powell GJ, Southby J, Danks JA, et al. Localization of Parathyroid Hormone-related Protein in Breast Cancer Metastases: Increased Incidence in Bone Compared with Other Sites. Cancer Res. 1991;51:3059–61. [PubMed] [Google Scholar]
- 8.Guise TA, Yin JJ, Taylor SD, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest. 1996;98:1544–9. doi: 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 10.Birkedal-Hansen H. Catabolism and turnover of collagens: collagenases. Methods Enzymol. 1987;144:140–71. doi: 10.1016/0076-6879(87)44177-3. [DOI] [PubMed] [Google Scholar]
- 11.Lynch CC, Hikosaka A, Acuff HB, et al. MMP-7 promotes prostate cancer-induced osteolysis via the solubilization of RANKL. Cancer Cell. 2005;7:485–96. doi: 10.1016/j.ccr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Stetler-Stevenson WG. The role of matrix metalloproteinases in tumor invasion, metastasis, and angiogenesis. Surg Oncol Clin N Am. 2001;10:383–92. x. [PubMed] [Google Scholar]
- 13.Djonov V, Cresto N, Aebersold DM, et al. Tumor cell specific expression of MMP-2 correlates with tumor vascularisation in breast cancer. Int J Oncol. 2002;21:25–30. [PubMed] [Google Scholar]
- 14.Ruokolainen H, Paakko P, Turpeenniemi-Hujanen T. Expression of matrix metalloproteinase-9 in head and neck squamous cell carcinoma: a potential marker for prognosis. Clin Cancer Res. 2004;10:3110–6. doi: 10.1158/1078-0432.ccr-03-0530. [DOI] [PubMed] [Google Scholar]
- 15.Freije JM, Diez-Itza I, Balbin M, et al. Molecular cloning and expression of collagenase-3, a novel human matrix metalloproteinase produced by breast carcinomas. J Biol Chem. 1994;269:16766–73. [PubMed] [Google Scholar]
- 16.Mauviel A. Cytokine regulation of metalloproteinase gene expression. J Cell Biochem. 1993;53:288–95. doi: 10.1002/jcb.240530404. [DOI] [PubMed] [Google Scholar]
- 17.Johansson N, Airola K, Grenman R, Kariniemi AL, Saarialho-Kere U, Kahari VM. Expression of collagenase-3 (matrix metalloproteinase-13) in squamous cell carcinomas of the head and neck. Am J Pathol. 1997;151:499–508. [PMC free article] [PubMed] [Google Scholar]
- 18.Cazorla M, Hernandez L, Nadal A, et al. Collagenase-3 expression is associated with advanced local invasion in human squamous cell carcinomas of the larynx. J Pathol. 1998;186:144–50. doi: 10.1002/(SICI)1096-9896(1998100)186:2<144::AID-PATH147>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Johansson N, Vaalamo M, Grenman S, et al. Collagenase-3 (MMP-13) is expressed by tumor cells in invasive vulvar squamous cell carcinomas. Am J Pathol. 1999;154:469–80. doi: 10.1016/S0002-9440(10)65293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu CP, Shen GH, Ko JL. Matrix metalloproteinase-13 expression is associated with bone marrow microinvolvement and prognosis in non-small cell lung cancer. Lung Cancer. 2006;52:349–57. doi: 10.1016/j.lungcan.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Aslakson CJ, Miller FR. Selective Events in the Metastatic Process Defined by Analysis of the Sequential Dissemination of Subpopulations of a Mouse Mammary Tumor. Cancer Res. 1992;52:1399–405. [PubMed] [Google Scholar]
- 22.Wilson TJ, Nannuru KC, Futakuchi M, Sadanandam A, Singh RK. Cathepsin G Enhances Mammary Tumor-Induced Osteolysis by Generating Soluble Receptor Activator of Nuclear Factor-{kappa}B Ligand. Cancer Res. 2008;68:5803–11. doi: 10.1158/0008-5472.CAN-07-5889. [DOI] [PubMed] [Google Scholar]
- 23.Futakuchi M, Nannuru KC, Varney ML, et al. Transforming growth factor-beta signaling at the tumor-bone interface promotes mammary tumor growth and osteoclast activation. Cancer Sci. 2009;100:71–81. doi: 10.1111/j.1349-7006.2008.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 25.Miyazono K, Ten DP, Heldin CH. TGF-beta signaling by Smad proteins. Adv Immunol. 2000;75:115–57. doi: 10.1016/s0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- 26.Bennett CF, Cowsert LM. Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim Biophys Acta. 1999;1489:19–30. doi: 10.1016/s0167-4781(99)00144-x. [DOI] [PubMed] [Google Scholar]
- 27.Henry SP, Geary RS, Yu R, Levin AA. Drug properties of second-generation antisense oligonucleotides: how do they measure up to their predecessors? Curr Opin Investig Drugs. 2001;2:1444–9. [PubMed] [Google Scholar]
- 28.Nannuru KC, Futakuchi M, Sadanandam A, et al. Enhanced expression and shedding of receptor activator of NF-kappaB ligand during tumor-bone interaction potentiates mammary tumor-induced osteolysis. Clin Exp Metastasis. 2009 doi: 10.1007/s10585-009-9279-2. [DOI] [PubMed] [Google Scholar]
