Abstract
The recent discovery of human T-lymphotropic virus type 3 (HTLV-3) in Cameroon highlights the importance of expanded surveillance to better understand the prevalence and public health impact of this new retrovirus. HTLV diversity was investigated in 402 persons in rural Cameroon who reported simian exposures. Plasma from 29 persons (7.2%) had reactive serology. HTLV tax sequences were detected in 3 persons. Phylogenetic analysis confirmed HTLV-1 infection in two individuals, and HTLV-3 infection in a third person (Cam2013AB). The complete proviral genome from Cam2013AB shared 98% identity and clustered tightly in distinct lineage with simian T-lymphotropic virus type 3 (STLV-3) subtype D recently identified in two guenon monkeys near this person’s village. These results document a fourth HTLV-3 infection with a new and highly divergent strain we designate HTLV-3 (Cam2013AB) subtype D demonstrating the existence of a broad HTLV-3 diversity likely originating from multiple zoonotic transmissions of divergent STLV-3.
Keywords: retrovirus, zoonoses, HTLV, STLV, emergence, nonhuman primates, hunters, evolution, diversity
In 2005, two new human T-lymphotropic viruses (HTLV) were discovered in Cameroon, types 3 and 4 (HTLV-3 and HTLV-4), doubling the number of known HTLVs, which previously included only HTLV-1 and HTLV-2 (Blattner et al., 1984; Calattini et al., 2005; Gessain, Meertens, and Mahieux, 2002; Khabbaz, Fukuda, and Kaplan, 1993; Manns and Blattner, 1991; Wolfe et al., 2005b). All four HTLV groups are believed to have originated from cross-species transmission of simian T-lymphotropic viruses (STLVs) to humans, though an STLV counterpart of HTLV-4 has yet to be identified (Calattini et al., 2005; Gessain and de The, 1996; Gessain and Mahieux, 2000; Gessain, Mahieux, and de The, 1996; Gessain, Meertens, and Mahieux, 2002; Mahieux et al., 1998; Nerrienet et al., 2001; Van Brussel et al., 1998; Vandamme, Salemi, and Desmyter, 1998; Wolfe et al., 2005b). Recently, more robust phylogenetic analysis of an STLV strain from a Macaca arctoides (MarB43) has shown that this is a highly divergent virus distinct from other STLV-1/HTLV-1 and has contingently been re-classified as STLV-5 (Liegeois et al., 2008; Switzer et al., 2009; Van Dooren et al., 2005). Regardless of suspected zoonotic origin, STLV in humans is typically called HTLV and both STLV and HTLV are known as the primate T-lymphotropic viruses (PTLV). While HTLV-1 and -2 have spread globally to infect millions of persons and cause disease in about 5% of infected persons (Blattner et al., 1984; Gessain, Meertens, and Mahieux, 2002; Khabbaz, Fukuda, and Kaplan, 1993; Mahieux and Gessain, 2003; Manns, Hisada, and La Grenade, 1999; Roucoux and Murphy, 2004), very little is known about the epidemiology and public health significance of HTLV-3 and HTLV-4. Thus, additional studies are required to determine the prevalence, geographic distribution, and disease potential of these emerging retroviruses.
STLV-3 has a wide geographic and simian host range across Africa and is composed of four subtypes based on phylogeographic clustering of sequences obtained from infected simians (Meertens and Gessain, 2003; Meertens et al., 2002; Meertens et al., 2003; Sintasath et al., 2009a; Sintasath et al., 2009b). Subtype A consists of STLV-3 from Papio hamadryas and Theropithecus gelada baboons from East Africa (Eritrea and Ethiopia, prototype strains Ph969 and Tge2117) (Goubau et al., 1994; Van Dooren et al., 2004b). Subtype B contains STLV-3 from monkeys (Cercopithecus, Cercocebus, and Papio species) from West Central Africa (Cameroon, Nigeria, Senegal; prototype strains CTO604, CTO-NG409, PPAF3) (Goubau et al., 1994; Meertens and Gessain, 2003; Meertens et al., 2002; Meertens et al., 2003; Van Dooren et al., 2004b). Partial genomic sequences from additional subtype B strains have been found in C. agilis, C. cephus, and Lophocebus albigens from Cameroon (Courgnaud et al., 2004; Liegeois et al., 2008; Sintasath et al., 2009a). Subtype C contains partially characterized viruses from C. nictitans from Cameroon (strains Cni217, Cni227, Cni3034, and Cni3038) (Liegeois et al., 2008; Sintasath et al., 2009b; Van Dooren et al., 2001). Highly divergent STLV-3 recently found in two guenons (C. mona and C. nictitans) from Cameroon form subtype D (strains Cmo8699AB, Cni7867AB) (Sintasath et al., 2009a; Sintasath et al., 2009b). Currently three HTLV-3 strains (2026ND, Pyl43, Lobak18) have been identified and all three are genetically related to the STLV-3 subtype B found in Cameroon (Calattini et al., 2009; Calattini et al., 2005; Wolfe et al., 2005b). HTLV-3(Pyl43) and HTLV-3(Lobak18) are nearly identical sharing about 99% nucleotide identity to each other and to STLV-3(CTO604) suggesting a common and recent ancestor (Calattini et al., 2009). In contrast, HTLV-3(2026ND) is about 10% divergent from other subtype B viruses (Switzer et al., 2006b).
