Abstract
Although the past decade has witnessed the sequencing from an increasing number of parasites, modern high-throughput DNA sequencing technologies have the potential to generate complete genome sequences at even higher rates. Along with the discovery of genes that might constitute potential targets for chemotherapy or vaccination, the need for novel protein expression platforms has become a pressing matter. In addition to reviewing the advantages and limitations of the currently available and emerging expression systems, we discuss novel approaches that could overcome the current limitations, including the ‘pseudoparasite’ concept, an expression platform in which the choice of the surrogate organism is based on its phylogenetic affinity to the target parasite, while taking advantage of the whole engineered organism as a vaccination adjuvant.
Proteins from protozoan parasites as targets for diagnosis and intervention
The availability of the genome sequences of several parasites of medical importance has led to exponential progress in our understanding of their biology, and enabled the identification of potential targets for intervention [1]. Further, the continued advances in genome sequencing technologies holds great promise for the accomplishment of similar goals for virtually any parasite of interest [2]. When compared to other pathogens of human and veterinary importance, such as viruses and bacteria, large gaps exist in our knowledge of the virulence and pathogenesis mechanisms of protozoan parasites, and methodologies for eradication or management of parasitic diseases through vaccination or treatment are still in the distant future. Characterization of genes of interest for potential intervention identified by mining these genomes has been hindered because of the lack of suitable protein expression systems. A clear example is Plasmodium falciparum, the etiological agent of malaria, for which the lack of an effective vaccine, and the rapid emergence of drug-resistant strains, have made most intervention attempts extremely challenging [3]. Therefore, the development of innovative and efficient systems for the expression of recombinant proteins from protozoan parasites has become an urgent public health matter.
Applications of recombinant proteins from protistan parasites
Recombinant proteins from parasites are required for numerous applications:
(i) Development of diagnostic tools
A rapid and accurate diagnosis of the etiological agent is the key for effective management of diseases caused by protozoan parasites, while avoiding the unnecessary use of therapeutic agents that might result in the selection of drug-resistant strains. The development of diagnostic tools requires substantial amounts of either the parasite at particular life cycle stages, or selected proteins expressed at those stages [4, 5]. For selected proteins, the recombinant proteins should accurately represent the native equivalents; otherwise, the field performance of the diagnostic test could be compromised [6]. Currently the only US-FDA approved kit uses the HRP-2 and aldolase antigens (BinaxNow, Inverness Medical, USA) [4]. Diagnostics for trypanosomiasis, babesiosis, and leishmaniasis also rely on recombinant antigens [7-9].
(ii) Immunogens for vaccination
Vaccination is the most desirable prophylactic method for any infectious disease. The use of proteins isolated directly from parasites is advantageous over recombinant proteins in that all structural and immunogenic characteristics that are native to the organism are displayed in the vaccine. However, their availability at the required purity standard and quantity has been the main limiting factor. In addition, polymorphism of the protein of interest can result in vaccines with poor reproducibility. Most subunit vaccines rely on the industrial production of the recombinant antigen of choice (Table 1). Further, as compared to whole-organism vaccines, subunit vaccines have low inherent safety risks associated with their manufacture processes [10].
Table 1.
Systems used for producing vaccine candidates against protozoan parasitesa
| Organism | Disease | Candidateb | System |
|---|---|---|---|
| Plasmodium | Malaria | ABRA | Escherichia coli |
| MSP1 | Escherichia coli | ||
| AMA-1 | Escherichia coli | ||
| MSP1(42) | Escherichia coli | ||
| LSA-1 | Escherichia coli | ||
| RTSS/AS02A | Saccharomyce cerevisiae | ||
| Pfs25 | Pichia pastoris | ||
| Pvs25 | Saccharomyces cerevisiae | ||
| CSP | Saccharomyces cerevisiae | ||
| Toxoplasma | Toxoplasmosis | SAG1, -2 | Salmonella |
| OP2 | Plasmid DNA | ||
| Cryptosporidium | Cryptosporidiosis | P23, CP15 | Plasmid DNA |
| Theileria | East Coast fever | p67 | Baculovirus |
| Neospora | - | NcSAG1, -2 | Escherichia coli |
| NcGRA7, NcsHSP33 | Plasmid DNA | ||
| Babesia | Babebiosis | 12D3, 11C5 | Escherichia coli |
| Bd37 | Escherichia coli | ||
| Eimeria | - | EtMIC2 | Plasmid DNA |
This is not a comprehensive list. Not all of these proteins are mentioned in the main text.
ABRA, acidic basic repeat antigen; AMA, apical membrane antigen; EtMIC2, Eimeria tenella microneme protein 2; GRA, tachyzoite-dense granules; HSP, heat-shock protein; Pfs25, P. falciparum surface 25; Pvs25, P. vivax surface 25.
(iii) Structure–function analysis of proteins from parasites
Novel approaches for effectively and specifically targeting essential parasite enzymes are currently being developed [11]. However, the rigorous proof of function requires the biochemical, biophysical, and functional characterization of the protein of interest. Protozoan parasites represent the ultimate challenge in this regard as most of them are obligate intracellular parasites for which no in vitro culture methods have been developed to date. Nevertheless, homology modeling and molecular-docking experiments have enabled the design of specific inhibitory drugs that can be subsequently tested experimentally as potential therapeutic agents [12].
(iv) Screening and profiling of candidate drugs
The development of therapeutics is a viable alternative to mitigate the effects of a disease, which is particularly useful where a vaccine is not available, or, in those cases where the vaccine is not sufficiently effective, as a complement to the vaccine. Along these lines, great efforts have been invested in studies leading to the production of drugs against apicomplexan parasites, based on interference with metabolic pathways associated with the apicoplast [13]. In some cases, cultured parasites provide enough material for drug screening (i.g. using Leishmania tarentolae 20 × 1011 cells, 50 mg of tubulin at 10–30 mg ml−1 is enough for screening approximately 1600 compounds) [14]; however, given the availability of extensive combinatorial chemical libraries, current drug-discovery screens require large amounts of proteins (i.e. PfMetAP1b 222 nM for 175,000 compounds arrayed in 384-well plates) [15], a fact that also underscores the urgent need of suitable expression systems.
Production of recombinant proteins from parasites: challenges and limitations
(i) Scale of production
Although improved technologies have reduced the amount of protein required for extensive screenings, the application of high-throughput methodologies still requires relatively high protein quantities (i.e. 5–25 mg ml−1 for screening 500–1000 crystallization conditions; 10 nM pfDHOD for screening a 208,000 compound library) [16, 17], virtually unattainable from parasites isolated from the host. By contrast, recombinant proteins can be produced in the relatively large amounts. In some cases, however, the industrial-scale production of recombinant proteins necessary for a selected application may require a complex infrastructure. For example, production of the P. falciparum liver-stage antigen 1 (LSA-1) for the pre-erythrocyte-stage protein-based vaccine, required 300 liters of broth to produce 8.0 kg of Escherichia coli paste, which yielded 8 mg LSA per gram of paste (<0.005 endotoxin units per 50 μg of protein) [18]).
