Abstract
KSHV is implicated in the pathogenesis of KS, a chronic inflammation associated malignancy. COX-2 and its metabolite PGE2, two pivotal proinflammatory/oncogeneic molecules, are proposed to play roles in the expression of major KSHV latency associated nuclear antigen-1 (LANA-1). Microsomal prostaglandin E2 synthase (mPGES), PGE2 and its receptors (EP1, EP2, EP3, and EP4) were detected in KS lesions with the distinct staining of EP2/EP4 in KS lesions. In latently infected endothelial TIVE-LTC cells, EP receptor antagonists down-regulated LANA-1 expression as well as Ca2+, p-Src, p-PI3K, p-PKCζ/λ, and p-NF-κB, which are also some of the signal molecules proposed to be important in KS pathogenesis. Exogenous PGE2 and EP receptor agonists induced the LANA-1 promoter in 293 cells, and YY1, Sp1, Oct-1, Oct-6, C/EBP and c-Jun transcription factors appear to be involved in this induction. PGE2/EP receptor induced LANA-1 promoter activity was down-regulated significantly by the inhibition of Ca2+, p-Src, p-PI3K, p-PKCζ/λ, and p-NF-κB. These findings implicate the inflammatory PGE2/EP receptors and the associated signal molecules in herpes virus latency and uncover a novel paradigm that demonstrates the evolution of KSHV genome plasticity to utilize inflammatory response for its survival advantage of maintaining latent gene expression. This data also suggests that potential use of anti-COX-2 and anti-EP receptor therapy may not only ameliorate the chronic inflammation associated with KS but could also lead to elimination of the KSHV latent infection and the associated KS lesions.
Keywords: cyclooxygenase-2, prostaglandin E2, EP receptors, KSHV latency, signaling
INTRODUCTION
KSHV (HHV-8) is etiologically associated with KS, the most common and aggressive AIDS-defining malignancy (1-3). KS is characterized by a pro-inflammatory microenvironment (1-3). Thus, unraveling the biology of KSHV pathogenesis, and therefore KS, is closely tied to understanding the chronic inflammatory conditions that set the stage for KS.
Previous reports had demonstrated that cyclooxygenase-2 (COX-2), a proinflammatory molecule, was highly up-regulated in vitro by KSHV infection (4, 5). The tumorigenic properties of COX-2 are attributed to its metabolite prostaglandin E2 (PGE2) that exerts its effect through eicosonoid (EP) receptors (EP1-4) (6-12). COX-2 inhibition significantly abrogated expression of the major KSHV latent gene LANA-1 during de novo KSHV infection of fibroblast (HFF) and endothelial (HMVEC-d) cells and exogenous PGE2 reversed this down-regulation (5). These studies have indicated that COX-2/PGE2 mediated inflammation is crucial for KSHV latency program. Although, the role of COX-2 and PGE2 in herpes viral lytic cycle is demonstrated, their role in viral latency has been observed only in KSHV.
However, the mechanistic aspects of how COX-2/PGE2 mediates KSHV latent gene expression is not known and the role of EP receptors is unexplored in herpes virus biology. Our study shows that Ca2+, Src, PI3K, PKCζ/λ, and NFkB signal molecules are regulated by EP receptors in latently infected cells and blocking EP receptors down-regulated LANA-1 and COX-2 gene expression. PGE2 stimulated the LANA-1 promoter via a network of Ca2+, Src, PI3K, PKCζ/λ, and NFkB activation. Collectively, these studies demonstrate that KSHV utilizes the host proinflammatory COX-2/PGE2/EP receptor pathway for its own advantage of establishing and maintaining latent gene expression.
Materials and Methods
Cells and KSHV
TIVE-LTC (long-term-infected telomerase immortalized umbilical vein endothelial cells) TIVE cells, a gift from Dr. Rolf Renne (University of Florida), and 293 cells were cultured as described before (13). KSHV was prepared and assessed for its infectivity, mycoplasma, and LPS as described before (5).
Plasmids
LANA-1 promoter sequence (pGL3.6 or p-LANA-1-Luc) and the LANA-1 promoter deletion sequences (pGL3.4, pGL3.3, pGL3.2, and pGL3.1) cloned in pGL3.0 vector (Promega, Madison, WI) with the reporter gene Firefly luciferase were gifts from Dr. Yuan Chang, University of Pittsburgh (14).
Reagents
Akt 1/2 inhibitor, TMB-8, PD98059, Wortmannin, Ly290042, U0126, and LPA were from Sigma, St. Louis, Mo. GFX, GO:6976, PP2, and Bay11-7085 were from Calbiochem, La Jolla, CA. PGE2, EP1-4 agonists, AH6809, and GW627368X were from Cayman chemical, Ann Arbor, MI. Fura-2AM was from Invitrogen, Carlsbad, CA. SC-51322 was from Enzo Life Sciences, Plymouth Meeting, PA.
Antibodies
Anti-mouse (COX-1 and COX-2) antibodies as well as anti-rabbit (mPGES, EP1, EP2, EP3, and EP4) antibodies were from Cayman chemicals. Anti-mouse (PI3K, α-tubulin, and p-Src) antibodies were from BD Biosciences, San Jose, CA, Sigma, and Calbiochem, respectively. Anti-mouse (p-NFκB, p-Akt, and p-ERK 1/2) and anti-rabbit (Akt, Src, NFκB p65, p-PKCζ/λ, and p-PI3K) antibodies were from Cell Signaling Technology, Inc., Denver, CO. Anti-rabbit PGE2 was from Abcam, Cambridge, MA. Anti-rabbit (PKCζ and ERK2) antibodies were from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. LANA-1 antibody (15).
Transfection and luciferase reporter Assay
Transfections on 293 cells were conducted as described before (5). The luciferase assays were conducted as per the manufacturer’s guidelines (Promega). The relative LANA-1 promoter activity or relative luciferase units (RLU) were normalized to Renilla luciferase protein levels.
Fluorescent activated cell sorting (FACS)
Samples for FACS analysis were prepared as per manufacturer’s guidelines (BD Biosciences). The data was collected using FACSCalibur flow cytometer (Becton Dickinson, Bedford, MA) and analyzed with CellQuest Pro software (Becton Dickinson) at the RFUMS flow cytometry core facility.