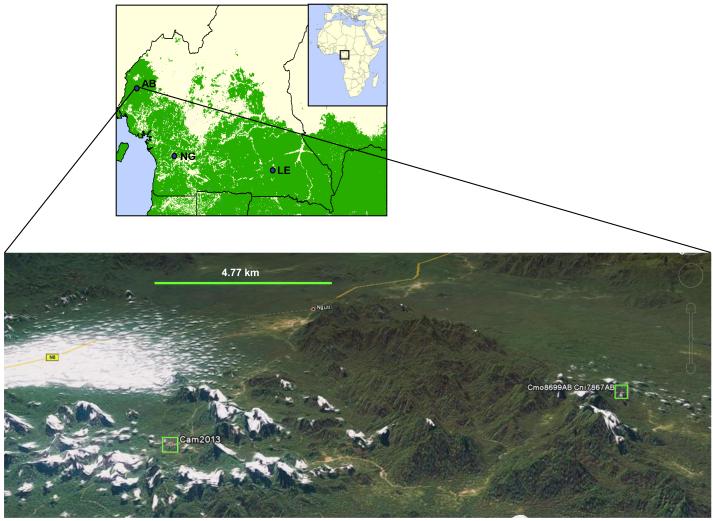
To investigate further the distribution of HTLV we conducted a prevalence study in Cameroon approved by local ethics committees, the Cameroon Ministry of Public Health, and the CDC Institutional Review Board. In total, 405 persons living in eight villages located in three separate forested areas of western and southeastern Cameroon (Fig. 1) volunteered for participation in the study and completed a questionnaire designed to collect information about nonhuman primate (NHP) exposure and demographic data. From this population, 402 persons donated blood specimens. Global positioning systems (GPS) were used to map locations of the villages.
Figure 1.
Geographic location of study sites in Cameroon. Upper panel, location of three major sites in Cameroon where the eight villages are located. Lower panel, topographical map showing proximity of the village of person Cam2013AB to the location where hunters collected specimens from STLV-3D-infected monkeys. Small boxes indicate locations of HTLV-3D-infected person’s village and STLV-3D-infected NHPs. White areas on map are cloud cover. Topomap courtesy of Google Maps.
Plasma samples from these persons were tested for antibodies to HTLV using the Vironostika HTLV-1/2 EIA (Biomerieux, Durham, N.C.) that contains purified HTLV-1 and -2 viral lysates and a recombinant HTLV-1 p21 Envelope (Env) protein. All reactive samples were tested further by Western blot (WB) using a kit (HTLV Blot 2.4, Genelabs Diagnostics, Singapore) that contains disrupted HTLV-1 virions, a gp21 recombinant envelope (Env) protein (GD21) common to both HTLV-1 and HTLV-2, and two HTLV-type specific recombinant Env peptides, MTA-1 and K55, which allow differentiation between HTLV-1 and HTLV-2, respectively. Specimens that were reactive to the Gag (p24) and Env (GD21) proteins were considered seropositive. Seropositive specimens that were reactive to MTA-1 or K55 were considered HTLV-1–like or HTLV-2–like, respectively. Seropositive samples not reactive to either the MTA-1 or K55 peptides were considered HTLV positive, but untypeable. Specimens that were reactive to either p24 or GD21 alone or in combination with other HTLV proteins were considered indeterminate. These EIA and WB assays have been shown previously to be capable of detecting antibodies to a broad range of PTLVs (Meertens et al., 2002; Van Dooren et al., 2004b; Wolfe et al., 2005b).
