Abstract
Hepatitis C virus (HCV) directly induces oxidative stress and liver injury. Bach1, a zipper (bZip) mammalian transcriptional repressor, negatively regulates heme oxygenase 1 (HMOX1), a key cytoprotective enzyme that has anti-oxidant and anti-inflammatory activities. microRNAs are small non-coding RNAs (~22 nt) that are important regulators of gene expression. Whether and how microRNAs regulate Bach1 or HCV are largely unknown. The aims of this study were to determine whether miR-196 regulates Bach1, HMOX1, and/or HCV gene expression. HCV replicon cell lines (Con1 and 9-13) of the Con1 isolate and J6/JFH1-based HCV cell culture system were used in this study. The effects of miR-196 mimic on Bach1, HMOX1 and HCV RNA and protein levels were measured by qRT-PCR and Western blots, respectively. The Dual Glo™ Luciferase Assay System was used to determine reporter activities. miR-196 mimic significantly down-regulated Bach1 and up-regulated HMOX1 gene expression, and inhibited HCV expression. Dual luciferase reporter assays demonstrated that transfection of miR-196 mimic resulted in a significant decrease in Bach1 3′-UTR-dependent luciferase activity but not in mutant Bach1 3′-UTR-dependent luciferase activity. Moreover, there was no detectable effect of mutant miR-196 on Bach1 3′-UTR-dependent luciferase activity.
Conclusions
miR-196 directly acts on the 3′-UTR of Bach1 mRNA and translationally represses the expression of this protein, and up-regulates HMOX1. miR-196 also inhibits HCV expression in HCV replicon cell lines (genotype 1b) and in J6/JFH1 (genotype 2a) HCV cell culture system. Thus, miR-196 plays a role in both HMOX1/Bach1 expression and the regulation of HCV expression in human hepatocytes. Over-expression of miR-196 holds promise as a potential novel strategy to prevent or ameliorate hepatitis C infection, and to protect against liver injury in chronic HCV infection.
Keywords: microRNA, heme oxygenase-1, hepatitis C virus, 3′-UTR, reporter gene assay
Hepatitis C virus (HCV) infection is a world-wide health problem. The current standard therapy for chronic hepatitis C (CHC) is a combination of pegylated interferon (IFN) and ribavirin, but only ~50% of patients respond to such treatment, which is expensive, prolonged, and attended by numerous unpleasant side-effects (1, 2). There is a continuing need to develop new anti-viral therapies. The molecular mechanisms underlying HCV-associated liver injury remain imperfectly understood. However, studies have indicated that HCV directly induces oxidative stress in hepatocytes through multiple mechanisms that include chronic inflammation, increases in iron, and liver injury. Therefore oxidative stress has emerged as an important pathogenetic mechanism in chronic hepatitis C (3–5), and antioxidant and cytoprotective therapy has been proposed to treat hepatitis C.
Heme oxygenase 1 (HMOX1) is a key cytoprotective enzyme, catalyzing heme degradation and generating ferrous iron, carbon monoxide and biliverdin, the latter two of which have anti-oxidant and anti-inflammatory activities in vivo (6–8). Bach1, a basic leucine zipper (bZip) mammalian transcriptional repressor of HMOX1, negatively regulates HMOX1 gene expression. Bach1 forms antagonizing heterodimers with the Maf-related oncogene family. These heterodimers bind to Maf recognition elements (MAREs) and suppress expression of genes [e.g., HMOX1 and NAD(P)H:quinone oxidoreductase (NQO1)] (9–12).
microRNAs (miRNAs) are a class of small non-coding RNAs (~22 nt) that regulate gene expression primarily through translational repression (13, 14). The exact mechanism by which target genes are down-regulated remains unclear. Sequence complementarity in the 6–8 base pair ‘seed regions’ at the end of miRNA-mRNA heteroduplexes seem to determine the specificity of miRNA-target RNA interactions (15). miR-196 was first recognized to have extensive and evolutionarily conserved complementarity to homeobox clusters, groups of related transcription factor genes crucial for numerous developmental programs in animals, and to regulate homeobox gene expression (16, 17). A recent report indicated one perfect match between miR-196 and non-structural (NS) 5A coding region of the HCV JFH1 genome (genotype 2a), and IFN β treatment resulted in significant induction of miR-196 in the human hepatoma cell line Huh-7 and in primary murine hepatocytes (18). A search of the TargetScan 4.2 data base revealed that at least two miR-196 seed match sites occur in the 3′-UTR of Bach1 mRNA. This led us to propose that miR-196 is a miRNA that targets the HCV genome and the 3′-UTR of Bach1 mRNA, the latter leading to up-regulation of the HMOX1 gene. These dual effects are analogous to effects of miR-122, recently reported from our laboratory (19).
In the work reported here we experimentally validated the computational prediction of miR-196 binding in the 3′-UTR of Bach 1 mRNA by luciferase reporter assay. miR-196 mimic significantly down-regulated Bach1 and up-regulated HMOX1 gene expression, and inhibited HCV expression. These results suggest that up-regulation of miR-196 may be an additional new therapeutic approach to prevent or ameliorate hepatitis C infection, and to protect against liver injury in chronic HCV infection. The findings indicate that miR-196 plays a role in the regulation of HMOX1/Bach1 expression and HCV replication in hepatocytes. They also add to the growing evidence that up-regulation of HMOX1 may be beneficial in HCV infection (5, 19, 20).
MATERIALS AND METHODS
Cell Culture
9-13 cells were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). 9-13 cells harbor a replicating HCV non-structural region with the use of the NS3 -NS5B gene regions from the Con1 isolate (21). Huh-7.5 and Con1 subgenomic genotype 1b HCV replicon cell lines were from Apath LLC (St, Louis, MO). Huh-7.5 is a highly permissive, alpha interferon-cured Huh-7 human hepatocellular carcinoma cell line derivative. The Con1 cell line is a Huh-7.5 cell population containing the full-length HCV genotype 1b replicon. The 9-13, Huh.7.5 and Con1 cells were maintained in DMEM supplemented with 10% (v/v) FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and selection antibiotic 500 μg/mL G418 for 9-13 cells, or 750 μg/mL G418 for Con1 cells.
microRNAs, siRNAs and Reporter Constructs
The miRIDIAN microRNA mimics for has-miR-196, has-miR-16, customized mutant has-miR-196, miRNA mimic negative control (MMNC), and Bach1-siRNA were obtained from Dharmacon (Lafayette, CO). pRL-TK vector was from Promega. The pRL-TK reporter vector contains a cDNA (Rluc) encoding Renilla luciferase as an internal control reporter. pGL3-Bach1 luciferase reporter construct, containing an 1837 bp fragment of Bach1 3′-UTR, was a kind gift of Dr. Rolf Renne (University of Florida, Gainesville, FL) (22). Mutant pGL3-Bach1 was generated by GENEWIZ, Inc. (South Plainfield, NJ). pLSV40-Rluc and pLSV40-GL3/Bach1 reporter vectors were kindly provided by B. R. Cullen (Duke University, Durham, NC). pLSV40-Rluc contains a cDNA (Rluc) encoding Renilla luciferase as an internal control reporter and pLSV40-GL3/Bach1 firefly luciferase reporter construct contains the full-length 3′-UTR of Bach1 mRNA (23). Constructs were confirmed by restriction enzyme digestion and sequencing.