Western blotting and measurement of PGE2
Total cell lysates prepared from cells after respective treatments were used for western blotting and quantified as described before (5). α-tubulin was used as the loading control for all the blots. Secreted amounts of PGE2 were measured using a PGE2 ELISA Kit as per the manufacturer’s guidelines (R and D, Minneapolis, MN).
Real-time reverse transcription PCR (RT-PCR)
LANA-1, COX-2, and COX-1 transcripts were detected by real-time RT-PCR as described before (5).
Confocal microscopy and immunohistochemistry
Confluent TIVE and TIVE-LTC cells were used for confocal microscopy using EP1-4 antibodies as before (13). Tissue sections from 3 healthy subjects and 3 KS+ patients were obtained from the AIDS and Cancer Specimen Resource (ACSR), National Cancer Institute (NCI). Immunohistochemistry in Fig. 1 was done by similar method as described before (13).
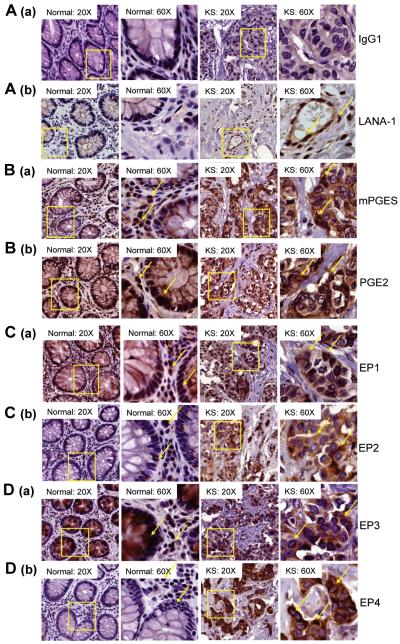
Figure 1.
Characterization of mPGES, PGE2 and EP receptors in KS Lesions. (A-D) Serial sections of normal and KS lesions from 3 healthy subjects and 3 KS+ patients were stained by immunohistochemistry for rabbit IgG1 (A (a)), LANA-1 (A (b)), mPGES (B (a)), PGE2 (B (b)), EP1 (C (a)), EP2 (C (b)), EP3 (D (a)), and EP4 (D (b)).
Measurement of Ca2+
293 or TIVE-LTC cells plated onto coverslips and placed in six well plates were mounted on the stage of an inverted microscope (Olymous IX71) and incubated with 5μM of Ca2+ indicator fura-2AM (Invitrogen) for 30 minutes at 37°C in Hanks balanced salt solution (Sigma), pH 7.4. Cells were then continuously perfused with Ca2+ free Hanks balanced salt solution (BSS) pH 7.4. When cytoplasmic Ca2+ levels rise after respective treatments, if any, Ca2+ will bind to fura-2AM in the cytoplasm inducing a fluorescence signal that can be used as a measure of the level of cytoplasmic calcium. The calcium induced fluorescence signal of fura-2AM was measured by alternatively exciting at 340 and 380nm and collecting the emitted fluorescence at 510nm using a CCD-based imaging system running SimplePCI software (Hamamatsu Corporation, Bridgewater, NJ). Changes in cytoplasmic Ca2+ ([Ca2+]i) are reported as the ratio of 340/380 emissions. Data analysis was performed using Origin Pro software (Origin Lab Corporation, Northampton, MA). The Ca2+ studies were conducted at RFUMS Ca2+ imaging facility.
Statistical analysis
In Fig. 2, the statistical significance (t-test) was conducted with respect to untreated or uninfected cells. In Fig. 3, the statistical significance (t-test) was conducted with respect to supernatant treatment. In Fig. 4 the statistical significance (t-test) was conducted similar to Fig. 2. In Fig. 5, the statistical significance (t-test) was calculated with respect to PGE2 alone treatment. In Fig. 6, the statistical significance (t-test) was conducted similar to Fig. 2. (*)p<0.01,(**)p<0.001,(***)p<0.0001.
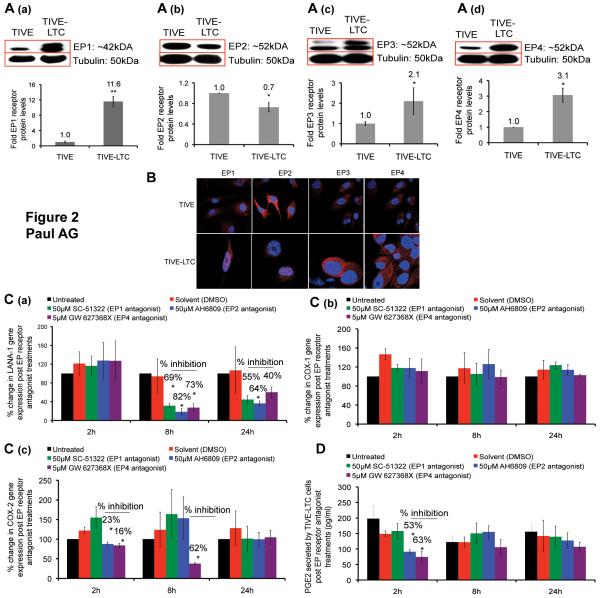
Figure 2.
Effect of EP receptor antagonists on LANA-1, COX-1, COX-2, gene expression and PGE2 secretion in TIVE-LTC cells. (A) Cells were serum starved for 48h, immunoblotted for EP1 (A (a)), EP2 (A (b)), EP3 (A (c)), and EP4 (A (d)) receptor levels with respect to tubulin to find the relative difference between TIVE and TIVE-LTC cells. (B) Permeabilized TIVE and TIVE-LTC cells were stained for EP1-4 receptors and analyzed by confocal microscopy. (C-D) TIVE-LTC cells were serum starved for 48h and treated with EP1, EP2, and EP4 antagonists. Total RNA and supernatant were collected at 2h, 8h and 24h post treatment to examine the gene expression levels of LANA-1 (C (a)), COX-1 (C (b)), and COX-2 (C (c)) by real time RT-PCR and PGE2 by ELISA (D). The percentage change was calculated with respect to untreated cells for each time point.
