Abstract
Synthetic peptides offer an attractive option for development of a V3-directed vaccine. However, immunization with flexible linear peptides may result in an immune response to multiple conformations, many of which differ from the native conformation of the corresponding region in the protein. Here we show that optimization of the location of a disulfide bond in peptides constrained to mimic the β-hairpin conformation of the V3, yields an immunogen that elicits a 30-fold stronger HIV-1 neutralizing response in rabbits compared with the homologous linear V3 peptide. The HIV-1 neutralizing response elicited by the optimally constrained peptide is also significantly stronger than that elicited by a gp120 construct in which the V3 is exposed. Neutralization of an HIV-1 strain that shares only 72% identity with the immunizing peptide was demonstrated. The most effective immunogen was also able to neutralize primary isolates that are more resistant to neutralization such as SS1196 and 6535.
Keywords: HIV-1, Vaccine, neutralizing antibodies, V3 loop, constrained peptides, gp120
Introduction
Induction of broadly neutralizing antibodies (bnAb) against diverse HIV-1 strains is critical for the development of a successful AIDS vaccine. Unfortunately, attainment of such a broadly neutralizing response by active immunization has proven to be an extremely difficult task because of the high mutation rate of the virus and the occlusion of target epitopes prior to the binding of the virus to its target cells. The recent success of the human trial carried out in Thailand indicates that despites these obstacles some degree of a protective immune response can be achieved by active immunization in humans(Rerks-Ngarm et al., 2009).
The V3 region is a major target for neutralizing antibodies. A recent immunization study with bivalent trimeric-gp120 demonstrated that most of the neutralizing activity elicited in rabbits was against V3 and V4 (Burke et al., 2009). High levels of V3 directed antibodies were also present in HIV-1 infected patients and in healthy individuals following gp120 based vaccination (Davis et al., 2009a; Davis et al., 2009b). These antibodies were broadly reactive, i.e. were able to neutralize a wide variety of chimeric HIV viruses with different V3 sequences presented on a HIV-2 scaffold. This is in agreement with previous data showing that when the different V3 sequences of neutralization-resistant isolates were presented in the context of the neutralization sensitive strain SF-162, these chimeric viruses became sensitive to V3 antibodies(Davis et al., 2009a; Zolla-Pazner et al., 2008)
Among HIV-1 neutralizing antibodies, 447-52D derived from an HIV-1 infected donor is one of the most potent and most broadly neutralizing human monoclonal antibodies, and in a study of HIV-1 broadly neutralizing antibodies 447-52D neutralized 45% of the clade-B isolates tested and was capable of neutralizing both ×4 and R5 viruses as well as some primary isolates (Binley et al., 2004).To gain insight into the conformation of the V3 region presented to the immune system by HIV-1, the structures of V3 peptides bound to HIV-1 neutralizing antibodies were determined by X-ray crystallography (Bell et al., 2008; Dhillon et al., 2008; Stanfield et al., 2003; Stanfield et al., 2004; Stanfield et al., 2006) and by NMR (Rosen et al., 2005; Sharon et al., 2003; Tugarinov et al., 1999; Tugarinov et al., 2000). All bound V3 structures revealed a β-hairpin conformation with an RMSD between the hairpin regions of any two V3 structures in the different complexes ranging between 1.2 and 2.5 Å. In some cases differences in the pairing of residues and in the hydrogen bond network were observed in different antibody-bound V3-peptides. Our working hypothesis is that the hairpin structure of the V3 region represents the structure of this domain in the viral envelope proteins and that antibodies targeting elements of this hairpin will be effective anti-viral agents.
The critical role for V3 in HIV-1 co-receptor binding restricts its sequence variation (Rosen et al., 2005; Rosen et al., 2006; Sharon et al., 2003; Stanfield et al., 2004; Stanfield et al., 2006). Only those sequences that are recognized by the cognate co-receptors, CCR5 or CXCR4 will result in an infective virus. Therefore, the conserved structural and sequence motifs in the V3 loop could be used, in principle, to induce a broadly neutralizing immune response. Indeed, this is consistent with recent data suggesting that a V3-directed antibody response both in infected patients and following vaccination can be more broadly reactive than initially assumed(Davis et al., 2009b). However, in most primary isolates the V3 is concealed and as a result these strains are resistant to V3-directed antibodies.
Linear V3 peptides can in principle be used as a component of an anti-HIV-1 vaccine (Haynes et al., 2006). However, such peptides, and even peptides representing the entire 35-residues V3 region with its native disulfide bond at its base, are mostly flexible in solution and except for a transient β-turn formed by the GPGR segment do not have a well-defined secondary structure (Chandrasekhar, Profy, and Dyson, 1991; Mor et al., 2009; Zvi, Hiller, and Anglister, 1992). As a result of this flexibility, unconstrained V3 peptides used as immunogens likely induce a broad distribution of antibodies, many of which will not recognize the native conformation of the corresponding region in gp120. Nevertheless, in a very comprehensive study Haynes and coworkers demonstrated neutralization of a spectrum of neutralization sensitive HIV-1 isolates by immune sera obtained after immunization with a panel of 29 linear V3 peptides in the form of C4-V3 peptides (Haynes et al., 2006).
Many attempts have been made to constrain the V3 structure using backbone cyclization and disulfide bond constraints. These focused mostly on short segments centered at the GPGR sequence (Cabezas et al., 2000; Tolman et al., 1993) or used sequences from HIVIIIB or HIVMN laboratory adapted ×4 strains containing 8 to 23 residues (Conley et al., 1994). Significantly, the anti-sera were primarily selective for neutralization of the strain harboring the immunogenic sequence and exhibited weak to moderate HIV-1 neutralization. Anti-sera obtained after immunization with a 23-residue disulfide-cyclized V3MN peptide neutralized also AL-1, SF-2 and HIVIIIB at higher concentrations but not four other strains that were tested. Shorter disulfide constrained peptides were less effective immunogens. Later attempts to use V3 peptides constrained with a hydrazone link for immunization (Cabezas et al., 2000) yielded considerably poorer neutralization results in comparison with the experiments of Conley and co-workers (Conley et al., 1994).
A number of laboratories have created V3-immunogens by inserting short V3 segment, centered on the GPGR sequence, into loops of proteins (Bryder et al., 1999; FitzGerald et al., 1998; Fontenot et al., 1995). In some cases moderate levels of V3 directed antibodies were generated and weak cross-clade neutralization was observed (Chakraborty et al., 2006). Stabilization of a β-hairpin conformation in the chimeric protein by introducing a disulfide bond connecting residues 307 and 318 resulted in a decrease in V3-specific antibody levels and loss of neutralization (Chakraborty et al., 2006). A more successful recent study used the entire V3, with the native disulfide bond at its base, fused to a carrier protein, to immunize rabbits (Zolla-Pazner et al., 2008). Cross-clade HIV-1 neutralization has been demonstrated using a clade-C gp120 DNA prime with a boost of a fusion protein containing the entire 35 residue clade-B cyclic V3 (Zolla-Pazner et al., 2009). Thus although mixed results have been reported, it is clear that properly designed constrained V3 antigens can lead to a potent HIV-1 neutralizing antibody response.
In previous studies we used disulfide bonds to constrain the V3 to the conformation recognized by the 447-52D antibody (Mor et al., 2009). We showed that some of the constrained peptides exhibited higher affinity to 447-52D Fab in comparison with linear V3 peptides (Mester et al., 2009). In the present communication, we report on a series of immunogens designed using the previous NMR and biochemical data. The constrained V3 peptides were linked to an eighteen-residue segment of the gp120 C4 region, which is known to be a helper T-cell epitope (Haynes et al., 2006; Sha et al., 2004; Zinckgraf, Winchell, and Silbart, 1999) and is required in order to elicit a robust antibody response. The C4-V3 peptides were synthesized by solid phase approaches, cyclized and used to immunize rabbits. Our results clearly show that a constrained V3 peptide can elicit a more potent HIV-1 neutralizing response in comparison with a linear peptide and gp120 immunogens. However, the location of the disulfide bond must be chosen carefully.
Material and methods
Chain assembly and purification of the linear (SH) containing C4-V3 peptides
The C4-V3 peptides were synthesized on a 443A peptide synthesizer (Applied Biosystems) using 0.1 mM fast Fmoc chemistry. Approximately 160 mg (0.1 mM) of preloaded Fmoc-Ile Wang resin (0.6 mmol/g) or Fmoc-Cys(Trt) Wang resin (0.5 mmol/g) were used. The first 10 amino acids were attached using single coupling, the remaining amino acids were assembled into the peptide chain using double coupling. The unreacted amino group was capped with acetic anhydride at the end of each coupling step. The resin weight gain was about 90% of theoretical and peaks with expected m/z values were observed in the mass spectrum of the crude peptide. The peptides were cleaved from the resin using a water/TFA cocktail with appropriate scavengers. In a representative procedure, to a vial containing 220 mg of the resin was added a solution of water (0.50 mL), phenol (500mg), 1,2-ethanedithiol (0.25 mL), thioanisole (0.50mL), triisopropylsilane (0.10mL) and trifluoroacetic acid (TFA; 10 mL) and the resulting mixture was stirred at 0 °C for 1 hour and at room temperature for 2 hours. Resin was separated from solution by filtration and washed two times with TFA. The combined filtrate was concentrated on a rotary evaporator under vacuum at a temperature below 30 °C and the resulting residue was treated with cold diethyl ether (40 mL). Precipitated peptide was isolated by centrifugation and ether was removed by decantation. After washing the solid precipitate was dried, dissolved in 2 mL acetonitrile (0.1%TFA), 4 mL of water (0.1%TFA) was added, the solution was frozen in dry ice and lyophilized for 48 hours to give final crude peptide in about 70% yield by weight.
For the synthesis of biotinylated homologs used in binding studies (Table 1) the resin was split into two parts (1:3) after completion of the assembly of the V3 epitope. A spacer sequence, mainly Gly-Ser-Gly, was built at the N-terminus of 25% of the peptide-resin followed by incorporation of biotin at the N-terminus. The couplings were carried out on the peptide synthesizer using the normal HOBT/HBTU activation method.
Table 1.