(ii) Immunogenicity of the recombinant product
Immunogenicity is of crucial importance when evaluating expression systems for production of recombinant vaccine antigens. Immunogenicity is inherent to the protein, and based mostly on structural features that determine its solubility, degradability, and interactions with multiple components of the innate and adaptive immune responses. For example, differences in the expression constructs for production of proteins evaluated as malaria vaccine candidates resulted in different immunogenicity, even when rabbits were immunized with equimolar amounts of each recombinant protein [19]. A standard method to enhance immunogenicity of the recombinant protein is to conjugate it to a carrier, usually a highly glycosylated protein or polysaccharide, although the outcomes are sometimes unexpected [20]. The P. falciparum apical membrane antigen 1 conjugated to bacterial ExoProteinA resulted in a 1000-fold higher antibody titer, as compared to the threefold increase in Pfs25 conjugated to the same carrier [20]. To date, the most promising vaccine against malaria uses the circumsporozoite protein (CSP) region, and a T-cell immunogenic epitope fused to the hepatitis B antigen, and formulated with AS02A [10]. Alternatively, multiple protective epitopes can be combined in a single recombinant immunogen [21].
(iii) Post-translational modifications
Recombinant proteins should reflect their authentic counterparts as faithfully as possible, including their post-translational modifications. Among these, glycosylation can be critical, not only for immunogenicity in that it may enhance or conceal immunogenic epitopes, but also to modulate biological activity. For example, the hyperglycosylated recombinant Toxoplasma surface antigen (SAG2) produced in yeast reacted strongly with pooled anti-Toxoplasma human serum, pooled anti-Toxoplasma mouse serum, and a SAG2-specific monoclonal antibody [22]. Toxoplasma tachyzoites treated with tunicamycin, which inhibits N-glycosylation, were mostly incapable of invading new host cells, thus revealing its relevance for virulence, and possibly for immunogenicity [23]. By contrast, the Trypanosoma brucei recombinant transferrin receptors expressed in insect cells in the presence of tunicamycin bind ligand effectively [24]. Other post-translational modifications typical from eukaryotes that have been described in apicomplexan proteins and might be required for production of recombinant proteins include the removal of N-terminal methionine and acylation [15, 25, 26].
Surrogate systems for expression of proteins from protozoan parasites
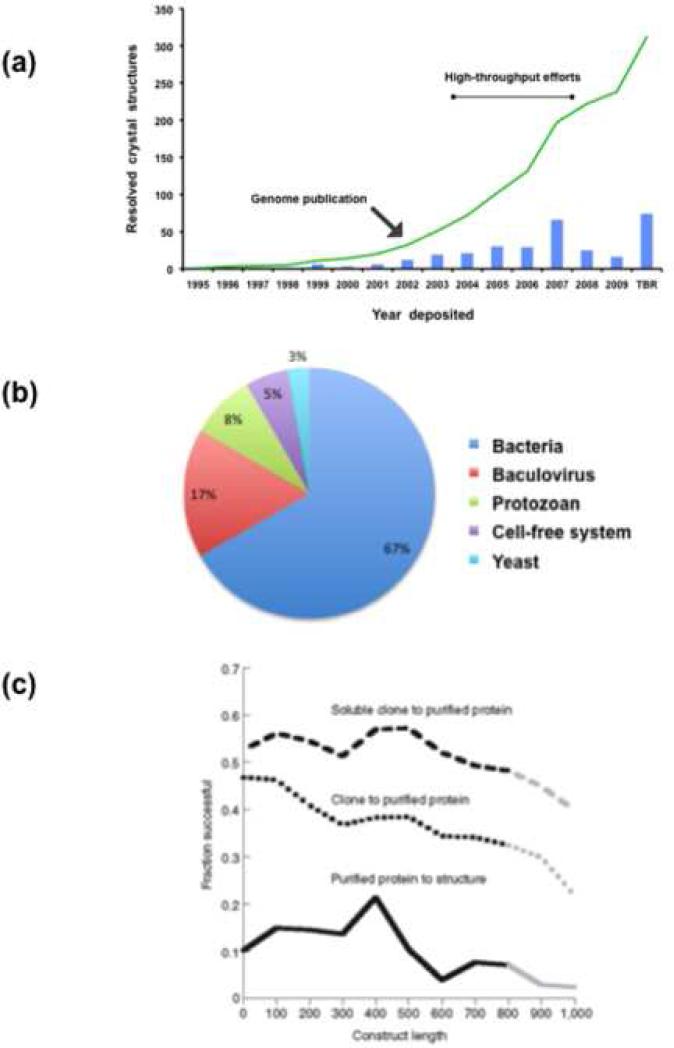
The selection of heterologous expression systems for proteins from protozoan parasites has been determined, at least in part, by the specific objective(s) pursued, production cost, and availability of the required facilities. To date, most efforts have been directed to the production of Plasmodium recombinant proteins (reviewed in Ref. [27]). A search of the PDB (Protein Data Bank) database (www.pdb.org, May 2009) yielded a limited number of matches to protozoan protein sequences for which the structures have been resolved (Figure 1a), Although an increase in the number of structures resolved during the past eight years is evident, the pattern illustrates the limited success attained in the available heterologous expression systems. In the following section, we focus on the systems that have been tested (successfully and unsuccessfully) for expression of parasite proteins (Table 2), those that are currently under development, and potentially useful innovative platforms.
Figure 1.
(a) Resolved crystal structures of Plasmodium genes. PDB database (www.pdb.org, May 2009) was searched for Plasmodium protein resolved structures and plotted against the year deposited (bars: number of structures deposited in a given year; line: cumulative structure deposited numbers). After the sequence of the genome, there were two high-throughput efforts that resulted in a limited increase in the number of structures resolved. Overall, recombinant protein production remains a bottleneck for many scientific endeavors. TBR: to be released. (b) Heterologous protein expression systems for parasites. A PubMed search of recombinant proteins from parasites in the past seven years reveals that prokaryotic systems are still the workhorse, followed by insect cells/baculovirus. Interestingly, protozoan organisms are used more often than yeast. Cell-free systems are just starting to take off. (c) Solubility as a function of construct length; Ref. [32] is the original source. The overall success rate for soluble recombinant proteins remains very low, especially for protein targets with lengths > 800 amino acids [32]. Dotted line, fraction of cloned targets resulting in successful large-scale purifications. Dashed line, fraction of soluble clones at a 1 ml scale resulted in pure protein at large scale. Solid line, fraction of purified proteins resulting in successful crystal structure determinations.
Table 2.
Performance comparison of most common heterologous expression systems.