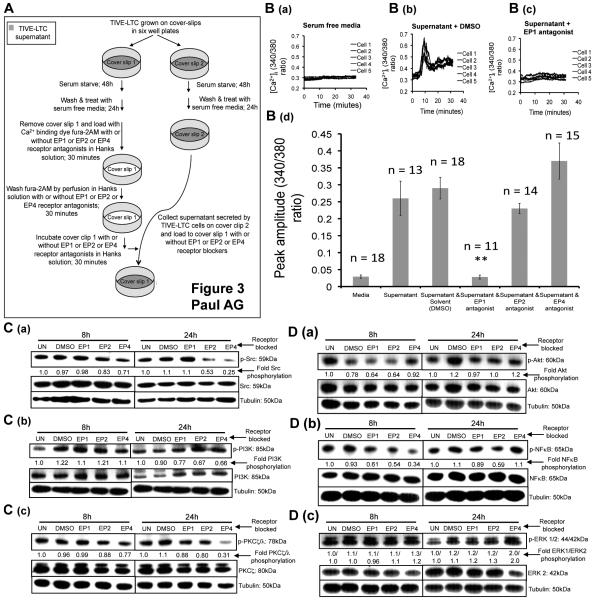
Figure 3.
Effect of EP receptor antagonists on Ca2+, Src, PI3K, PKCζ/λ, Akt, NFκB, and ERK 1/2 in TIVE-LTC cells. (A) Schematic of the experimental design for Ca2+ experiments. (B) TIVE-LTC cells were grown on cover slips placed in six-well plates and serum starved for 48h. The cells were washed and treated with fresh serum free media for 24h. Coverslip 1 was removed, loaded with fura-2AM (5μM) with or without different EP receptor antagonists. The cells were then washed and incubated with serum free media (B (a)) or with the supernatant produced by coverslip 2 TIVE-LTC cells with or without EP1, EP2, and EP4 antagonists or DMSO (B (b)). Transient elevation in [Ca2+]i was measured for 35 minutes. For each graph, 5 representative TIVE-LTC cells were selected to portray the transient [Ca2+]i signal. (B (d)) Summary results showing the peak amplitude (mean±SEM) of transient elevation in [Ca2+]i observed in different treatments. ‘n’ refers to the number of cells that demonstrated a change in [Ca2+]i signal within the field studied. (C-D) TIVE-LTC cells were serum starved for 48h and treated with EP1, EP2, and EP4 antagonists or DMSO. 8h and 24h post treatment, total cell lysates were prepared and immunoblotted for p-Src (C (a)), p-PI3K (C (b)), p-PKCζ/λ (C (c)), p-Akt (D (a)), p-NFκB (D (b)), and p-ERK 1/2 (D (c)), and normalized with respect to the levels of total protein levels. The fold change was calculated with respect to the signal intensity for untreated (UN) cells for each time point.
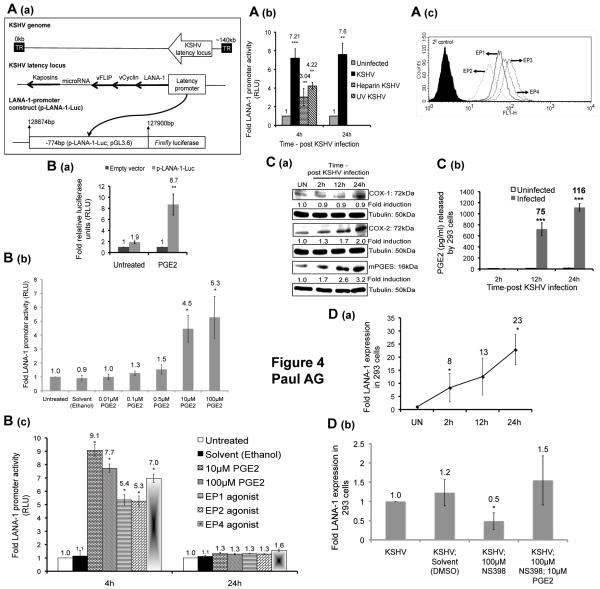
Figure 4.
Effect of KSHV infection and exogenous PGE2 treatment on LANA-1 promoter activity and the effect of de novo KSHV infection on 293 cells. (A) Schematic of the LANA-1 promoter constructs (p-LANA-1-Luc or pGL3.6). (A (b)) 293 cells transfected with p-LANA-1-Luc were infected with live KSHV or heparin treated KSHV, UV-KSHV, or left uninfected for 2h, washed with PBS, and supplemented with serum free DMEM. (A (c)) Permeabilized 293 cells were stained for EP1, EP2, EP3, and EP4 receptors by FACS. (B) 293 cells transfected with p-LANA-1-Luc or empty vector (pGL3.0) were treated with 10μM PGE2 (B (a)) or different concentrations of PGE2 for 4h (B (b)) or PGE2 (10μM and 100μM), 10μM of EP1-4 agonists or left untreated for 4h and 24h (B (c)). (A (b) and B) At the indicated times, the cells were harvested, lysed, and assayed for relative luciferase units (RLU). (C and D (a)) 293 cells, serum starved for 24h, were infected with KSHV for 2h or left uninfected (UN), washed and supplemented with serum free DMEM throughout the experiment. At 2h, 12h, and 24h p.i., whole cell lysate, supernatant, and total RNA were collected to measure for COX-1, COX-2, mPGES protein levels (C (a)) PGE2 by ELISA (C (b)), and LANA-1 expression (D (a)), respectively. (D (b)) 293 cells serum starved for 24h were pretreated with NS-398 (100μM) for 24h, washed, infected with KSHV for 2h in the presence of NS-398 (100μM) or NS-398 (100μM) and PGE2 (10μM), washed and supplemented with serum free DMEM in the presence of NS-398 (100μM) or NS-398 (100μM) and PGE2 (10μM). 24h p.i. Total RNA was prepared to analyze the gene expression levels of LANA-1 by real time RT-PCR. The fold change and corresponding statistics (t-test) were calculated with respect to untreated or uninfected (UN) 293 cells for each time point.
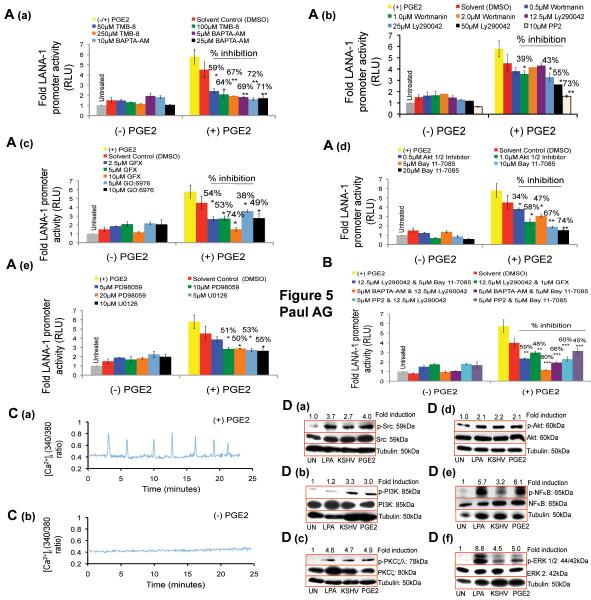
Figure 5.
