Summary
The amino acid sequence of viral capsid proteins contains information about their folding, structure and self-assembly processes. While some viruses assemble from small preformed oligomers of coat proteins, other viruses such as phage P22 and herpesvirus assemble from monomeric proteins (Fuller and King, 1980; Newcomb et al., 1999). The subunit assembly process is strictly controlled through protein:protein interactions such that icosahedral structures are formed with specific symmetries, rather than aberrant structures. dsDNA viruses commonly assemble by first forming a precursor capsid that serves as a DNA packaging machine (Earnshaw, Hendrix, and King, 1980; Heymann et al., 2003). DNA packaging is accompanied by a conformational transition of the small precursor procapsid into a larger capsid for isometric viruses. Here we highlight the pseudo-atomic structures of phage P22 coat protein and rationalize several decades of data about P22 coat protein folding, assembly and maturation generated from a combination of genetics and biochemistry.
Keywords: virus assembly, protein folding, bacteriophage, capsid protein, HK97-fold, suppressor
Morphogenesis of bacteriophage P22
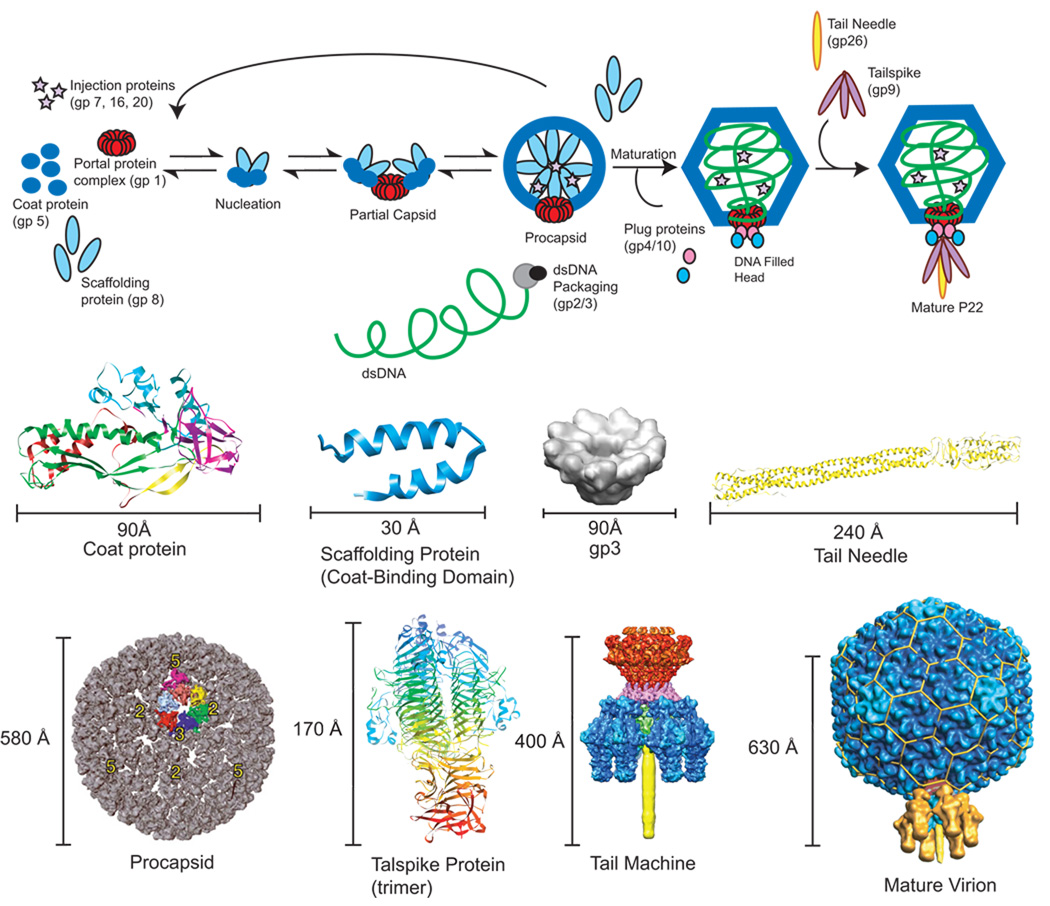
The morphogenic pathway of the T=7 Salmonella bacteriophage P22 involves the co-assembly of 415 molecules of monomeric coat protein (product of gene 5; gp5) with about 60–300 molecules of scaffolding protein (gp8) as well as some minor proteins (gp7, 16 and 20, referred to as ejection proteins) and the portal protein complex (gp1) into a procapsid (King et al., 1976; Prevelige and King, 1993) (Figure 1, Table 1). Scaffolding protein directs the assembly of the procapsid but is not found in mature phage, and so acts as a true assembly chaperone. Without scaffolding protein, coat protein assembles into T= 4 capsids and aberrant spiral structures. Spirals have pentameric capsomers located inappropriately, and not at icosahedral 5-fold vertices, so that closed procapsid structures do not form (Earnshaw and King, 1978; Lenk et al., 1975; Thuman-Commike et al., 1998). The minor ejection proteins are incorporated early in assembly, also through interactions with scaffolding protein (Greene and King, 1996). The dsDNA genome is actively packaged in an ATP-dependent fashion into the procapsid through the unique portal vertex (Bazinet and King, 1988) by the terminase complex (gp2 and gp3). Concomitant with DNA packaging, scaffolding protein exits from the procapsid to take part in additional rounds of assembly, and the capsid matures, resulting in a 10–15% increase in volume (Earnshaw, Casjens, and Harrison, 1976; Prasad et al., 1993). The dsDNA is stabilized by the addition of plug proteins (gp4, 10 and 26) that close the portal vertex, and finally tailspikes (gp9), the cell recognition and attachment proteins, are added (King, Lenk, and Botstein, 1973). None of the proteins are covalently modified or proteolyzed during folding and assembly.
Figure 1. P22 assembly pathway including known structures.
Shown in the top panel is the morphogenic pathway of phage P22. Below are the corresponding structures as follows: P22 coat protein in the procapsid state determined by cryo-EM homology modeling (PDB entry 3IYI) (Parent et al., 2010); Scaffolding protein C-terminal coat-protein binding domain NMR structure (PDB entry 2GP8) (Sun et al., 2000); Procapsid cryo-EM density map (Jiang et al., 2003); gp3 terminase cryo-EM density map (modified from Nemecek et all 2008, JMB) (Nemecek et al., 2008); Tail Needle (gp26 trimer) crystal structure (PDB entry 2POH) (Olia, Casjens, and Cingolani, 2007); Tailspike trimer crystal structure (PDB entry 1TSP) (Steinbacher et al., 1994); asymmetric cryo-reconstruction of tail machine complex (modified from Lander et al 2009) (Lander et al., 2009); asymmetric cryo-reconstruction of full virion (modified from Lander et al 2006, Science) (Lander et al., 2006).
Table 1.
Composition of P22 Procapsids and Capsids
| Component | Role | Location† | # Copies | Mass (kDa)* | Structure†† | |
|---|---|---|---|---|---|---|
| gp1 | Portal | P | C | 12 | 82.7 | Cryo-EM |
| gp4 | Tail plug | C | 12 | 18.0 | Cryo-EM | |
| gp5 | Major capsid | P | C | 415 | 46.7 | Cryo-EM |
| gp7 | Ejection (pilot) | P | C | 12–20 | 23.4 | N.A. |
| gp8 | Scaffolding | P | 60–300 | 33.6 | NMR** | |
| gp9 | Tailspike | C | 18 | 71.9 | X-ray/CryoEM | |
| gp10 | Tail plug | C | 6 | 52.5 | Cryo-EM | |
| gp16 | Ejection (pilot) | P | C | 12–20 | 64.4 | N.A. |
| gp20 | Ejection (pilot) | P | C | 12–20 | 50.1 | N.A. |
| gp26 | Tail Needle | C | 3 | 24.7 | X-ray/Cryo-EM | |
| dsDNA | Genome | C | 1 | ~26,500a | Cryo-EM | |
| Procapsid | Precursor capsid | P | 1 | ~30,300 | Cryo-EM | |
| Capsid | Infectious virion | C | 1 | ~50,600 | Cryo-EM | |
Location of component in either the procapsid (P) or the capsid (C)
The mass calculated from the translated amino acid sequence (Pedulla et al., 2003).
N.A. = information not available
Scaffolding protein capsid-binding domain (residues 238–303).
the total genome size is 41724 base pairs
Phage P22 has long been a model used for understanding assembly mechanisms of dsDNA viruses (Capen and Teschke, 2000; Casjens, 1979; Casjens et al., 1985; Fuller and King, 1980; Fuller and King, 1981; Fuller and King, 1982; Prevelige, Thomas, and King, 1993; Prevelige, Thomas, and King, 1988). An important advantage of phage P22 as an assembly model is its simplicity. The proteins needed for assembling procapsid-like particles (PLP) of P22 can be purified and are active for assembly (Fuller and King, 1981). In vitro, only coat and scaffolding proteins are required to assemble a PLP (Fuller and King, 1980; Prevelige, Thomas, and King, 1988). Both in vivo and in vitro, the number of scaffolding protein molecules incorporated into a procapsid (or a PLP) varies, depending on the conditions of assembly (Casjens et al., 1985; Parent, Ranaghan, and Teschke, 2004; Prevelige, Thomas, and King, 1993). In vitro assembled PLPs have the same morphology and size as in vivo generated procapsids (Fuller and King, 1980). Thus, data generated from in vitro assembly experiments can be readily compared to in vivo experiments, allowing the rationalization of phage phenotypes due to coat variants with observable differences in folding and assembly of coat protein. A nearly complete structure of the coat protein is now available (Parent et al., 2010), which allows us to examine the structure/function relationship of the coat protein during the entire phage life cycle.