| System | Advantages | Disadvantages |
|---|---|---|
| Prokaryotic | • Rapid growth at high densities | • Absence of post-transcriptional modifications |
| • Inexpensive growth media | • Amino acid substitutions or modifications | |
| • Suitable for large scale production | ||
| • Heterogeneous products | ||
| • Well known genetics | • Contamination with endotoxin | |
| • Easy to transform | • Biased codon usage | |
| • Multiple cloning vectors, purification tags, and host strains | • Low level of secretion | |
| • Failure to fold in the native active state | ||
| • Abundant literature (troubleshooting) | ||
| • Accumulation as insoluble inclusion bodies | ||
| • Multiple commercial brands | ||
| • High protein yields | • Lack of complexity for functional organelle targeting | |
|
Eukaryotic | ||
| Yeast | • Inexpensive | • Glycosylation patterns different from those in the native parasite protein |
| • Suitable for large-scale production | ||
| • Multiple cloning vectors, purification tags, and host strains | • Hyper-glycosylation | |
| • Low yield derived from codon bias, depletion of precursors and energy, proteolysis | ||
| • Secretion into the culture medium | ||
| • Unfolded proteins shooting down the secretory pathway | ||
| • Trafficking through the secretory pathway favors disulphide bounds formation and protein folding | ||
| • Truncated proteins | ||
| • Easy genetic manipulation | ||
| • Post-transcriptional modifications | ||
| • Endotoxin-free | ||
| • Abundant literature (troubleshooting) | ||
| • Several commercial brands | ||
| • High protein yields | ||
|
Mammalian cell lines |
• Post-transcriptional modifications (glycosylation, phosphorylation, and the addition of fatty acid chains) |
• Labor intensive and expensive |
| • Increased biosafety (BL2) when using some virus for gene delivery | ||
| • Transient or stable transfection | • Slowly growth | |
| • Limited large scale production | ||
| • Abundant literature (troubleshooting) | • Low protein yield | |
| • Contamination with mammalian pathogens | ||
|
Baculovirus expression vector system |
• Basic cell culture equipment |
• Labor intensive |
| • Minimal safety precautions | • Time consuming (up to 3 weeks to obtain enough viruses) | |
| • Large inserts (up to 15 Kb) | ||
| • Post-transcriptional modifications | • Expensive methodology | |
| • No easy amenable for automation | ||
| • Abundant literature (troubleshooting) | ||
| • Suitable only for batch production | ||
| • Glycosylation patterns different from those in the native parasite protein (lack of multiantennary glycans) | ||
|
D. discoideum |
• Easy culture at high densities |
• Still under development |
| • Easy genetic manipulation | • No commercially available | |
| • Multiple cloning vectors | • Preferential codon usage | |
| • Eukaryotic organelle complexity for functional studies | • No FDA-approved SOPsa | |
| • AT rich genome | ||
| • Abundant literature (model organism) | ||
|
T. thermophila |
• Post-transcriptional modifications |
• Still under development |
| • AT rich genome | • No commercially available | |
| • GPI anchored proteins | • No FDA-approved SOPs | |
|
Cell-free systems |
• Free of translation inhibitors |
• Absence of post-transcriptional modifications |
| • Non-biased codon usage | • Expensive methodology | |
| • Non-specialized training | • Low yield | |
| • Simple equipment required | • Not amenable for automation and parallelization | |
| • Specially suitable for integral membrane proteins | • No FDA-approved SOPs | |
Abbreviations: FDA, Federal Drug Administration; SOP, Standard Operating Procedures.
Prokaryotic systems
The E. coli expression system is the most commonly used for industrial production of recombinant proteins for pharmaceutical applications (reviewed in Ref. [28]) (Figure 1b). Prokaryotic organisms grow rapidly and at high densities in relatively inexpensive growth media, are easy to transform, and can produce high quantities of soluble recombinant product; in addition, multiple cloning vectors and host strains are available. Limitations of prokaryotic expression systems include the absence of post-transcriptional modifications, amino acid substitutions or modifications, heterogeneous products, contamination with endotoxin, and accumulation of the recombinant product as inclusion bodies. For proteins from protozoan parasites, the overall success rate for obtaining soluble, immunogenic recombinant products in prokaryotic systems remains low [29–32] (Figure 1c). For example, from 303 Plasmodium genes, only 7% of the recombinant proteins induced antibody titers [31]. A high-throughput study targeting apicomplexan parasites resulted in 30.2% purified soluble recombinant proteins, with only 3.8% of their structures resolved [29]. Similarly, from a total of 1000 open reading frames (ORFs) from Plasmodium tested, <7% were expressed as soluble proteins [30]. Current approaches to improve expression in prokaryotic systems include the production of temperature-induced recombinant protein [33], and the use of thermal melt and/or thermal shift assays for identifying ligands that stabilize recombinant proteins [34]. However, Plasmodium proteins of large size or with high pI remain difficult to produce in bacteria, and no plastid-targeted recombinant proteins have yet been expressed as soluble products [30]. This has been attributed to the codon bias derived from the high frequency at which a particular codon is used preferentially by the Plasmodium AT-rich genome. Although the impact of codon bias on heterologous protein expression has been questioned by a recent large-scale study [30], alternative strategies have been developed to overcome this problem (reviewed in Ref. [35]). These include plasmids or host cells containing tRNA that recognize rare codons [36], and the use of harmonized codon usage frequencies [18]. As it becomes increasingly affordable, gene synthesis represents a viable option for codon usage harmonization, even for large-scale initiatives [37]. Recently, focus has been placed on the identification of sequence features that may modulate protein expression in heterologous systems. For example, the recombinant protein yield in E. coli was strongly dependent on the codons used for only a subset of amino acids, predominantly favoring codons read by tRNAs that are most highly charged during amino acid starvation, instead of codons that are most abundant for the highly expressed E. coli proteins [38]. In addition to codon bias, features that may be unique to any given protein determine success in expression yields and correct folding in prokaryotic systems. For example, cryptopain-1 from Cryptosporidium does not require its pro-domain for proper folding when expressed in E. coli [39]. In spite of the above-mentioned limitations, a minor subset of protozoan proteins have been produced in prokaryotic systems as soluble, active, and immunogenic recombinant products, without any major difficulties. Among these, the Entamoeba hystolica cysteine proteinase EhCP1 [40], the recombinant dihydrofolate reductase-thymidylate synthase (DHFR-TS) from Babesia gibsoni [41], and the dominant circumsporozoite CD8 T-cell epitope from Plasmodium berghei [42].
Eukaryotic systems
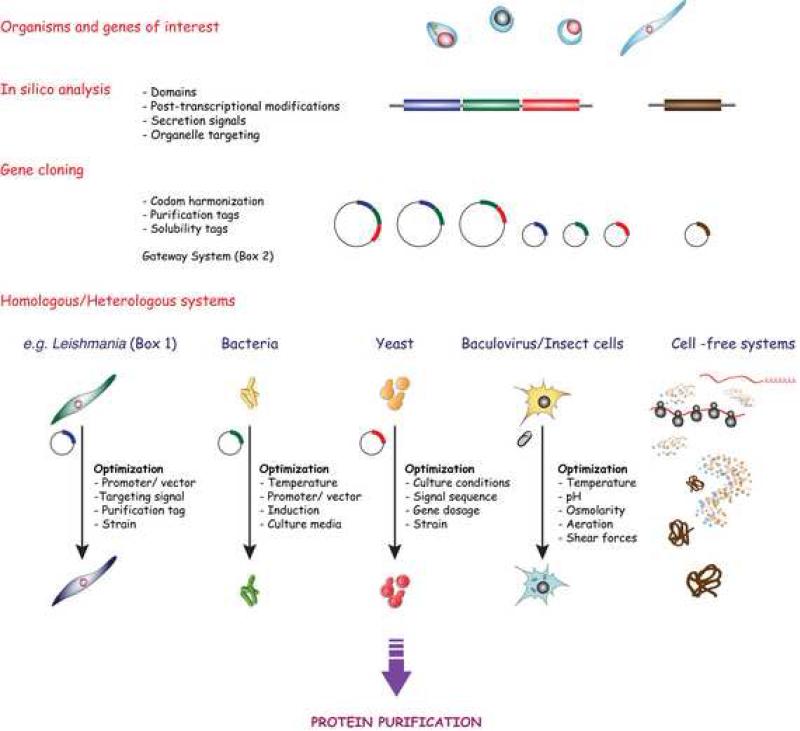
The main advantage of these systems derives from their closer phylogenetic relationship with the parasites of interest, presumably sharing to some extent with the parasites of interest post-translational modifications, such as glycosylation, acylation, ability to form disulfide linkages, interaction with eukaryotic chaperones, and proteolytic processing, together with sub-cellular compartmentalization, secretion mechanisms that avoid accumulation, and low toxicity [43] (Figure 2). The most frequently used eukaryotic expressions systems include:
Figure 2.