DNA was prepared from uncultured peripheral blood mononuclear cells (PBMCs) of all WB reactive specimens, and its integrity was confirmed by ß-actin polymerase chain reaction (PCR) as previously described (Wolfe et al., 2005b). All DNA preparation and PCR assays were performed in a laboratory where only human specimens are processed and tested according to recommended precautions to prevent contamination. DNA specimens were first screened with a generic PTLV PCR assay capable of detecting and differentiating partial tax sequences from each of the four major PTLV groups via phylogenetic analysis (Sintasath et al., 2009a; Vandamme et al., 1997; Wolfe et al., 2005b). Further resolution within each PTLV group was obtained by phylogenetic analysis of LTR sequences PCR-amplified from PBMC DNA of persons with detectable tax sequences using established primers and conditions (Meertens et al., 2001; Sintasath et al., 2009a; Wolfe et al., 2005b). The primers SPL2Fn (5′ ACC WTG AGC CCS ARR TAT CCC C 3′) and LTRU5E (5′ CGC AGT TCA GGA GGC ACC AC 3′) and SPL3Fn (5′ AGA GCC TYC CRI TGA MAA ACA TTT C 3′) and 420PLTR (5′ GAA CGC GAC TCA ACC GGC GTG GAT 3′) were also used to amplify PTLV-1 LTR sequences using standard nested PCR conditions and a 45°C annealing temperature. Additional phylogenetic and molecular dating analyses using Bayesian Markov Chain Monte Carlo (MCMC) and maximum likelihood (ML) inference were performed as described recently (Sintasath et al., 2009b; Switzer et al., 2009).
Zoonotic Retroviral Infection of Primate Hunters
The numbers of men and women in the study were almost equal (male=200, women=205) and their ages ranged from 18 – 64 (average = 36). Of the 405 persons, 336 (83%) reported contact with NHP, including hunting, butchering, and keeping NHP pets. Of the 336 primate-exposed persons, 151 (45%) reported hunting, 332 (99%) reported butchering, and 53 (16%) kept NHP pets. None of the participants reported bite wounds. 382 participants identified themselves as being from Bantu ethnic groups, 23 individuals identified themselves as being from non-Bantu ethnic groups including 21 participants who identified themselves as Baka, and two as Hausa. Plasma samples from 29 persons (7.2%) were HTLV-1/2 EIA and Western blot (WB) reactive with the following WB profiles: HTLV-1 (n=3), HTLV-2 (n=2), HTLV-positive but untypeable (n=6), and indeterminate (n=18).
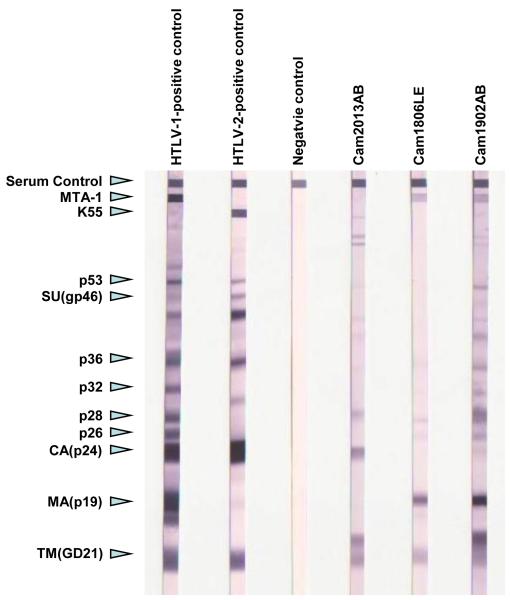
PTLV tax sequences were detected in PBMC DNA of two HTLV-1-seropositive persons (Cam1806LE and Cam1902AB) and one HTLV-untypeable person (Cam2013AB) with moderate WB reactivity to only gag (p24, p28) and recombinant envelope (GD21) proteins (Fig. 2). Plasma from persons CAM1806LE and Cam1902LE reacted to the HTLV-1 p19, p24, GD21 proteins and the MTA-1 peptides typical of HTLV-1/STLV-1 infection (Fig. 2). Interestingly, both Cam2013AB and Cam1902AB also showed reactivity to a protein slightly above the GD21 band not seen in the negative or positive controls (Fig. 2). Of the 26 PCR negative persons, 14 and 20 individuals reported hunting and butchering NHPs, respectively. Their ages ranged from 19 – 63 years old and 69% were males. It is not uncommon to find HTLV-seroreactive and PCR negative test results using specimens from Africans, possibly due to cross-reactivity with malaria or other antigens (Calattini et al., 2009; Calattini et al., 2005; Mahieux et al., 2000b; Wolfe et al., 2005b). Alternatively, the negative PCR results could be due to low sensitivity of the generic primers to detect infections with low viral loads or viral strains with sequence heterogeneity in the PCR primer regions.