Similarities and differences in the morphogenesis of phages P22 and HK97
HK97 is currently the only example of a bacteriophage where near-atomic structures (better than 4 Å resolution, solved by X-ray crystallography) are available for both precursor and mature particles, allowing for a detailed understanding of the mechanisms of assembly and maturation (Gertsman et al., 2009; Wikoff et al., 2000). P22 (family Podoviridae) and HK97 (family Siphoviridae) have much in common in terms of capsid morphology, assembly and maturation (Johnson and Chiu, 2007). Although the classification into separate families suggests a distant evolution (especially evident in the dramatic differences in tail appendages), their capsid assembly pathways are generally analogous. Both phage have nearly icosahedral lattices that exhibit T=7 laevo symmetry, with a portal complex at a unique vertex. The structures of their respective coat proteins are highly homologous (Parent et al., 2010) and the “HK97-fold” is often considered to be a common ancestor for many dsDNA phage, including P22 (Bamford, Grimes, and Stuart, 2005).
In HK97, the formation of the precursor particle (Prohead I) occurs by co-polymerization of the major capsid protein (gp5), which exists in equilibrium between pre-assembled hexons and pentons, and a protease (gp4)(Duda et al., 1995a; Xie and Hendrix, 1995). Once assembly of Prohead I is complete, proteolysis of the first 103 residues (the Δ-domain, which functions as a scaffold for assembly) converts gp5 to gp5*, forming Prohead II (Duda et al., 1995b). Maturation is triggered by dsDNA packaging and proceeds through at least three expansion intermediates (EI I, II, and III) (Lata et al., 2000). HK97 morphogenesis culminates in formation of Head II, characterized by extensive chainmail crosslinking (Popa et al., 1991). The presence of these multiple, discrete structures has made HK97 a model system, as detailed information about maturation has been gleaned (Conway et al., 1995; Gan et al., 2006; Ross et al., 2006).
Although many general features of bacteriophage assembly are common to both systems, there are some important distinctions between P22 and HK97. The major differences include: 1) P22 assembles from monomeric coat proteins added one at a time through the aid of a separately expressed scaffolding protein, gp8 (Casjens and King, 1974; Prevelige, Thomas, and King, 1988); 2) P22 maturation does not involve proteolytic cleavage of coat protein; and 3) P22 capsids are not stabilized by chemical crosslinks. Though the mechanisms of subunit and capsid stabilization differ between P22 and HK97 (Parent et al., 2010), the common HK97-fold indicates that this building block is well suited to promote effective assembly of a capsid. In addition, as covered in detail in this review, P22 coat protein can tolerate variation and can adapt under various environmental pressures. The widespread functionality, and ability to tolerate insertions and adaptations may be the reason this protein fold is so prevalent in dsDNA tailed bacteriophage.
P22 capsid structures-- a brief history
The first complete description of P22 head assembly used amber mutations in genes that code for head assembly products to determine which products would accumulate in the absence of each head protein (Botstein, Waddell, and King, 1973). Procapsids, filled phage heads, empty heads, and some aberrant structures were observed. These particles were further characterized using small angle X-ray scattering (SAXS) (Earnshaw, Casjens, and Harrison, 1976), which showed that procapsids are smaller in diameter (584 Å) than phage or empty heads (628 Å). Treatment of procapsids with 0.8% SDS caused them to expand to the diameter of phage heads. Expansion was postulated to be an important regulatory step for DNA packaging and scaffolding protein release. For the first time, in vitro assembly from purified subunits was shown to recapitulate in vivo procapsid assembly, resulting in PLPs with the same morphology and dimensions, but lacking the portal complex (Fuller and King, 1981; Prevelige, Thomas, and King, 1988). The first structures of empty procapsid shells (procapsids isolated from phage infected cells that are stripped of scaffolding protein, portal, and minor proteins) and DNA filled heads (phage without the tail machinery) examined by three-dimensional (3D) image reconstruction of vitrified samples to 28 Å, using icosahedral averaging, confirmed the differences in size and morphology that had been observed earlier (Prasad et al., 1993). The procapsid reconstruction revealed large holes at the center of the capsomers, where scaffolding protein was hypothesized to exit during DNA packaging. In addition, the reconstruction of the head showed clear conformational rearrangements that occurred during expansion, which closed the holes primarily through large shifts of coat protein mass.
Reconstruction of in vivo generated procapsids, which contain scaffolding protein, revealed that most of the scaffolding protein was not icosahedrally ordered within the procapsid (Thuman-Commike et al., 1996). However, a small region of scaffolding protein density could be seen on the interior surface of procapsids at the tips of some coat subunits. Scaffolding dimers were proposed to link coat subunits across the strict and quasi-three fold axes of symmetry, thereby causing interactions between subunits in adjacent capsomers and driving icosahedral assembly (Thuman-Commike et al., 1996; Thuman-Commike et al., 1999).
The first identification of P22 coat protein secondary structure was accomplished with subnanometer reconstructions of DNA filled heads (9.5 Å) and empty procapsid shells (8.5 Å), which had delineated subunit interfaces so that each coat subunit could be distinguished in the capsid lattices (a lattice refers to the arrangement of coat protein in the icosahedron) (Jiang et al., 2003). Several α-helices were assigned based on the density and revealed significant conformational changes and coat protein refolding during expansion. In these reconstructions, P22 coat protein was shown to have a fold similar to the HK97 capsid protein, in spite of the low sequence identity between the proteins (<10 %). Though this was a major improvement in resolution, and added to the growing knowledge that the HK97 fold is common to dsDNA tailed phage and viruses, threading the complete P22 protein sequence through the electron density was not yet possible.
More recently, two asymmetric reconstructions of phage P22 were completed (Chang et al., 2006; Lander et al., 2006). Asymmetric reconstructions differ from icosahedrally-averaged reconstructions in that no impositions of symmetry are made. This technique allows the visualization of nucleic acids and proteins that are not icosahedrally localized in a capsid such as dsDNA, the portal complex, ejection proteins and the tail machinery. Furthermore, these reconstructions revealed subunit details such as the symmetry mismatch between the portal complex and the capsid, highlighting structural interactions between P22 coat protein and portal subunits that were not visualized in maps produced from icosahedrally averaged reconstructions. However, these reconstructions (~17 Å resolution) were not at a high enough resolution to trace the backbone of coat protein.
P22 coat protein subunit pseudo-atomic structure
One important prerequisite to attaining a pseudo-atomic structure by cryo-TEM is to have a pure, homogenous sample. A conformational transition similar to the expansion caused by DNA packaging in vivo can be induced in vitro (Earnshaw, Casjens, and Harrison, 1976; Galisteo and King, 1993). During heat expansion the pentons are released but the overall structure of the particle corresponds to the mature form (Teschke, McGough, and Thuman-Commike, 2003). We took advantage of the high stability of the heat-expanded heads, which allowed us to remove all the minor proteins leaving a very pure sample (Parent et al., 2010). The heat-expanded capsid is structurally simple, having only coat protein only in hexon positions. Recently structures of in vitro heat-expanded heads and empty procapsid shells were reported at a resolution of 7.0 Å and of 9.1 Å, respectively (Parent et al., 2010).
The majority of the backbone of P22 coat protein (~91 %) was separately traced through electron density maps of both structures. To accurately guide the threading of the amino acid sequence through the electron density, landmarks were required. The amino-terminus (amino acids 1–42) was identified by difference map analysis of protease-treated compared to untreated expanded heads. Protease treatment removes the first 42 amino acids of coat protein (Parent et al., 2010). Other threading landmarks were generated via engineered cysteine variants within coat protein. Procapsids of these variants were labeled with 1.4 nm gold beads covalently modified with maleimide. The location of electron dense gold beads was readily observed by difference map analysis for three procapsid shell reconstructions. Together, these guideposts allowed us to use the density maps from expanded heads and procapsids along with iterative homology modeling to produce a pseudo-atomic model of coat protein before and after maturation (PDB entries 3IYI, and 31YH respectively) (Parent et al., 2010).