Transfection and Luciferase Activity Assays
Transfection of miR-196 mimic or Bach-siRNA was performed as described previously (24). Co-transfection of microRNA mimics and reporters were carried out using Lipofectamine 2000 from Invitrogen (Carlsbad, CA) according to manufacturer’s protocol. Briefly, cells were co-transfected with 0.4 ug/mL of pGL3-Bach1 or mutant pGL3-Bach, with 0.4 ug/mL of pRL-TK, and with 10–50 nM tested miRNAs. 24–48 h after transfection, cells were harvested and lysed, and the luciferase reporter activities were measured using the Dual-Glo® Luciferase Assay System. Firefly luciferase activity was normalized to Renilla luciferase activity and total protein, determined using the BCA protein assay kit. For ease of comparison, values for cells with pGL3-Bach1 and pRL-TK transfection were set equal to 1.
In Vitro Transcription, Transfection and Infection
The HCV infectious clone pJ6/JFH1, the full-length chimeric genome with the core-NS2 regions from the infectious J6 (genotype 2a) and NS3-NS5B regions from the infectious JFH1 (genotype 2a), was generously provided by C. M. Rice (the Rockefeller University, New York, NY). The production of J6/JFH1-based cell culture-generated HCV (HCVcc) has been reported previously (25). In brief, the pJ6/JFH1 plasmid was linearized with XbaI, and purified by ethanol precipitation, digestion with proteinase K, and phenol-chloroform extraction. The linearized plasmid was used as a template for in vitro transcription using the MEGAscript T7 kit (Ambion, Austin, TX). For HCV RNA transfection, Huh-7.5 cells were plated in 24-well plates one day prior to transfection and transfected at 70~80% confluence. Cells were transfected at an RNA/lipofectamine ratio of 1:2 by using 2 μg/well of HCV RNA and 4 uL/well Lipofectamine 2000 for 48 h. To infect naïve Huh-7.5 cells, cell culture supernatants from the cells transfected with HCV RNA for 48 h were collected and filtered through a 0.20 μm filter, and added to cultures of naïve Huh-7.5 cells.
The pFK-Con1 construct (genotype 1b) (21) was a kind gift of Dr. R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). Mutant pFK-Con1 (pFK-Con1-Mut) with mutations in four nucleotides in the putative binding sites for miR-196 was generated by GENEWIZ, Inc. (South Plainfield, NJ) and confirmed by sequencing. pFK-Con1-WT and pFK-Con1-Mut were linearized with AseI and ScaI. In vitro transcription and transfection were performed as described previously.
Western Blots
Western blots were performed using the standard protocols of our laboratory as reported previously (10). The method is described in detail in Supplemental Materials and Methods (see Supporting Information online).
Quantitative RT-PCR
Total RNA from tested cells was extracted and cDNA was synthesized (10). Real time quantitative RT-PCR was carried out using primers as reported previously (19, 26), and as described in detail in Supplemental Material and Methods (see Supporting Information online).
Statistical Analysis
Initial analysis showed that results were normally distributed. Therefore, parametric statistical procedures were used. The Student’s t-test (for comparison of two conditions) or analysis-of-variance (for comparisons among more than two) was used to analyze the differences among means. Values of P <0.05 were considered statistically significant. Experiments were repeated at least three times with similar results. All experiments included at least triplicate samples for each treatment group. Representative results from single experiments are presented. Statistical analyses were performed with JMP 4.0.4 software from SAS Institute (Cary, NC).
RESULTS
Analysis in silico predicts two seed region matches of miR-196 and the 3′-UTR of Bach1 mRNA
An online search of the TargetScan 4.2 data base (http://www.targetscan.org/vert_42/) demonstrated that at least two putative miR-196 seed match sites were harbored in the 3′-UTR of Bach1 mRNA. As shown in Figure 1A & B, one of the predicted binding sites (2280–2286 nt) was highly conserved in human, mouse, rat, chicken and dog, whereas the other putative site (2161–2166 nt) was poorly conserved across species. No predicted miR-196 binding sites were found in the Nrf2 and HMOX1 gene, and no putative miR-196 binding sites were found in the coding region of Bach1 gene (data not shown).
Figure 1. In silico prediction of putative binding sites for miR-196 in the 3′-UTR of Bach1.
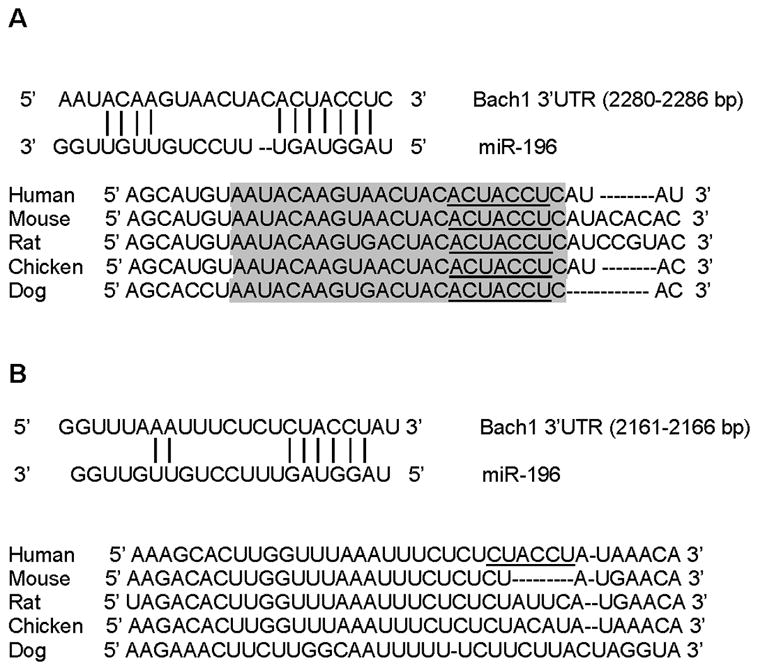
(A) Schematic of seed region match between miR-196 and the first putative Bach1 3′-UTR site targeted. The first site is highly conserved in the human, mouse, rat, dog and pig. (B) Schematic of second seed region match between miR-196 and the putative Bach1 3′-UTR site targeted. The second site is poorly conserved in the human, mouse, rat, dog and pig.
miR-196 down-regulates the transcriptional repressor Bach1 and up-regulates HMOX1 in 9-13 cells expressing HCV non-structural proteins
To experimentally verify that the putative miR-196 binding sites are functional, we transfected 9-13 cells with miR-196 specific mimic and measured Bach1 protein and mRNA levels by Western blots and qRT-PCR, respectively. 9-13 cells transfected with miR-196 mimic showed a significant reduction in the expression of Bach1 protein levels (~55% after 24 h transfection and ~64% after 48 h transfection), compared with miRNA mimic negative control (MMNC), whereas no effects on Bach1 protein levels were detectable in cells transfected with miRNA mimic negative control compared with mock transfection (Figure 2A). However, no significant effect of miR-196 on Bach1 mRNA levels was observed in 9-13 cells (Figure 2B). These results demonstrate that the regulation of miR-196 on Bach1 occurs at a translational level in human hepatoma 9-13 cells.
Figure 2. miR-196 mimic down-regulates Bach1 protein levels and up-regulates expression of the HMOX1 gene in 9-13 cells.