Most frequently used systems for the production of recombinant protein from parasites. After the selection of the gene of interest, an extensive in silico analysis should corroborate the presence of protein domains, secretion signals, organellar targeting, and post-translational modifications. These characteristics and the use of the recombinant protein will help guide the selection of the cloning vector and expression system. The gene of interest can be directly cloned, often under a strong promoter, into a suitable vector to express in a homologous system or in a surrogate system, ideally, the closest related species that can be grown in large quantities and is amenable for genetic manipulation. When no in vitro culture is available or the scale needed precludes the use of the homologous system, the bacterial system is often the first choice. General guidelines include removing predicted membrane-spanning regions, avoiding disrupting predicted secondary structural elements, respecting the boundaries of globular domains, if known; and avoiding the inclusion of low-complexity regions or hydrophobic residues at the N- and/or C-termini. When post-translational modifications in the recombinant protein are needed for function or antigenicity or the bacterial systems are not performing as expected, eukaryotic systems are available either using a surrogate organism or cell-free system. In either case, and especially for systems that have been in the market for long time, there is a suite of clones, strains, culture conditions, and tags that can be tested during the optimization process.
(i) Yeast
Pichia pastoris and Saccharomyces cerevisiae are usually the first choice expression systems due to their suitability for large-scale fermentors, and their high protein yields. Their main disadvantage resides in the N- and O-linked glycosylation patterns of the recombinant proteins, which, in some cases, might be qualitatively and quantitatively different from those in the native parasite protein, resulting in inactive products. Nevertheless, PfCP-2.9 and Pvs25 are two good examples of recombinant proteins produced in Pichia [27]. Further improvements on the expression of Plasmodium recombinant proteins in S. cerevisiae are based on the identification of Plasmodium protein–protein interactions using a high-throughput version of the yeast two-hybrid assay [44].
(ii) Mammalian cell lines
Although sharing numerous features with yeasts, the use of mammalian cells to produce recombinant proteins from parasites has been hindered by the labor-intensive and expensive methodology required for establishing stable recombinant cells. Thus, the use of mammalian cell lines has been limited to the identification and characterization of Plasmodium surface ligands involved in host–parasite interactions, binding assays, expression of membrane transporters, and testing of DNA-based vaccines [27].
(iii) Baculovirus-mediated expression systems in insect cells
As a transient expression system, the virus-mediated synthesis of heterologous proteins in insect cells and larvae (reviewed in Ref. [45]) can yield milligram quantities of recombinant protein, with some required post-transcriptional modifications. Insect cell culture is simpler and requires less equipment than does the culture of mammalian cells; it can be easily upgraded to bioreactor scale, and several genes of interest can be expressed simultaneously for the production of multiprotein complexes. Drawbacks include the time-consuming generation of recombinant baculovirus, their inability to synthesize branched multiantennary glycans, and their lethal effects on the host cells, which makes the system suitable only for batch production. The full-length Crithidia fasciculata mitochondrial topoisomerase II, a promising target for anti-trypanosomatid drugs, was successfully expressed in the Bac-to-Bac baculovirus expression system [46]. Viruses represent one of the most frequently used systems for developing vaccines, and have proved suitable both as delivery systems and as adjuvants. These include, recombinant poxvirus FP9 and a modified vaccinia virus Ankara [47, 48], the malaria protein B in yellow fever 17D virus [49], the Plasmodium merozoite surface protein (MSP) -1(42) in Bombyx mori nuclear polyhedrosis virus-silkworm [50], and the malaria MSP-1 in a replication-defective virus [51]. Toxoplasma surface antigens expressed in replication-deficient recombinant adenoviruses induce a protective immune response in mice [52]. Further, the recombinant MVA rhoptry protein 2 vaccinia virus was developed as a vaccine candidate for toxoplasmosis [53].
(iv) Tetrahymena thermophila
Based on its high cell densities with short generation times and suitability for culture in bioreactors, the ciliate T. thermophila was proposed about a decade ago as an alternative heterologous expression system [54]. Since then, it has been used successfully to express the I-antigen of the parasitic ciliate of fish Ichthyophthirius multifiliis [55], and the glycosylphosphatidylinositol (GPI)-anchored full-length Plasmodium CSP [56]. This constitutes the first report of the recognition of targeting and GPI anchoring signals in a heterologous expression host. This successful initiative led to further development of Tetrahymena as an expression system [57], although limited progress has been reported since then. Publication of the Tetrahymena genome should renew such interest by identifying secretion and organellar signals for secretion and targeting, regulatory elements for constructing transfection vectors, among others, in spite of a potential limitation of this system concerning the expression of plastid-derived genes, since no compelling evidence for its presence was found in the genome [58].
(v) Dictyostelium discoideum
This slime mold is recognized as a useful model organism for studies ranging from ameboid motility to multicellular morphogenesis [59]. Because of its multiple advantages, which include axenic in vitro culture in synthetic media, Dictyostelium has been recently proposed as an alternative expression system for eukaryotic proteins (reviewed in Ref. [60]). As an alternative subunit vaccine against malaria, the Plasmodium recombinant CSP anchored to the surface of Dictyostelium, elicited antibodies against two different regions of the target protein [61]. In this case, replacement of the CSP-C-terminal segment by the Dictyostelium contact site A glycosyl-phosphatidylinositol anchor signal sequence was essential for surface display [61, 62]. After these early successful studies [61, 62], no further attempts have been reported and, as for Tetrahymena, the availability of the genome [63] should facilitate the exploration of this system for the production of recombinant proteins from parasites.
(vi) Homologous and heterologous parasite expression systems
As indicated above, proteins directly isolated from parasites are advantageous over recombinant proteins produced in bacteria because they faithfully display all desirable characteristics, such as immunogenicity and biological activity. Thus, the enhanced production of proteins of interest in the particular parasite, either by over-expression or by increasing the parasite numbers by culture scale-up would represent the approach of choice. Recently, a chemically defined medium has been developed for continuous growth of the P. falciparum intraerythrocytic stage [64], which should facilitate the development of a homologous system for production of Plasmodium recombinant proteins. For parasites that cannot be cultured in vitro, alternative animal models can be used to overcome this limitation, at least in part. For example, rats have been proposed to substitute ruminants for enhancing propagation of Cryptosporidium spp. [65].
Protozoan parasites have also been used successfully as surrogate systems for expression of proteins of other parasites (Figure 3), mostly for functional studies, and developed into commercial heterologous expression systems (Box 1). Neospora caninum can express genes of the phylogenetically related T. gondii [66], whereas engineered Toxoplasma ts-4 mutants can express the Leishmania antigen kinetoplastid membrane protein-11 and elicit a specific immune response in BALB/c mice [67]. Trichomonas foetus can express functional Trichomonas vaginalis AP65 adhesin [68]. Plasmodium vivax chloroquine-resistance transmembrane protein was expressed in both P. falciparum and Dictyostelium for chloroquine drug tests [69]. Similarly, Trypanosoma cruzi, the etiological agent of Chagas disease, has been used to screen for drugs against Crithidia fasciculata ornithine decarboxylase [70].