Effect of Ca2+, p-Src, p-PI3K, p-PKCζ/λ, p-Akt 1/2, p-NFκB, and p-ERK 1/2 inhibition on PGE2 mediated LANA-1 promoter activity. (A-B) 293 cells transfected with p-LANA-1-Luc was pretreated with indicated pharmacological signal inhibitors for 2h and supplemented with 10μM PGE2 for 4h or left untreated and assayed for RLU. The fold change was calculated with respect to untreated cells. (C) 293 cells serum starved for 24h were loaded with fura-2AM, washed by perfusion, and treated with 10μM PGE2 or Hanks BSS (pH 7.4) and the [Ca2+]i levels were measured for 25 minutes. Shown are representative cells from a field of approximately 50-60 cells. (D) 293 cells serum starved for 24h were treated with 100ng of LPA, 10μM PGE2, infected with 20 DNA copies per cell of KSHV, or left untreated (UN) for 4h. Total cell lysate was prepared and immunoblotted for p-Src, p-PI3K, p-PKCζ/λ, Akt 1/2, NFκB, and ERK 1/2, normalized with respect to the levels of total protein levels and the fold change was calculated with respect to (UN) cells for each time point.
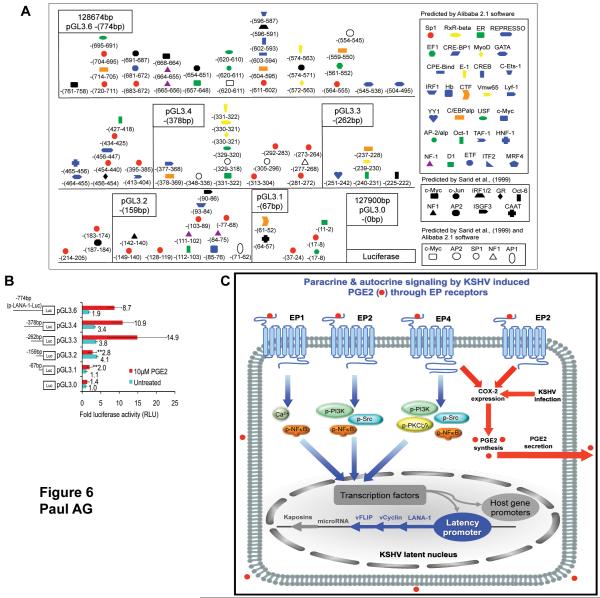
Figure 6.
PGE2 responsive regions of LANA-1 promoter. (A) PGE2 responsive TF binding sites on p-LANA-1-Luc. The LANA-1 promoter sequence (14) were analyzed by Alibaba 2.1 software. The schematic shows the different TFs predicted by Alibaba 2.1 software, the TF binding sites predicted by (14), and by both. The numbers for each TF refers to the beginning and end sequence of the binding site within the 774bp LANA-1 promoter sequence studied, with 128674bp and 127900bp as the start and end sites of the LANA-1 promoter, respectively, in the KSHV genome. The site of origin of each LANA-1 promoter deletion construct is marked. (B) Identification of PGE2 responsive regions of LANA-1 promoter. Sequential series of LANA-1 promoter deletion constructs with luciferase reporter or empty vector (pGL3.0) were transfected into 293 cells and treated with10μM PGE2 for 4h or left untreated. The cells were harvested, lysed, and assayed for RLU. The fold change and corresponding statistics (t-test) were calculated with respect to untreated cells for each time point. (C) Schematic model depicting the potential mechanism by which PGE2 and EP receptor play roles in KSHV pathogenesis. During latency, KSHV induces proinflammatory COX-2 gene expression and PGE2 secretion (5) resulting in the activation of EP receptors in an autocrine and paracrine fashion. Activated EP receptors influence specific signal cascades that in turn influence specific transcription factors converging on the LANA-1 promoter. EP2 and EP4 mediated signaling might also be involved in the regulation of this process by initiating a positive feedback loop to maintain COX-2 gene expression and therefore PGE2 synthesis/secretion resulting in the sustenance of the characteristic chronic proinflammatory environment created by KSHV infection.
RESULTS
PGE2, mPGES, and EP1-4 receptors are detected in KS lesions
We used immunohistochemistry to first investigate the presence of mPGES, PGE2 and EP1-4 in serial sections of biopsies from 3 healthy subjects and 3 KS+ patients. The presence of KSHV in KS lesions was confirmed by the detection of characteristic nuclear staining of LANA-1 (Fig. 1A (b)). Strong cytoplasmic mPGES, PGE2, and EP1-4 reactivity were detected in KS lesions (Fig. 1B-D). However, KS lesion samples exhibited distinct staining for EP2 and EP4 receptors compared to normal samples (Fig. 1C (b) and 1D (b)). Collectively, these results for the first time demonstrate the presence of inflammation associated EP receptors in KS lesions.
KSHV infection up-regulates EP1, EP3, and EP4 receptors and down-regulates EP2 receptors
We next examined the role of EP receptors in maintaining latent gene expression in TIVE-LTC cells that sustains expression of latency genes (16). In a separate study, we have observed the up-regulation of COX-2 and mPGES proteins, and PGE2 secretion in TIVE-LTC cells compared to control TIVE cells and down-regulation of LANA-1 expression in TIVE-LTC cells treated with COX-2 inhibitor NS-398 (13). Western blot analysis demonstrates that compared to TIVE cells, EP1, EP3, and EP4 receptors were significantly up-regulated in TIVE-LTC cells, whereas EP2 receptor was down-regulated in TIVE-LTC cells (Fig. 2A; a-d). Confocal microscopy also confirmed the presence and cellular localization of all 4 EP receptors in TIVE and TIVE-LTC cells (Fig. 2B).