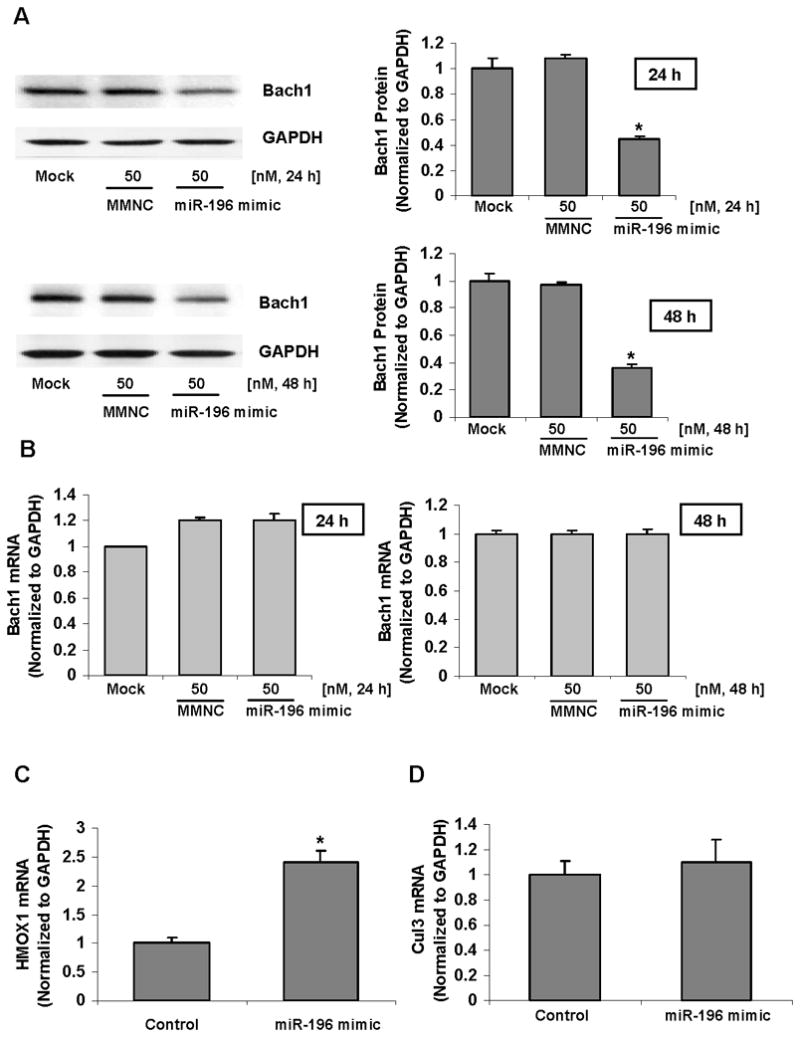
9-13 cells were transfected with 50 nM miR-196 mimic or miRNA mimic negative control (MMNC) by Lipofectamine 2000 as indicated. 24–48 h after transfection, the cells were harvested. Bach1 protein levels were assessed by Western blots with anti-Bach1 or GAPDH specific antibodies. The levels of Bach1 mRNA were quantified by qRT-PCR as described in Materials and Methods. The amounts of Bach1 protein and mRNA were normalized to GAPDH which did not vary with treatment. Values for cells with a mock transfection were set equal to 1. The levels of HMOX1 mRNA were quantified by qRT-PCR. The amounts of HMOX1 mRNA levels were normalized to GAPDH which did not vary with transfection. Values for cells with mimic negative control transfection were set equal to 1. Data are presented as means ± SE, n=3. * differs from negative control only, P<0.05. (A) miR-196 mimic down-regulated Bach1 protein levels. (B) The levels of Bach1 mRNA were not altered with miR-196 mimic transfection. (C) miR-196 mimic up-regulated HMOX1 mRNA levels. (D) miR-196 did not alter Cullin 3 mRNA levels.
Bach1 is a well-established transcriptional repressor of the HMOX1 gene (10, 11), therefore we next determined whether down-regulation of Bach1 protein by miR-196 could increase HMOX1 gene expression. 9-13 cells were transfected with miR-196 mimic or miRNA mimic negative control for 48 h, after which the levels of HMOX1 and Cullin 3 (Cul 3, non-specific gene control) mRNA were quantified by qRT-PCR. As expected, miR-196 mimic significantly up-regulated HMOX1 mRNA levels by ~2.4 fold (Figure 2C), but not Cul 3 mRNA levels (Figure 2D), compared with the same amount of miRNA mimic negative control.
miR-196 directly Interacts with the 3′-UTR of Bach1 mRNA in 9-13 cells expressing HCV non-structural proteins
To further establish that miR-196 targets the 3′-UTR of Bach1 mRNA, which contains two predicted seed match sites for miR-196 (Figure 3A), [rather than exerting a less direct and specific regulation], a reporter construct, which we called pGL3-Bach1, with Bach1 3′-UTR downstream of the firefly luciferase (f-luc) open reading frame (Figure 3B), was used. 9-13 cells were co-transfected with pGL3-Bach1 (f-luc), pRL-TK (renilla, to normalize for transfection efficiencies), and with miRNA-negative controls, miR-196 mimic or miR-16 (a “negative” miR with no predicted binding sites in the 3′-UTR of Bach1 mRNA). 48 h after transfection, the luciferase reporter activity was assayed. miR-196 mimic transfection significantly decreased reporter activity by ~53%, whereas miRNA mimic negative control and miR-16 mimic had no effect on reporter luciferase activity (Figure 3C). A similar effect was observed when we co-transfected 9-13 cells with a reporter construct pLSV40-GL3/Bach1 containing the full length 3′-UTR of Bach1 mRNA and miR-196 mimic. Specifically, 50 nM of miR-196 mimic decreased the firefly luciferase activity by ~59% (Suppl. Figure 1B), whereas miRNA mimic negative control (MMNC) had no significant effect on reporter luciferase activity (Suppl. Figure 1C).
Figure 3. miR-196 mimic represses the expression of a luciferase reporter containing Bach1 3′-UTR.
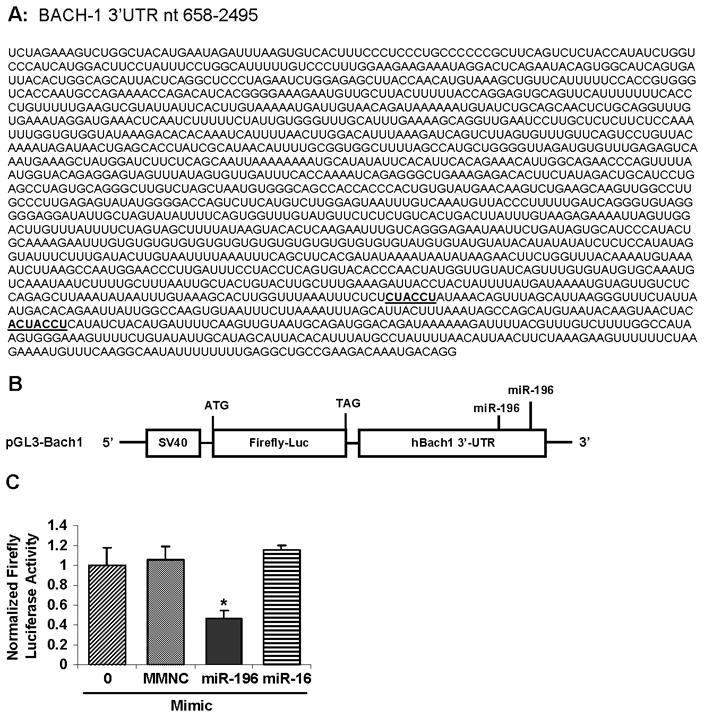
(A) The Bach1 3′-UTR contains two seed match sites (highlighted) for miR-196. (B) Schematic representation of pGL3-Bach1, the firefly luciferase (f-luc) reporter construct utilized. (C) Inhibition of the f-luc activities of pGL3-Bach1 3′-UTR reporter by miR-196 mimic. 9-13 cells were co-transfected with 0.4 μg/mL of pGL3-Bach1, 0.4 μg/mL of pRL-TK (renilla) and with 50 nM miR-196 mimic, miR-16 mimic or miRNA mimic negative control (MMNC) by Lipofectamine 2000 as indicated. 48 h after transfection, the luciferase reporter activities were measured using Dual Luciferase Assay System from Promega. Firefly luciferase activities were normalized to renilla luciferase activities and total protein, as indicated in Materials and Methods. Values for cells with a mock transfection were set equal to 1. Data are presented as means ± SE, n=3. * differs from negative control only, P<0.05.