Comparing capsid and procapsid subunit structures
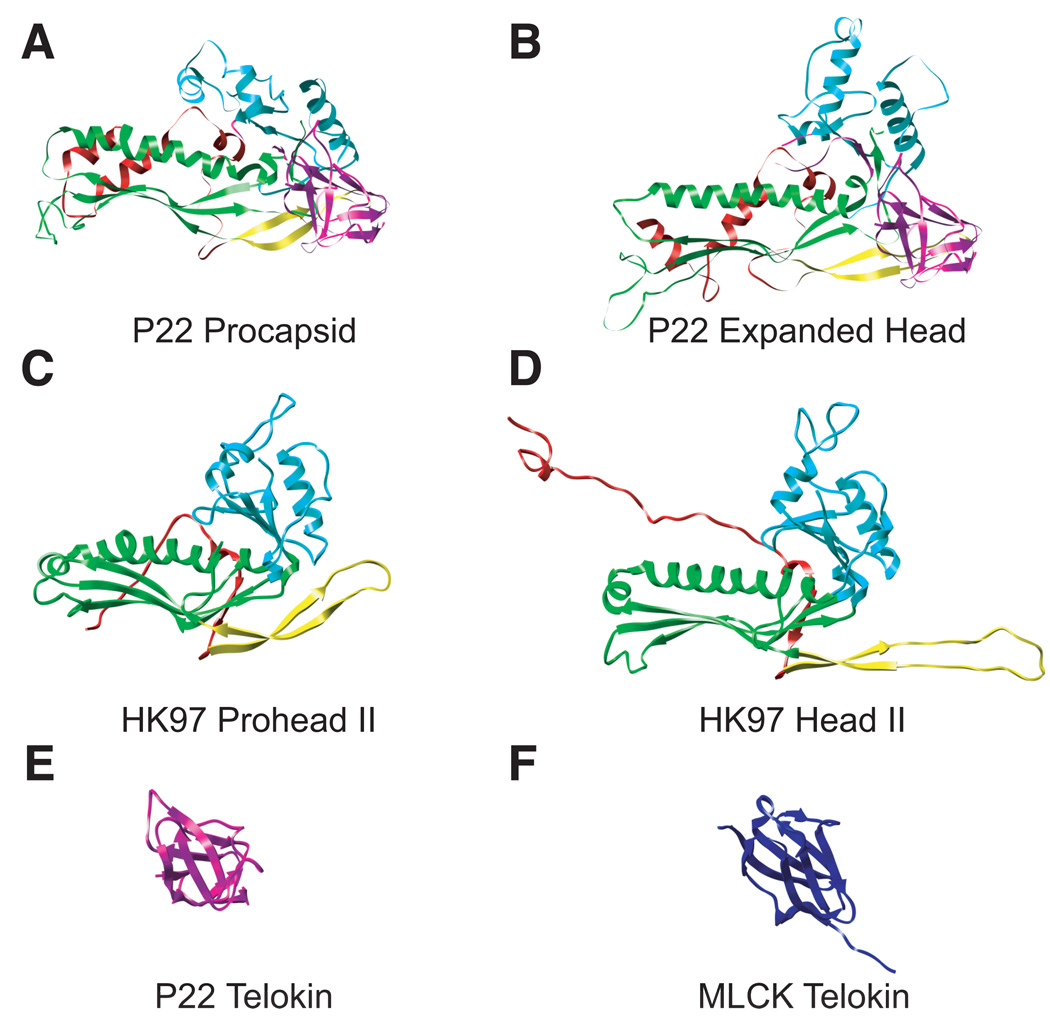
The P22 capsid and procapsid subunit backbone structures are shown as ribbon diagrams next to HK97 subunits (Head II PDB entry 1OHG and Prohead II PDB entry 3E8K) (Figure 2a–d). In HK97, the domains are named according to their location in the capsomer. The A domain is axial to the center, the P domain is found at the periphery, the N-arm at the amino terminus, and the E-loop is the ‘elongated’ loop where the subunits undergo cross-linking (Wikoff et al., 2000). The corresponding regions of P22 coat protein are labeled in the convention established for the HK97 capsid protein (Gertsman et al., 2009); however, the cross-linking reaction does not occur in P22, and the E-loop is shorter. P22 coat protein has the HK97 fold for ~68% of the protein, which we will refer to as the “core” of P22 coat protein. The major structural difference between P22 and HK97 is a telokin domain inserted in the P22 coat protein (residues 223–359 as determined through homology modeling), which is the protrusion called the “extra density” domain in Jiang et al., 2003. Telokin is a member of the immunoglobulin-like (Ig) family of domains, which are found in many phage proteins, primarily as additions to either the capsid or tail proteins (Fraser, Maxwell, and Davidson, 2007; Fraser et al., 2006). We show the telokin domain of P22 coat protein next to the telokin domain from myosin light chain kinase (PDB entry 1FHG) (Figure 2e and 2f).
Figure 2. P22 has an HK97-like body and a telokin-like addition.
P22 coat protein in both the immature state “procapsid” (A) and mature state “expanded head” (B) is compared to HK97 in comparable states, “Prohead II” (C), or “Head II” (D). The common structural elements are colored correspondingly; N-arm (red), E-loop (yellow) P-domain (green), A-domain (cyan). P22 has a unique telokin domain colored in magenta. The P22 telokin domain (E) is compared to the telokin domain from myosin light chain kinease “MLCK” (PDB entry 1FHG)(Holden et al., 1992)(F).
As mentioned above, several lines of evidence have indicated that there are conformational rearrangements and coat protein refolding during expansion (Jiang et al., 2003; Kang and Prevelige, 2005), and now we can see that these rearrangements primarily occur in the A-domain, the P-loop that connects the strands in the P-domain, and the N-arm. In the supplementary movie we compare the structure of the N-arm plus the P-loop and the A domain to highlight the conformational rearrangements that occur during expansion. During expansion, the first helix in the N-arm (residues 3 – 11) and the P-loop (residues 376 – 398) are drawn closer together by a combination of events where the helix moves to a higher radius, and the P-loop moves to a lower radius. The radii described are relative to the particle origin, where a lower radius refers to a position closer to the particle interior, and a higher radius is toward the outside of the particle. Together these make significant inter-subunit contacts, which likely contribute to the increased stability of the expanded head. In the A-domain (residues 127 – 222) there is a significant movement that closes the hole at the center of the capsomers, along with some refolding of the protein from coil to helix.
The observed conformational rearrangements must be sufficient to stabilize P22 capsids at strict and quasi three-fold axes since P22 coat protein does not crosslink in the fashion of HK97 (Wikoff et al., 2000) nor require accessory capsid proteins, as seen with Herpesviruses, T4 and lambda phage, as well as (Ishii, Yamaguchi, and Yanagida, 1978; Spencer et al., 1998; Yang et al., 2000). Stabilization upon maturation is likely needed in P22, since procapsids are labile enough that they slowly dissassemble at low concentration over a period of days (Parent, Suhanovsky, and Teschke, 2007a). Additionally, monomers can exchange with assembled coat subunits, which suggests a relatively weak intrasubunit interaction energy. In fact, the energy is only −6 to −7 kcal/mol (Parent, Zlotnick, and Teschke, 2006). Addition of all of the coat subunit interactions in the procapsid leads to a robust lattice, with an overall energy of ~ −3000 kcal/mol (Parent, Zlotnick, and Teschke, 2006). Expansion locks the subunits at the quasi and strict three fold axes and dramatically increases capsid stability as dissassociation of expanded heads has not been observed. The expanded capsid lattice even resists disruption in 7 M urea, indicating strong hexon:hexon interactions (Parent et al., 2010). However, the interaction energy between the penton subunits and their neighboring hexon subunits can be overcome with sufficient thermal energy, resulting in loss of pentons during heat expansion (Teschke, McGough, and Thuman-Commike, 2003). Complete heat denaturation of P22 empty capsids requires only slighty less energy than for empty HK97 heads; the Tm for WT P22 capsids is 82.5 °C (Capen and Teschke, 2000). Therefore, P22 is able to achieve high stability through maturation even in the absense of intersubunit crosslinks.
The capsid maturation reaction
Significant step-wise stabilization occurs as coat protein assembles from monomers to form procapsids, and then matures (Anderson and Teschke, 2003; Galisteo, Gordon, and King, 1995; Galisteo and King, 1993). One initial conformational change involves the A domain (residues 127–222). Amino acids 156–207 of coat protein are protease accessible in monomers and procapsids. The time required for digestion is much longer for procapsids indicating a change in accessibility to proteases and H/D exchange upon assembly (Capen and Teschke, 2000; Lanman, Tuma, and Prevelige, 1999; Teschke, 1999). However, the A domain becomes protease resistant and inaccessible to H/D exchange only once expanded to the mature form (Capen and Teschke, 2000; Kang and Prevelige, 2005; Lanman, Tuma, and Prevelige, 1999). The subunit structure in both procapsid and expanded heads agrees with Raman spectroscopy, which indicates only small changes in coat protein secondary structure during maturation (Tuma, Prevelige, and Thomas, 1998). The large hole at the center of the capsomers becomes occluded by the A-domain rearrangements in expansion (Jiang et al., 2003; Prasad et al., 1993).
Procapsids expand when they have been cleaved in the A-domain with protease at a lower temperature than uncleaved procapsids (Lanman, Tuma, and Prevelige, 1999). These data show that the resulting decrease in conformational constraint of the A domain allows expansion to occur more readily by lowering the activation energy for this process. Thus, we conclude that movement and refolding of the A domain may be the rate-limiting step for capsid maturation.