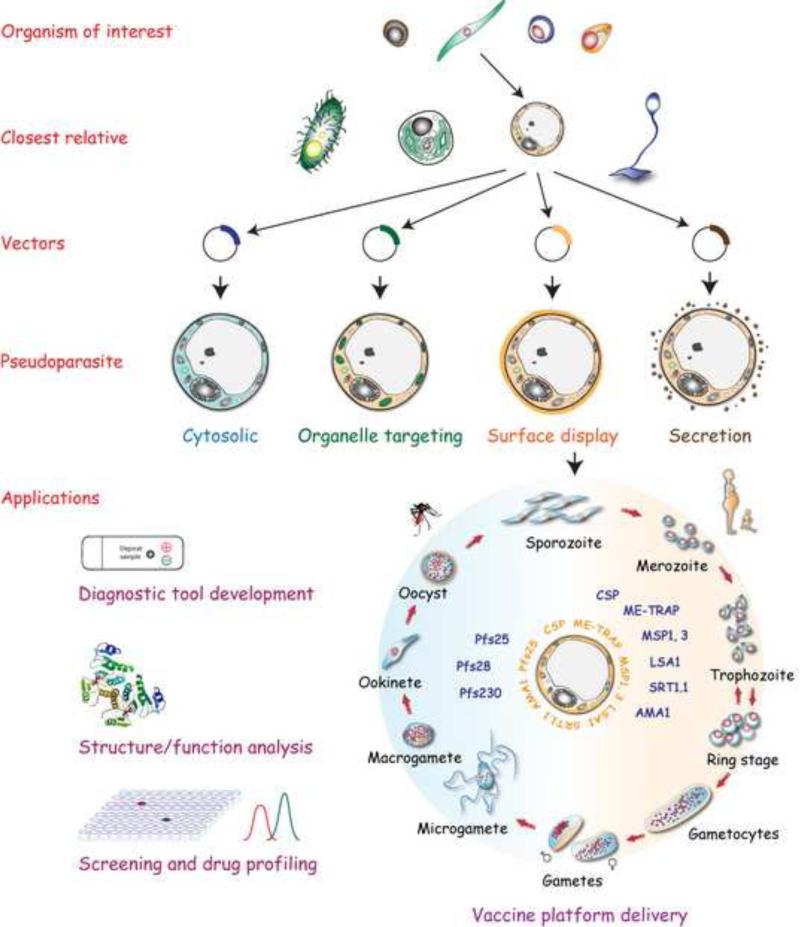
Figure 3.
The ‘pseudoparasite’ concept. The evolutionary relationships between organisms provide us with a ‘magnifying glass’ to select those with shared structures, mechanisms, and perhaps shared processes required for protein function. Therefore, ‘phylogenetically tailored’ heterologous expression systems might lead to the development of novel alternative platforms for expressing recombinant proteins from parasites. The first step would involve the selection of the closest relative to our organism of interest that shares the targeted structure/pathway/gene. Ideally, this organism would be amenable for genetic manipulation and for large-scale culture in axenic conditions and in a cell-free defined medium. In the second step, the cloning vector (cytosolic, organelle targeting, surface, secretion) will be selected depending on the predicted native targeting, and the potential applications of the recombinant protein, ranging from diagnostic tool development to screening and drug profiling. Taking this phylogenetic relationship further, the engineering of a pseudoparasite might be carried out in an organism that expresses genes that are homologous to those in the parasite of interest. These will enable the correct folding and post-translational modifications that take place in the target organism, and that are necessary to elicit a specific, effective immune response, thus, providing a multi-antigen, up-scalable vaccine delivery platform. This approach would enable the identification and selection of the best combination of multiple immunogens to be expressed in their immunologically relevant conformation by the pseudoparasite, while taking advantage of the whole engineered organism as an adjuvant. Hence, the pseudoparasite displaying the selected parasite antigens can be directly tested for the ability to induce an effective immune response in the absence of adjuvants. Gateway systems and alike can also be adopted to incorporate any gene of the organism of interest to the pseudoparasite platform. The example depicted focuses on multiple Plasmodium stages (intra-erythrocytic stage omitted from the diagram) from which candidate genes selected for a malaria vaccine are integrated into the pseudoparasite. Other genes of interest identified in future studies could be incorporated to develop a platform expressing as many genes as necessary to provide an effective vaccine.
Cell-free protein synthesis systems
The wheat-germ cell-free expression system was used to produce a functional PfDHFR-TS, known to have resisted multiple attempts in other expression systems [71]. Its application to malaria vaccine candidates [72] required no prior optimization or harmonization of the P. falciparum AT-biased codon usage, and yielded soluble, highly immunogenic proteins. A small-scale format expression analysis of 124 P. falciparum genes resulted in the production of 93 recombinant proteins, suggesting that this system may represent a suitable alternative for proteins from genes with codon bias and when post-translational modifications are not required.
The ‘next door neighbor’ as the optimal expression system
Phylogeny and systematics are powerful tools to establish evolutionary relationships between organisms, and they can contribute significantly to select species that may exhibit the highest potential to produce heterologous recombinant proteins. Similar structures and mechanisms present in distinct organisms might underscore unknown shared processes required for production of functional proteins [73]. Therefore, the identification of close relatives to the parasites of interest might lead to the development of novel alternative platforms for expressing recombinant proteins from protozoan parasites. Perkinsus marinus is a protozoan parasite of mollusks within the dinoflagellate lineage, still close to the divergence point from the apicomplexans [74]. In addition to the multiple morphological features that Perkinsus shares with the Apicomplexa, the availability of cell-free, fully defined media formulations for its culture, the 16–24 h doubling time reaching densities up to 108 cells per ml, and the ease of culture scale-up [75, 76], suggest that Perkinsus might constitute an optimal expression system for producing recombinant proteins from apicomplexan parasites, that meet the necessary qualitative and quantitative requirements. Further, Perkinsus is non-pathogenic to humans, and available resources include a fully sequenced genome and transfection methodology [77].
The alga Chromera velia has been dubbed a ‘bridge over troublesome plastids’ by providing a strong case for a single endosymbiotic origin of the dinoflagellates and apicomplexan plastids derived from a red algal endosymbiont [78, 79]. Because Chromera can also be easily grown in culture, it has been deemed to have considerable potential for the successful expression of apicomplexan plastid-encoded proteins, as well as for high-throughput screening for drugs targeting the plastid [79]. Nevertheless, considerable characterization and optimization of the system will be required before it can become useful for these and other applications. On the basis of the extensive body of knowledge on the use of bioengineered algae for industrial applications [80], however, it could be anticipated that its potential may be realized in the near future.
Conclusions and future directions
The biological roles of most annotated genes of protozoan parasites remain unknown, and the availability of their products, which is key to addressing questions about their structure–function relationships remains hindered by the lack of optimal expression platforms for producing the recombinant proteins that would facilitate structural studies. From the above discussion, it becomes evident that the available systems are far from optimal. The development of novel expression systems (Box 1), together with the continued optimization of the currently available systems (Box 2) should contribute to rapidly increase the number of functional immunogenic recombinant proteins from protozoan parasites. Greater understanding of the cell biology of parasites and their close relatives by the comprehensive use of the existing resources will be key to advancing the field, and enable the development of ‘phylogenetically tailored’ heterologous expression systems. Hence, strategies for producing parasite recombinant proteins should be designed on an individual basis, and gradually developed to higher levels of complexity (Figure 2). ‘Systems biotechnology’ is still in its early stages, however, and novel disciplines (such as evolutionary engineering and inverse metabolic engineering) illustrate the emergence of innovative concepts (reviewed in Ref. [81]) that may spearhead progress in addressing parasitic diseases. In this context, the parasite P. marinus could be engineered as a suitable surrogate for production of apicomplexan proteins. Thus, designing a non-pathogenic, albeit phylogenetically closely related ‘pseudoparasite’ (Figure 3), which expresses multiple parasite genes with the correct folding and post-translational modifications, while taking advantage of the whole engineered organism as an adjuvant, might elicit a specific effective immune response and thus, constitute a promising alternative to the currently available platforms for vaccine design and delivery.