The amino acid sequence of peptides used for immunization and for binding measurements a.
| Immunogenic peptides | |
| C4-V3L | KQIINMWQEVGKAMYA-RPNNNTRKSIHIGPGRAFYTTGEI |
| C4-V3 K305C-T320C | KQIINMWQEVGKAMYA-RPNNNTRCSIHIGPGRAFYTCGEI |
| C4-V3 K305C-G321C | KQIINMWQEVGKAMYA-RPNNNTRCSIHIGPGRAFYTTCEI |
| C4-V3 R304C-G321C | KQIINMWQEVGKAMYA-RPNNNTCKSIHIGPGRAFYTTCEI |
| C4-V3 T303C- E322C | KQIINMWQEVGKAMYA-RPNNNCRKSIHIGPGRAFYTTGCG |
| C4-V3 T303C- I323C | KQIINMWQEVGKAMYA-RPNNNCRKSIHIGPGRAFYTTGEC |
| C4-V3 N302C-I323C | KQIINMWQEVGKAMYA-RPNNCTRKSIHIGPGRAFYTTGECG |
| C4-V3 N301C-G325C | KQIINMWQEVGKAMYA-RPNCNTRKSIHIGPGRAFYTTGEIIC |
| Peptides used for ELISA | |
| V3L | Biotin- GSGTRKSIHIGPGRAFYTTGEI |
| V3 K305C-T320C | Biotin- GSGTRCSIHIGPGRAFYTCGEI |
| V3 K305C-G321C | Biotin- GSGTRCSIHIGPGRAFYTTCEI |
| V3 R304C-G321C | Biotin- GSGCKSIHIGPGRAFYTTC |
| V3 T303C-E322C | Biotin- SGSCRKSIHIGPGRAFYTTGC |
| V3 T303C-I323C | Biotin- GSGCRKSIHIGPGRAFYTTGEC |
| V3N302C-I323C | Biotin- GSGCTRKSIHIGPGRAFYTTGECG |
| V3 N301C-G325C | Biotin- GSGCNTRKSIHIGPGRAFYTTGEIIC |
| C4 | Biotin- GSGKQIINMWQEVGKAMYA |
Peptide are referred to by the residues that were replaced by cysteines; the postulated conformation of the V3 loop that was mimicked is stated in brackets; residues replaced by cysteines are underlined.
Preparative HPLC purification of linear peptide was carried out on a Waters DeltaPak column 19 × 300 mm using a 20-50% acetonitrile/water gradient in 80 min with a flow rate of 5 ml/min and detection at 220 nm and 280 nm. Both solvent reservoirs contained 0.1% TFA. Usually about 15 mg of crude peptide were dissolved in 1 ml of acetonitrile/water (1:4) and injected onto the column. Analytical HPLC of the linear peptides was carried out on a Zorbax Eclipse XDB-C8 column 4.6 × 150 mm using detection at 220 nm detection and 10-60% acetonitrile/water gradient over 20 min with a flow-rate of 1 ml/min. The linear peptides used for cyclization were >90% homogeneous.
Disulfide bond formation
Peptides were cyclized using either ferricyanide mediated oxidation, DMSO mediated oxidation or glutathione mediated oxidation depending on the sequence and length of the peptide. In general the reaction progress was monitored by electron spray ionization mass spectrometry. In most of the cases the linear C4-V3 peptides were cyclized using oxidized glutathione (GSSG) as the oxidant. In a typical procedure, a solution of linear C4-V3 peptide (2.7 mg) dissolved in 3 mL of water containing 0.1% of TFA was added dropwise to a solution of GSSG (14 mg) in 80 mL of ammonium acetate (0.1 M, pH7.9). The combined solution was stirred overnight at room temperature. HPLC analysis showed almost no change in retention time and the progress of the reaction was monitored by MS analysis. After completion of the reaction, the solution was acidified to pH 2.0 by dropwise addition of 0.5 mL of TFA, filtered and the filtrate was loaded onto a preparative Waters C18 Delta Pak column. Product was eluted using a 10-50% acetonitrile/water gradient over 80 min. Both solvents contained 0.1% TFA. The pure fractions were combined and lyophilized to result in about 1.3 mg of pure cyclic peptide. The cyclic C4-V3 peptide was >95% homogeneous as analyzed by HPLC and the MS difference between linear and cyclic was 2 Da as expected.
Immunization of rabbits with V3 peptides
Twelve week old female New Zealand white rabbits were purchased from the animal breeding center at the Weizmann Institute of Science (Rehovot, Israel). Animals were treated according to the guidelines and under the supervision of the Animal Care and Use Committee. All the work was done under the supervision of the veterinary resources department. Animals were immunized up to five times at weeks 1, 4 8, 13 and 37 with 250 μg of HPLC purified peptide in phosphate buffered saline (PBS) or 250 μg gp120 in 50 mM Tris-HCl, 300 mM NaCl administrated intramuscularly. Peptide was mixed at 1:1 volume:volume ratio with Complete Freund's Adjuvant (CFA; 1 mL) at the first injection and Incomplete Freund's Adjuvant (IFA) in the second injection; further boosts were administered with no adjuvant. Animals were bled 10 days after each boost starting from the 3rd immunization.
Expression and purification of gp120
The vector pSyn gp120 that encodes gp120JR-FL was kindly provided by the NIH AIDS reagent program (http://www.aidsreagent.org). From this vector, a gene encoding residues 88-492 of gp120JR-FL i.e. 88-492gp120 was constructed and inserted into the pIRES vector which was developed for high level expression in HEK293 (Clontech, Mountain-View, CA). This vector encodes the IgK secretion signal, enabling secretion of the gp120 protein to the growth medium and a 6× histidine tag followed by a Tobacco Etch Virus (TEV) protease recognition site at the N-terminus of gp120. Additionally the segments coding for the V1 and V2 variable loops were deleted and replaced by a segment coding for Gly-Ala-Gly (GAG) and two glycosylation sites were modified (N301Q and T388A). These modifications have been reported to increase susceptibility to neutralization by CD4-binding site antibodies (Koch et al., 2003). This gp120 construct,88-492gp120ΔV1/V2, N301Q, T388A, (referred to as gp120) was stably transfected into a mutated HEK293 cell line lacking the gene for N-acetylglucosaminyltransferase I (Reeves et al., 2002). The expressed proteins are homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans. The protein was purified initially on a 50 ml Cibacron Blue Sepharose column (GE Healthcare), followed by a 5 ml HisTrap HP column purification (GE Healthcare). The eluted fraction was cleaved by TEV protease, followed by an additional purification on a Ni column to remove the TEV and uncleaved gp120. Finally the protein was purified on a superdex 200 16/60(GE Healthcare). The homogenously glycosylated 45 kD protein was identified by SDS Polyacrylamide Gel Electrophoresis and superdex 200 10/300 analytical gel filtration.
Determining peptide binding titers by ELISA
To test the binding of the resulting antibodies to the immunizing peptide, Reacti-Bind™ Streptavidin High Binding Capacity Coated Plates clear, 96-wells were used (PIERCE Cat No 15500). All procedures were done at room temperature. Plates were washed three times with PBS, 0.1% BSA, and 0.05% Tween-20 (wash buffer). 100 μl of the V3 or C4 biotinylated peptide (Table 1) at 1 μg/ml in wash buffer were added to each well and incubated for 2 hours with shaking. After rinsing the ELISA wells, serial dilutions of the serum in wash buffer were added to each well and incubated for 1.5 h. This was followed by several washes and 45 minutes incubation with 1:2500 dilutions of secondary antibodies in wash buffer (HRP-conjugated donkey anti-rabbit-Jackson 711-035-152). The plates were washed and HRP substrate (TMB/E by Chemicon International) was added. The reaction was stopped by adding 100 μl of 0.1% sodium fluoride and OD was read at 650 nm in a VersaMax microplate reader.
Alternatively Ni-column purified His-tagged gp120 (see above) was coated onto HisGrab™ Nickel Coated, High Binding Capacity Plates clear, 96-well (PIERCE Cat No 15142) for 2 hours with shaking with 100 μl of 88-492gp120ΔV1/V2 at 10μg/ml in PBS. Subsequent steps are as described above for peptide ELISA.
In order to determine half-max binding values, OD values were plotted against the serum dilution and fitted using Origin software to a one-site binding model. Data is presented as the reciprocal of the serum dilution at half maximum binding, values were rounded to two significant figures
When testing for binding to reduced V3 peptides, the peptides were incubated overnight in wash buffer supplemented with 10 mM DTT, followed by incubation on the plate with 10 mM DTT. Serum dilutions and washes were done in wash buffer supplemented with 2 mM DTT. Linear peptide was treated the same as the control.
Neutralization assay
Pseudoviruses single round of infection-based neutralization assay was carried out by Monogram Biosciences, Inc. South San Francisco as previously described (Richman et al., 2003). Virus particles containing virus envelope proteins were produced by co-transfecting HEK293 cells with a plasmid expressing HIV-1 primary isolates Env plus an HIV genomic vector that contains the luciferase indicator gene. Murine leukemia virus (MLV) Env plasmid was used as a negative control to assess non-specific neutralization. Recombinant pseudotyped viruses were harvested 48 h post-transfection and incubated for 1 h at 37 °C with serial two-fold dilutions of heat-inactivated rabbit sera starting at 1:10. The virus/serum dilutions were incubated with U87 CD4+, CCR5+ and CXCR4+ cells. Virus infectivity was determined 72 h post-inoculation by measuring the amount of luciferase activity expressed in infected cells. Neutralizing activity is displayed as the percent inhibition of viral replication (luciferase activity) at each antibody dilution compared with no antibody sample, % inhibition = {1 - [luciferase+Ab/luciferase-Ab]} *100. Titers were calculated as the reciprocal of the serum dilution conferring 50% inhibition.
Statistical analysis
In order to assess the difference in binding to the immunizing V3 peptide and gp120 a one sample T-test for a hypothetical mean value of 1 was used. Unpaired T-test to compare two means was used to evaluate differences in binding to cyclic vs. reduced V3 peptide. Analysis was performed using the GraphPad QuickCalc Internet tools (http://www.graphpad.com/quickcalcs/). P-value<0.05 was considered significant.