Coat protein monomers
Assembly of phage P22 procapsids occurs via the addition of coat monomers to the growing edge of the lattice (Fuller and King, 1982; Prevelige, Thomas, and King, 1988). Coat monomers have been extensively studied (Anderson and Teschke, 2003; Fuller and King, 1981; Teschke and King, 1993; Tuma et al., 2001). Assembly-competent monomers are generated by denaturation of empty procapsid shells, followed by extensive dialysis to refold the protein (Anderson and Teschke, 2003; Fuller and King, 1982; Prevelige, Thomas, and King, 1988). In the absence of scaffolding protein, coat protein mostly remains monomeric at low concentrations, though a very slow, uncontrolled assembly reaction does occur with over of days at 20° C (Suhanovsky, unpublished). The overall thermodynamic stability of native WT coat monomers, determined by equilibrium urea denaturation experiments, is only ~ −7 kcal/mol (Anderson and Teschke, 2003), which is rather unstable for a protein the size of coat protein (Fersht, 1999; Finn et al., 1992)
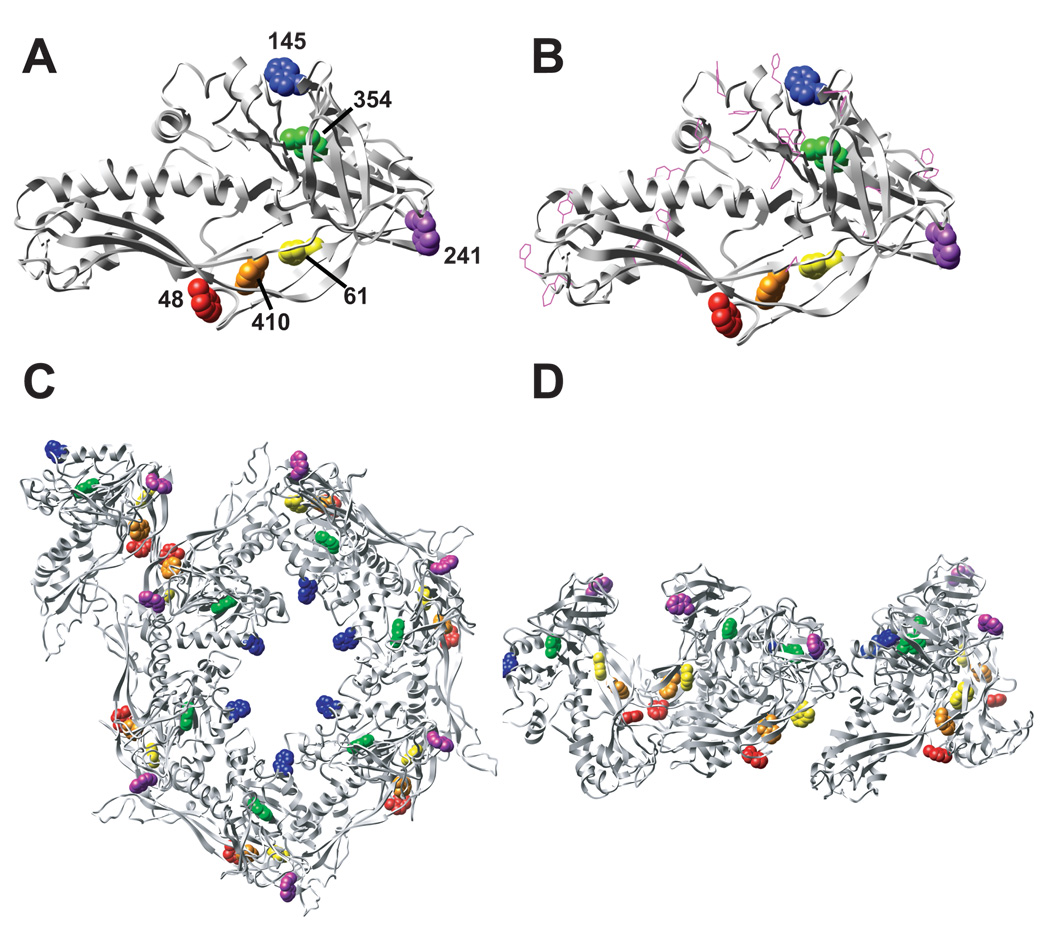
The pseudo-atomic model (Parent et al., 2010) provides an excellent approximation of the structure, but it is not an accurate, full atomic description of the coat protein. Nevertheless, it can be used to rationalize previous biochemical experiments. For example, four of the six tryptophans in the P22 subunit appear to be in quite similar environments, likely interacting with phenylalanines or other tryptophans (Figure 3b). Trps W48, W61 and W410 may cluster in the P-domain. Trp W354 (P-domain) is potentially in a hydrophobic environment with several phenylalanine and tryrosine residues in close proximity. W145 (A-domain) and W241 (telokin domain) are probably solvent exposed in monomers, therefore quenched and do not contribute to the fluorescence. Coat protein tryptophans have been shown by fluorescence lifetime measurements and Raman spectroscopy to reside in equivalent environments in folded monomers (Prevelige, King, and Silva, 1994; Tuma et al., 2001), which is consistent with the positions of the tryptophans in the assembled subunit. W48 and W410 appear be involved in contacts between capsomers (Figure 3c and d) and we hypothesize that this explains the observed increase in fluorescence upon assembly (Teschke and King, 1993). W48 contributes to the entropic stabilization of procapsids, but not monomers, consistent with its location at a subunit interface (Prevelige, King, and Silva, 1994).
Figure 3. Location of tryptophans in P22 coat protein.
One subunit of P22 coat protein (procapsid state) with the 6 tryptophan residues shown as spheres and color-coded based on location (B). Same view of the coat protein as in panel A, but including phenylalanines and tyrosines as magenta-colored ball and stick atoms. The location of all of the tryptophans in one asymmetric unit (6 hexon subunits and one penton subunit) in the procapsid state from a top down view. Tryptophans are color-coded according to A. (D) Same as C, but a side view.
SAXS experiments revealed that coat protein monomers are ~ 100 Å long (Tuma et al., 2001). Since the end-to-end distance of the coat subunit in procapsids is ~ 87 Å (Parent et al 2010), coat protein is only slightly more compact upon assembly. In addition, only 4 % of peptide backbone is altered when monomers assemble, indicating that assembly does not induce major folding changes (Tuma et al., 2001). Together, these data suggest that the coat protein subunit structure as determined from procapsids is an approximation of the monomer structure, therefore, we will use it to analyze previously collected data on coat monomer folding and assembly.
“Let the phage do the work”
“Let the phage do the work” is a quote from Professor Jonathan King, an expert in phage P22 genetics and biochemistry. By this he meant that results from phage genetics could guide biochemical experiments, and lead to an understanding of phage protein function. For example, over the last several decades many P22 coat protein variants have been investigated for their effects on folding and assembly. The first coat protein mutants were isolated during searches for mutants in all phage P22 genes and were used to elucidate the sequence of events that occur during phage morphogenesis (Botstein, Waddell, and King, 1973; Jarvik and Botstein, 1973; Jarvik and Botstein, 1975; King, Lenk, and Botstein, 1973). Phage were generally mutagenized with nitrosoguanosine or ultraviolet light and screened for conditional-lethal and lethal phenotypes: temperature-sensitive (conditional lethal, “ts”), cold-sensitive (conditional lethal, “cs”), and amber mutants (lethal “am”). The position of each mutation was mapped by complementation to the P22 circular genome (Gough and Levine, 1968; Kolstad and Prell, 1969; Lew and Roth, 1970). Mutated phage were used to determine which assembly products accumulated in infections using conditional-lethal or lethal mutants. Thus, with phage carrying mutations in the late genes, which are involved in capsid assembly and completion, procapsids were shown to be precursors to DNA filled heads, followed by the addition of head completion proteins.
Temperature-sensitive-folding (tsf) variants of coat protein
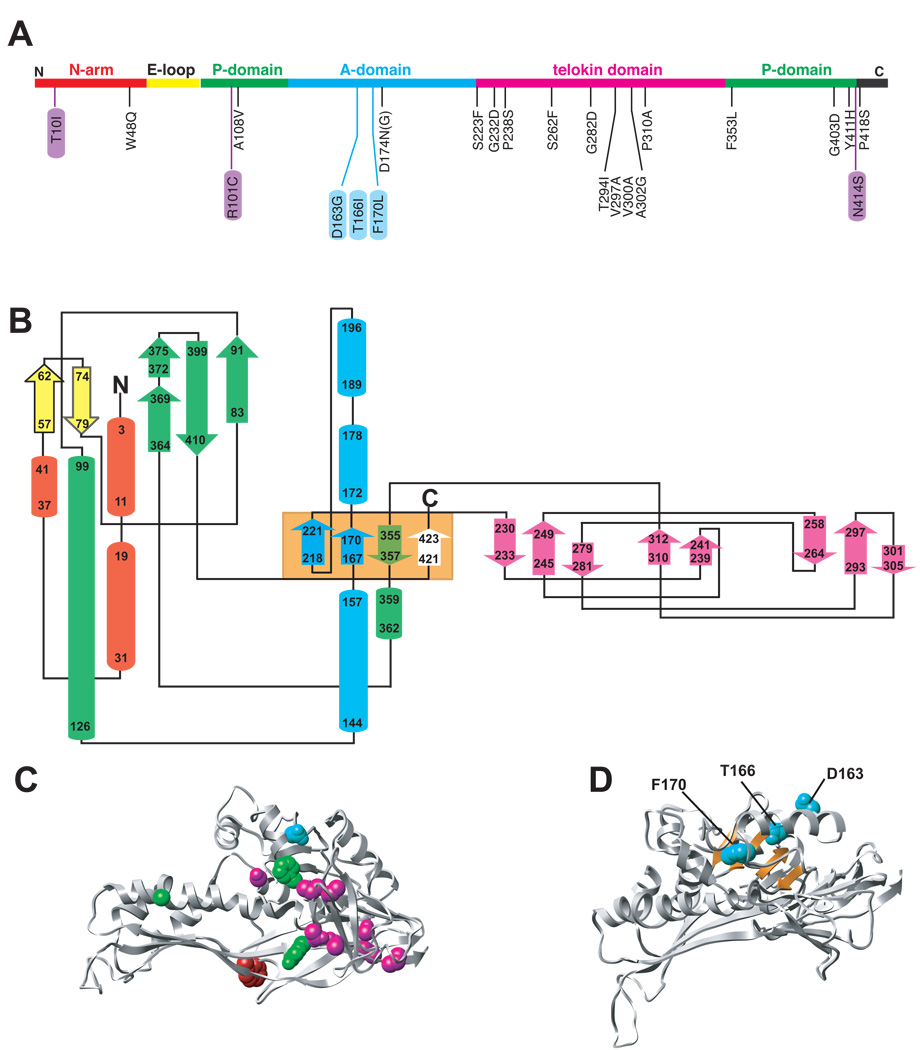
Eighteen amino acid substitutions in P22 coat protein result in a tsf phenotype (Figure 4a). In vivo, tsf substitutions significantly reduce the yield of soluble coat protein at high temperatures because the newly synthesized tsf coat polypeptides aggregate and form inclusion bodies prior to reaching an assembly-competent state (Gordon et al., 1994; Nakonechny and Teschke, 1998). Thus, these positions are crucial for the proper folding of coat protein. Such tsf defects lead to a decrease in the rate and yield of capsid assembly both in vitro and in vivo. The folding and assembly of the tsf coat proteins can be rescued at high temperature in vivo by growing phage in cells that overproduce GroEL/S (Gordon et al., 1994; Nakonechny and Teschke, 1998).
Figure 4. Location of conditional lethal variants in P22.