Box 1. Commercial eukaryotic protein expression systems.
The protozoan Leishmania tarentolae, which was isolated from the Moorish gecko Tarentola mauritanica [82], has been developed into a eukaryotic protein expression system (LEXSY, Jena Bioscience). Its eukaryotic gene expression machinery, which includes full glycosylation and disulfide bond formation, represents a potential advantage over other expression systems. In addition, the system offers a suite of inducible or constitutive vectors, and the choice to target the protein to intracellular compartments or for secretion. Moreover, the strains expressing the protein of interest are stable, easy to culture, and amenable of production at fermentation scale. The system is relatively new in the market, and its thorough implementation should, in the future, reveal to what extent it can contribute to resolve the current limitations in the production of recombinant protein from protozoan parasites.
Box 2. Gateway recombination strategy.
The actual strategies for production of recombinant proteins include the screening of multiple host systems that require building multiple constructs with different purification tags. To reduce the pathway defined as ‘linear’: build an expression clone for a particular host, if the recombinant protein is not suitable for the application, go back and prepare another vector for the same or a new host [83], Gateway (Invitrogen) (for a review on this method see [84]) developed a method that relies on a multistep recombinational cloning eliminating the need for the traditional enzymatic restriction and DNA ligase [85]. This system can be applied to both ‘low-throughput’ test a single protein of interest in different hosts or ‘high-throughput’ to test a large number of proteins in the same heterologous system and under the same conditions, specially if a ORF library is available with each ORF as individual clone and the target vector has all the elements necessary for driving the expression and purifying the recombinant protein. This system has been applied successfully to P. falciparum single-exon genes, with an 84% cloning efficiency and a 100% success rate when subcloned into three expression vectors [31]. The same approach was used to identify novel malaria antigens for DNA vaccines [86] and it is being adopted for cloning and expressing genes in Entamoeba histolytica [87]. Similarly, this system can be quickly adopted for any parasite of interest to build a battery of expression vectors designed specifically to express and produce recombinant proteins in any parasite specific heterologous system developed [88].
Box 3. Source of the currently available expression systems.
For a Review of widely used prokaryotic and eukaryotic systems, see Ref. [28]. Invitrogen commercializes the Pichia Expression Kit and Bac-to-Bac baculovirus expression system (www.invitrogen.com). FlashBAC™ (Oxford Expression Technologies, UK) offers the potential for automation and parallelization. With the same objective, BioSciences Inc. (USA) and Invitrogen (USA) have introduced the InsertDirect™ and HEK 293FreeStyle ™ systems, respectively. The protein expression system using the protozoan Leishmania tarentolae LEXSY has been recently commercialized by Jena Bioscience (www.jenabioscience.com). The wheat germ cell-free protein expression system (WREPO®) was developed by Cell Free Sciences (www.cfsciences.com). Non-commercial systems under development or with potential for became alternative systems for the expression of recombinant proteins are available from the laboratories where it is been developed. T. thermophila is the most advanced [54]. In the case of Dictyostelium, which has recently been proposed as an alternative expression system for eukaryotic proteins ([60]) it is available at the Dictyostelium consortium (http://www.nih.gov/science/models/d_discoideum/). The P. marinus system is under development at the corresponding author laboratory. The alga Chromera has also considerable potential for the successful expression of apicomplexan proteins [79]. Most of these organisms are available at ATCC (www.ATCC.org).
Acknowledgments
Supported by Grant 1R21AI076797-01A2 from the NIH, by NOAA-MD Sea Grant SA7528068-I, and by NSF/USDA-CSREES NSF/USDA 0333240.
Glossary of Terms
- Codon harmonization (synonymous translational attenuation)
During the translation in the ribosomes, low frequency codons may cause translational pausing, favoring the correct folding of the proteins. Codon harmonization consists in matching the usage frequencies, specially the low usage frequencies, of the native codons from the gene within the native organism as nearly as possible to codons recognized as low usage frequency codons by the surrogate host. This ensures that the positional codon frequency of low/intermediate and high usage codons remains unaltered in the surrogate host, allowing the needed halts in the translational processes to match that of the organism of interest (natural host). Differences in synonymous codon usage between the natural host and surrogate cells may lead to low protein expression and the formation of insoluble aggregates.
- Codon optimization
Most of the 20 genetically determined amino acids are encoded by multiple codons, which each species use at a distinct frequency. Protein synthesis in protozoan parasites relies on codons that are rarely employed by the surrogate organism in a heterologous expression system. Codon optimization consists in matching the frequencies at which different codons are used in the natural host (donor) and the surrogate organism.
- Homologous protein expression system
a system in which production of the protein of interest takes place in the natural host under a specific (inducible or not) promoter, often different from the native promoter.
- Heterologous protein expression system
a system in which production of the protein of interest is carried out in an organism different from the organism source of the target protein.
- Inclusion bodies
insoluble aggregates of denatured/unfolded protein produced in the cytoplasm or nucleus of bacteria.
- Surrogate system
organism or cellular components used to study a particular mechanism or express a protein naturally produced by a different organism.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Azevedo WF, Jr., Soares MB. Selection of targets for drug development against protozoan parasites. Curr. Drug Targets. 2009;10:193–201. doi: 10.2174/138945009787581186. [DOI] [PubMed] [Google Scholar]
- 2.Coombs A. The sequencing shakeup. Nat. Biotechnol. 2008;26:1109–1112. doi: 10.1038/nbt1008-1109. [DOI] [PubMed] [Google Scholar]
- 3.Greenwood BM, et al. Malaria: progress, perils, and prospects for eradication. J. Clin. Invest. 2008;118:1266–1276. doi: 10.1172/JCI33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray CK, Bennett JW. Rapid diagnosis of malaria. Interdiscip. Perspect. Infect. Dis. 2009;2009:415953. doi: 10.1155/2009/415953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seitzer U, et al. Evaluation of Theileria annulata recombinant immunodominant proteins for the development of ELISA. Transbound. Emerg. Dis. 2008;55:244–248. doi: 10.1111/j.1865-1682.2008.01030.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee N, et al. Effect of sequence variation in Plasmodium falciparum histidine- rich protein 2 on binding of specific monoclonal antibodies: Implications for rapid diagnostic tests for malaria. J. Clin. Microbiol. 2006;44:2773–2778. doi: 10.1128/JCM.02557-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anon International meeting: new diagnostic tests are urgently needed to treat patients with Chagas disease. Rev. Soc. Bras. Med. Trop. 2008;41:315–319. doi: 10.1590/s0037-86822008000300020. [DOI] [PubMed] [Google Scholar]
- 8.Hunfeld KP, et al. Babesiosis: recent insights into an ancient disease. Int. J. Parasitol. 2008;38:1219–1237. doi: 10.1016/j.ijpara.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Neuber H. Leishmaniasis. J. Dtsch Dermatol. Ges. 2008;6:754–765. doi: 10.1111/j.1610-0387.2008.06809.x. [DOI] [PubMed] [Google Scholar]