Results
Design and synthesis of disulfide-constrained V3-peptide immunogens
The constrained peptides used as immunogens in the present study were based on the consensus sequence for clade-B R5 viruses and included the entire epitope recognized by the 447-52D antibody (K305-T320) (Table 1) with seven additional residues at the N-terminus (298RPNNNTR304) and 2 or 4 residues at the C-terminus (322EI323 or 322EIIC325, respectively). The elongation of the sequence added residues T303 and R304 that were found to interact extensively with 2219 (Stanfield et al., 2006). R304 and K305 have been found to be critical for the interactions of several HIV-1 neutralizing antibodies with V3(Pantophlet et al., 2008) and therefore it was important to include them in our immunogen design. To constrain a peptide to the 447-52D bound conformations of V3, a disulfide bond could be inserted at positions 301, 303, 305 or 307 of the N-terminal strand of the V3 loop (Mester et al., 2009; Mor et al., 2009). Residue I307 is part of the V3 epitope recognized by 447-52D, and interacts extensively with this antibody (Sharon et al., 2003). Therefore, its replacement could be detrimental to obtaining antibodies cross-reactive with native gp120. Moreover, a disulfide bond involving residue 307 (and either 318 or 319 in the C-terminus) would not constrain N-terminal residues found upstream of 307 (i.e. 305 and 306), and C-terminal residues found downstream of 318/319 (i.e. 319 and 320) to a β-hairpin conformation (Mor et al., 2009). We therefore decided to test peptides constrained using disulfide bonds involving residues 301, 303 or 305. The peptides constrained at positions 303 and 305 bound more strongly to 447-52D in comparison with the linear V3 peptides with V3K305C-T320C exhibiting the highest affinity to 447-52D (Mester et al., 2009). The peptides were designed to mimic the conformation of either V3MN or the V3consensus bound to 447-52D which differ in the pairing of the residues in the V3 β-hairpin (Mester et al., 2009; Mor et al., 2009; Rosen et al., 2006; Sharon et al., 2003). Furthermore two additional V3 peptides were designed to mimic the conformation of V3IIIB bound to the 0.5β antibody. This antibody is a strain specific antibody and V3 peptides constrained to the 0.5β bound conformation exhibited a significant reduction in affinity to 447-52D antibody (Mester et al., 2009).
The 0.5β-bound V3 conformation is different from that of the 447-52D bound conformation in the hydrogen bond forming residues in the N-terminal strand of V3. In order to constrain a peptide to the 0.5β bound conformation a disulfide bond could be inserted at positions 302, 304 or 306 of the V3 loop (Mester et al., 2009; Mor et al., 2009). In this study we included two V3 peptides constrained to the above conformation using a single disulfide bond involving replacement of either N302 or R304 by cysteine (i.e C4-V3N302C-I323C and C4-V3R304C-G321C respectively) Table 1. The immunogens used in this investigation contained 40 or 42 residues and regions with a significant tendency to assume β-sheet structures. β-Sheet formation during solid phase peptide synthesis often buries the chain ends and prevents chain elongation. Although all peptides were synthesized using automated solid phase peptide synthesizer with double coupling and capping after each step, the final crude product had significant heterogeneity (Fig. 1A). Moreover, in several syntheses we originally failed due to the early termination of the chain assembly. Truncation sequences formed in these syntheses were identified by mass spectrometry and the difficult couplings were circumvented by changing the coupling conditions or lowering the substitution on the resin to eliminate intermolecular interactions of growing peptide chains. Despite the synthetic challenges, using ESI MS we were able to identify the linear disulfhydryl precursor product and using preparative reversed phase HPLC we purified this to >90% homogeneity (Fig. 1B). The linear sulfhydryl containing C4-V3 peptides or their biotinylated homologs were oxidized to form the constrained immunogens using a variety of procedures (See experimental procedures). No change in retention was discerned on reversed phase HPLC and the cyclization was monitored by mass spectrometry. In all cases, the final cyclic peptide was highly homogeneous (Fig. 1C and 1 E. Notice a 2 Da difference in mass between 1D and 1E). The degree of cyclization was also ascertained by slowly scanning the M/Z peaks and monitoring the isotope distribution (data not shown). The MS results allowed us to distinguish small amounts of linear peptide in the cyclic product. All peptides used in binding or neutralization studies were >95% cyclic.
Fig. 1.

Chromatographic and Mass-Spectra (MS) evaluation of synthetic constrained immunogens. Analytical HPLC profile of crude linear (A), purified linear (B) and purified cyclic (C) C4-V3 N301C-G325C. ESI-MS spectrum of purified linear (D) and purified cyclic (E) C4-V3 N301C-G325C. The HPLC was run using a 10-60% acetonitrile/water gradient (containing 0.1%TFA) over 20 min; Column: Zorbax-Eclipse XDB- C8, 150×4.6 mm; Flow-rate: 1.0 mL/min; Detection at 220nm; Product Rt 11.799 min; The average molecular weight of the linear was 4798.1 while the average molecular weight of the cyclic was 4796.0.
Reactivity of the C4-V3 immune sera with V3 and C4 peptides
A list of the constrained peptides used for immunization is presented in Table 1. For comparison we tested also a gp120 and a linear C4-V3 immunogen. Initially rabbits immunized with C4-V3L, C4-V3T303C-I323C and C4-V3K305C-T320C were subjected to a 4th and a 5th inoculation at weeks 13 and 37, respectively. The post-4 sera displayed a small reduction in binding titer to the corresponding V3 immunogen compared to the post-3 sera. A test bleed done at week 24 in order to follow the V3 titer also indicated some reduction in antibody levels. The animals were rested and injected for the fifth time at week 37. Serum drawn after the 5th inoculation had lower levels of V3 antibodies than post-3 sera (data not shown). Therefore, sera drawn after the 3rd inoculation were considered optimal, and tested for gp120 binding and neutralization of HIV-1 clade B viral strains, and three injections were adopted as the standard protocol for all further immunizations. High levels of V3 specific antibodies were observed following three injections for all C4-V3 immune sera, with geometric-mean-titer (GMT) values for half-maximal binding to the homologous V3 peptides ranging from 3100 to 20000 (Table 2 and Fig. 2) with the exception of C4-V3R304C-G321C, mimicking the 0.5β bond conformation, which elicited unusually low titers of anti-V3 antibodies (half-max GMT=1000). The highest titers were obtained for C4-V3N301C-G325C and C4-V3T303C-E322C. To verify that most of the antibody response was directed against the V3 epitope, the reactivity of several of the sera was tested for V3 and C4 separately. Significantly lower C4 binding titers were observed compared with V3 titers ranging from 150-1900. This reflects in most cases a 4-30 fold reduction in the C4 titer in comparison with the V3-titer with the exception of C4-V3R304C-G321C which elicited low V3 titers (Table 2).
Table 2.
summary of binding data obtained by ELISA a.
| Rabbit | Immunogen | V3 | gp120 | C4 |
|---|---|---|---|---|
| B712 | C4-V3L | 1800 | 660 | 420 |
| B707 | C4-V3L | 1800 | 840 | 150 |
| B702 | C4-V3L | 12500 | 7700 | ND |
| B722 | C4-V3L | 4200 | 2900 | ND |
| GMT | 3600 | 1880 | ||
| B717 | C4-V3K305c-T320c | 8300 | 8300 | 1400 |
| B715 | C4-V3K305C-T320C | 2400 | 1400 | 220 |
| B719 | C4-V3K305C-T320C | 11000 | 7700 | ND |
| B723 | C4-V3K305C-T320C | 4300 | 2000 | ND |
| GMT | 5500 | 3700 | ||
| B866 | C4-V3K305C-G321C | 4350 | 4000 | |
| B871 | C4-V3K305C-G321C | 1600 | 1600 | |
| B872 | C4-V3K305C-G321C | 2400 | 1500 | |
| B873 | C4-V3K305C-G321C | 5300 | 3300 | |
| GMT | 3100 | 2400 | ||
| B961 | C4-V3T303C-E322C | 20000 | 25000 | 1500 |
| B962 | C4-V3T303C-E322C | 10000 | 10000 | 330 |
| B963 | C4-V3T303C-E322C | 14000 | 14000 | 850 |
| B964 | C4-V3T303C-E322C | 50000 | 50000 | 1700 |
| GMT | 19000 | 20000 | ||
| B966 | C4-V3R304C-G321C | 510 | 820 | 312 |
| B967 | C4-V3R304C-G321C | 1200 | 1500 | 450 |
| B968 | C4-V3R304C-G321C | 660 | 690 | 317 |
| B975 | C4-V3R304C-G321C | 2700 | 2300 | 549 |
| GMT | 1000 | 1200 | ||
| B716 | C4-V3T303C-I323C | 2400 | 2800 | 260 |
| B720 | C4-V3T303C-I323C | 6250 | 7700 | 460 |
| B714 | C4-V3T303C-I323C | 5900 | 5000 | ND |
| B718 | C4-V3T303C-I323C | 12500 | 14000 | ND |
| GMT | 5800 | 6200 | ||
| C018 | C4-V3N302C-I323C | 12500 | 12500 | |
| C020 | C4-V3N302C-I323C | 3300 | 2300 | |
| C022 | C4-V3N302C-I323C | 5300 | 4200 | |
| C024 | C4-V3N302C-I323C | 17000 | 17000 | |
| GMT | 7800 | 6700 | ||
| B889 | C4-V3N301C-G325C | 50000 | 50000 | 1900 |
| B890 | C4-V3N301C-G325C | 11000 | 8300 | 960 |
| B892 | C4-V3N301C-G325C | 14000 | 10000 | 1300 |
| GMT | 20000 | 16000 | ||
| B955 | gp120 | 5900 | 17000 | |
| B958 | gp120 | 3700 | 12500 | |
| B959 | gp120 | 2700 | 14000 | |
| B960 | gp120 | * | * | |
| GMT | 3900 | 14000 | ||
Binding of the C4-V3 and gp120 elicited immune sera to V3 peptides, gp120 and the C4 peptide. Immune-sera were obtained after three immunizations and half-maximal values of the binding reaction were determined. ND-not determined. Geometric mean titer (GMT) is presented.
Undetectable levels of binding (excluded from calculation). Values are rounded to two significant figures.
Fig. 2.