Next, we co-transfected 9-13 cells with Luc reporter construct containing a four nucleotide mutant Bach1 3′-UTR, which we called pGL3-Bach1-Mut (Figure 4A), and with pRL-TK, and with miR-196 mimic or miR-155 mimic (22) (a “negative” miR, with no changes of predicted miR-155 binding sites in pGL3-Bach1-WT and pGL3-Bach1-Mut), and assayed the luciferase reporter activity. miR-196 mimic transfection significantly decreased luciferase activity in a dose-dependent fashion, whereas miR-196 did not change reporter activity in cells transfected with the reporter construct containing mutant binding sites for miR-196 (Figure 4B). miR-155 significantly decreased reporter activity in both pGL3-Bach1-WT and pGL3-Bach1-Mut (Figure 4C). These results demonstrate that miR-196 directly regulates Bach1 gene expression and miR-196 mediates down-regulation of Bach1 though the 3′-UTR of Bach1 mRNA.
Figure 4. miR-196 does not inhibit luciferase activity of reporter with mutant Bach1 3′-UTR.
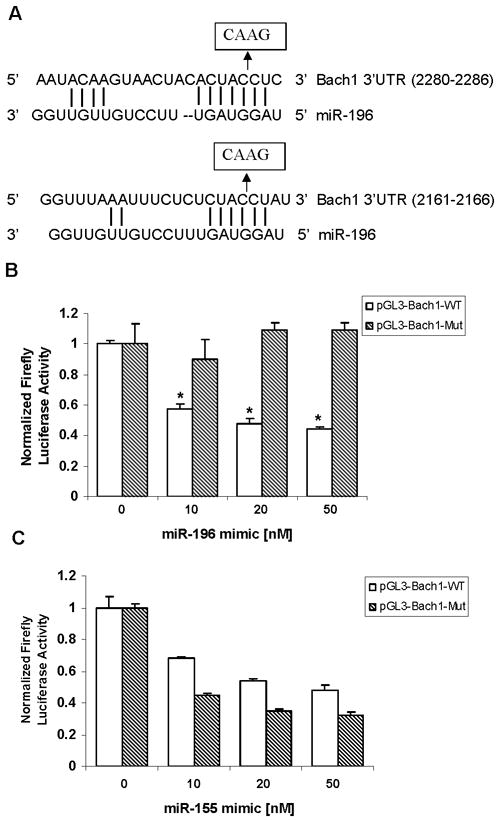
(A) Four nucleotides in two seed match sites of Bach1 3′-UTR were replaced. (B) miR-196 mimic decreased the f-luc activity of pGL3-Bach1-WT but not pGL-Bach1-Mut reporter. 9-13 cells were co-transfected with 0.4 μg/mL of mutant pGL3-Bach1 or 0.4 μg/mL of pGL3-Bach1 and with 0.4 μg/mL of pRL-TK (renilla), and with miR-196 mimic (10–50 nM) as indicated in Materials and Methods, 48 h after transfection, the luciferase reporter activities were measured using Dual Luciferase Assay System from Promega, firefly luciferase activities were normalized to renilla luciferase activities and total protein. (C) miR-155 mimic decreased the f-luc activities of both pGL3-Bach1-WT and pGL-Bach1-Mut reporter. 9-13 cells were co-transfected with 0.4 μg/mL of mutant pGL3-Bach1 or 0.4 μg/mL of pGL3-Bach1 and with 0.4 μg/mL of pRL-TK (renilla), and with miR-155 mimic (10–50 nM). 48 h after transfection, the luciferase reporter activities were assayed as described in Materials and Methods. Values for cells with a mock transfection were set equal to 1. Data are presented as means ± SE, n=3. * differs from negative control only, P<0.05.
To further establish the direct interaction between miR-196 and Bach1-3′-UTR, we created mutant miR-196 (Figure 5A) in which seed match sites for the 3′-UTR of Bach1 mRNA were abolished. Cells were co-transfected with pGL3-Bach1-WT, and with pRL-TK, and with increasing concentrations of miRNA mimic negative control, miR-196 mimic or mutant miR-196 mimic, and luciferase reporter activity was assayed. As anticipated, miR-196 resulted in a significant reduction in luciferase activity in a dose-dependent fashion, which was consistent with our previous observations, whereas miRNA negative controls and mutant miR-196 did not affect luciferase activity (Figure 5B), further indicating the direct interaction between miR-196 and the 3′-UTR of Bach1 mRNA.
Figure 5. Mutant miR-196 mimic does not alter the luciferase activity of Bach1 3′-UTR reporter but inhibits the luciferase activity of mutant Bach1 3′-UTR reporter.
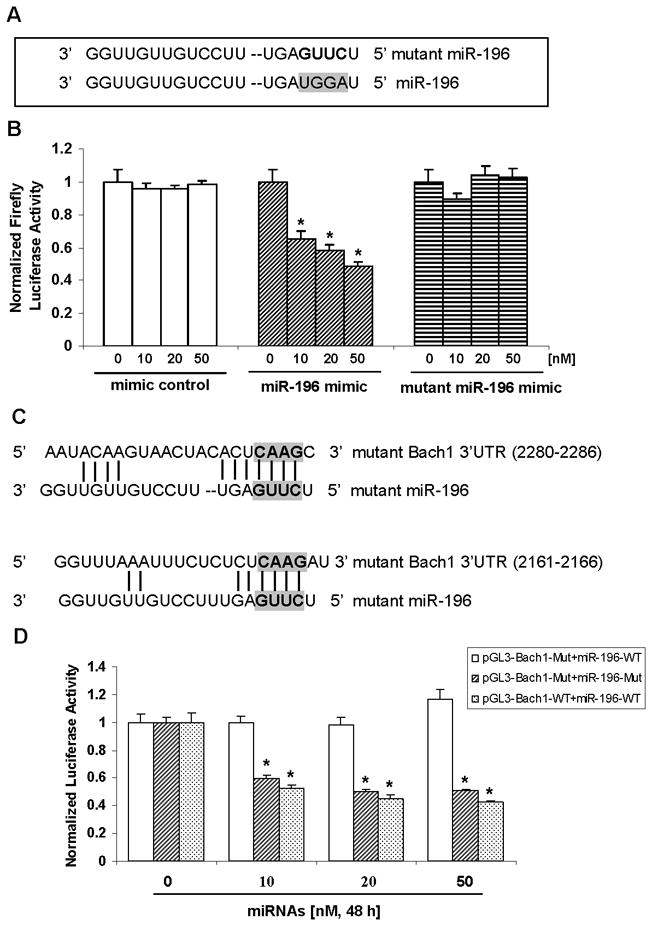
(A) Four nucleotides mutation were introduced to the seed match sites of miR-196. (B) Effect of mutant miR-196 on Bach1 3′-UTR reporter activity. 9-13 cells were co-transfected with 0.4 μg/mL of pGL3-Bach1, 0.4 μg/mL of pRL-TK and with 0, 10, 20 or 50 nM mimic negative control, miR-196 mimic or mutant miR-196 for 48 h, firefly and renilla luciferase activities were measured using Dual Luciferase Assay System from Promega. Firefly luciferase activities were normalized to renilla luciferase activities and total protein, as indicated in Materials and Methods. Values for cells without microRNA transfection were set equal to 1. (C) Restoration of seed match between mutant miR-196 and mutant Bach1 3′-UTR are shown. (D) Inhibition of mutant Bach1 3′-UTR reporter activity by mutant miR-196 mimic. 9-13 cells were co-transfected with 0.4 μg/mL of mutant reporter (pGL3-Bach1-Mut), 0.4 μg/mL of pRL-TK, and with 0, 10, 20 or 50 nM mutant miR-196 or wild type miR-196 mimic for 48 h, firefly and renilla luciferase activities were measured. Firefly luciferase activities were normalized to renilla luciferase activities and total protein. Values for cells without microRNA transfection were set equal to 1. Data are presented as means ± SE, n=3. * differs from mimic negative control, P<0.05.