- 10.Ripley Ballou W. Obstacles to the development of a safe and effective attenuated pre-erythrocytic stage malaria vaccine. Microbes Infect. 2007;9:761–766. doi: 10.1016/j.micinf.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Fatumo S, et al. Estimating novel potential drug targets of Plasmodium falciparum by analysing the metabolic network of knock-out strains in silico. Infect. Genet. Evol. 2009;9:351–358. doi: 10.1016/j.meegid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Awale M, et al. Homology modeling and atomic level binding study of Leishmania MAPK with inhibitors. J. Mol. Model. 2009 doi: 10.1007/s00894-009-0565-3. [Epub. ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Wiesner J, et al. The plastid-like organelle of apicomplexan parasites as drug target. Curr. Pharm. Des. 2008;14:855–871. doi: 10.2174/138161208784041105. [DOI] [PubMed] [Google Scholar]
- 14.Morgan RE, et al. Inhibitors of tubulin assembly identified through screening a compound library. Chem. Biol. Drug Des. 2008;72:513–524. doi: 10.1111/j.1747-0285.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, et al. Inhibitors of Plasmodium falciparum methionine aminopeptidase 1b possess antimalarial activity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14548–14553. doi: 10.1073/pnas.0604101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joachimiak A. High-throughput crystallography for structural genomics. Curr. Opin. Struct. Biol. 2009;19:573–584. doi: 10.1016/j.sbi.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel V, et al. Identification and characterization of small molecule inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J. Biol. Chem. 2008;283:35078–35085. doi: 10.1074/jbc.M804990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillier CJ, et al. Process development and analysis of liver-stage antigen 1, a preerythrocyte-stage protein-based vaccine for Plasmodium falciparum. Infect. Immun. 2005;73:2109–2115. doi: 10.1128/IAI.73.4.2109-2115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnot DE, et al. Comparative testing of six antigen-based malaria vaccine candidates directed toward merozoite-stage Plasmodium falciparum. Clin. Vaccine Immunol. 2008;15:1345–1355. doi: 10.1128/CVI.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian F, et al. Conjugating recombinant proteins to Pseudomonas aeruginosa ExoProtein A: a strategy for enhancing immunogenicity of malaria vaccine candidates. Vaccine. 2007;25:3923–3933. doi: 10.1016/j.vaccine.2007.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Q, et al. Immunogenicity and in vitro protective efficacy of a polyepitope Plasmodium falciparum candidate vaccine constructed by epitope shuffling. Vaccine. 2007;25:5155–5165. doi: 10.1016/j.vaccine.2007.04.085. [DOI] [PubMed] [Google Scholar]
- 22.Lau YL, Fong MY. Toxoplasma gondii: serological characterization and immunogenicity of recombinant surface antigen 2 (SAG2) expressed in the yeast Pichia pastoris. Exp. Parasitol. 2008;119:373–378. doi: 10.1016/j.exppara.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Luk FC, et al. N-linked glycosylation of proteins in the protozoan parasite Toxoplasma gondii. Mol. Biochem. Parasitol. 2008;157:169–178. doi: 10.1016/j.molbiopara.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier A, Steverding D. Expression and purification of non-glycosylated Trypanosoma brucei transferrin receptor in insect cells. Exp. Parasitol. 2008;120:205–207. doi: 10.1016/j.exppara.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Miao J, et al. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Saksouk N, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birkholtz LM, et al. Heterologous expression of plasmodial proteins for structural studies and functional annotation. Malar. J. 2008;7:197. doi: 10.1186/1475-2875-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 29.Vedadi M, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Mehlin C, et al. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol. Biochem. Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Aguiar JC, et al. High-throughput generation of Plasmodium falciparum functional molecules by recombinational cloning. Genome Res. 2004;14:2076–2082. doi: 10.1101/gr.2416604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graslund S, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspeta L, et al. The effect of heating rate on Escherichia coli metabolism, physiological stress, transcriptional response, and production of temperature-induced recombinant protein: a scale-down study. Biotechnol. Bioeng. 2009;102:468–482. doi: 10.1002/bit.22084. [DOI] [PubMed] [Google Scholar]
- 34.Crowther GJ, et al. Buffer optimization of thermal melt assays of Plasmodium proteins for detection of small-molecule ligands. J. Biomol. Screen. 2009;14:700–707. doi: 10.1177/1087057109335749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahdev S, et al. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol. Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 36.Baca AM, Hol WG. Overcoming codon bias: a method for high-level overexpression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int. J. Parasitol. 2000;30:113–118. doi: 10.1016/s0020-7519(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 37.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 38.Welch M, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Na BK, et al. Cryptopain-1, a cysteine protease of Cryptosporidium parvum, does not require the pro-domain for folding. Parasitology. 2009;136:149–157. doi: 10.1017/S0031182008005350. [DOI] [PubMed] [Google Scholar]
- 40.Quintas-Granados LI, et al. Purification, refolding and autoactivation of the recombinant cysteine proteinase EhCP112 from Entamoeba histolytica. Protein Expr. Purif. 2009;63:26–32. doi: 10.1016/j.pep.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Aboge GO, et al. Cloning, expression, and characterization of Babesia gibsoni dihydrofolate reductase-thymidylate synthase: inhibitory effect of antifolates on its catalytic activity and parasite proliferation. Antimicrob. Agents Chemother. 2008;52:4072–4080. doi: 10.1128/AAC.00384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt NW, et al. CD8 T cell immunity to Plasmodium permits generation of protective antibodies after repeated sporozoite challenge. Vaccine. 2009;27:6103–6106. doi: 10.1016/j.vaccine.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaCount DJ, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 45.Shrestha B, et al. Baculovirus expression vector system: an emerging host for high-throughput eukaryotic protein expression. Methods Mol. Biol. 2008;439:269–289. doi: 10.1007/978-1-59745-188-8_19. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Expression, purification and characterization of recombinant mitochondrial topoisomerase II of kinetoplastid Crithidia fasciculata in High-five insect cells. Protein Expr. Purif. 2008;58:122–131. doi: 10.1016/j.pep.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 47.Birkett A, et al. A modified hepatitis B virus core particle containing multiple epitopes of the Plasmodium falciparum circumsporozoite protein provides a highly immunogenic malaria vaccine in preclinical analyses in rodent and primate hosts. Infect. Immun. 2002;70:6860–6870. doi: 10.1128/IAI.70.12.6860-6870.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walther M, et al. Safety, immunogenicity, and efficacy of prime-boost immunization with recombinant poxvirus FP9 and modified vaccinia virus Ankara encoding the full-length Plasmodium falciparum circumsporozoite protein. Infect. Immun. 2006;74:2706–2716. doi: 10.1128/IAI.74.5.2706-2716.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonaldo MC, et al. Surface expression of an immunodominant malaria protein B cell epitope by yellow fever virus. J. Mol. Biol. 2002;315:873–885. doi: 10.1006/jmbi.2001.5258. [DOI] [PubMed] [Google Scholar]