- 29.Dougall WC, Chaisson M. The RANK/RANKL/OPG triad in cancer-induced bone diseases. Cancer Metastasis Rev. 2006;25:541–9. doi: 10.1007/s10555-006-9021-3. [DOI] [PubMed] [Google Scholar]
- 30.Nakao A, Imamura T, Souchelnytskyi S, et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–62. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellahcene A, Merville MP, Castronovo V. Expression of Bone Sialoprotein, a Bone Matrix Protein, in Human Breast Cancer. Cancer Res. 1994;54:2823–6. [PubMed] [Google Scholar]
- 32.Bellahcene A, Kroll M, Liebens F, Castronovo V. Bone sialoprotein expression in primary human breast cancer is associated with bone metastases development. J Bone Miner Res. 1996;11:665–70. doi: 10.1002/jbmr.5650110514. [DOI] [PubMed] [Google Scholar]
- 33.Bellahcene A, Menard S, Bufalino R, Moreau L, Castronovo V. Expression of bone sialoprotein in primary human breast cancer is associated with poor survival. Int J Cancer. 1996;69:350–3. doi: 10.1002/(SICI)1097-0215(19960822)69:4<350::AID-IJC19>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Leygue E, Snell L, Dotzlaw H, et al. Expression of Lumican in Human Breast Carcinoma. Cancer Res. 1998;58:1348–52. [PubMed] [Google Scholar]
- 35.Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 36.Bondareva A, Downey CM, Ayres F, et al. The lysyl oxidase inhibitor, beta-aminopropionitrile, diminishes the metastatic colonization potential of circulating breast cancer cells. PLoS One. 2009;4:e5620. doi: 10.1371/journal.pone.0005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kansara M. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J Clin Invest. 2009;119:837–51. doi: 10.1172/JCI37175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a Novel Fusion Oncokinase Identified by an Immunohistochemistry-based Diagnostic System for ALK-positive Lung Cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 39.Lafleur MA, Drew AF, de Sousa EL, et al. Upregulation of matrix metalloproteinases (MMPs) in breast cancer xenografts: a major induction of stromal MMP-13. Int J Cancer. 2005;114:544–54. doi: 10.1002/ijc.20763. [DOI] [PubMed] [Google Scholar]
- 40.Ellsworth RE, Seebach J, Field LA, et al. A gene expression signature that defines breast cancer metastases. Clin Exp Metastasis. 2009;26:205–13. doi: 10.1007/s10585-008-9232-9. [DOI] [PubMed] [Google Scholar]
- 41.Ohshiba T, Miyaura C, Inada M, Ito A. Role of RANKL-induced osteoclast formation and MMP-dependent matrix degradation in bone destruction by breast cancer metastasis. Br J Cancer. 0 AD;88:1318–26. doi: 10.1038/sj.bjc.6600858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uria JA, Balbin M, Lopez JM, et al. Collagenase-3 (MMP-13) Expression in Chondrosarcoma Cells and Its Regulation by Basic Fibroblast Growth Factor. Am J Pathol. 1998;153:91–101. doi: 10.1016/S0002-9440(10)65549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein A, Olendrowitz C, Schmutzler R, et al. Identification of brain- and bone-specific breast cancer metastasis genes. Cancer Letters. 2009;276:212–20. doi: 10.1016/j.canlet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Uria JA, Stahle-Backdahl M, Seiki M, Fueyo A, Lopez-Otin C. Regulation of collagenase-3 expression in human breast carcinomas is mediated by stromal-epithelial cell interactions. Cancer Res. 1997;57:4882–8. [PubMed] [Google Scholar]
- 45.Blair JM, Zhou H, Seibel MJ, Dunstan CR. Mechanisms of disease: roles of OPG, RANKL and RANK in the pathophysiology of skeletal metastasis. Nat Clin Pract Oncol. 2006;3:41–9. doi: 10.1038/ncponc0381. [DOI] [PubMed] [Google Scholar]
- 46.Leeman MF, Curran S, Murray GI. The Structure, Regulation, and Function of Human Matrix Metalloproteinase-13. Critical Reviews in Biochemistry and Molecular Biology. 2002;37:149. doi: 10.1080/10409230290771483. [DOI] [PubMed] [Google Scholar]
- 47.Knauper V, Smith B, Lopez-Otin C, Murphy G. Activation of progelatinase B (proMMP-9) by active collagenase-3 (MMP-13) Eur J Biochem. 1997;248:369–73. doi: 10.1111/j.1432-1033.1997.00369.x. [DOI] [PubMed] [Google Scholar]
- 48.Engsig MT, Chen QJ, Vu TH, et al. Matrix Metalloproteinase 9 and Vascular Endothelial Growth Factor Are Essential for Osteoclast Recruitment into Developing Long Bones. J Cell Biol. 2000;151:879–90. doi: 10.1083/jcb.151.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–76. [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson TJ, Nannuru KC, Singh RK. Cathepsin G-Mediated Activation of Pro-Matrix Metalloproteinase 9 at the Tumor-Bone Interface Promotes Transforming Growth Factor-B Signaling and Bone Destruction. Mol Cancer Res. 2009;7:1224–33. doi: 10.1158/1541-7786.MCR-09-0028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.