We predicted that mutant miR-196 (miR-196-Mut), containing base complementarity with mutant pGL3-Bach1 (pGL3-Bach1-Mut), should restore its effect on mutant reporter (Bach1-3′-UTR-Mut) activity, because they again match perfectly in their seed regions (Figure 5C). As we predicted, mutant miR-196 mimic significantly inhibited luciferase activity in cells transfected with mutant pGL3-Bach1, which was mutated to “fit” mutant miR-196, whereas no significant effects of miR-196 wild type on mutant reporter (pGL3-Bach1-Mut) luciferase activity were observed, further establishing the direct interaction between miR-196 and the 3′-UTR of Bach1 mRNA (Figure 5D).
miR-196 represses HCV RNA and protein expression in HCV replicon cells
To determine whether miR-196 represses HCV mRNA and protein expression, HCV replicon cell lines Con1 and 9-13 cells were transfected with miR-196 mimic or microRNA mimic negative control. As shown in Figure 6A & B, 50 nM of miR-196 mimic resulted in a significant reduction of HCV replication by ~50% in Con1 cells and NS5A protein levels by ~52%, compared with the same concentration of miRNA mimic negative control. When Bach1 gene expression was silenced by Bach1-siRNA (Figure 6C), the levels of HCV replication were significantly decreased by ~40% (Figure 6D).
Figure 6. miR-196 mimic represses HCV expression in HCV replicon cells.
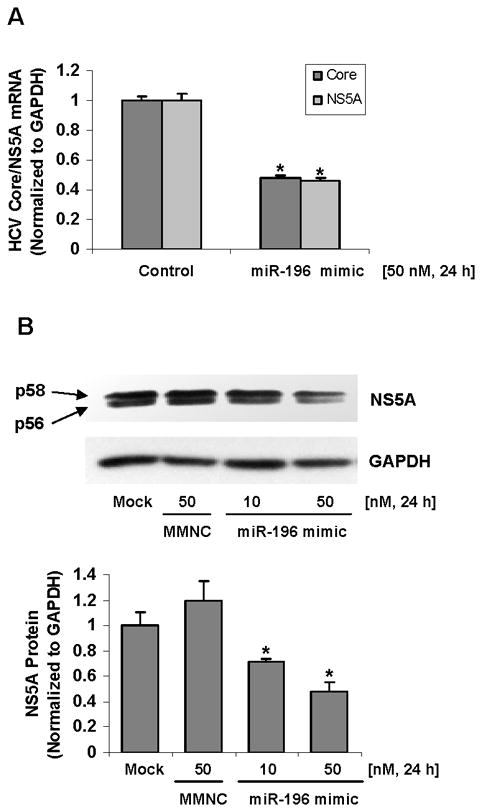
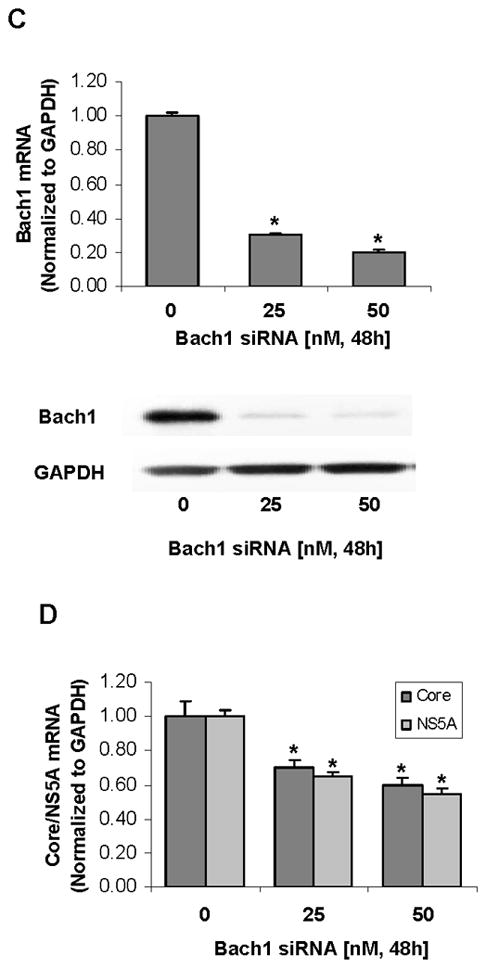
Cells were transfected for 24-48 h with 10–50 nM miR-196 mimic or Bach1-siRNA by Lipofectamine 2000, after which cells were harvested and total RNA and protein were extracted. The levels of HCV core and NS5A and host cell GAPDH mRNA and protein levels were measured by quantitative RT-PCR and Western blots as described in Materials and Methods. Data are presented as means ± SE, n=3. * differs from negative control only, P<0.05. (A) Down-regulation of HCV core and NS5A mRNA expression by miR-196 mimic in Con1 cells. (B) Down-regulation of HCV NS5A protein expression by miR-196 mimic in 9-13 cells. (C) Silencing Bach1 gene expression by Bach1-siRNA in Con1 cells. (D) Down-regulation of HCV core and NS5A mRNA expression by Bach1-siRNA in Con1 cells.
Comparative studies of miR-196 on HCV expression in Con1-WT- vs Con1-Mut-transfected Huh-7.5 cells
Analysis in silico predicts two 6-nt match sites within HCV Con1 genome to a region of miR-196 (nucleotides 2–7) (Figure 7A), which are not considered as perfect matches of a 7-nt match to the seed region of the miRNA (nucleotides 2–8), and different from the perfect seed match between miR-196 and HCV JFH1 genome (18). However there is a possibility that two 6-nt complementary matches could lead to an effect. Therefore, we investigated whether the inhibition of HCV expression by miR-196 is the result of an effect through Bach1 and HMOX1 or by directly targeting the HCV genome, or both. We introduced four nucleotide mutants to generate mutant pFK-Con1-Mut, in which two match sites for the HCV Con1 genome were abolished (Figure 7A). 50 nM of miR-196 transfection still resulted in a significant reduction of HCV core and NS5A expression in Huh-7.5 cells transfected with Con1-Mut RNA, as observed in Huh-7.5 cells transfected with Con1-WT RNA (Figure 7B & C), but the effect of miR-196 on HCV core and NS5A in Con1-Mut-transfected cells was slightly less than that in Con-WT-transfected cell (Figure 7D). These findings, together with data shown in Figure 6C & D, suggested that the inhibition of HCV expression by miR-196 is the result of an effect through Bach1 as well as targeting the HCV Con1 genome, and the indirect effect of miR-196 on HCV expression though Bach1 and HMOX1 is a major contribution to inhibition of HCV expression.