KSHV utilizes EP receptors to maintain LANA-1 and COX-2 gene expression and PGE2 secretion
We measured LANA-1, COX-2, and COX-1 gene expression in TIVE-LTC cells treated with non-cytotoxic concentrations (Supplemental data Fig.1, Panels A to I) of well-characterized competitive EP receptor blockers SC-51322 (EP1 antagonist; 50μM) or AH6809 (EP2 antagonist; 50μM) or GW 627368X (EP4 antagonist; 5μM) at 2h, 8h and 24h post-treatment and observed no significant change in COX-1 gene expression (Fig. 2C (b)). COX-1 gene expression was conducted as a control, since COX-1 promoter is constitutively active. EP1, EP2, and EP4 antagonists down-regulated LANA-1 gene expression significantly by 69%, 82%, and 73%, and by 55%, 64%, and 40% at 8h and 24h post-treatments, respectively (Fig. 2C (a)). COX-2 gene expression was down-regulated by EP2 and EP4 antagonists at 2h post-treatment and by EP4 antagonist at 8h post-treatment (Fig. 2C (c)). At 2h post-treatment, EP2 and EP4 antagonists down-regulated PGE2 secretion significantly with no significant changes at 8h and 24h post treatments (Fig. 2D).
KSHV utilizes EP1 receptor mediated cytoplasmic Ca2+ ([Ca2+]i) signaling to maintain LANA-1 gene expression
From the above results demonstrating the down-regulation of LANA-1 gene expression by EP1 blockade (Fig. 2C (a)) together with the fact that EP1 receptor is a well-characterized Ca2+ inducing GPCR (10), we hypothesized that PGE2 secreted in the supernatant of TIVE-LTC cells might be inducing Ca2+ signaling via the EP1 receptor. Therefore, we predicted that if we block the EP1 receptor, the potential of the PGE2 in the supernatant to induce Ca2+ signaling via the EP1 receptor, if any, would also be blocked. The purpose of our experiment was not to test the effect of down regulating PGE2 secretion in the supernatant by EP receptor antagonists and consequently the calcium signal because even at 2h post treatment with EP2 and EP4 antagonists, PGE2 is present with in the range of 80-100pg/ml (Fig. 2D). However, our goal was to investigate the effect of blocking EP receptors on the supernatant-induced calcium signal, if any. By doing so, we are answering the role of PGE2 in the supernatant in inducing calcium signal through EP receptors.
To test our hypothesis, TIVE-LTC cells were grown on coverslips and [Ca2+]i was measured as outlined in Fig. 3A. To measure [Ca2+]i, cover slip 1 was removed and loaded with the Ca2+ indicator fura-2AM for 30 minutes with and without EP receptor antagonists in Hanks solution. However, while being loaded with fura-2AM, the cells in coverslip 1 are not exposed to the physiological supernatant of TIVE-LTC cells. Therefore, to test whether the supernatant of TIVE-LTC can induce Ca2+ signaling in coverslip 1 cells, we used the supernatant produced by cells grown in parallel on cover slip 2. The supernatant of coverslip 2 TIVE-LTC cells incubated in the presence of DMSO (Fig. 3B (b)) induced a significant transient elevation in [Ca2+]i. We did not observe any significant change between the observed peak amplitude of transient elevation in [Ca2+]i induced by the supernatant alone or with solvent control (DMSO) (Fig. 3B (d)). In contrast, serum free medium used as a negative control had no significant effect on [Ca2+]i (Fig. 3B (a)) indicating that factors present in the physiological supernatant of TIVE-LTC cells can induce Ca2+ signaling.
Next, we examined whether the transient [Ca2+]i induced by TIVE-LTC cell supernatant is due to activation of EP1 receptor by the supernatant PGE2. Therefore, we treated the cells on coverslip 1 with EP1 or EP2 or EP4 antagonist, while being treated with fura-2AM and the supernatant from coverslip 2 cells, as outlined in Fig. 3A. Treatments with EP1 but not EP2 or EP4 antagonists significantly abolished the supernatant mediated Ca2+ signal in TIVE-LTC cells (Fig. 3B (c) and 3B (d)). These observations clearly demonstrated that PGE2 present in the supernatant of TIVE-LTC cells could induce a Ca2+ signal through the EP1 receptor.
Inhibition of EP receptors down regulates p-Src, p-PI3K, p-PKCζ/λ, p-Akt and p-NFκB and up-regulates p-ERK in KSHV infected TIVE-LTC cells
We next examined the signal cascades regulated by EP receptors. TIVE-LTC cells serum starved for 48h were treated with EP1, EP2, and EP4 antagonists or DMSO. EP2 and EP4 antagonists down-regulated p-Src by 17% and 47% and 29% and 75% at 8h and 24h post-treatments, respectively (Fig. 3C (a)). At 24h post-treatment, EP1, EP2, and EP4 antagonists down-regulated PI3K phosphorylation by 23%, 33% and 34%, respectively with no effect at 8h post-treatment (Fig. 3C (b)). EP2 and EP4 antagonists down-regulated p-PKCζ/λ by 12% and 20% and 23% and 69% at 8h and 24h post-treatments, respectively (Fig. 3C (c)). Compared to DMSO treatment, we did not observe any changes on p-Akt by EP receptor antagonists at 8h and 24h post-treatment (Fig. 3D ()). At 8h post-treatment, p-NFκB was down-regulated by 39%, 46%, and 66% with EP1, EP2, and EP4 antagonists, respectively, and by 41% with EP2 antagonist 24h post-treatment (Fig. 3D (b)). With EP4 antagonist, p-ERK 1 and p-ERK 2 was up-regulated by 1.3 and 2.0-folds and 1.2 and 2.0-folds, at 8h and 24h post-treatment, respectively (Fig. 3D (e)).
KSHV infection and exogenous PGE2 activates the LANA-1 promoter
Based on our data from TIVE-LTC cells, we hypothesized that PGE2 mediated signaling can up-regulate LANA-1 promoter activity. To test this, 293 cells were transfected with a luciferase reporter gene under the control of 774bp LANA-1 promoter (p-LANA-1-Luc, Fig. 4A (a)) (14). The efficacy of our system was first demonstrated by the induction of p-LANA-1-Luc activity by primary KSHV infection whereas less induction of p-LANA-1-Luc activity was observed with entry incompetent heparin treated and UV inactivated virus (Fig. 4A (b)).