Shown in panel A is the primary sequence of P22 with the “backbone” color-coded by domain as in Figure 2 highlighting the 18 tsf mutations (black), 3 cs mutants (purple) and 3 global su mutations (blue). Panel B shows a topology diagram of P22 coat protein obtained through PDBsum (Laskowski et al., 1997) with secondary structural elements represented as cylinders (helices) and arrows (sheets) color coded according to domain. The starting and ending residues in each element are indicated as well as the N and C termini. The orange box designates four β-strands that comprise the β-hinge. C) Ribbon diagram of one subunit with the 17 positions of tsf mutations shown as spheres and color coded according to domain. D) Ribbon diagram of one subunit showing the β-hinge (orange) and the WT residues at the positions of the 3 global su substitutions as cyan spheres.
In vitro, tsf coat protein monomers have altered secondary and tertiary structure, as well as increased surface hydrophobicity, as compared to WT coat protein (Doyle et al., 2003). The major effect of the tsf substitutions is a destabilization of the monomers, which rapidly “flicker” between the folded state and unfolding intermediates. Because of the increased population of intermediate species, when tsf coat protein monomers are incubated with GroEL in vitro they are bound efficiently (Doyle et al., 2003). The coat proteins in the binary complex showed conformations with substantial secondary and tertiary structure, similar to a late folding intermediate (de Beus, Doyle, and Teschke, 2000).
The relative positions of the known conditional lethal mutations are highlighted in the primary sequence, and in topology and ribbon diagrams (Figure 4a–d). The substitutions appear to be well distributed in the primary sequence (Figure 4a), but close inspection of the topology diagram and the folded subunit clearly shows that the majority of tsf substitutions cluster in the telokin domain (Figure 4b and 4c). Three other substitutions are found in either the P- or A-domains. In addition, three tsf substitutions near the C-terminus reside in a ‘β-hinge’ that connects the A- and telokin domains. This region comprises four anti-parallel β-sheets (Figure 4c, center light orange box).
Suppressors of coat protein folding defects
Intragenic second site suppressors (su) of the tsf gene 5 mutations were isolated by plating tsf phage at elevated temperatures to identify additional residues involved in coat protein folding (Aramli and Teschke, 1999). Three global su substitutions (D163G, T166I, F170L) were repeatedly and independently isolated from different gene 5 tsf parents, scattered throughout the gene. The tsf:su coat proteins demonstrate both stabilized native and intermediate states as compared to their tsf parents, so that these proteins remain GroEL substrates even though their folding is less defective (Doyle et al., 2004; Parent and Teschke, 2007). The three global su substitutions are located in the β-hinge, which implicates this region as important for folding (Figure 4d).
Coat protein folding
WT coat protein
The folding of WT coat protein monomers has been thoroughly investigated (Anderson and Teschke, 2003; Teschke and King, 1993). In equilibrium urea denaturation experiments, coat protein is incubated in 0 – 7 M urea until equilibrium is established, and the unfolding transition is monitored by circular dichroism (CD) at 222 nm for secondary structure and tryptophan fluorescence (trp) at 340 nm as a marker of tertiary structure. A three state model best describes the reversible folding transition of coat protein: native (N) ⇔ intermediate ensemble (I) ⇔ unfolded ensemble (U). The equilibrium intermediate ensemble is characterized by the loss of secondary structure but native-like tryptophan fluorescence, which would typically suggest the existence of tertiary structure in the intermediate ensemble (Anderson and Teschke, 2003). Here this is more likely the result of clustering of tryptophans.
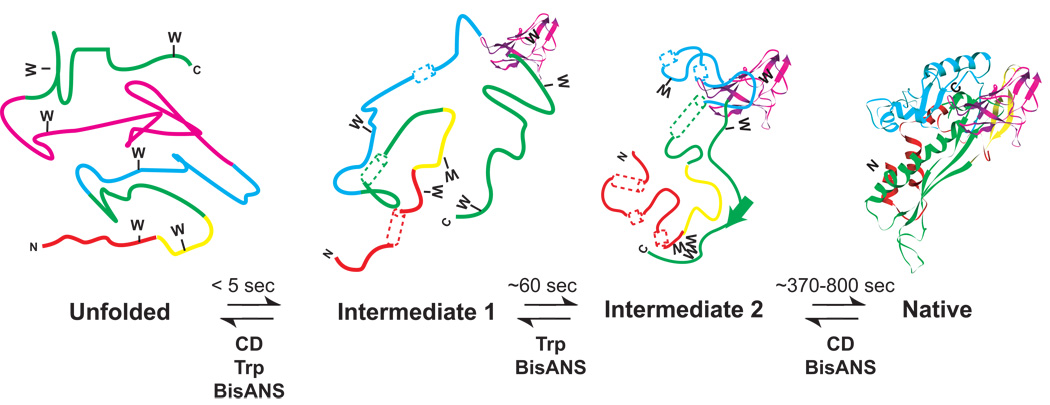
When unfolded coat protein is diluted into buffer that promotes folding, 80% of the coat protein molecules follow a slow kinetic path. Kinetic folding experiments have demonstrated the existence of two kinetic intermediates (Doyle et al., 2003), so that coat protein must pass through four distinct states during folding (Figure 5). The U ⇔ I1 transition is observed in both trp fluorescence and CD experiments; 65% percent of the total trp fluorescence change and 33% percent of secondary structure is formed in this ‘burst phase’ (< 5 sec) leading from U to I1. I2 is a later kinetic intermediate, characterized by fully buried tryptophans but incomplete secondary structure, and is similar to the equilibrium intermediate ensemble. The I1 to I2 transition, which is observed by trp fluorescence, has a relaxation time of 60 sec at 20 °C. The I2 to N transition is characterized by a slow increase in secondary structure, with a relaxation time of ~ 370 sec (Anderson and Teschke, 2003). When bisANS, a hydrophobic dye, is used to monitor burial of hydrophobic patches during folding, there is a large burst (U ⇒ I1), followed by a slower continued increase in bisANS fluorescence (I1 ⇒ I2), and finally a very slow loss in bisANS binding (~800 sec), after which the native state (N) is formed. The I1 to I2 transition rate is consistent between bisANS and trp fluorescence, whereas the hydrophobic burial during I2 to N is slower than secondary structure formation (Anderson and Teschke, 2003; Doyle et al., 2003; Teschke and King, 1993).
Figure 5. Kinetic folding model for P22 coat protein.
A proposed model for the slow folding reaction is shown. Secondary structure that is not fully formed is represented as dashed tubes for helices and dashed ribbons for sheets. Fully formed secondary structure is represented as solid ribbons and tubes. The unfolded protein rapidly folds to intermediate 1, which has some secondary structure formed and some clustering of tryptophans. During the transition from intermediate 1 to intermediate 2 the tryptophans get fully buried. The intermediate 2 to native state has the formation of remaining secondary structure and complete folding to the native state.
The telokin domain has only one tryptophan residue (W354) that is probably solvent exposed, which suggests that changes in tryptophan fluorescence are not related to telokin domain folding (Figure 3). Three out of the six tryptophans (241, 354, and 410) in coat protein are included in the C-terminal half, which is largely composed of β-sheets and flexible loops. Conversely, the HK97-like core of the P22 coat protein has all of the α-helices and should contribute most strongly to the CD signal at 222 nm. Refolding experiments using separately expressed halves of P22 coat protein (N-terminal 1–190, and C-terminal 191–430) showed that only the C-terminal half, which contains the telokin domain, was capable of refolding independently, although it formed dimers rather than monomers (Kang and Prevelige, 2005). Based on this result, the C-terminal half was suggested to be the folding scaffold for the entire subunit (Kang and Prevelige, 2005), which is consistent with our folding experiments.
Tsf coat protein folding
The majority of the tsf mutants are located in the telokin domain. The tsf substitutions generally affect the conformation of the folded monomers so that they appear to have more β-sheet or random coil than WT coat protein monitored by CD (Doyle et al., 2004; Galisteo, Gordon, and King, 1995; Parent, 2007; Teschke and King, 1995). A three-state equilibrium folding model best describes the folding of the tsf coat variants, like WT coat protein. The major effect of the tsf substitutions on folding is to dramatically destabilize the I ⇔ N transition as measured by equilibrium experiments, but the stability for U ⇔ I transition is also affected, so that the native state is not well-populated (Doyle et al., 2004; Doyle et al., 2003; Parent, 2007).
Folding kinetics of A108V, D174N, and F353L tsf coat proteins, which are located in the HK97 core of the coat subunit, showed that the only secondary structure generated occurs in the burst phase, though even this is destabilized by the substitutions. (Homology modeling assigned F353 to the telokin domain, but closer inspection of the folding data presented here, more logically places F353 in the β-hinge.) The I2 to N transition is so destabilized for these protein variants that the N state is only transiently populated and flickers between N, I2, and I1 (Doyle et al., 2004). Thus, these proteins are very aggregation prone and dependent on GroEL/S for folding (Gordon et al., 1994; Nakonechny and Teschke, 1998; Teschke and King, 1995).
S223F and G232D are the only two tsf mutants in the telokin domain that have been thoroughly investigated (Doyle et al., 2004; Doyle et al., 2003). Their native states are destabilized compared to WT coat protein in equilibrium denaturation experiments. However, helix formation in the I2 to N transition for S223F is observed as folding kinetics measured by CD. This is unlike tsf mutants in the core where only the CD change in the burst phase is observable. Tsf mutants in the telokin domain, P238S, S262F, and V300A, show CD folding kinetics, as does G232D in a recent reanalysis of folding data (Teschke, unpublished data). However, these proteins still rapidly unfold from N back to intermediates, consistent with a destabilized native state. These data suggest that if the tsf substitutions are in the telokin domain, the ability to fold the core is retained, but if the amino acid substitutions are in the core there is a more severe folding defect.