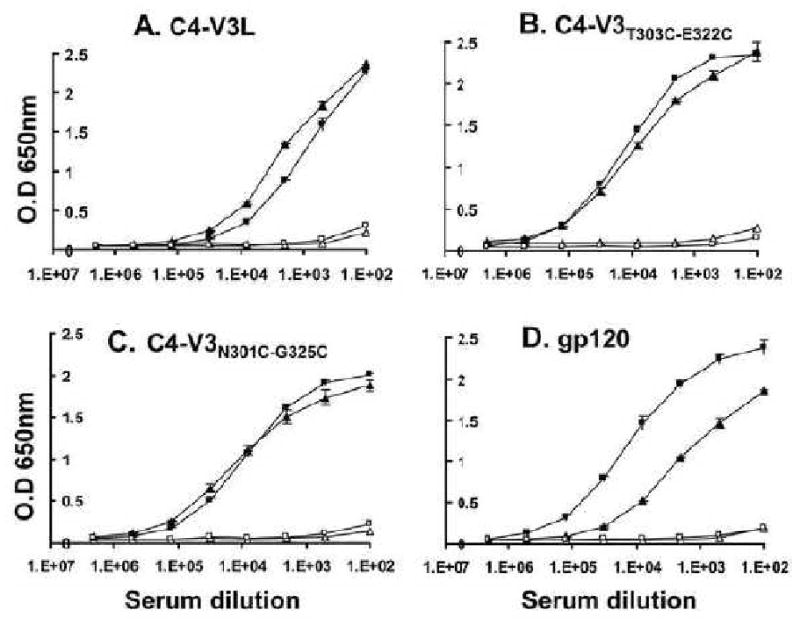
Binding of antibodies elicited by C4-V3 peptides or gp120 to the corresponding V3 peptide and gp120. A) Rabbit B707 serum, immunized with C4-V3L, B) Rabbit B963 serum, immunized with C4-V3T303C-E322C, C) Rabbit B892 immunized with C4-V3N301C-G325C and D) Rabbit B959 serum, immunized with gp120. Binding to the homologous peptide is shown in triangles (▲ post-immune, Δ pre-immune). Binding to gp120 in is shown in squares (■ post-immune, □ pre-immune). Y-axis represents OD at 650nm; X-axis represents the reciprocal of serum dilution. Standard deviation is for duplicates on plate.
Cross-reactivity of the C4-V3 immune sera with gp120
An essential prerequisite for neutralization by a vaccine-elicited sera is that it will be cross-reactive with a gp120 protein in which the V3 is exposed. Therefore, the pre-immune and the immune sera were tested for binding to the ΔV1/V2 gp120 construct in which the V3 is fully exposed (Fig. 3, Table 2) by a simple ELISA test. For each serum the relative binding to the corresponding V3 peptide and gp120 molecule was calculated and the data is presented as the average of the ratio between gp120 and the immunizing-peptide binding (Fig. 3). The two peptides constrained by a disulfide bond involving residue 303 elicited sera that bound gp120 as strongly as they bound the V3-peptide immunogen, while the sera from the rabbits immunized with the linear V3 (V3L) peptide was the least cross reactive to gp120 with an average ratio of 0.54 (p=0.0078) between gp120 and V3L binding (Fig. 3). The average ratios between gp120 and constrained V3 peptide binding of the serum induced by C4-V3K305C-T320C and C4-V3K305C-G321C, which were constrained to the V3MN (Sharon et al., 2003) and V3consensus conformation (Rosen et al., 2006), respectively, were 0.69 and 0.79, respectively, while the sera elicited by C4-V3N301C-G325C had a ratio of gp120/constrained V3 peptide binding of 0.82. For these last three peptides the reduced binding to gp120 was less consistent than for the linear peptide as evidenced by the larger standard deviation which did not reach statistical significance as judged by the T-test (P value ranging from 0.07-0.19). Of the two peptides designed to mimic the 0.5β-bound V3-conformation C4-V3N302C-I323C had a small reduction in affinity to gp120 compared with it homologous V3 peptide, and C4-V3R304C-G321C unexpectedly had a little higher affinity to gp120 than to its homologue V3 peptide. For both peptides this observation was not statistically significant (P value of 0.193 and 0.327 respectively).
Fig. 3.
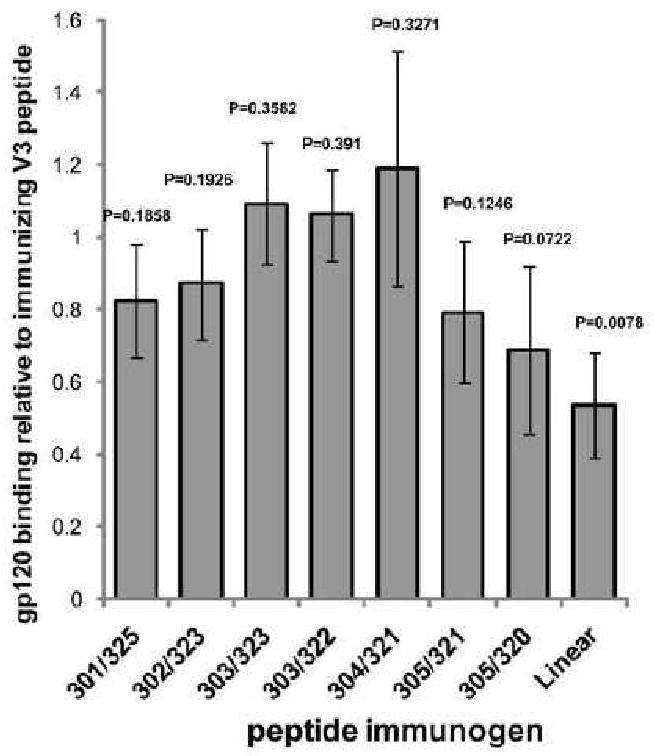
Relative cross reactivity of immune sera with gp120 and V3 peptide immunogens. The ratio between the immune-sera binding to gp120 and binding to the homologous peptide used for immunization is represented. The ratio is obtained by dividing the half maximal titer for gp120 by the half maximal titer to the V3 peptide used as immunogen. Shown is the average and standard deviation for each of the four rabbits immunized with each of the peptides. Peptide immunogens are listed by the positions replaced by cysteine according to Table1. P-value for one sample T-test for a hypothetical mean of 1 is shown above the histograms.
Due to the low V3 titer in this later peptide we believe that this observation is not significant. Thus, although the differences in gp120 cross-reactivity among the different peptides' immune-sera are not large, it is clear that peptides constrained by a disulfide bond involving residue 303 to the conformation of the V3 bound to 447-52D exhibited higher cross-reactivity with gp120 in comparison with the sera elicited by the flexible linear peptide and the other C4-V3 peptides in which the V3 was constrained to the conformations recognized by the 447-52D antibody.
The reactivity of the peptide immune-sera is conformation dependent
To further characterize the conformational specificity of the immune sera we tested the binding of the C4-V3L and C4-V3T303C-I323C immune-sera to the V3T303C;I323C peptide in a reduced and cyclic form and as a control we tested binding to V3L in the presence and absence of reducing agent (Fig. 4). For the C4-V3L immune sera, the ratio between cyclic V3T303C-I323C binding and V3L binding was 0.64 and the ratio between reduced V3T303C-I323C and V3L binding was 0.89, indicating that most of the decrease in the binding of the C4-V3L immune sera to cyclic V3T303C-I323C compared with the V3L peptide is due to the disulfide-bond constraint (P=0.0004 for reduced vs. cyclic). The ratio between C4-V3T303C-I323C immune-sera binding to cyclic V3T303C-I323C and V3L was 1.16 while the ratio between reduced C4-V3T303C-I323C and V3L binding was 0.97. (P=0.0083 for reduced vs cyclic). These results indicate that the C4-V3T303C-I323C immune sera bind slightly better to the peptide constrained by a disulfide bond implying recognition of the conformation of this epitope. Moreover only a fraction of the C4-V3L immune sera bound gp120 and constrained V3T303C-I323C (Fig.3 and Fig. 4). The reduced reactivity of the C4-V3L induced sera with the cyclic V3T303C-I323C compared to the reduced V3T303C-I323C indicated that these immune sera elicited an antibody response that recognized a wider range of conformations than those represented by V3T303C-I323C.
Fig. 4.
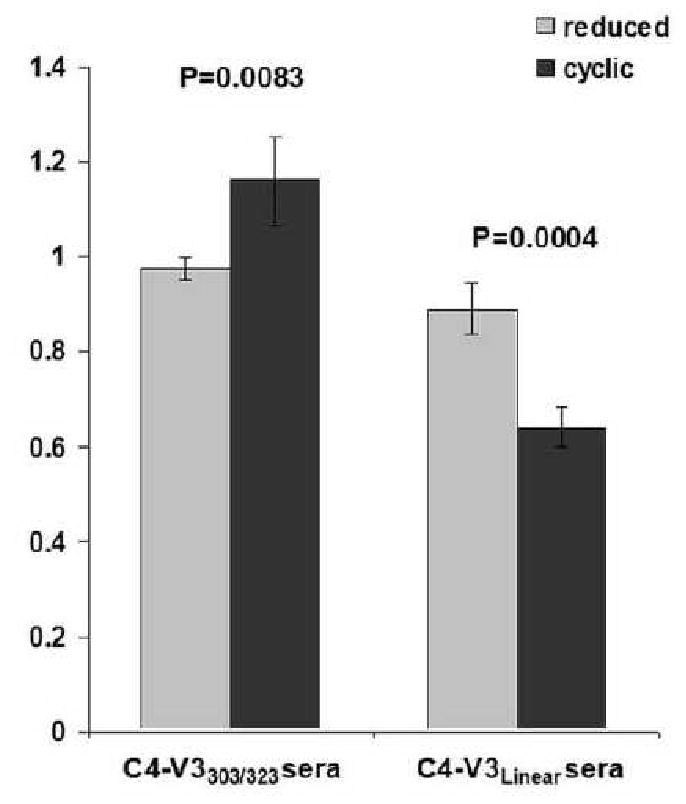
Influence of peptide conformation on the binding of immune sera to a cyclic V3 peptide. The binding of the immune-sera to cyclic vs. reduced V3 T303C/I323C is compared to binding to V3L for the C4V3L and C4-V3T303C/I323C induced sera. Half-maximal binding titer to cyclic and reduced V3T303C/I323C and to V3L with and without DTT was determined by ELISA for each V3T303C/I323C and V3L immune serum. In each experiment the binding of the serum to V3T303C/I323C in the cyclic (dark) or reduced state (light) was divided by the binding to V3L. Shown is the average and standard deviation for each of the four rabbits immune sera. P-value for two samples T-test for reduced vs. cyclic is shown.