Figure 7. Effect of miR-196 on HCV expression in Huh-7.5 cells transfected with Con1-WT or Con1-Mut RNA.
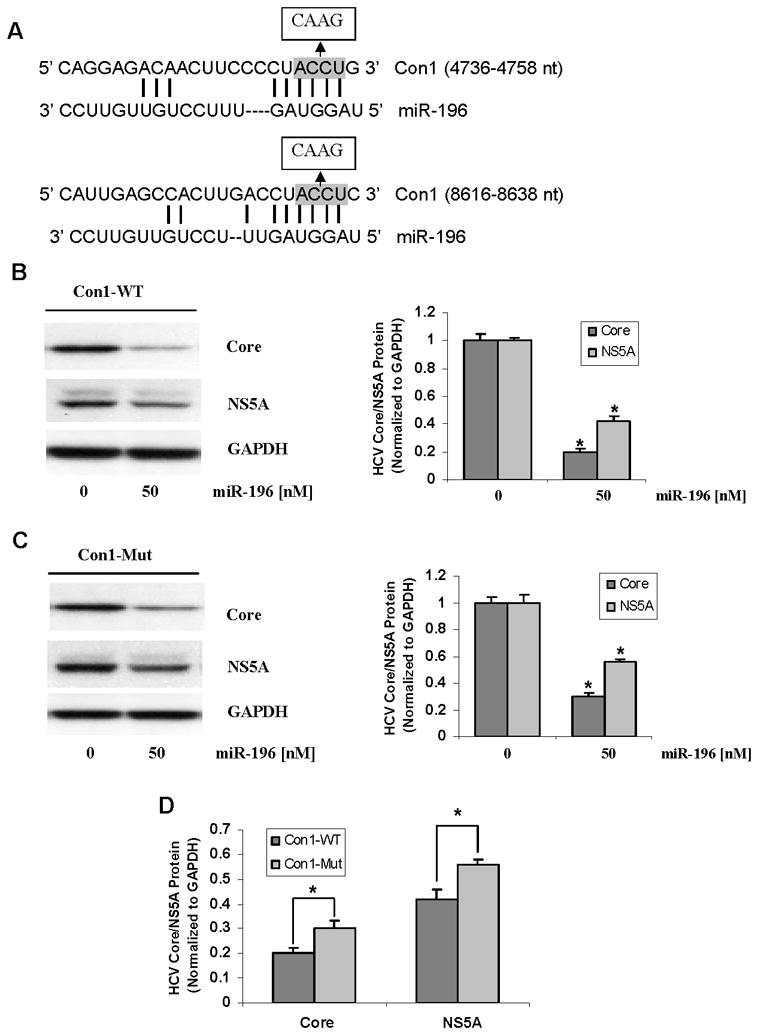
Huh-7.5 cells were transfected with 2 μg/well of HCV Con1-WT or Con-Mut RNA by Lipofectamine 2000. After 24 h, cells were transfected with 50 nM of miR-196 mimic, or miRNA mimic negative control (MMNC) as control. After 48 h, cells were harvested and total proteins were extracted. HCV core, NS5A and GAPDH protein levels were measured by Western blots. Data are presented as means ± SE, n=3. * differs from control P<0.05. (A) In silico prediction of putative binding sites for miR-196 in HCV genotype1 Con1 genome and the four nucleotides mutation introduced to the binding sites of miR-196. (B) Effects of miR-196 on HCV core and NS5A protein levels in Con1-WT-transfected Huh-7.5 cells. (C) Effects of miR-196 on HCV core and NS5A protein levels in Con1-Mut-transfected Huh-7.5 cells. (D) Comparison of the effects in Con1-WT- vs Con1-Mut- transfected Huh-7.5 cells.
miR-196 inhibits HCV expression in the HCV JFH1-based cell culture system
As reported recently, a perfect match for miR-196 was found in the coding region of the HCV NS5A gene in HCV JFH1 genome (18). As expected, we observed a similar down-regulatory effect of miR-196 on HCV expression in the HCV J6/JFH1 cell culture system. 50 nM of miR-196 led to a significant decrease of HCV J6/JFH1 RNA by nearly 70% in J6/JFH1 transfected Huh-7.5 cells (Figure 8A), ~50% in J6/JFH1 infected Huh-7.5 cells (Figure 8B) and ~60% reduction of HCV NS3 protein in J6/JFH1 infected Huh-7.5 cells (Figure 8C). These results were consistent with previous observations in the JFH1 cell culture system (18).
Figure 8. miR-196 mimic inhibits HCV expression in the HCV J6/JFH1-based cell culture system.
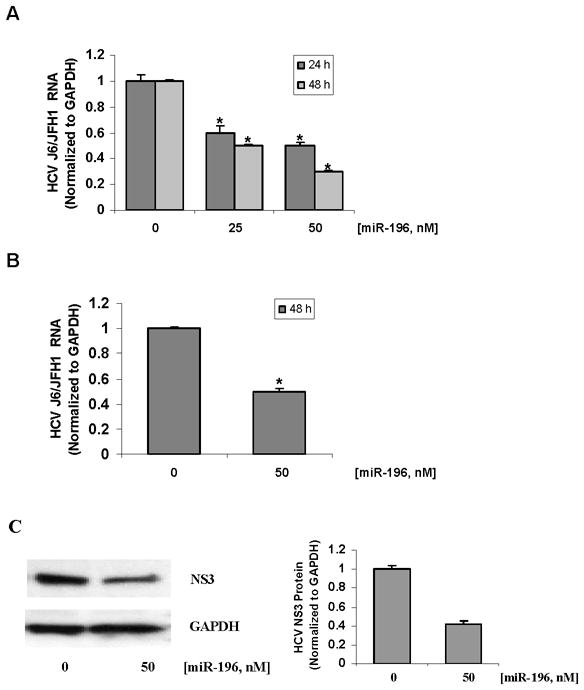
Huh-7.5 cells were transfected with 2 μg/well of HCV J6/JFH1 RNA by Lipofectamine 2000. After 48 h, cells were transfected with miR-196 mimic, or miRNA mimic negative control (MMNC). For HCV infection, naïve Huh-7.5 were cultured with 1 mL of cell culture supernatants harvested from J6/JFH1-transfected cells, as described in Materials and Methods. After 48 h of exposure to the supernatants, cells were transfected with miR-196 mimic, or miRNA mimic negative control (MMNC) for 48 h. Cells were harvested and total RNA and proteins were extracted. HCV RNA was quantified by qRT-PCR, and HCV NS3 and GAPDH protein levels were measured by Western blots. Data are presented as means ± SE, n=3. * differs from control P<0.05. (A) Down-regulation of HCV J6/JFH1 RNA levels by miR-196 mimic in Huh-7.5 cells transfected with J6/JFH1 RNA. (B) Down-regulation of HCV J6/JFH1 RNA and (C) protein levels by miR-196 mimic in Huh-7.5 cells infected with J6/JFH1 hepatitis C virus secreted into the culture supernatant.