All of our kinetic and equilibrium folding data of WT, tsf variants, and the tsf:su variants (discussed below) are consistent with the notion that coat protein folds as two autonomous domains: the telokin domain and the core (Figure 5). Based on our experiments with the tsf coat proteins, we propose the following model. First, our data suggest that the telokin domain is the folding nucleus, since this part of the protein has the majority of the tsf substitutions, it is able to fold autonomously, and amino acid substitutions in this domain still allow for final formation of helical secondary structure in the core of the protein. Our data are consistent with telokin folding in the burst phase of the folding reaction, along with burying of some of tryptophans in the core (48, 61, 354, 410), followed by formation of α-helices as the rate-limiting step in the reaction. When there are amino acid substitutions in the telokin domain, the rest of the protein has difficulty achieving the native state, causing the tsf variants to be aggregation-prone. These data are consistent with the observation that the tsf coat proteins induce GroEL/S and require GroEL/S for folding at high temperature (Gordon et al., 1994; Nakonechny and Teschke, 1998; Parent, Ranaghan, and Teschke, 2004).
Maturation of tsf procapsids
As mentioned previously, the major conformational changes that occur during expansion are reorganization of the A-domain and movement of the N-arm and P-loops. WT procapsids expand with what appears to be simple first order kinetics, without indication of any populated intermediate, which is consistent with a cooperative reaction (Capen and Teschke, 2000). However, much evidence suggests that there are multiple steps during expansion. For instance, some tsf variants exhibited sigmoidal expansion kinetics, which means that capsid expansion is at least a two step process (Capen and Teschke, 2000). The tsf mutants that affect the kinetics of expansion are distributed throughout the protein. These data suggest that conformational information must be communicated between different regions of the protein prior to expansion (step 1). Consistent with the data presented thus far is the possibility that the reorganization and movement of the A domain is the rate-limiting intermediate step of the reaction during expansion (step 2), which is followed by the physical expansion in volume (step 3).
Evidence for the second step in expansion comes from the observation that amino acid substitutions in the A domain both enhance and inhibit the capsid maturation reaction. Empty procapsid shells with tsf substitutions at one position, (D174N or D174G), show sigmoidal expansion kinetics and a decrease in the temperature required for complete expansion as compared to WT shells. Thus, there must be a lower activation energy for the rate-limiting step, which is consistent with our hypothesis that A domain movements predominantly affect the rate-limiting step in expansion. Conversely, procapsids of coat su variants, (D163G, T166I and F170L), do not display sigmoidal kinetics and expand slower than WT even at high temperatures. This suggests an increased energy requirement for expansion. Indeed, empty procapsid shells of these variants are sometimes disrupted rather than smoothly transition to the expanded state (Parent, Suhanovsky, and Teschke, 2007b). Though D174 occurs near the global su sites, it resides in one of the A-domain helices, whereas all three global su residues are in the β-hinge that connects the core of the protein and the telokin domain. This strand undergoes significant movement in response to the refolding of the A domain. We propose that this movement is a prerequisite for expansion. So these data suggest that there may be as many as three discrete events during expansion: communication between the domains that expansion is occurring, refolding of the A domain, and of the strands in the hinge. In WT procapsids, these events may occur concomitantly or in rapid succession.
Tsf:su coat proteins
The su substitutions, D163G, T166I, and F170L were determined to be global suppressors, i.e., each suppresses the phenotype of several tsf mutants in coat protein, which are distributed throughout sequence. All these su substitutions occur in a β-strand and turn in the β-hinge region (Figure 4d). Each global su substitution remedies the folding defects of multiple tsf parents by a common mechanism. Yet, the three global su substitutions do not function the same way. For example, T166I and D163G both fix the original tsf defects by a shared mechanism. However, F170L corrects tsf protein folding defects by a second mechanism (Aramli and Teschke, 2001; Doyle et al., 2004; Parent and Teschke, 2007).
T166I and F170L have been examined extensively in the context of several tsf substitutions (indicated by tsf:T166I). T166I is the most frequently isolated suppressor substitution (Aramli and Teschke, 1999). Although, stabilization of the native state is the obvious and most frequently observed mechanism by which su substitutions work (Sekijima et al., 2006; Van der Schueren, Robben, and Volckaert, 1998), surprisingly the T166I substitution actually increases monomer aggregation (Aramli and Teschke, 2001). In equilibrium and kinetic folding experiments, the T166I substitution increases the population of the intermediates. Consistent with these data, the tsf:T166I proteins increase expression of GroEL/S even compared to the tsf parents (Parent, Ranaghan, and Teschke, 2004). However, the S223F:T166I coat protein more readily folds to N (Doyle et al., 2004). We hypothesized that the increase in the population of intermediate species allowed for more effective interactions with GroEL/S in vivo. Not only is GroEL/S essential for successful in vivo folding and assembly of the tsf:T166I proteins, but also scaffolding protein (Parent, Ranaghan, and Teschke, 2004). The small population of tsf:T166I coat proteins that fold to the native state are trapped in procapsids by scaffolding protein. GroEL/S is unable to rescue the folding of the tsf:T166I coat proteins when there are decreased levels of intracellular scaffolding protein. The D163G substitution appears to function by the same mechanism as T166I in vivo (Parent and Teschke, 2007).
The F170L su substitution functions via a completely different mechanism than the T166I substitution. The addition of F170L to tsf coat proteins causes stabilization of the native state (Parent and Teschke, 2007). Unlike the tsf:T166I coat proteins, expression of tsf:F170L coat proteins does not induce GroEL/S, though there remains a requirement for the chaperone complex. Tsf:F170L coat proteins are resistant to unfolding at elevated temperatures, where tsf:T166I coat proteins completely aggregate (Aramli and Teschke, 2001).
The price you pay
If the su substitutions (D163G, T166I, F170L) promote folding, and these mutants can assemble into infectious phage, then why haven’t these amino acids become part of the WT coat protein sequence? We investigated this question by generating the su substitutions on a plasmid that expresses coat protein (Parent, Suhanovsky, and Teschke, 2007b). The individual, double (D163G:T166I, D163:F170L, and T166I:F170L), and triple (D163G:T166I:F170L) mutants were all generated. The phenotypes were determined by trans complementation using a phage that is unable to produce coat protein. The single su substitutions showed no phenotype, but the double and triple variants had cs phenotypes, with the triple mutant being extremely cs and unable to support phage growth even at 25 °C. Hence, su substitutions likely failed to become part of the WT sequence owing to a negative consequence for phage production in certain conditions.
The products of in vivo assembly of the su coat proteins were visualized by electron microscopy. All mutants led to the formation of aberrant products (primarily spiral forms and broken procapsids) at 16 °C. Spirals result when the placement of the five-fold vertices is inaccurate (Earnshaw and King, 1978). In vitro assembly reactions recapitulate in vivo results except that the F170L coat variant also forms tubes, called “polyheads” (Parent, Suhanovsky, and Teschke, 2007b). In the absence of scaffolding protein, more F170L coat protein forms polyheads (Suhanovsky, et al., unpublished data). Image reconstruction of vitrified F170L polyheads demonstrates that they are solely composed of hexons (Parent et al., unpublished data). We propose that interaction with scaffolding protein during assembly can alter the conformation of F170L coat protein monomers so that subunits at the pentameric vertices can be added properly.
Cs mutants in coat protein
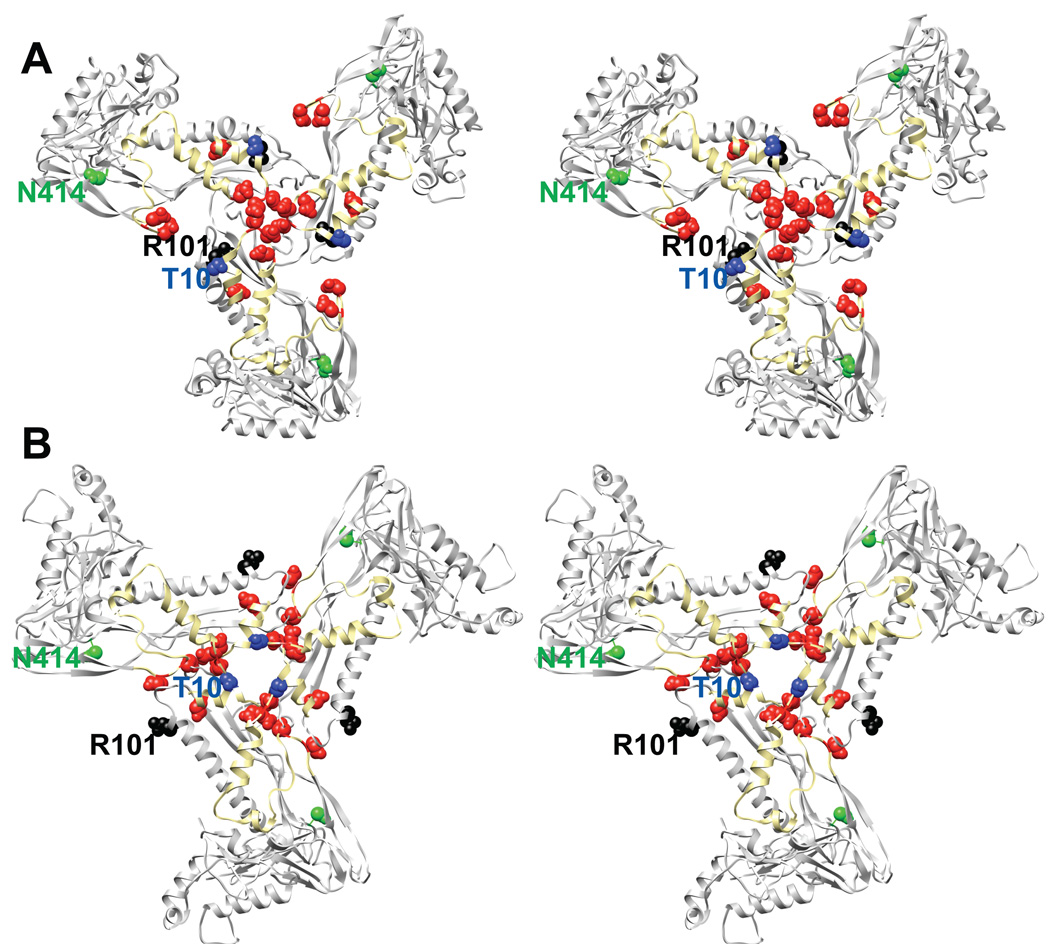
Three P22 cs coat mutants, T10I, R101C, and N414S, have been well characterized. R101C and N414S were isolated in general searches for cs mutants, whereas T10I was isolated as a suppressor of the coat tsf mutant, F353L (Jarvik and Botstein, 1973; Jarvik and Botstein, 1975). The cs mutants cause assembly problems, but do not negatively affect folding (Fong, Doyle, and Teschke, 1997; Gordon, 1993; Teschke and Fong, 1996). Cs coat proteins are able to fold and assemble at low temperatures but the infections are unable to progress beyond procapsids, owing to altered interaction with scaffolding protein (Gordon, 1993; Teschke and Fong, 1996). In vivo, N414S binds scaffolding protein tightly enough that DNA packaging is precluded, as scaffolding protein cannot exit the procapsids. Conversely, T10I and R101C coat proteins interact weakly with scaffolding protein, observed both as scaffolding protein leaking from procapsids and the inability to assemble PLPs in vitro. However, these mutants interact with scaffolding at least on a basal level to incorporate the portal protein complex (Gordon, 1993; Teschke and Fong, 1996). Weak interaction with scaffolding protein does not explain the inability of these mutants to package DNA in vivo. We propose that these substitutions affect the ability of the N-arm to move appropriately during maturation (Figure 6).