Antibody response to the gp120 based immunogen
We compared the immune response against the C4-V3 peptide immunogens with that obtained against gp120 to determine whether there is any advantage in using constrained V3 peptides to obtain a potent HIV-1 neutralizing response. The gp120 construct used as immunogen in this study contained the full length V3 loop, lacked the first and second variable loops as well as the first 86 and last 19 N- and C-terminal residues, respectively, and was homogenously glycosylated with Man5GlcNAc2 glycans at sites normally occupied by complex or hybrid glycans (see Material and Methods). Three out of the four rabbits raised a strong anti-gp120 immune response; however one rabbit had undetectable antibody levels against both gp120 and V3 (B960) and therefore was excluded from the GMT calculation and from the analysis (Table 2). The GMT for half-maximal gp120 binding of the gp120-elicited immune sera was 14,000. This was comparable to the GMT values measured for binding to the homologous V3 immunogen of immune sera from constrained V3 peptides that elicited strong antibody responses such as C4-V3N301C-G325C and C4-V3T303C-E322C. The V3 directed antibody responses of the gp120-immune sera, as measured by V3T303C-I323C binding, were 2.8-5.3 fold lower than that against gp120, with GMT for half-maximum binding of 3,900. The peptide V3T303C-I323C was used in this experiment to assess the V3-directed response of the gp120 immune-sera because a very good correlation between gp120 and peptide binding was observed for the sera elicited by this peptide (see above). The results indicate that gp120 elicited an antibody response to other epitopes in addition to the V3 crown (residues R304-E322), and that a moderate level of V3 directed antibodies was obtained using the gp120 construct in which the V3 loop is fully exposed by deletion of the V1 and V2 loops and by reducing the size of the carbohydrate chains in the protein.
Neutralization of clade-B neutralization sensitive HIV-1 isolates by C4-V3 and gp120 immune sera
A pseudovirus based neutralization assay was used to evaluate the potency and breadth of the neutralizing antibody response elicited by the different C4-V3 and gp120 immunogens against a spectrum of clade-B viruses. Initially we focused on strains that are highly sensitive to neutralization. All sera were tested against Bal, BX08, MN, SF-162 and NL-43 strains and all sera except C4-V3K305C-G321C were tested also against NSC. JR-CSF, which is known to be a neutralization resistant strain, was also included in all the tests. The V3 sequences and phenotype of these HIV-1 strains are presented in Table 3. All strains are R5 viruses with the exception of MN and NL-43 which utilize CXCR4. aMLV (Murine Leukemia Virus) was used as a negative control. As a further control, representative pre-immune sera were tested for neutralization of SF-162 and no neutralization activity was detected (data not shown). The neutralization results are summarized in Table 4. The most effective immunogens were peptides constrained at position 303, of which C4-V3T303C-E322C, representing the conformation of a V3MN peptide bound to 447-52D, induced the strongest neutralizing response demonstrated by the effective neutralization of SF-162 (Fig. 5). Both C4-V3T303C-E322C and C4-V3T303C-I323C immune sera neutralized the four R5 neutralization sensitive strains as well as MN which is an ×4 strain. C4-V3T303C-E322C immune sera neutralized also the ×4 neutralization-sensitive strain NL-43 and these were the only immune sera that neutralized NL-43 significantly. It should be noted that NL-43 contains a rare two residue insertion preceding the GPGR segment in the V3 loop. Importantly, both peptides induced a much more potent neutralizing response then the linear C4-V3 peptide. Only one linear-peptide immune-serum neutralized 5 sensitive strains and two other C4-V3L immune-sera neutralized only the two most neutralization sensitive strains, SF162 and NSC. The neutralization titers were on average significantly lower for C4-V3L immune sera than those elicited by peptide immunogens constrained at T303. For example, the IC50 GMT of C4-V3L immune sera for NSC neutralization was more than one order of magnitude lower than that of the C4-V3T303C-I323C immune sera and two orders of magnitude lower than that of C4-V3T303C-E322C immune sera. Peptides constrained at K305 were found to be even less effective than the linear peptide in inducing antibodies capable of neutralizing HIV-1 isolates; none of the sera in the two groups of rabbits immunized by these peptides neutralized all sensitive strains tested, only one serum in each group neutralized more then two strains and the neutralization titers were much lower in most cases than those observed for the other constrained C4-V3 peptides. The peptide C4-V3N301C-G325C is constrained to assume the same V3consensus conformation as C4-V3T303C-I323C. However, its disulfide bond is removed further away from the GPGR loop and the ring size enclosed by the disulfide bond is therefore four-residues larger. This peptide was included in order to test whether T303 is the optimal position for the disulfide constraint. C4-V3N301C-G325C induced a potent anti-gp120 and anti-V3 response which was more effective than that elicited by either the linear peptide or the peptides constrained at K305, nevertheless it was not as potent as C4-V3T303C-I323C and C4-V3T303C-E322C in eliciting an HIV-1 neutralizing response. For example, the IC90 GMT for SF-162 is 2 and 3 fold higher for the C4-V3T303C-I323C and C4-V3T303C-E322C immune sera in comparison with the C4-V3N301C-G325C immune sera. Only one out of three immune sera reached 90% inhibition for MN in the group immunized with C4-V3N301C-G325C compared with three out of four and four out of four for C4-V3T303C-I323C and C4-V3T303C-E322C immune sera, respectively (data not shown). This demonstrates that T303 is indeed the optimal position for the disulfide bond when V3 peptides are constrained to the 447-52D bound conformations. The modified gp120 molecule elicited a relatively modest neutralizing response that resembles the antibody response elicited by C4-V3N301C-G325C and although it neutralized the two most sensitive strains only one serum neutralized all 5 sensitive strains that we tested. Both peptides designed to mimic the 0.5β conformation elicited poor neutralizing responses (Table 4 and Figure 5), None of the C4-V3N302C-I323C induced sera neutralized all 5 sensitive strains and only two sera neutralized more than one strain. These results demonstrate a drastic decline in neutralization potency compared with the immune sera of peptides constrained to the 447-52D conformation at positions 303 or 301. C4-V3R304C-G321C elicited an even poorer neutralizing response with only two sera capable of neutralizing SF-162. Due to the low V3 titer in C4-V3R304C-G321C this group of rabbits was subjected to two additional boosts. 2-3 fold higher V3 titer was detected after the fifth inoculation and improved neutralization titers were also achieved with IC:50 GMT of 630 and 111 (for 3/4 available) for SF-162 and MN, respectively). Nevertheless the neutralization was limited to SF-162 and MN. None of the sera neutralized BX08 and only one serum neutralized Bal (data not shown).
Table 3.
V3 sequence of the viral strains tested for neutralization by the immune sera a.
| Strain | Tropism | V3 Sequence |
|---|---|---|
| JR-FL | R5 | TRKSIHI - - GPGRAFYTTGEII |
| SF-162 | R5 | . . . . . T. - - . . . . . . .A. .D. . |
| MN | ×4 | K. .R. . . - - . . . . . . . . .KN. . |
| NSC | R5 | . .R. .TM - - . . . . . . . . . . . . . |
| BX08 | R5 | . . . . . . . - - . . . . . . . . . .D. . |
| Bal | R5 | . . . . . . . - - . . . . . . . . . . . . . |
| SS1196 | R5 | . . . . . . . - - . . . . . . . . . .GV. |
| 6535 | R5 | . . . . .NL - - . . . . . . . . . .D. . |
| QH0692 | R5 | . . . . . . . - - . . . . . . . . . .D. . |
| JR-CSF | R5 | . . . . . . . - - . . . . . . . . . . . . . |
| NL-43 | ×4 | . . . . .R. QR. . . . . .V.I.K-. |
V3 sequence (residues 303-324) of viral strains compared to JR-FL representing the clade-B consensuses, · identical to consensuses; - (deletion).
Table 4.
Neutralization of clade-B isolates by the C4-V3 and gp120 immune sera a.
| Rabbit | Immunogen | BaL | BX08 | MN | NSC | SF162 | JRCSF | NL-43 | aMLV |
|---|---|---|---|---|---|---|---|---|---|
| B712 | C4-V3L | <10 | <10 | <10 | 53 | 53 | <10 | <10 | <10 |
| B707 | C4-V3L | <10 | <10 | <10 | 11 | 63 | <10 | <10 | <10 |
| B702 | C4-V3L | 12 | 60 | 244 | 32 | 15 | <10 | 19 | <10 |
| B722 | C4-V3L | 15 | 31 | 38 | 62 | 964 | <10 | <10 | <10 |
| GMT | <12 | <21 | <31 | 33 | <83 | N.N | N.N | N.N | |
| B717 | C4-V3K305C-T320C | <10 | 64 | <10 | 13 | 111 | <10 | <10 | <10 |
| B715 | C4-V3K305C-T320C | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| B719 | C4-V3K305C-T320C | <10 | 27 | 15 | 165 | 191 | <10 | 32 | <10 |
| B723 | C4-V3K305C-T320C | <10 | <10 | <10 | <10 | 14 | <10 | <10 | <10 |
| GMT | N.N | <20 | <11 | <22 | <42 | N.N | <13 | N.N | |
| B866 | C4-V3K305C-G321C | 12 | 15 | <10 | N.D | <10 | <10 | <10 | <10 |
| B871 | C4-V3K305C-G321C | <10 | <10 | <10 | N.D | <10 | <10 | <10 | <10 |
| B872 | C4-V3K305C-G321C | <10 | <10 | <10 | N.D | 25 | <10 | <10 | <10 |
| B873 | C4-V3K305C-G321C | 19 | 17 | 51 | N.D | 3379 | <10 | <10 | <10 |
| GMT | <12 | N.N | <15 | N.D | <54 | N.N | N.N | N.N | |
| B966 | C4-V3R304C-G321C | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| B967 | C4-V3R304C-G321C | <10 | <10 | <10 | <10 | 79 | <10 | <10 | <10 |
| B968 | C4-V3R304C-G321C | <10 | <10 | 22 | 500 | 246 | <10 | <10 | 15 |
| B975 | C4-V3R304C-G321C | <10 | <10 | 17 | 14 | 15 | <10 | <10 | <10 |
| GMT | N.N | N.N | N.N | <29 | <41 | N.N | N.N | N.N | |
| B961 | C4-V3T303C-E322C | 162 | 135 | 1958 | >5120 | >5120 | <10 | 597 | <10 |
| B962 | C4-V3T303C-E322C | 17 | 25 | 93 | 775 | 359 | <10 | 44 | <10 |
| B963 | C4-V3T303C-E322C | 60 | 61 | 1018 | >5120 | >5120 | <10 | 747 | <10 |
| B964 | C4-V3T303C-E322C | 57 | 46 | 474 | >5120 | 4939 | <10 | 269 | <10 |
| GMT | 56 | 55 | 544 | >3194 | >2612 | N.N | 269 | N.N | |
| B716 | C4-V3T303C-I323C | 12 | 27 | 30 | 1647 | 557 | <10 | <10 | <10 |
| B720 | C4-V3T303C-I323C | 111 | 285 | 1178 | 350 | >5120 | 22 | 10 | <10 |
| B714 | C4-V3T303C-I323C | 21 | 52 | 115 | 276 | 1150 | <10 | 42 | <10 |
| B718 | C4-V3T303C-I323C | 16 | 55 | 133 | 190 | 1900 | <10 | <10 | <10 |
| GMT | 26 | 68 | 152 | 417 | >1580 | N.N | <14 | N.N | |
| C018 | C4-V3N302C-I323C | 23 | 33 | 33 | 20 | 832 | <10 | <10 | <10 |
| C020 | C4-V3N302C-I323C | <10 | 13 | <10 | 12 | 12 | <10 | <10 | <10 |
| C022 | C4-V3N302C-I323C | <10 | 13 | <10 | 18 | 39 | <10 | <10 | <10 |
| C024 | C4-V3N302C-I323C | 17 | 31 | 44 | 1737 | 1391 | <10 | <10 | <10 |
| GMT | N.N | 20 | <19 | 52 | 152 | N.N | N.N | N.N | |
| B889 | C4-V3N301C-G325C | 28 | 33 | 30 | 274 | 637 | <10 | <10 | <10 |
| B890 | C4-V3N301C-G325C | 39 | 38 | 582 | >5120 | 2412 | <10 | <10 | <10 |
| B892 | C4-V3N301C-G325C | 12 | 15 | 44 | 36 | 253 | <10 | <10 | <10 |
| GMT | 24 | 26 | 92 | >368 | 730 | N.N | N.N | N.N | |
| B955 | gp120 | 45 | 40 | 131 | 902 | 727 | <10 | <10 | <10 |
| B958 | gp120 | 14 | 18 | 64 | 438 | 540 | <10 | 15 | <10 |
| B959 | gp120 | 16 | 15 | 158 | 79 | 632 | <10 | <10 | <10 |
| B960* | gp120 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| GMT | 22 | 22 | 110 | 315 | 628 | N.N | N.N | N.N | |
Titers, calculated as the serum dilution conferring 50% inhibition (IC50) of pseudovirus infection are presented. The first measurement was done at 1:10 dilution and was followed by consecutive 2 fold dilutions. Highlighted in gray are samples in which the IC50 was at least 3× higher than any inhibition of the aMLV negative control. Geometric mean titer (GMT) is presented (<10 was calculated as 10) .N.D=not determined. N.N= non-neutralizing.