DISCUSSION
The major novel findings of this work are that (1) miR-196 has functional binding sites in the 3′-UTR of Bach1 mRNA and (2) down-regulation of Bach1 by either miR-196 or Bach1-siRNA represses HCV expression. Functionality of miR-196 in regulation of Bach1 is demonstrated by several lines of evidence, including initial in silico analysis (Figure 1), by expected effects of miR-196 mimics (Figures 2), and by the expected effects of miR-196 mimics on luciferase reporter constructs containing the wild type and mutant Bach1 3′-UTR (Figures 3, 4 and 5, and Suppl. Figure 1). In addition, we demonstrate that down-regulation of Bach1 by either miR-196 or Bach1-siRNA leads to the up-regulation of the HMOX1 gene (Figure 2C & D) and to down-regulation of HCV expression in replicon cells (the genotype 1b Con1 strain) (Figure 6) and the HCV J6/JFH1-generated cell culture system (Figure 8). These results suggest that over-expression or up-regulation of miR-196 represents a potential new additional therapeutic strategy for chronic HCV infection, as well as other conditions in which up-regulation of HMOX1 may be desirable.
Based upon a large number of experimental and clinical studies carried out during the past several years, it is now generally accepted that HCV infection produces an increase in oxidative stress in infected hepatocytes. One important mediator of such increased oxidative stress is the HCV core protein (27, 28). In parallel with these observations are a series of observations in numerous systems, including experimental systems with expression of HCV, showing that HMOX1 helps to protect numerous cells and tissues against the potentially damaging effects of excess oxidative stress. These actions are based upon the ability of HMOX1 to decrease “free” or loosely-bound heme, which can act as a potent prooxidant, and to increase production of carbon monoxide, biliverdin, and bilirubin, which have potent antioxidant and anti-inflammatory and antifibrogenic effects (6–8, 29, 30). HMOX1 has also emerged as an important anti-apoptotic enzyme (31). Over-expression or induction of HMOX1 suppresses HCV replication and increases resistance of hepatocytes to oxidant injury (19, 20).
Regulation of expression of the HMOX1 gene is complex. However, we and others have shown that among the important sites for regulation are a series of expanded AP-1 sites, also called antioxidant responsive elements (31), Maf protein responsive elements [MARE], and metalloporphyrin-responsive elements [MPRE] in the 5′ untranslated region of HMOX genes, across many species (32–36). Bach1 plays a key role in tonic repression of expression of the HMOX1 gene. It does so by forming heterodimers with small Maf proteins and blocking transcriptional activation of the gene. Bach1 contains several consensus binding sites (all containing CP motifs), when they bind heme, lead to a change in conformation of the protein with marked reduction in affinity for Maf proteins and subsequent derepression and increase in activity of HMOX1 gene expression (9, 10, 12).
In view of the above, it is not surprising that HMOX1 activity might be increased in HCV infection, and, indeed, we and others have shown this to be the case (5). Nevertheless, in some other experimental systems and also in some clinical studies, a decrease in expression of HMOX1 has been observed in the setting of chronic hepatitis C (37, 38). These findings suggest the possibility that patients with genetic or other factors that lead to lower levels of HMOX1 gene expression may be at increased risk for development of chronic hepatitis C infection after acute HCV exposure and/or with greater risks of development of more rapidly progressive liver disease due to HCV infection. In this regard, there are at least two known genetic factors that influence levels of expression of HMOX1, namely, the length of GT repeats in the 5′-untranslated region and the presence of a single nucleotide polymorphism at position -413 (A/T, rs2071746) in the HMOX1 promoter region (39, 40).
The discovery of miRNAs in C. elegans ushered in a new and exciting area of study. The numbers of miRNAs continues to grow, and additional mRNAs and candidate genes regulated by them continue to be identified. With respect to the liver, miR-122 was identified as the most abundant miRNA expressed in hepatocytes (accounting for ~70% of total miRNA’s) and shown to have major effects on several enzymes of cholesterol metabolism (41, 42). Unexpectedly, miR-122 was also shown to be required for HCV expression (19, 43), at least in cell culture systems. More recent work has shown that the effects of miR-122 depend upon the context and location of its cognate seed sequence binding sites. The sites in the 5′ region are mostly associated with up-regulation of expression, whereas those in the 3′ untranslated region are mostly associated with repression of expression (44). Our current work adds miR-196 as a down regulator of HCV expression (Figure 6 & 8) and an attractive candidate as new therapeutic agents for chronic HCV infection.
Our study has limitations. Effects of miR-196 on, Bach1, HMOX1, and HCV thus far have been shown only in cell culture models, and the suppression of HCV expression has been moderate, not extremely high. The field of HCV research has been stymied by the lack of simple and robust animal models. Among non-human species, only chimpanzees have thus far been capable of being infected with HCV, and the disease in them is generally relatively mild. They are also extraordinarily difficult and expensive to maintain. Recently, murine models have been developed, based upon immune deficient animals into which human hepatocytes can be implanted without rejection, and these hepatocytes then infected with the hepatitis C virus (45, 46). Another recent model has been able to establish this in non-immunodeficient mice in which the host animal hepatocytes undergo necrosis and apoptosis and can be rescued with human hepatocytes (47). Thus, over-expression of a combination of miR-196 and other selected miRNAs in order to decrease the viral output further in cell cultures and murine models are currently under study in our laboratory.
In summary, we demonstrate functional miR-196 binding sites in the 3′-UTR of Bach1, which lead to down-regulation of Bach1 gene expression, up-regulation of HMOX1 gene expression, and down-regulation of HCV expression. These findings add to the growing panoply of miRNAs that influence expression of genes and proteins of the hepatitis C virus and of HMOX1, a key cytoprotective enzyme. They suggest potential new additional therapies for chronic HCV infection and, perhaps, for other diseases characterized by increased oxidative stress.
Supplementary Material
Acknowledgments
We thank Dr. Rolf Renne (University of Florida, Gainesville, FL) for the generous gift of luciferase reporter construct pGL3-Bach1 and Dr. Bryan R. Cullen (Duke University, Durham, NC) for providing pLSV40-Rluc, pLSV40-GL3 and pLSV40-GL3/Bach1 reporter vectors. We are grateful to Dr. Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for kindly supplying the HCV 9-13 cells and pFK-Con1 plasmid, and Dr. Charles M. Rice (the Rockefeller University, New York, NY) for providing the J6/JFH1 molecular clone.
Financial Support: This work was supported by National Institutes of Health (NIH) grant R01-DK38825.
Abbreviations
- BCA
Bicinchoninic acid
- bZip
Basic leucine zipper
- CHC
chronic hepatitis C
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HMOX
Heme oxygenase
- HCV
Hepatitis C virus
- IFN-α
Interferon-alpha
- IFN-β
Interferon-beta
- MARE
Maf recognition element
- miRNA
microRNA
- MMNC
miRNA mimic negative control
- MINC
miRNA inhibitor negative control
- Nrf2
Nuclear regulatory factor erythroid 2-related factor 2
- NS
Nonstructural protein (s) of HCV
- NS5A
Nonstructural protein 5a of HCV
- NQO1, NAD(P)H
quinone oxidoreductase
- UTR
untranslated region
- PAGE
polyacrylamide gel electrophoresis
- PVDF
Polyvinylidene fluoride
- qRT-PCR
Quantitative real time polymerase chain reaction
- SDS
Sodium dodecyl sulfate
Contributor Information
Weihong Hou, Email: weihong.hou@carolinashealthcare.org.
Qing Tian, Email: qing.tian@carolinashealthcare.org.
Jianyu Zheng, Email: jianyuzheng@hotmail.com.