Figure 6. Cs mutants and the electrostatic residues in coat protein that likely dictate scaffolding protein binding.
A) Stereo image of one subunit per neighboring hexamer at a true three-fold axis of symmetry shown as a ribbon diagram viewed from inside the procapsid. The N-arm is colored in yellow, with the acidic residues in the N-arm represented as red spheres. The 3 cs mutations are also shown as spheres (T10I in blue, R101C in black, N414S in green). B) Same as panel A, except for the mature state (expanded head).
The pseudo-atomic model of the coat subunit can easily explain T10I and R101C phenotypes (Figure 6). The N-arm and P-loop have been hypothesized to be important for scaffolding protein binding (Parent et al., 2010). The coat-binding domain of scaffolding protein is rich in basic amino acids (Tuma et al., 1998). Intriguingly the N-arm of P22 coat protein is very acidic, which suggests this may be a site to which scaffolding protein binds. Conformational changes in the N-arm and P-loop during maturation conceal some of the electrostatic residues and probably encourage scaffolding protein release through the holes at the center of the capsomers. The N414S substitution is more difficult to rationalize since the location of this residue is quite distant from the suggested sites of scaffolding interaction.
Conclusions
The recently published pseudo-atomic model of P22 coat protein has allowed us to interpret our folding and assembly data. The telokin domain appears to have been added and maintained during evolution because it stabilizes monomers. Indeed, it is likely the folding nucleus and it is the site of many tsf mutants, which cause protein aggregation at high temperature. The C-terminal half of the protein, containing the telokin domain, can fold autonomously. It will be revealing to investigate the folding and stability of the isolated telokin domain to determine if it’s folding is rapid compared to the whole protein.
The su substitutions identify an additional region of coat protein that is important for folding. We propose that the formation of the β- hinge is the final “lock” for stabilizing the folded state and that is why substitutions in that region affect folding, both positively and negatively. The β-hinge is composed of regions N- and C-terminal to the telokin domain, by which we infer that the whole protein must be post-translationally folded. This is consistent with a hypothesis that the C-termini of phage proteins are likely to be critical for folding to prevent misassembly of defective or incomplete proteins, i.e., only completely synthesized proteins can fold and assemble (Casjens et al., 1991). Additionally, the regions in coat protein without known mutants may also indicate areas that are important for folding. For instance, the carboxyl terminal half of coat protein has 14 tsf sites, while the amino terminal half has only three, which may indicate that the formation of the helices in the P-domain is crucial for folding. In the past, we’ve use phage genetics to identify crucial sites in the protein, but now the recent structures will provide the information to directly test how different regions of coat protein affect folding or assembly.
Finally, the mutants that have been isolated over the years have provided a framework to validate the newly released structures, which have proven consistent with the biochemical and genetic data of many labs. To ‘Let the phage do the work’ in determining the important regions in coat protein has truly been an instructive and productive approach.
Supplementary Material
This movie was created with the Yale Morph Server (http://molmovdb.org/) (Flores et al., 2006), which creates by linear interpolation a series of hypothetical snap-shots of the capsid structure from the known procapsid and expanded head models (Parent et al., 2010). “D” subunits from neighboring hexons are distinguished by different colors, with the N-arms of each chain in darker shades of the same colors. The P-loops are shown in black. The top panel represents a view from outside the capsid, along a strict 3-fold axis of symmetry. The bottom panel shows a side view of what is displayed in the top panel.
Acknowledgements
We would like to thank Dr. Jonathan King for inspiring much of the work presented in this paper, and the rest of the P22 community. We also thank Dr. Timothy S. Baker and members of the Teschke lab for critical reading of the manuscript and helpful discussion. This work was supported by R01 GM76661 to CMT and by NIH fellowship F32A1078624 to KNP. KNP was also supported in part by R37 GM33050 to TSB.
Abbreviations
- gp
gene product
- ts
temperature-sensitive
- cs
cold-sensitive
- tsf
temperature-sensitive-folding
- su
suppressor substitution
- am
amber mutation
- 3D
three-dimensional
- PLP
procapsid-like particles
- SAXS
small angle X-ray scattering
- H/D exchange
hydrogen-deuterium exchange
- N
native state
- I
intermediate state
- U
unfolded state
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson E, Teschke CM. Folding of Phage P22 Coat Protein Monomers: Kinetic and Thermodynamic Properties. Virology. 2003;313:184–197. doi: 10.1016/s0042-6822(03)00240-x. [DOI] [PubMed] [Google Scholar]
- Aramli L, Teschke C. Alleviation of a defect in protein folding by increasing the rate of subunit assembly. J Biol Chem. 2001;276(27):25372–25377. doi: 10.1074/jbc.M101759200. [DOI] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J. Biol. Chem. 1999;274(32):22217–22224. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–63. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 1988;202(1):77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Capen CM, Teschke CM. Folding defects caused by single amino acid substitutions in a subunit are not alleviated by assembly. Biochemistry. 2000;39(5):1142–1151. doi: 10.1021/bi991956t. [DOI] [PubMed] [Google Scholar]
- Casjens S. Molecular organization of the bacteriophage P22 coat protein shell. J. Mol. Biol. 1979;131:1–19. doi: 10.1016/0022-2836(79)90298-5. [DOI] [PubMed] [Google Scholar]
- Casjens S, Adams MB, Hall C, King J. Assembly-controlled autogenous modulation of bacteriophage P22 scaffolding protein gene expression. J Virol. 1985;53:174–179. doi: 10.1128/jvi.53.1.174-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Eppler K, Sampson L, Parr R, Wyckoff E. Fine structure genetic and physical map of the gene 3 to 10 region of the bacteriophage P22 chromosome. Genetics. 1991;127(4):637–647. doi: 10.1093/genetics/127.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 1974;2(2–4):202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM Asymmetric Reconstruction of Bacteriophage P22 Reveals Organization of its DNA Packaging and Infecting Machinery. Structure. 2006;14:1–10. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253(1):86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- de Beus MD, Doyle SM, Teschke CM. GroEL binds a late folding intermediate of phage P22 coat protein. Cell Stress Chaperones. 2000;5(3):163–173. doi: 10.1379/1466-1268(2000)005<0163:gbalfi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Anderson E, Parent KN, Teschke CM. A concerted mechanism for the suppression of a folding defect through interactions with chaperones. J. Biol. Chem. 2004;279(17):17473. doi: 10.1074/jbc.M400467200. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Anderson E, Zhu D, Braswell EH, Teschke CM. Rapid unfolding of a domain populates an aggregation-prone intermediate that can be recognized by GroEL. J. Mol. Biol. 2003;332:937–951. doi: 10.1016/s0022-2836(03)00955-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol. Biol. 1995a;247(4):618–35. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Xie Z, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995b;247:636–47. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison SC. Assembly of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 1976;104(2):387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Hendrix R, King J. Structural studies of bacteriophage lambda heads and proheads by small angle X-ray diffraction. J. Mol. Biol. 1980;134:575–594. doi: 10.1016/0022-2836(79)90368-1. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- Fersht A. In: Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. Julet MR, editor. New York: W.H. Freeman and Company; 1999. [Google Scholar]
- Finn BE, Chen X, Jennings PA, Saal'au-Bethell SM, Matthews CR. Principles of protein stability. Part 1 - reversible unfolding of proteins: kinetic and thermodynamic analysis. In: Rees AR, Sternberg MJE, Wetzel R, editors. Protein Engineering: A practical approach. Oxford: Oxford University Press; 1992. pp. 168–189. [Google Scholar]
- Flores S, Echols N, Milburn D, Hespenheide B, Keating K, Lu J, Wells S, Yu EZ, Thorpe M, Gerstein M. The Database of Macromolecular Motions: new features added at the decade mark. Nucleic Acids Res. 2006;34:D296–D301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong DG, Doyle SM, Teschke CM. The folded conformation of phage P22 coat protein is affected by amino acid substitutions that lead to a cold-sensitive phenotype. Biochemistry. 1997;36(13):3971–3980. doi: 10.1021/bi962188y. [DOI] [PubMed] [Google Scholar]
- Fraser JS, Maxwell KL, Davidson AR. Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol. 2007;10(4):382–7. doi: 10.1016/j.mib.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359(2):496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Fuller MT, King J. Regulation of coat protein polymerization by the scaffolding protein of bacteriophage P22. Biophys. J. 1980;32(1):381–401. doi: 10.1016/S0006-3495(80)84963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, King J. Purification of the coat and scaffolding protein from procapsids of bacteriophage P22. Virology. 1981;112:529–547. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Fuller MT, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J. Mol. Biol. 1982;156(3):633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, Gordon CL, King J. Stability of wild-type and temperature-sensitive protein subunits of the phage P22 capsid. J. Biol. Chem. 1995;270(28):16595–16601. doi: 10.1074/jbc.270.28.16595. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys. J. 1993;65:227–235. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, Hendrix RW, Duda RL, Liljas L, Johnson JE. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–65. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee KK, Speir JA, Duda R, Hendrix RW, Komives E, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–50. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CL. Ph.D. thesis in Biochemistry. Cambridge, MA: Massachusettes Institute of Technology; 1993. [Google Scholar]