To determine the effect of exogenous PGE2 on p-LANA-1-Luc, we first confirmed the presence of EP1-4 receptors by FACS (Fig. 4A (c)). Exogenous PGE2 (10μM) induced p-LANA-1-Luc activity by 8.7-fold at 4h post-treatment with no significant effect on the empty vector (Fig. 4B (a)). Treatments with varying concentrations of PGE2 demonstrates that 10μM of PGE2 or more were necessary to activate p-LANA-1-Luc significantly with no significant difference between the effects of 10μM and 100μM of PGE2 (Fig. 4B (b-c)). Well-characterized agonists for EP1-4 receptors were also able to induce p-LANA-1-Luc activity significantly at 4h but not at 24h post-treatment (Fig. 4B (c)).
De novo KSHV infection of 293 cells induces the COX-2/PGE2 pathway
To validate whether the 293 cells used above were ideal to study the paradigm, we demonstrated that de novo KSHV infection of 293 cells, induces COX-2 and mPGES proteins (Fig. 4C (a)), PGE2 secretion (Fig. 4C (b)), and LANA-1 expression (Fig. 4D(a)). To determine whether the COX-2/PGE2 pathway is important for maintaining LANA-1 gene expression in 293 cells, we demonstrated that 10μM PGE2 can restore the reduction in LANA-1 expression caused by COX-2 specific inhibitor NS-398 (100μM) (Fig. 4D (b)).
Inhibition of Ca2+, p-Src, p-PI3K, p-PKCζ/λ, p-Akt 1/2, p-NFκB, and p-ERK 1/2 blocks PGE2 mediated LANA-1 promoter
We next examined the role of Ca2+, Src, PI3K, PKCζ/λ, Akt 1/2, NFkB, and ERK 1/2 in PGE2 mediated LANA-1 transcriptional regulation by measuring the LANA-1 promoter activity in 293 cells pretreated with specific inhibitors for 2h followed by PGE2 (10μM) treatment for 4h and then incubated with PGE2. We used pharmacological inhibitors of Ca2+ (BAPTA-AM and TMB-8), PI3K (Wortmannin and Ly290042), Src kinase (PP2), PKC (GFX and GO:6976), Akt 1/2 (Akt 1/2 inhibitor), NFκB (Bay 11-7085), and ERK 1/2 (PD98059 and U0126), at the indicated non-cytotoxic concentrations.
Ca2+ chelation (Fig. 5A (a)), Src inhibition (Fig. 5A (b)) and PKC inhibition (Fig. 5A (c), decreased PGE2 mediated p-LANA-1-Luc activity significantly (Fig. 5A (a-c)). Similarly, PI3K inhibitors wortmannin (1.0μM) and Ly290042 (25μM and 50μM) reduced PGE2 induced p-LANA-1-Luc activity significantly (Fig. 5A (b)). Akt 1/2 and NFκB inhibition down-regulated PGE2 mediated p-LANA-1-Luc activity significantly (Fig. 5A (d)). Though, PGE2 mediated LANA-1 promoter activity was down-regulated by Akt 1/2 inhibitor, we did not observe any effect on Akt phosphorylation by EP receptor antagonists (Fig. 3D (a)). Furthermore, exogenous PGE2 was able to induce Akt 1/2 in 293 cells. The dichotomy between TIVE-LTC and 293 cells indicates that in KSHV latent TIVE-LTC cells, Akt phosporylation might also be under the control of EP2/EP4 receptor independent mechanisms. However, in serum starved 293 cells, PGE2 might be acting as a powerful signal inducer through EP receptors to induce Akt that may subsequently activate the LANA-1 promoter. ERK inhibition by PD98059 (10μM and 20μM) and U0126 (5μM and 10μM) reduced PGE2 induced p-LANA-1-Luc activity significantly by 50-55% with no significant inhibition on the basal activity (Fig. 5A (e)). Our promoter studies using ERK 1/2 inhibitors are further validated by the observation that de novo KSHV infection and exogenous PGE2 activate ERK 1/2 in 293 cells (Fig. 5D (f)). This is in contrast to the EP4 antagonist induced up-regulation of ERK1/2 phosphorylation in TIVE-LTC cells. This could be due to the differences in the cell systems used as the determining factors regulating ERK phosphorylation in TIVE-LTC could be different from that of 293 cells. Under serum-starved conditions, PGE2 might be acting as a power signal inducer in 293 cells through EP receptors (Fig. 5D (f)). Therefore, PGE2 induced LANA-1 promote activity is inhibited by ERK inhibitors. However, in TIVE-LTC cells, the presence of viral proteins and a cytokine/chemokine rich supernatant might be altering the signal transduction profile of the cell to such an effect that EP2 and EP4 receptors might be responsible for inhibiting ERK phosphorylation.
To explore further the signal molecules studied here, next we used different combinations of signal inhibitors (Fig. 5B) at non-cytotoxic concentrations (supplemental data Fig.1, panel J). Blocking of PI3K and Ca2+ simultaneously demonstrated a significant additive effect of 80% (Fig. 5B) on the decrease in LANA-1 promoter activity compared with 5μM of BAPTA-AM (Fig. 5A (a)) and 12.5μM of Ly290042 (Fig. 5A (b)), when used alone.
Induction of Ca2+, p-Src, p-PI3K, p-PKCζ/λ, p-Akt 1/2, p-NFκB, and p-ERK 1/2 by PGE2 in 293 cells
To validate the capacity of PGE2 to induce LANA-1 promoter activity through the signal molecules that were blocked in Fig. 5A, we examined whether exogenous PGE2 (10μM) and KSHV infection can induce them. To test whether PGE2 can induce Ca2+, we treated serum starved 293 cells loaded with fura-2AM with PGE2 (10μM) that evoked an oscillatory Ca2+ signal for 25 minutes (Fig. 5C (a)). As a negative control, we also measured the basal levels of Ca2+ in 293 cells treated with Ca2+ free Hanks BSS and did not observe any intracellular Ca2+ signals (Fig. 5C (b)). Compared to untreated cells, LPA treatment (positive control), KSHV infection and PGE2 (10μM) increased the phosphorylation of p-Src, p-PI3K, p-PKCζ/λ, Akt 1/2, NFκB, and ERK 1/2 (Fig. 5D).