excluded from calculation due to lack of immune response in that rabbit
Fig. 5.
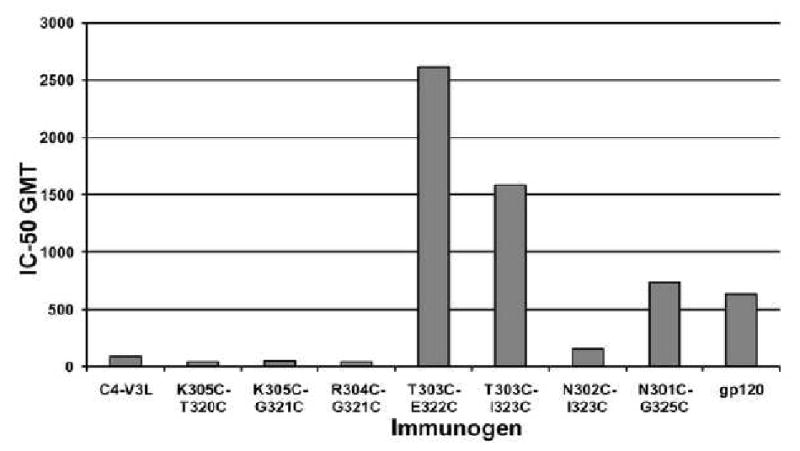
comparison of the neutralization of SF-162. GMT for serum dilution inflicting 50% inhibition (IC:50) of SF-162 is presented for the different immunogens used in the study.
Altogether our results support the use of peptides based immunogens over gp120 based immunogens and demonstrate the benefit of optimally constraining the V3 loop to induce a more potent and cross-reactive neutralizing response.
Neutralization of clade-B moderately resistant HIV-1 primary isolates
In order to further assess the neutralizing potential of the immune sera, the sera for C4-V3L, C4-V3T303C-E322C, C4-V3T303C-I323C and gp120 were tested against the more resistant primary isolates SS1196, 6535 and QH0692. As shown in Table 5 no neutralization of these strains was detected for the C4-V3T303C-I323C, and gp120 sera and only one sera from rabbits immunized with C4-V3L weakly neutralized SS1196. However, C4-V3T303C-E322C immune sera were able to neutralize SS1196 with GMT of 64 (average of 4/4 rabbits), and 6535 with GMT of less than 26 (37 when average is calculated for 3 out 4 rabbits that showed neutralization) but not QH0692 which is more difficult to neutralize in comparison with 6535. This experiment clearly demonstrates that C4-V3T303C-E322C immune sera can neutralize several primary isolates that are more resistant to neutralization and the immunogenic potential of properly constrained V3-peptides over linear peptides and monomeric-gp120 based constructs
Table 5.
Neutralization of clade-B modestly resistant strainsa.
| Rabbit | Immunogen | SS1196 | 6535 | QH0692 |
|---|---|---|---|---|
| B961 | C4-V3T303C-E322C | 82 | 64 | 14 |
| B962 | C4-V3T303C-E322C | 51 | <10 | 13 |
| B963 | C4-V3T303C-E322C | 80 | 32 | <10 |
| B964 | C4-V3T303C-E322C | 49 | 24 | 12 |
| GMT | 64 | <26 | N.N | |
| B716 | C4-V3T303C-I323C | <10 | <10 | <10 |
| B720 | C4-V3T303C-I323C | 18 | 15 | 12 |
| B714 | C4-V3T303C-I323C | <10 | <10 | <10 |
| B718 | C4-V3T303C-I323C | <10 | <10 | <10 |
| GMT | N.N | N.N | N.N | |
| B712 | C4-V3L | <10 | <10 | <10 |
| B707 | C4-V3L | <10 | <10 | <10 |
| B702 | C4-V3L | 22 | <10 | <10 |
| B722 | C4-V3L | 10 | <10 | <10 |
| GMT | <12 | N.N | N.N | |
| B955 | gp120 | 25 | <10 | <10 |
| B958 | gp120 | <10 | <10 | <10 |
| B959 | gp120 | 11 | <10 | <10 |
| B960* | gp120 | <10 | <10 | <10 |
| GMT | N.N | N.N | N.N | |
Titers, calculated as the plasma dilution conferring 50% inhibition (IC50) are shown. Geometric mean titer (GMT) is presented (<10 was calculated as 10). Highlighted in gray are samples in which the IC50 was at least 3× higher than any inhibition of the aMLV negative control. Geometric mean titer (GMT) is presented (<10 was calculated as 10). N.N= non-neutralizing.
excluded from calculation due to lack of immune response in that rabbit.
Discussion
This study evaluated the hypothesis that antibodies elicited by V3 peptides constrained to a conformation that mimics the β-hairpin conformation of the V3 loop, will be more cross-reactive with monomeric gp120 and more potent in neutralizing HIV-1 compared with antisera elicited by the homologous linear V3 peptide. To test this hypothesis we used structural data acquired on constrained V3 peptides (Mor et al., 2009) together with the 447-52D binding affinity of these peptides to guide the design of V3 peptide immunogens constrained by a single disulfide bond. Our design considered the hydrogen bond network and the cross-strand alignment of the β-hairpin conformation observed in the NMR structures of V3 peptides bound to antibody 447-52D (Rosen et al., 2006). Based on the above we systematically changed the location of the disulfide bond and compared the gp120 cross reactivity and HIV-1 neutralizing response obtained by immunization with the constrained-peptide to that obtained by an analogous linear V3-immunogen and by monomeric-gp120. This analysis has lead to the following observations.
Peptides constrained by a disulfide bond involving residue 303 were more effective immunogens for eliciting sera with gp120 cross-reactivity
Of the peptide mimicking the 447-52D conformation, C4-V3T303C-E322C and C4-V3T303C-I323 induced immune sera showed the best cross-reactivity with monomeric-gp120 in comparison with C4-V3L with ratios of 1.06 and 1.1 between binding to gp120 and the corresponding V3 peptide, respectively (Fig. 3). These values indicate that the immune sera recognize monomeric-gp120 as well as they recognize the V3 immunogen. The C4-V3K305C-T320C and C4-V3K305C-G321C immune sera exhibited slightly poorer cross-reactivity with gp120 (ratios of 0.69 and 0.79, respectively). This is despite the fact that the affinity to 447-52D of V3K305C-T320C is slightly higher than that of V3T303C-E322C and the affinity of V3K305C-G321C is higher than that of V3T303C-I323 (Mester et al., 2009). Unlike 447-52D which is tolerant to replacements of amino acids in the β-strand (Keller et al., 1993; Stanfield et al., 2004), and to replacements of K305 and I307 by cystine (Mester et al., 2009) it is possible that a subpopulation of the antibodies elicited by V3K305C-T320C and V3K305C-G321C interact with the side chain of the cystine residue at position 305 and are sensitive to its replacement by other residues.
The immune sera of peptides constrained to the 0.5β bond conformation cross-reacted very well with monomeric-gp120. C4-V3N302C-I323C immune sera had a small reduction in binding to gp120, and C4-V3R304C-G321C immune sera had a slightly higher antibody titer to gp120 than to its homologous V3 peptide. Nevertheless C4-V3R304C-G321C was a poor immunogen inducing low V3 peptide antibody titers (GMT 3-20 fold lower compared with other peptides), that are comparable to the C4-directed antibody titer. Due to the flexible nature of the gp120 used we can not exclude the possibility that C4 binding played a role in the slight increase in gp120 binding compared with the V3 titer. Notably, several studied have demonstrated that antibodies raised against linear C4 peptide have limited gp120 binding and no neutralization activity (Kelker et al.,; Robey et al., 1995). Therefore, the observation of C4-binding antibodies is not relevant for neutralization.