- 50.Pang AL, et al. In vivo expression and immunological studies of the 42-kilodalton carboxyl-terminal processing fragment of Plasmodium falciparum merozoite surface protein 1 in the baculovirus-silkworm system. Infect. Immun. 2002;70:2772–2779. doi: 10.1128/IAI.70.6.2772-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Draper SJ, et al. Recombinant viral vaccines expressing merozoite surface protein-1 induce antibody- and T cell-mediated multistage protection against malaria. Cell Host Microbe. 2009;5:95–105. doi: 10.1016/j.chom.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caetano BC, et al. Vaccination with replication-deficient recombinant adenoviruses encoding the main surface antigens of Toxoplasma gondii induces immune response and protection against infection in mice. Hum. Gene Ther. 2006;17:415–426. doi: 10.1089/hum.2006.17.415. [DOI] [PubMed] [Google Scholar]
- 53.Roque-Resendiz JL, et al. MVA ROP2 vaccinia virus recombinant as a vaccine candidate for toxoplasmosis. Parasitology. 2004;128:397–405. doi: 10.1017/s0031182003004761. [DOI] [PubMed] [Google Scholar]
- 54.Gaertig J, et al. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotechnol. 1999;17:462–465. doi: 10.1038/8638. [DOI] [PubMed] [Google Scholar]
- 55.Clark TG, et al. The I-antigens of Ichthyophthirius multifiliis are GPI-anchored proteins. J. Eukaryot. Microbiol. 2001;48:332–337. doi: 10.1111/j.1550-7408.2001.tb00322.x. [DOI] [PubMed] [Google Scholar]
- 56.Peterson DS, et al. The circumsporozoite protein of Plasmodium falciparum is expressed and localized to the cell surface in the free-living ciliate Tetrahymena thermophila. Mol. Biochem. Parasitol. 2002;122:119–126. doi: 10.1016/s0166-6851(02)00079-8. [DOI] [PubMed] [Google Scholar]
- 57.Weide T, et al. A recombinase system facilitates cloning of expression cassettes in the ciliate Tetrahymena thermophila. BMC Microbiol. 2007;7:12. doi: 10.1186/1471-2180-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller-Taubenberger A, Bozzaro S. From cell-cell adhesion and cellular oscillations to spectacular views inside the cell--50 years of research with Dictyostelium. Eur. J. Cell Biol. 2006;85:851–858. doi: 10.1016/j.ejcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Arya R, et al. Dictyostelium discoideum--a promising expression system for the production of eukaryotic proteins. FASEB J. 2008;22:4055–4066. doi: 10.1096/fj.08-110544. [DOI] [PubMed] [Google Scholar]
- 61.Reymond CD, et al. Anchoring of an immunogenic Plasmodium falciparum circumsporozoite protein on the surface of Dictyostelium discoideum. J. Biol. Chem. 1995;270:12941–12947. doi: 10.1074/jbc.270.21.12941. [DOI] [PubMed] [Google Scholar]
- 62.Fasel N, et al. Dictyostelium discoideum as an expression host for the circumsporozoite protein of Plasmodium falciparum. Gene. 1992;111:157–163. doi: 10.1016/0378-1119(92)90683-g. [DOI] [PubMed] [Google Scholar]
- 63.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asahi H. Plasmodium falciparum: Chemically defined medium for continuous intraerythrocytic growth using lipids and recombinant albumin. Exp. Parasitol. 2009;121:22–28. doi: 10.1016/j.exppara.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Suresh P, Rehg JE. Comparative evaluation of several techniques for purification of Cryptosporidium parvum oocysts from rat feces. J. Clin. Microbiol. 1996;34:38–40. doi: 10.1128/jcm.34.1.38-40.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howe DK, et al. Expression of Toxoplasma gondii genes in the closely-related apicomplexan parasite Neospora caninum. Mol. Biochem. Parasitol. 1997;86:29–36. [PubMed] [Google Scholar]
- 67.Ramirez JR, et al. Attenuated Toxoplasma gondii ts-4 mutants engineered to express the Leishmania antigen KMP-11 elicit a specific immune response in BALB/c mice. Vaccine. 2001;20:455–461. doi: 10.1016/s0264-410x(01)00341-3. [DOI] [PubMed] [Google Scholar]
- 68.Kucknoor AS, et al. Heterologous expression in Tritrichomonas foetus of functional Trichomonas vaginalis AP65 adhesin. BMC Mol. Biol. 2005;6:5. doi: 10.1186/1471-2199-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sa JM, et al. Expression and function of pvcrt-o, a Plasmodium vivax ortholog of pfcrt, in Plasmodium falciparum and Dictyostelium discoideum. Mol. Biochem. Parasitol. 2006;150:219–228. doi: 10.1016/j.molbiopara.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Carrillo C, et al. Trypanosoma cruzi as a model system to study the expression of exogenous genes coding for polyamine biosynthetic enzymes. Induction of DFMO resistance in transgenic parasites. Biochim. Biophys. Acta. 2007;1770:1605–1611. doi: 10.1016/j.bbagen.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 71.Mudeppa DG, et al. Cell-free production of functional Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Mol. Biochem. Parasitol. 2007;151:216–219. doi: 10.1016/j.molbiopara.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 72.Tsuboi T, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect. Immun. 2008;76:1702–1708. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leander BS, Keeling PJ. Morphostasis in alveolate evolution. Trends Ecol. Evol. 2003;18:395–402. [Google Scholar]
- 74.Saldarriaga JF, et al. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int. J. Syst. Evol. Microbiol. 2003;53:355–365. doi: 10.1099/ijs.0.02328-0. [DOI] [PubMed] [Google Scholar]
- 75.Gauthier JD, et al. Effect of fetal bovine serum glycoproteins on the in vitro proliferation of the oyster parasite Perkinsus marinus: Development of a fully defined medium. J. Eukaryot. Microbiol. 1995;42:307–313. doi: 10.1111/j.1550-7408.1995.tb01585.x. [DOI] [PubMed] [Google Scholar]
- 76.La Peyre JF, Faisal M. Development of a protein-free chemically defined culture medium for the propagation of the oyster pathogen Perkinsus marinus. Parasite. 1997;4:67–73. [Google Scholar]
- 77.Fernández-Robledo JA, et al. Transfection of the protozoan parasite Perkinsus marinus. Mol. Biochem. Parasitol. 2008;157:44–53. doi: 10.1016/j.molbiopara.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 78.Keeling PJ. Evolutionary biology: bridge over troublesome plastids. Nature. 2008;451:896–897. doi: 10.1038/451896a. [DOI] [PubMed] [Google Scholar]
- 79.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 80.Raja R, et al. A perspective on the biotechnological potential of microalgae. Crit. Rev. Microbiol. 2008;34:77–88. doi: 10.1080/10408410802086783. [DOI] [PubMed] [Google Scholar]
- 81.Graf A, et al. Yeast systems biotechnology for the production of heterologous proteins. FEMS Yeast Res. 2009;9:335–348. doi: 10.1111/j.1567-1364.2009.00507.x. [DOI] [PubMed] [Google Scholar]
- 82.Wallbanks KR, et al. The identity of Leishmania tarentolae Wenyon 1921. Parasitology. 1985;90(Pt 1):67–78. doi: 10.1017/s0031182000049027. [DOI] [PubMed] [Google Scholar]
- 83.Esposito D, et al. Gateway cloning for protein expression. Methods Mol. Biol. 2009;498:31–54. doi: 10.1007/978-1-59745-196-3_3. [DOI] [PubMed] [Google Scholar]
- 84.Hartley JL. Use of the gateway system for protein expression in multiple hosts. Curr. Protoc. Protein Sci. 2003 doi: 10.1002/0471140864.ps0517s30. Chapter 5, Unit 5 17. [DOI] [PubMed] [Google Scholar]
- 85.Hartley JL, et al. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haddad D, et al. Novel antigen identification method for discovery of protective malaria antigens by rapid testing of DNA vaccines encoding exons from the parasite genome. Infect. Immun. 2004;72:1594–1602. doi: 10.1128/IAI.72.3.1594-1602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abhyankar MM, et al. Development of the Gateway system for cloning and expressing genes in Entamoeba histolytica. Parasitol. Int. 2009;58:95–97. doi: 10.1016/j.parint.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delbecq S, et al. Hydrophobic moeties in recombinant proteins are crucial to generate efficient saponin-based vaccine against apicomplexan Babesia divergens. Vaccine. 2006;24:613–621. doi: 10.1016/j.vaccine.2005.08.073. [DOI] [PubMed] [Google Scholar]