Identification of candidate PGE2 responsive elements on LANA-1 promoter
To determine the minimal LANA-1 promoter region responsive to exogenous PGE2, a sequential series of LANA-1 promoter deletion constructs were assayed in a luciferase reporter experiment in 293 cells (Fig. 6A and Fig.6B). We then examined the p-LANA-1-Luc sequence using Alibaba 2.1 TF software to characterize the transcription factor (TF) binding site profile (Fig. 6A). Exogenous PGE2 (10μM) activated pGL3.6, pGL3.4, and pGL3.3 promoter constructs at a similar level while, the pGL3.2 and pGL3.1 promoter constructs had significantly lower activities (Fig. 6B). Taken together, these results suggested that the promoter region located between −262bp and −159bp with candidate TFs such as YY1, Sp1, Oct-1, Oct-6, C/EBP, and c-Jun is required for PGE2 mediated LANA-1 promoter activity (Fig. 6A and 6B).
DISCUSSION
The novelty of our comprehensive study is the demonstration for the first time that a proinflammatory lipid metabolite, such as PGE2 and its receptors, plays a crucial role in herpes virus latency. Previous reports have indicated the role of Ca2+ in KSHV lytic cycle (17-20). In contrast, our studies showing the down-regulation of LANA-1 expression by EP1 receptor antagonist, the blockage of supernatant induced [Ca2+]i signal by EP1 antagonist, and the down-regulation PGE2 induced LANA-1 promoter activity by calcium chelators are the first demonstration of a role for [Ca2+]i in KSHV latency program. Unlike calcium, previous reports have shown the role of Src, PI3K, PKCζ/λ, and NFκB in KSHV latency program (21-23). However, the novelty of our study is that the data linking PGE2/EP receptors with KSHV LANA-1 expression and LANA-1 promoter through PGE2 via Src, PI3K, PKCζ/λ, and NFκB signal induction provides a new framework to understand the host mechanisms utilized by KSHV to induce these signal cascades. Furthermore, the promoter region we studied accounts for the transcripts of LANA-1, vFLIP, vCyclin, and some of the viral microRNAs (24, 25), and its induction by PGE2 together suggests that KSHV might be utilizing PGE2 via EP receptors for regulating the expression of other latency genes.
The down-regulation of COX-2 gene expression by EP2 and EP4 antagonist could either be due to the direct effect of signal cascades on the COX-2 gene, which has an inducible promoter (26) or due to the down-regulation of COX-2 mRNA transcript half-life that has been shown to be mediated by p38/MK2 dependent signaling acting on the ARE sequences in the 3′ UTR region of the COX-2 mRNA (27) The absence of any effect on COX-2 gene expression by EP receptor antagonism 24h post-treatment could be due to COX-2 promoter induction by other factors and suggest that KSHV utilizes multiple pathways with specific levels of temporal hierarchy to ensure the sustained activity of COX-2/PGE2/EP axis of inflammation including a positive feedback loop mediated through EP2 and EP4 receptor signaling.
The effect of exogenous PGE2 on LANA-1 promoter is the eventuality of numerous distinct yet related signal cascades converging on specific TFs (Fig. 6H), which are also probably used to the maintenance of host gene expression, such as COX-2, that is crucial for KSHV survival. Besides COX-2/PGE2, KSHV must be also utilizing several signature proinflammatory and angiogenic molecules that are induced during infection for the sustained induction of these signal networks (2, 28-32). Nevertheless, the present study demonstrating the PGE2/EP receptor utilization for latent gene expression is unique due to the fact that PGE2 is a lipid signal inducer that functions by autocrine and paracrine fashion at the site of synthesis with a circulating half-life of approximately 30 seconds and normal plasma levels varying from 3-15pg/ml (33). Despite the short half life, signaling events initiated by PGE2 through EP receptors are proposed to initiate an avalanche of temporal effects on cellular signaling such as Src, PI3K, PKCζ/λ, NFkB, and Ca2+ with immense oncogenic potential (6, 34, 35), which are the same signal pathways that are identified here to be regulated by PGE2/EP receptors in KSHV latency program.
The hallmark of KSHV interaction in human host, like in other herpes viruses, is the establishment of life long latency with periodic reactivation and reinfection. Successful reactivation and reinfection, however, is the consequence of the continuous tug of war between KSHV and the host immune system. Regardless of the outcome of this battle, maintenance and establishment of latency is crucial for herpes virus survival. Therefore, in the course of evolution, KSHV might have recalibrated the very purpose of inflammation from being a host response to eliminate the virus to the host mechanism that enables viral survival through the continuous production of inflammatory cytokines and growth factors. Pirating the pro-inflammatory lipid metabolite PGE2 and EP receptors for maintaining latency gene expression is a hallmark of such a phenomenon and thus gives a glimpse of the plasticity of the KSHV genome and also a novel paradigm shift in comprehending host mechanisms underlying KSHV latency. More excitingly, it adds a new paradigm in the understanding of the pathogenic mechanisms underlying chronic inflammation of KSHV associated KS lesions. Furthermore, the study also exposes a potential “Achilles heel” of KSHV pathogenesis wherein the use of anti-COX-2 and anti-EP receptor therapy could ameliorate KS by simultaneously controlling latency gene expression and chronic inflammation.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by Public Health Service grant CA 099925 and the RFUMS H. M. Bligh Cancer Research Fund to B.C and CA128560 to NSW. We thank Keith Philibert for critically reading the manuscript, Rita Levine for help in FACS analyses, Terri Li of University of Chicago for immunohistochemistry staining and Dr. Daniel Peterson and Scott Surridge of the RFUMS Core Confocal facility for help in immunohistochemistry imaging. We thank the NIH (NCI) AIDS Research and Reference Reagent Program for normal and KS specimens. We thank Dr. Rolf Renne (University of Florida) for the TIVE-LTC and TIVE cells and Dr. Yuan Chang (University of Pittsburgh) for LANA-1 promoter constructs.