The linear C4-V3L peptide elicited antibodies that were less cross-reactive with monomeric-gp120 in comparison with the V3T303C-E322C or V3T303C-I323 immune sera. Nevertheless the significant cross-reactivity of the C4-V3L immune sera with gp120 is surprising (binding ratio gp120/V3L = 0.54). This ratio is only a factor of two lower than that observed for the immune-sera of peptides constrained by a disulfide bond involving residue 303. It is known that the GPGR segment in V3 peptides transiently populates a β-turn conformation (Chandrasekhar, Profy, and Dyson, 1991; Zvi, Hiller, and Anglister, 1992). Thus, although it does not form a β-hairpin like conformation, the V3L peptide is not completely flexible. The β-turn forming GPGR segment is the core epitope for many V3-directed HIV-1 neutralizing antibodies and, for example, in the V3MN complex with 447-52D the GPGR segment occupies a central pocket in the antibody binding site. The significantly populated β-turn conformation of V3L, a conformation that most likely characterizes the corresponding region in native gp120, may be the reason why this peptide elicits a relatively high proportion of gp120-cross-reactive antibodies which neutralize HIV-1. In this study the monomeric gp120 immunogen was designed to present an exposed V3 in order to gain data about the reactivity of the sera with V3-containing gp120 without the effect of epitope masking, The most significant finding was the relatively low cross reactivity of the linear peptide compared with other constrained peptides and that peptides constrained at T303 are the most potent immunogens. The conclusion that the T303 constrained-V3 peptides are viable immunogens is supported by the neutralization of different HIV-1 strains with different levels of V3 masking by sera raised against C4-V3T303C-I322C.
Peptides constrained to mimic the 0.5β-bound conformation elicited considerably poorer HIV-1 neutralizing responses compared with peptides mimicking the 447-52D bound conformation
In this study we focused on identifying the most effective disulfide constrained V3 peptide, using the 447-52D bound conformation of the V3 as a template for constraining V3 peptide immunogens. Additionally two peptides were constrained to the 0.5β bound conformation of the V3. Our data clearly shows that V3 peptides constrained to the 0.5β-bound conformation resulted in sera which exhibited a drastic decline in HIV-1 neutralizing capability. Even C4-V3N302C-I323C which retains the entire 447-52D epitope unchanged, elicited significantly poorer neutralization responses compared with C4-V3T303C-I323C C4-V3T303C-E322C and C4-V3N301C-G325C. This observation indicates that the hydrogen-bond network and the resulting side-chain conformation governs the nature of the elicited antibody response. C4-V3R304C-G321C elicited low levels of V3 directed antibodies and only sporadic HIV-1 neutralizating responses. Taken together the comparison of sera raised against peptides constrained to mimic 0.5β- and 447-52D bound V3 conformations clearly shows that the 447-52D-bound conformation is indeed a useful model for designing peptide immunogens that elicit an HIV-1 neutralizing antibody response.
A peptide constrained by a disulfide bond linking residues 303 and 322 is the most effective V3 immunogen for eliciting an HIV-1 neutralizing response
Comparison of the neutralization efficiency of a panel of HIV-1 strains by the immune sera elicited by the different peptide immunogens reveals that the peptide V3T303C-E322C is optimally constrained and that it elicited a considerably more potent HIV-1 neutralization in comparison with the linear peptide and with peptides constrained by a disulfide bond containing both shorter and longer ring sizes (i.e. V3K305C-T320C, V3K305C-G321C and V3N301C-G325C). This was reflected in a significantly higher neutralizing titer when comparing neutralization sensitive strains. For example, when neutralization of the neutralization-sensitive strain SF162 was compared among the immune-sera elicited by the different constrained-peptide immunogens, we found that the neutralization efficiency of the C4-V3T303C-E322C sera was at least 30-fold higher than that of C4-V3L immune sera, and more than 48-fold higher than the V3K305C-T320C, V3K305C-G321C immune sera (Fig. 5). When the disulfide bond was located further away from the GPGR segment, i.e. involving position 301 and 325, the neutralization efficiency dropped by more than three fold suggesting that position 303 is optimal for the disulfide bond location.
As shown in Table 5 C4-V3T303C-E322C immune sera were capable of neutralizing SS1196 and 6535 primary isolates, of which 6535 is considered difficult to neutralize. These strains were not neutralized by C4-V3T303C-I323 immune sera. This immunogen encloses a ring that is only one residue longer than that of C4-V3T303C-EI322 and that has the same hydrogen forming network at the N-terminus. This demonstrates the need to fine tune the location of the disulfide bond in order to achieve the most effective antibody response. Taken together with its ability to neutralize NL-43 the neutralization of strains SS1196 and 6535 indicated that C4-V3T303C-E322C elicited an antibody response that is different then that of C4-V3T303C-I323C and can neutralize a wider range of viral strains including those that are more resistant to neutralization.
It is surprising that the C4-V3L, C4-V3K305C-T320C and C4-V3K305C-G321C elicited much poorer HIV-1 neutralizing responses in comparison with C4-V3T303C-E322C and C4-V3T303C-I323C although the differences in gp120 cross-reactivity were not pronounced. A possible explanation is that both C4-V3T303C-E322C and C4-V3T303C-I323C elicited a larger fraction of the high-affinity antibodies that are crucial for HIV-1 neutralization. Another possibility is that binding to the modified monomeric gp120 does not reflect exactly the binding to V3 on the virus and that it could be that C4-V3T303C-E322C elicits antibodies that can better access the V3 region on the virus.
C4-V3T303C-E322C elicits a better HIV-1 neutralizing response than monomeric-gp120
The HIV-1 neutralization elicited by monomeric-gp120 in the present study is considerably poorer than that elicited by C4-V3T303C-E322C. When neutralization of SF-162 or NSC is compared, the C4-V3T303C-E322C immune sera was 5-fold or 10-fold, respectively more potent than the gp120 immune sera (Table 4 and Fig. 5), and the gp120 induced sera did not neutralize NL-43 at all. The poorer HIV-1 neutralization by the gp120 immune sera was obtained despite the fact that the V3 region was fully exposed in the truncated gp120 construct used in the present study (Huang et al., 2005). Two conclusions can be reached on the basis of the observed poorer neutralization by the gp120 immune sera. First, the gp120 construct used in this study which is expected to elicit enhanced V3 response in comparison with gp120 containing the V1 and V2 loop as well as longer carbohydrate chains was a poorer immunogen in comparison with C4-V3T303C-E322C. Second, if gp120 elicited neutralizing antibodies that targeted regions other than the V3, their potency in HIV-1 neutralization was substantially weaker in comparison with the C4-V3T303C-E322C immune sera. This is an important conclusion in view of the fact that monomeric gp120 immunogens were used in the Thai vaccine trial (Rerks-Ngarm et al., 2009).
The C4-V3T303C-E322C immune sera are broadly neutralizing
The panel of the HIV-1 strains neutralized by C4-V3T303C-E322C includes eight strains, some of which are considerably different in sequence from the immunizing consensuses sequence (Table 3). Variations are observed at position T303, K305, S306, H308, I309, Y318, T319, G321 and E322. The NL-43 strain that resembles HIV-1IIIB is the most distant from the immunizing peptide and in addition to four mutations (H308R, Y318V, T320I and E322K) it contains a two residue insertion (Q310-R311) and a one residue deletion (I323). Interestingly 447-52D neutralizes NL-43 and IIIB very efficiently while other broadly neutralizing V3 directed antibodies fail to do so (Binley et al., 2004; Eda et al., 2006) This implies that C4-V3T303-E322C is the most effective in inducing an 447-52D-like antibody response. Altogether these differences in the V3 sequences of the tested HIV-1 strains and especially the large differences between the NL-43 strain and the immunizing peptide indicate that the immune sera generated by the C4-V3T303C-E322C peptide are highly cross-reactive with clade-B viruses which contain the GPGR motif and are capable of neutralizing both ×4 and R5 viruses.
Peptide C4-V3T303C-E322C is the most potent constrained V3 immunogen studied to date
Previously, the most comprehensive and successful study using constrained V3 peptides was carried out by Conley and co-workers (Conley et al., 1994), and despite more recent studies, the most potent HIV-1 neutralizing response remained the one induced using a 23-residue V3MN peptide cyclized by a disulfide bond at its termini (L-705,402 or V3N300C-T326C according to our nomenclature). The neutralization titer for the MN strain was 640 and that for IIIB (similar to NL43 tested in the present study) was more than 15-fold lower. Three additional clade-B strains were neutralized (AL-1, SF-2 and WMJ-2) while three others (Du 6587-5, Du 7887-7 and RF) were resistant to the anti-sera. Conley and co-workers concluded that the anti-V3 response was comparable to that obtained by gp120 immunogens (Conley et al., 1994). Shorter peptides either lacking the entire epitope recognized by 447-52D or which were poor mimics of the V3 β-hairpin conformation resulted in considerably weaker HIV-1 neutralizing responses.
Similar to the optimal V3 peptide discovered in the present study, the long V3MN peptide used by Conley and co-workers, L-705,402, contained the entire epitope recognized by 447-52D. The 25-residue disulfide enclosed ring in L-705,402 is slightly longer than the 23-residue ring of V3N301C-G325C which showed reduced efficiency in eliciting a potent HIV-1 neutralizing response compared to the V3T303C-E322C peptide (18-residue ring) which was found to be the optimal immunogen in our investigation. However, compared to the L-705,402 immune sera, V3T303C-E322C elicited immune-sera showed considerably improved neutralization of HIV-1 strains that differed in their V3 sequence from the consensus clade-B immunizing peptide. In addition, V3T303C-E322C elicited at least a four-fold stronger HIV-1 neutralizing response in comparison with gp120 while the peptides containing longer ring size V3N301C-G325C and 705,402 (V3MNN300C-T326C) elicited HIV-1 neutralizing responses comparable to that induced by gp120 (present study and Conley). Thus it appears that V3T303C-E322C has the optimal ring size. It is likely that this optimum reflects a compromise between the requirement to present the entire 447-52D epitope without any replacement to cysteine residues and to contain as short a ring size as possible to better mimic the β-hairpin conformation of the V3.
It should be noted that direct comparison of our results with those obtained by Haynes and co-workers (Haynes et al., 2006) and by Conley and co-workers (Conley et al., 1994) is not possible because of the different animals used for immunization, different peptide immunogens and different neutralization assays. Therefore, the goal of this study was to test the principle of using a linear V3 immunogen in comparison with a constrained V3-immunogen under the same conditions. We hypothesized that the constrained peptide found to be optimal in Conley's study contained a ring structure that was too long and too flexible and therefore the second goal of this study was to determine the optimal location of the disulfide bond constrained within the V3. Our data support our hypothesis because the strongest HIV-1 neutralizing response was elicited by a constrained peptide that contained a shorter ring size than that found in Conley's peptide. In addition we compared the sera elicited by gp120 to that elicited by the C4-V3 peptide under the same conditions and demonstrated that optimally constrained V3 peptide elicit more potent HIV-1 neutralizing response in comparison with monomeric gp120 molecule.