- Gordon CL, Sather SK, Casjens S, King J. Selective in vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J. Biol. Chem. 1994;269(45):27941–27951. [PubMed] [Google Scholar]
- Gough M, Levine M. The circularity of the phage P22 linkage map. Genetics. 1968;58(2):161–169. doi: 10.1093/genetics/58.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225(1):82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 2003;10(5):334–41. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Holden HM, Ito M, Hartshorne DJ, Rayment I. X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8Å resolution. J Mol Biol. 1992;227:840–851. doi: 10.1016/0022-2836(92)90226-a. [DOI] [PubMed] [Google Scholar]
- Ishii T, Yamaguchi Y, Yanagida M. Binding of the structural protein soc to the head shell of bacteriophage T4. J Mol Biol. 1978;120:533–44. doi: 10.1016/0022-2836(78)90352-2. [DOI] [PubMed] [Google Scholar]
- Jarvik J, Botstein D. A genetic method for determining the order of events in a biological pathway. Proc. Natl. Acad. Sci. USA. 1973;70(7):2046–2050. doi: 10.1073/pnas.70.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik J, Botstein D. Conditional-lethal mutations that suppress genetic defects in morphogenesis by altering structural proteins. Proc. Natl. Acad. Sci. USA. 1975;72(7):2738–2742. doi: 10.1073/pnas.72.7.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003;10(2):131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17:237–43. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kang S, Prevelige PE., Jr Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J Mol Biol. 2005;347(5):935–48. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos. Trans. R. Soc. London B. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22 II. Morphogenetic pathway. J. Mol. Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- Kolstad RA, Prell HH. An amber map of Salmonella phage P22. Mol Gen Genet. 1969;104:339–50. doi: 10.1007/BF00334233. [DOI] [PubMed] [Google Scholar]
- Lander GC, Khayat R, Li R, Prevelige PJ, Jr, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architechture of an infectious conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The Structure of an Infectious p22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006;312:1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lanman J, Tuma R, Prevelige PE., Jr Identification and characterization of the domain structure of bacteriophage P22 coat protein. Biochemistry. 1999;38(44):14614–14623. doi: 10.1021/bi9915420. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–90. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100(2):253–63. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- Lenk E, Casjens S, Weeks J, King J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology. 1975;68:182–199. doi: 10.1016/0042-6822(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Lew KK, Roth JR. Isolation of UGA and UAG nonsense mutants of bacteriophage P22. Virology. 1970;40:1059–62. doi: 10.1016/0042-6822(70)90153-4. [DOI] [PubMed] [Google Scholar]
- Nakonechny WS, Teschke CM. GroEL and GroES control of substrate flux in the in vivo folding pathway of phage P22 coat protein. J. Biol. Chem. 1998;273(42):27236–27244. doi: 10.1074/jbc.273.42.27236. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Trus BL, Cheng N, Steven AC, Booy FP, Brown JC. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–50. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007;14:1221–26. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- Parent KN. University of Connecticut Ph.D. thesis. 2007. Folding and assembly of phage P22 coat protein. [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links. Structure. 2010 doi: 10.1016/j.str.2009.12.014. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Ranaghan MJ, Teschke CM. A second site suppressor of a folding defect functions via interactions with a chaperone network to improve folding and assembly in vivo. Mol. Microbiol. 2004;54:1036–54. doi: 10.1111/j.1365-2958.2004.04326.x. [DOI] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Phage P22 procapsids equilibrate with free coat protein subunits. J Mol Biol. 2007a;365:513–22. doi: 10.1016/j.jmb.2006.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Polyhead formation in phage P22 pinpoints a region in coat protein required for conformational switching. Mol. Microbiol. 2007b;65(5):1300–10. doi: 10.1111/j.1365-2958.2007.05868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Teschke CM. GroEL/S substrate specificity based on substrate unfolding propensity. Cell Stress Chaperones. 2007;12(1):20–32. doi: 10.1379/CSC-219R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Zlotnick A, Teschke CM. Quantitative analysis of multi-component spherical virus assembly: Scaffolding protein contributes to the global stability of phage P22 procapsids. J. Mol. Biol. 2006;359:1097–1106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Karthikeyan T, Houtz bM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. Corrected sequence of the bacteriophage p22 genome. J. Bacteriol. 2003;185:1475–77. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa MP, McKelvey TA, Hempel J, Hendrix RW. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J Virol. 1991;65:3227–37. doi: 10.1128/jvi.65.6.3227-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Jr, Marieta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PE, Jr, King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- Prevelige PE, Jr, King J, Silva JL. Pressure denaturation of the bacteriophage P22 coat protein and its entropic stabilization in icosahedral shells. Biophys. J. 1994;66:1631–1641. doi: 10.1016/S0006-3495(94)80955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J. Mol. Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, Steven AC, Duda RL. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J Mol Biol. 2006;364(3):512–25. doi: 10.1016/j.jmb.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y, Dendle MT, Wiseman RL, White JT, D'Haeze W, Kelly JW. R104H may suppress transthyretin amyloidogenesis by thermodynamic stabilization, but not by the kinetic mechanism characterizing T119 interallelic trans-suppression. Amyloid. 2006;13:57–66. doi: 10.1080/13506120600722449. [DOI] [PubMed] [Google Scholar]
- Spencer J, Newcomb W, Thomsen D, Homa F, Brown J. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 1998;72:3944–51. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal Structure of P22 Tailspike Protein: Interdigitated Subunits in a Thermostable Trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PEJ, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000;297(5):1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- Teschke CM. Aggregation and assembly of phage P22 temperature-sensitive coat protein mutants in vitro mimic the in vivo phenotype. Biochemistry. 1999;38(10):2873–2881. doi: 10.1021/bi982739f. [DOI] [PubMed] [Google Scholar]
- Teschke CM, Fong DG. Interactions between coat and scaffolding proteins of phage P22 are altered in vitro by amino acid substitutions in coat protein that cause a cold-sensitive phenotype. Biochemistry. 1996;35(47):14831–14840. doi: 10.1021/bi960860l. [DOI] [PubMed] [Google Scholar]
- Teschke CM, King J. Folding of the phage P22 coat protein in vitro. Biochemistry. 1993;32(40):10839–10847. doi: 10.1021/bi00091a040. [DOI] [PubMed] [Google Scholar]
- Teschke CM, King J. In vitro folding of phage P22 coat protein with amino acid substitutions that confer in vivo temperature sensitivity. Biochemistry. 1995;34(20):6815–6826. doi: 10.1021/bi00020a028. [DOI] [PubMed] [Google Scholar]
- Teschke CM, McGough A, Thuman-Commike PA. Penton release from P22 heat-expanded capsids suggests importance of stabilizing penton-hexon interactions during capsid maturation. Biophys J. 2003;84:2585–2592. doi: 10.1016/S0006-3495(03)75063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jokana J, Prasad BVV, King J, Prevelige PE, Jr, Chiu W. Three-dimensional structure of scaffolding-containing phage P22 procapsids by electron cryo-microscopy. J. Mol. Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383. [DOI] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, Burbea M, McGough A, Chiu W, Prevelige PEJ. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys. J. 1999;76(6):3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys. J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R, Parker MH, Weigele P, Sampson L, Sun Y, Krishna NR, Casjens S, Thomas GJJ, Prevelige PEJ. A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J. Mol. Biol. 1998;281(1):81–94. doi: 10.1006/jmbi.1998.1916. [DOI] [PubMed] [Google Scholar]
- Tuma R, Prevelige PE, Jr, Thomas GJ., Jr Mechanism of capsid maturation in a double-stranded DNA virus. Proc. Natl. Acad. Sci. USA. 1998;95:9885–9890. doi: 10.1073/pnas.95.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R, Tsuruta H, Benevides JM, Prevelige PE, Jr, Thomas GJ., Jr Characterization of subunit structural changes accompanying assembly of the bacteriophage P22 procapsid. Biochemistry. 2001;40(3):665–74. doi: 10.1021/bi001965y. [DOI] [PubMed] [Google Scholar]
- Van der Schueren J, Robben J, Volckaert G. Misfolding of chloramphenicol acetyltransferase due to carboxy-terminal truncation can be corrected by second-site mutations. Protein Eng. 1998;11(12):1211–1217. doi: 10.1093/protein/11.12.1211. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289(5487):2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol. 1995;253(1):74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nature Struct. Biol. 2000;230:230–7. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This movie was created with the Yale Morph Server (http://molmovdb.org/) (Flores et al., 2006), which creates by linear interpolation a series of hypothetical snap-shots of the capsid structure from the known procapsid and expanded head models (Parent et al., 2010). “D” subunits from neighboring hexons are distinguished by different colors, with the N-arms of each chain in darker shades of the same colors. The P-loops are shown in black. The top panel represents a view from outside the capsid, along a strict 3-fold axis of symmetry. The bottom panel shows a side view of what is displayed in the top panel.