Figure 2.
HTLV Western blot profiles of HTLV-3D and HTLV-1 PCR-positive nonhuman primate hunters from Cameroon. Cam2013AB is infected with HTLV-3D and Cam1806LE and Cam1902AB are both infected with HTLV-1 variants. MTA-1 and K55 are the HTLV-1 and HTLV-2 type-specific envelope peptides, respectively. Reactivity to additional HTLV-1 proteins and the serum control are shown on the left. CA, Gag capsid; MA, Gag matrix; SU, Env surface protein; TM, Env transmembrane protein.
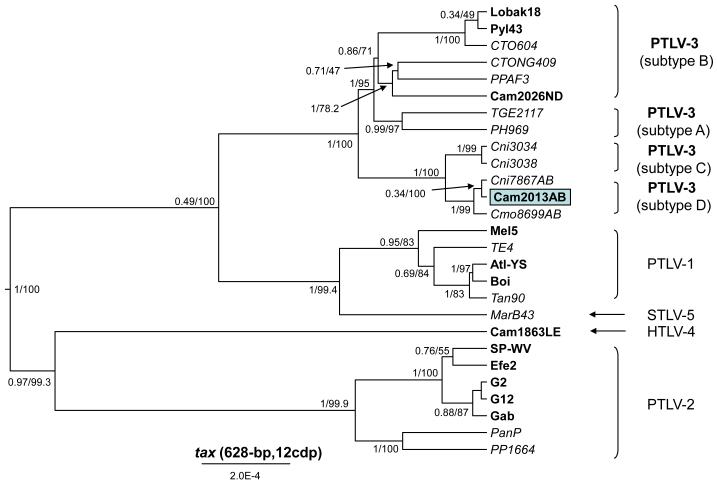
Remarkably, partial tax sequences from person Cam2013AB clustered strongly within the PTLV-3 clade and were 100% identical to those from a highly divergent STLV-3D we discovered recently in C. mona and C. nictitans monkeys in Cameroon (strains Cmo8699AB, Cni7867AB) (Sintasath et al., 2009a; Sintasath et al., 2009b) (Fig. 3a). Cam2013AB tax sequences clustered tightly with only the STLV-3D subtypes, and not other subtypes including STLV-3C for which only partial tax-LTR sequences are currently available for comparison (Fig. 3a). LTR sequences obtained from Cam2013AB shared 98% identity to the two STLV-3D sequences and all three sequences clustered together with high bootstrap support in a distinct lineage in the PTLV-3 phylogroup (Fig. 3b). Person Cam2013AB is a 32-year old Bantu male who reported using a gun and reported hunting crowned monkeys (C. pogonias), drills, and red-capped mangabeys (C. torquatus). He also reported butchering these same NHPs and is married with five children. Specimens were not available from spouses or children to investigate person-to-person transmission in this study.
Figure 3.
Identification of a new HTLV-3 subtype by phylogenetic analysis of (a) partial tax, and (b) LTR sequences. Bayesian and maximum likelihood (ML) trees inferred from 1st and 2nd codon positions (cdp) of partial (628-bp) tax sequences and 200 million MCMC chains and a relaxed molecular clock. Posterior probabilities over ML bootstrap values are provided at each node in the partial tax gene tree. (b) Neighbor joining (NJ) and ML trees inferred from 273-bp LTR sequence alignment and gave identical topologies. NJ over ML boostrap values are shown at the major nodes of the LTR derived tree. HTLV sequences are in bold, STLV sequences are italicized.