Conclusions
In the present study we used the structure of V3-peptides bound to antibody 447-52D to guide the design of constrained immunogens. We found that the C4-V3T303C-E322C immunogen elicited a potent HIV-1 neutralizing response that was more efficient in neutralization of sensitive strains, more broadly neutralizing, and more capable of neutralizing primary isolates that are difficult to neutralize than previously studied constrained V3-immunogens. Small changes in the location of the disulfide bond were shown to have a significant impact on the nature of the antibody response. The results support our working hypothesis that constrained V3-peptides can be better immunogens than their linear V3-peptide homologs and gp120.
Comparison of the different constrained V3 immunogens in view of their NMR structure (Mor et al., 2009) and their affinity to 447-52D (Mester et al., 2009) leads us to conclude that: a) In order to achieve an effective neutralizing response, V3 peptides must be designed to mimic the 447-52D-bound conformation of the V3 as opposed to the 0.5β bound conformation. b) It is important to include the intact K305-T320 segment of V3 without replacement to cysteine. c) High affinity binding to 447-52D can be misleading in selecting an optimal immunogen (Mester et al., 2009). d) It may not be necessary to achieve a rigid β-hairpin conformation in the peptide immunogen and some flexibility may be beneficial for eliciting a strong HIV-1 neutralizing response. e) The ring enclosed by the disulfide bond should be minimal while containing the entire 447-52D epitope to avoid too much flexibility.
Finally, the results suggest that a V3-directed antibody response can neutralize a broad subset of primary isolates which will be limited primarily by the occlusion of this epitope in primary isolates. Strategies to expose the V3 loop, focus the antibody response on exposed V3 segments and increase the titer of the elicited antibody response should, therefore, be vigorously pursued.
Acknowledgments
We are most grateful to Dr. Orith Leitner and Ziv Landau from the antibody unit at the Weizmann Institute for the immunizations, animal handling and good advice; to Dr. Osnat Rosen for most fruitful discussions, to Drs. Julie Goss, Terri Wrin and Pham Phung from Monogram Biosciences South San Francisco CA USA for there services in HIV-1 neutralization assay; to Dr. Hilary Voet from the Faculty of Agriculture, at the Hebrew University for advice on statistical analysis. We thank Dr. H. G. Khorana (Massachusetts Institute of Technology) for the gift of GnT I-deficient HEK 293S.
Footnotes
This study was supported by NIH grant GM53329 (JA) and GM22087 (FN), and by the Horowitz Foundation, Gurwin Foundation and the Kimmelman Center. J.A is the Dr. Joseph and Ruth Owades Professor of Chemistry and F.N. is the Leonard and Esther Kurtz Term Professor at the College of Staten Island at the City University of New York
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bell CH, Pantophlet R, Schiefner A, Cavacini LA, Stanfield RL, Burton DR, Wilson IA. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375(4):969–978. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryder K, Sbai H, Nielsen HV, Corbet S, Nielsen C, Whalen RG, Fomsgaard A. Improved immunogenicity of HIV-1 epitopes in HBsAg chimeric DNA vaccine plasmids by structural mutations of HBsAg. DNA Cell Biol. 1999;18(3):219–225. doi: 10.1089/104454999315439. [DOI] [PubMed] [Google Scholar]
- Burke B, Gomez-Roman VR, Lian Y, Sun Y, Kan E, Ulmer J, Srivastava IK, Barnett SW. Neutralizing antibody responses to subtype B and C adjuvanted HIV envelope protein vaccination in rabbits. Virology. 2009;387(1):147–156. doi: 10.1016/j.virol.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas E, Wang M, Parren PW, Stanfield RL, Satterthwait AC. A structure-based approach to a synthetic vaccine for HIV-1. Biochemistry. 2000;39(47):14377–14391. doi: 10.1021/bi0003691. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Durani V, Miranda ER, Citron M, Liang X, Schleif W, Joyce JG, Varadarajan R. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. Biochem J. 2006;399(3):483–491. doi: 10.1042/BJ20060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar K, Profy AT, Dyson HJ. Solution conformational preferences of immunogenic peptides derived from the principal neutralizing determinant of the HIV-1 envelope glycoprotein gp120. Biochemistry. 1991;30(38):9187–9194. doi: 10.1021/bi00102a009. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Conard P, Bondy S, Dolan CA, Hannah J, Leanza WJ, Marburg S, Rivetna M, Rusiecki VK, Sugg EE, et al. Immunogenicity of synthetic HIV-1 gp120 V3-loop peptide-conjugate immunogens. Vaccine. 1994;12(5):445–451. doi: 10.1016/0264-410x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol. 2009a;83(3):1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology. 2009b;387(2):414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AK, Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set. Acta Crystallogr D Biol Crystallogr. 2008;D64(Pt 7):792–802. doi: 10.1107/S0907444908013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda Y, Takizawa M, Murakami T, Maeda H, Kimachi K, Yonemura H, Koyanagi S, Shiosaki K, Higuchi H, Makizumi K, Nakashima T, Osatomi K, Tokiyoshi S, Matsushita S, Yamamoto N, Honda M. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J Virol. 2006;80(11):5552–5562. doi: 10.1128/JVI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald DJ, Fryling CM, McKee ML, Vennari JC, Wrin T, Cromwell ME, Daugherty AL, Mrsny RJ. Characterization of V3 loop-Pseudomonas exotoxin chimeras. Candidate vaccines for human immunodeficiency virus-1. J Biol Chem. 1998;273(16):9951–9958. doi: 10.1074/jbc.273.16.9951. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gatewood JM, Mariappan SV, Pau CP, Parekh BS, George JR, Gupta G. Human immunodeficiency virus (HIV) antigens: structure and serology of multivalent human mucin MUC1-HIV V3 chimeric proteins. Proc Natl Acad Sci U S A. 1995;92(1):315–319. doi: 10.1073/pnas.92.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Ma B, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia SM, Korber BT, Liao HX. Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology. 2006;345(1):44–55. doi: 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelker HC, Itri VR, Valentine FT. A strategy for eliciting antibodies against cryptic, conserved, conformationally dependent epitopes of HIV envelope glycoprotein. PLoS One. 5(1):e8555. doi: 10.1371/journal.pone.0008555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PM, Arnold BA, Shaw AR, Tolman RL, Van Middlesworth F, Bondy S, Rusiecki VK, Koenig S, Zolla-Pazner S, Conard P, et al. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology. 1993;193(2):709–716. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Mester B, Manor R, Mor A, Arshava B, Rosen O, Ding FX, Naider FR, Anglister J. HIV-1 peptide vaccine candidates: Selecting constrained V3 peptides with highest affinity to antibody 447-52D. Biochemistry. 2009 doi: 10.1021/bi900146g. [DOI] [PubMed] [Google Scholar]
- Mor A, Segal E, Mester B, Arshava B, Rosen O, Ding FX, Russo J, Dafni A, Schvartzman F, Scherf T, Naider F, Anglister J. Mimicking the structure of the V3 epitope bound to HIV-1 neutralizing antibodies. Biochemistry. 2009;48(15):3288–3303. doi: 10.1021/bi802308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Wrin T, Cavacini LA, Robinson JE, Burton DR. Neutralizing activity of antibodies to the V3 loop region of HIV-1 gp120 relative to their epitope fine specificity. Virology. 2008;381(2):251–260. doi: 10.1016/j.virol.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009 doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey FA, Kelson-Harris T, Roller PP, Robert-Guroff M. A helical epitope in the C4 domain of HIV glycoprotein 120. J Biol Chem. 1995;270(41):23918–23921. doi: 10.1074/jbc.270.41.23918. [DOI] [PubMed] [Google Scholar]
- Rosen O, Chill J, Sharon M, Kessler N, Mester B, Zolla-Pazner S, Anglister J. Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization. Biochemistry. 2005;44(19):7250–7258. doi: 10.1021/bi047387t. [DOI] [PubMed] [Google Scholar]
- Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc Natl Acad Sci U S A. 2006;103(38):13950–13955. doi: 10.1073/pnas.0606312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha BE, Onorato M, Bartlett JA, Bosch RJ, Aga E, Nokta M, Adams EM, Li XD, Eldridge J, Pollard RB. Safety and immunogenicity of a polyvalent peptide C4-V3 HIV vaccine in conjunction with IL-12. Aids. 2004;18(8):1203–1206. doi: 10.1097/00002030-200405210-00015. [DOI] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure. 2003;11(2):225–236. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Ghiara JB, Ollmann Saphire E, Profy AT, Wilson IA. Recurring conformation of the human immunodeficiency virus type 1 gp120 V3 loop. Virology. 2003;315(1):159–173. doi: 10.1016/s0042-6822(03)00525-7. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman RL, Bednarek MA, Johnson BA, Leanza WJ, Marburg S, Underwood DJ, Emini EA, Conley AJ. Cyclic V3-loop-related HIV-1 conjugate vaccines. Synthesis, conformation and immunological properties. Int J Pept Protein Res. 1993;41(5):455–466. doi: 10.1111/j.1399-3011.1993.tb00465.x. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Zvi A, Levy R, Anglister J. A cis proline turn linking two beta-hairpin strands in the solution structure of an antibody-bound HIV-1IIIB V3 peptide. Nat Struct Biol. 1999;6(4):331–335. doi: 10.1038/7567. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Zvi A, Levy R, Hayek Y, Matsushita S, Anglister J. NMR structure of an anti-gp120 antibody complex with a V3 peptide reveals a surface important for co-receptor binding. Structure. 2000;8(4):385–395. doi: 10.1016/s0969-2126(00)00119-2. [DOI] [PubMed] [Google Scholar]
- Zinckgraf JW, Winchell JM, Silbart LK. Antibody responses to a mucosally delivered HIV-1 gp120-derived C4/V3 peptide. J Reprod Immunol. 1999;45(2):99–112. doi: 10.1016/s0165-0378(99)00042-x. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen S, Pinter A, Krachmarov C, Wrin T, Wang S, Lu S. Cross-clade neutralizing antibodies against HIV-1 induced in rabbits by focusing the immune response on a neutralizing epitope. Virology. 2009 doi: 10.1016/j.virol.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008;372(2):233–246. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zvi A, Hiller R, Anglister J. Solution conformation of a peptide corresponding to the principal neutralizing determinant of HIV-1IIIB: a two-dimensional NMR study. Biochemistry. 1992;31(30):6972–6979. doi: 10.1021/bi00145a015. [DOI] [PubMed] [Google Scholar]