Herbert L. Bonkovsky, Email: herbert.bonkovsky@carolinashealthcare.org.
References
- 1.Fried MW. Side effects of therapy of hepatitis C and their management. Hepatology. 2002;36:S237–244. doi: 10.1053/jhep.2002.36810. [DOI] [PubMed] [Google Scholar]
- 2.Russo MW, Fried MW. Side effects of therapy for chronic hepatitis C. Gastroenterology. 2003;124:1711–1719. doi: 10.1016/s0016-5085(03)00394-9. [DOI] [PubMed] [Google Scholar]
- 3.Choi J, Ou JH. Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. Am J Physiol Gastrointest Liver Physiol. 2006;290:G847–851. doi: 10.1152/ajpgi.00522.2005. [DOI] [PubMed] [Google Scholar]
- 4.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 5.Ghaziani T, Shan Y, Lambrecht RW, Donohue SE, Pietschmann T, Bartenschlager R, Bonkovsky HL. HCV proteins increase expression of heme oxygenase-1 (HO-1) and decrease expression of Bach1 in human hepatoma cells. J Hepatol. 2006;45:5–12. doi: 10.1016/j.jhep.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 6.Bonkovsky HL, Elbirt KK. Heme Oxygenase: Its Regulation and Role. In: Cutler RG, Rodriguez H, editors. Oxidative Stress and Aging. River Edge, NJ: World Scientific; 2002. pp. 699–706. [Google Scholar]
- 7.Elbirt KK, Bonkovsky HL. Heme oxygenase: recent advances in understanding its regulation and role. Proc Assoc Am Physicians. 1999;111:438–447. [PubMed] [Google Scholar]
- 8.Lambrecht RW, Fernandez M, Shan Y, Bonkovsky HL. Ascites and Renal Dysfunction in Liver Disease. 2. Oxford: Blackwell Science; 2005. Heme oxygenase and carbon monoxide in cirrhosis and portal hypertension. [Google Scholar]
- 9.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Y, Lambrecht RW, Ghaziani T, Donohue SE, Bonkovsky HL. Role of Bach-1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAS. J Biol Chem. 2004;279:51769–51774. doi: 10.1074/jbc.M409463200. [DOI] [PubMed] [Google Scholar]
- 11.Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, Furuyama K, Nakayama M, et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J Biol Chem. 2003;278:9125–9133. doi: 10.1074/jbc.M209939200. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. Embo J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 17.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Wilson AT, Mathahs MM, Wen F, Brown KE, Luxon BA, Schmidt WN. Heme oxygenase-1 suppresses hepatitis C virus replication and increases resistance of hepatocytes to oxidant injury. Hepatology. 2008;48:1430–1439. doi: 10.1002/hep.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 22.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. Faseb J. 2006;20:2651–2653. doi: 10.1096/fj.06-6346fje. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 26.Hou W, Shan Y, Zheng J, Lambrecht RW, Donohue SE, Bonkovsky HL. Zinc mesoporphyrin induces rapid and marked degradation of the transcription factor Bach1 and up-regulates HO-1. Biochim Biophys Acta. 2008;1779:195–203. doi: 10.1016/j.bbagrm.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 28.Irshad M, Dhar I. Hepatitis C virus core protein: an update on its molecular biology, cellular functions and clinical implications. Med Princ Pract. 2006;15:405–416. doi: 10.1159/000095485. [DOI] [PubMed] [Google Scholar]
- 29.Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 30.Hill-Kapturczak N, Chang SH, Agarwal A. Heme oxygenase and the kidney. DNA Cell Biol. 2002;21:307–321. doi: 10.1089/104454902753759726. [DOI] [PubMed] [Google Scholar]
- 31.Brouard S, Otterbein LE, Anrather J, Tobiasch E, Bach FH, Choi AM, Soares MP. Carbon monoxide generated by heme oxygenase 1 suppresses endothelial cell apoptosis. J Exp Med. 2000;192:1015–1026. doi: 10.1084/jem.192.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbirt KK, Whitmarsh AJ, Davis RJ, Bonkovsky HL. Mechanism of sodium arsenite-mediated induction of heme oxygenase-1 in hepatoma cells. Role of mitogen-activated protein kinases. J Biol Chem. 1998;273:8922–8931. doi: 10.1074/jbc.273.15.8922. [DOI] [PubMed] [Google Scholar]
- 33.Shan Y, Lambrecht RW, Bonkovsky HL. Identification of key elements that are responsible for heme-mediated induction of the avian heme oxygenase-1 gene. Biochim Biophys Acta. 2004;1679:87–94. doi: 10.1016/j.bbaexp.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Lu TH, Shan Y, Pepe J, Lambrecht RW, Bonkovsky HL. Upstream regulatory elements in chick heme oxygenase-1 promoter: a study in primary cultures of chick embryo liver cells. Mol Cell Biochem. 2000;209:17–27. doi: 10.1023/a:1007025505842. [DOI] [PubMed] [Google Scholar]
- 35.Shan Y, Pepe J, Lu TH, Elbirt KK, Lambrecht RW, Bonkovsky HL. Induction of the heme oxygenase-1 gene by metalloporphyrins. Arch Biochem Biophys. 2000;380:219–227. doi: 10.1006/abbi.2000.1921. [DOI] [PubMed] [Google Scholar]
- 36.Shan Y, Pepe J, Lambrecht RW, Bonkovsky HL. Mapping of the chick heme oxygenase-1 proximal promoter for responsiveness to metalloporphyrins. Arch Biochem Biophys. 2002;399:159–166. doi: 10.1006/abbi.2001.2742. [DOI] [PubMed] [Google Scholar]
- 37.Wen F, Brown KE, Britigan BE, Schmidt WN. Hepatitis C core protein inhibits induction of heme oxygenase-1 and sensitizes hepatocytes to cytotoxicity. Cell Biol Toxicol. 2008;24:175–188. doi: 10.1007/s10565-007-9027-9. [DOI] [PubMed] [Google Scholar]
- 38.Abdalla MY, Britigan BE, Wen F, Icardi M, McCormick ML, LaBrecque DR, Voigt M, et al. Down-regulation of heme oxygenase-1 by hepatitis C virus infection in vivo and by the in vitro expression of hepatitis C core protein. J Infect Dis. 2004;190:1109–1118. doi: 10.1086/423488. [DOI] [PubMed] [Google Scholar]
- 39.Yamada N, Yamaya M, Okinaga S, Nakayama K, Sekizawa K, Shibahara S, Sasaki H. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am J Hum Genet. 2000;66:187–195. doi: 10.1086/302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Exner M, Minar E, Wagner O, Schillinger M. The role of heme oxygenase-1 promoter polymorphisms in human disease. Free Radic Biol Med. 2004;37:1097–1104. doi: 10.1016/j.freeradbiomed.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 44.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kneteman NM, Weiner AJ, O’Connell J, Collett M, Gao T, Aukerman L, Kovelsky R, et al. Anti-HCV therapies in chimeric scid-Alb/uPA mice parallel outcomes in human clinical application. Hepatology. 2006;43:1346–1353. doi: 10.1002/hep.21209. [DOI] [PubMed] [Google Scholar]
- 46.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 47.Vanwolleghem T, Meuleman P, Libbrecht L, Roskams T, De Vos R, Leroux-Roels G. Ultra-rapid cardiotoxicity of the hepatitis C virus protease inhibitor BILN 2061 in the urokinase-type plasminogen activator mouse. Gastroenterology. 2007;133:1144–1155. doi: 10.1053/j.gastro.2007.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