References
- 1.Ablashi DV, Chatlynne LG, Whitman JE, Jr., Cesarman E. Spectrum of Kaposi’s sarcoma-associated herpesvirus, or human herpesvirus 8, diseases. Clin Microbiol Rev. 2002;15:439–64. doi: 10.1128/CMR.15.3.439-464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas JL, Gustin JK, Dezube B, Pantanowitz JL, Moses AV. Kaposi’s sarcoma: a model of both malignancy and chronic inflammation. Panminerva Med. 2007;49:119–38. [PubMed] [Google Scholar]
- 3.Ganem D. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu Rev Pathol. 2006;1:273–96. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 4.Naranatt PP, Krishnan HH, Svojanovsky SR, Bloomer C, Mathur S, Chandran B. Host gene induction and transcriptional reprogramming in Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8)-infected endothelial, fibroblast, and B cells: insights into modulation events early during infection. Cancer Res. 2004;64:72–84. doi: 10.1158/0008-5472.can-03-2767. [DOI] [PubMed] [Google Scholar]
- 5.Sharma-Walia N, Raghu H, Sadagopan S, et al. Cyclooxygenase 2 induced by Kaposi’s sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J Virol. 2006;80:6534–52. doi: 10.1128/JVI.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55:115–22. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majima M, Amano H, Hayashi I. Prostanoid receptor signaling relevant to tumor growth and angiogenesis. Trends Pharmacol Sci. 2003;24:524–29. doi: 10.1016/j.tips.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282:11613–17. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 9.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005;310:1504–10. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 10.Fulton AM, Ma X, Kundu N. Targeting prostaglandin E EP receptors to inhibit metastasis. Cancer Res. 2006;66:9794–97. doi: 10.1158/0008-5472.CAN-06-2067. [DOI] [PubMed] [Google Scholar]
- 11.Casibang M, Moody TW. AH6809 antagonizes non-small cell lung cancer prostaglandin receptors. Lung Cancer. 2002;36:33–42. doi: 10.1016/s0169-5002(01)00476-7. [DOI] [PubMed] [Google Scholar]
- 12.Piazuelo E, Jimenez P, Strunk M, et al. Effects of selective PGE2 receptor antagonists in esophageal adenocarcinoma cells derived from Barrett’s esophagus. Prostaglandins Other Lipid Mediat. 2006;81:150–61. doi: 10.1016/j.prostaglandins.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Sharma-Walia N, Paul AG, Bottero V, et al. Kaposi’s Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion. PLoS Pathog. 2010;6:e1000777. doi: 10.1371/journal.ppat.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–46. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi’s sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J Virol. 2004;78:3601–20. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An FQ, Folarin HM, Compitello N, et al. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi’s sarcoma-associated herpesvirus latency in vitro and in vivo. J Virol. 2006;80:4833–46. doi: 10.1128/JVI.80.10.4833-4846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupu-Meiri M, Silver RB, Simons AH, Gershengorn MC, Oron Y. Constitutive signaling by Kaposi’s sarcoma-associated herpesvirus G-protein-coupled receptor desensitizes calcium mobilization by other receptors. J Biol Chem. 2001;276:7122–28. doi: 10.1074/jbc.M006359200. [DOI] [PubMed] [Google Scholar]
- 18.Lagunoff M, Majeti R, Weiss A, Ganem D. Deregulated signal transduction by the K1 gene product of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1999;96:5704–09. doi: 10.1073/pnas.96.10.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano K, Isegawa Y, Zou P, Tadagaki K, Inagi R, Yamanishi K. Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded vMIP-I and vMIP-II induce signal transduction and chemotaxis in monocytic cells. Arch Virol. 2003;148:871–90. doi: 10.1007/s00705-002-0971-7. [DOI] [PubMed] [Google Scholar]
- 20.Zoeteweij JP, Moses AV, Rinderknecht AS, et al. Targeted inhibition of calcineurin signaling blocks calcium-dependent reactivation of Kaposi sarcoma-associated herpesvirus. Blood. 2001;97:2374–80. doi: 10.1182/blood.v97.8.2374. [DOI] [PubMed] [Google Scholar]
- 21.Sadagopan S, Sharma-Walia N, Veettil MV, et al. Kaposi’s sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J Virol. 2007;81:3949–68. doi: 10.1128/JVI.02333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma-Walia N, Naranatt PP, Krishnan HH, Zeng L, Chandran B. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J Virol. 2004;78:4207–23. doi: 10.1128/JVI.78.8.4207-4223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naranatt PP, Akula SM, Zien CA, Krishnan HH, Chandran B. Kaposi’s sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity. J Virol. 2003;77:1524–39. doi: 10.1128/JVI.77.2.1524-1539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79:14457–64. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staudt MR, Dittmer DP. Promoter switching allows simultaneous transcription of LANA and K14/vGPCR of Kaposi’s sarcoma-associated herpesvirus. Virology. 2006;350:192–205. doi: 10.1016/j.virol.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Chandrasekharan NV, Simmons DL. The cyclooxygenases. Genome Biol. 2004;5:241–46. doi: 10.1186/gb-2004-5-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–74. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki Y, Jaffe ES, Chang Y, et al. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–43. [PubMed] [Google Scholar]
- 29.Carroll PA, Brazeau E, Lagunoff M. Kaposi’s sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology. 2004;328:7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ensoli B, Barillari G, Gallo RC. Cytokines and growth factors in the pathogenesis of AIDS-associated Kaposi’s sarcoma. Immunol Rev. 1992;127:147–55. doi: 10.1111/j.1600-065x.1992.tb01412.x. [DOI] [PubMed] [Google Scholar]
- 31.Sadagopan S, Sharma-Walia N, Veettil MV, et al. Kaposi’s sarcoma-associated herpesvirus upregulates angiogenin during infection of human dermal microvascular endothelial cells, which induces 45S rRNA synthesis, antiapoptosis, cell proliferation, migration, and angiogenesis. J Virol. 2009;83:3342–64. doi: 10.1128/JVI.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivakumar R, Sharma-Walia N, Raghu H, et al. Kaposi’s sarcoma-associated herpesvirus induces sustained levels of vascular endothelial growth factors A and C early during in vitro infection of human microvascular dermal endothelial cells: biological implications. J Virol. 2008;82:1759–76. doi: 10.1128/JVI.00873-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzpatrick FA, Aguirre R, Pike JE, Lincoln FH. The stability of 13,14-dihydro-15 keto-PGE2. Prostaglandins. 1980;19:917–31. doi: 10.1016/0090-6980(80)90126-4. [DOI] [PubMed] [Google Scholar]
- 34.Ansari KM, Rundhaug JE, Fischer SM. Multiple signaling pathways are responsible for prostaglandin E2-induced murine keratinocyte proliferation. Mol Cancer Res. 2008;6:1003–16. doi: 10.1158/1541-7786.MCR-07-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poligone B, Baldwin AS. Positive and negative regulation of NF-kappaB by COX-2: roles of different prostaglandins. J Biol Chem. 2001;276:38658–64. doi: 10.1074/jbc.M106599200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.