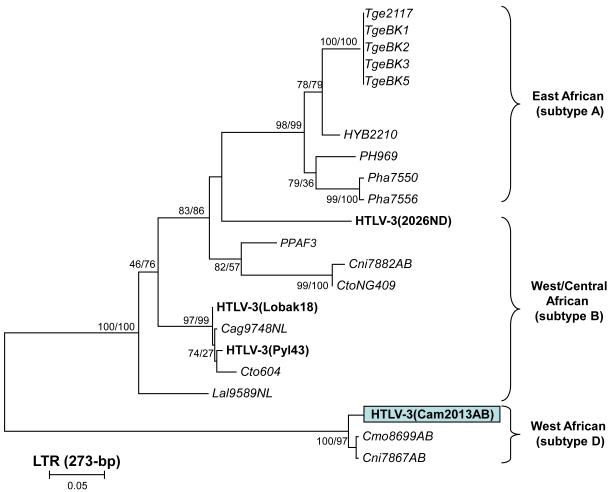
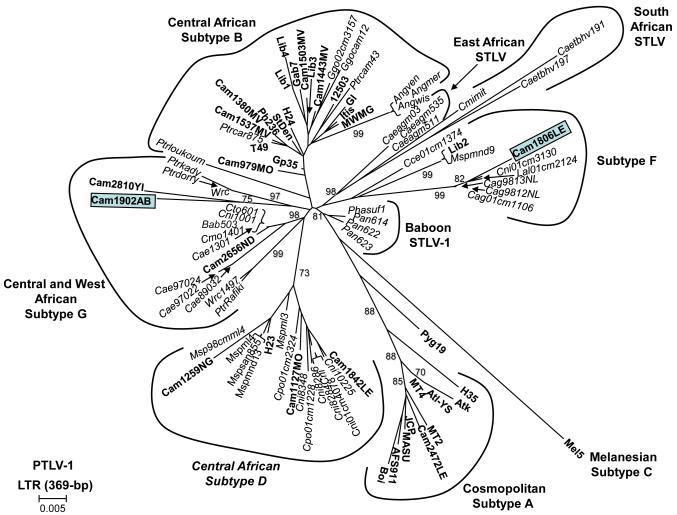
Phylogenetic analysis of tax (data not shown) and LTR sequences (Fig. 4) from Cam1806LE and Cam1902AB confirmed infection with HTLV-1 (GenBank accession numbers, tax = GU391297, GU391298, LTR = GU391299, GU3912300). Based on an alignment of 369-bp LTR sequences, Cam1806LE clustered within the PTLV-1 subtype F clade (Fig. 4) and shared approximately 99% sequence identity with STLV-1 from C. agilis, L. albigena and C. nicitans reported recently (Liegeois et al., 2008; Sintasath et al., 2009a). Person Cam1806LE is a 35-year old Bantu male that reported butchering and hunting of NHPs, including agile mangabeys (C. agilis), C. nictitans, C. pogonias, C. cephus, chimpanzee, gorilla, and Colobus guereza. Interestingly, LTR sequences from Cam1806LE clustered strongly with two sequences from C. agilis (Cag9812NL and Cag9813NL) identified recently (Sintasath et al., 2009a) that were located about 100 km south of Cam1806LE’s village, supporting further a phylogeographic clustering of STLV-3 and HTLV-3. Cam1806LE is married with five children. The Cam1902AB LTR sequence clustered in the newly identified PTLV-1G clade containing mostly STLV-1 (Fig. 4) and is closely related (97% identity) to HTLV-1(2810YI) also from Cameroon but whose villages are about 250 km apart (Wolfe et al., 2005b). Person Cam1902AB is a 48-year old male Bantu that also reported hunting and/or butchering NHPs, including gorilla, chimpanzee, drill (Mandrillus leucophaeus), Preuss’s colobus, Preuss’s cercopithecus, De Brazza monkeys, C. nictitans, and C. cephus. Cam1902AB also reported keeping a drill as a pet for 3 years. He is married and has seven children. Overall, our finding of sequences in Cam1806LE and Cam1902AB with high homology to previously characterized STLV-1 sequences supports a possible recent transmission of STLV-1 to these hunters. It is also possible that Cam1902AB is infected with an HTLV-1 that has crossed recently into humans from simians and is now circulating in the general population. This alternate hypothesis is based on the clustering of Cam1902AB LTR sequences with the only other HTLV-1 (Cam 2810YI) LTR sequence in clade G rather than with STLV-1 (Fig. 4). Identification and analysis of additional HTLV-1 subtype G sequences are needed to clarify this situation. Specimens were not available from close contacts to evaluate person-to-person transmission of these viruses
Figure 4.
Inferred phylogenetic relationships of PTLV-1 LTR sequences. Sequences from Cameroonian nonhuman primate hunters generated in the current study are boxed. Support for the branching order was determined by 1,000 bootstrap replicates using the neighbor-joining method; only values ≥ 60% are shown. HTLV sequences are in bold, STLV sequences are italicized.
Characterization of the Complete HTLV-3D Proviral Genome
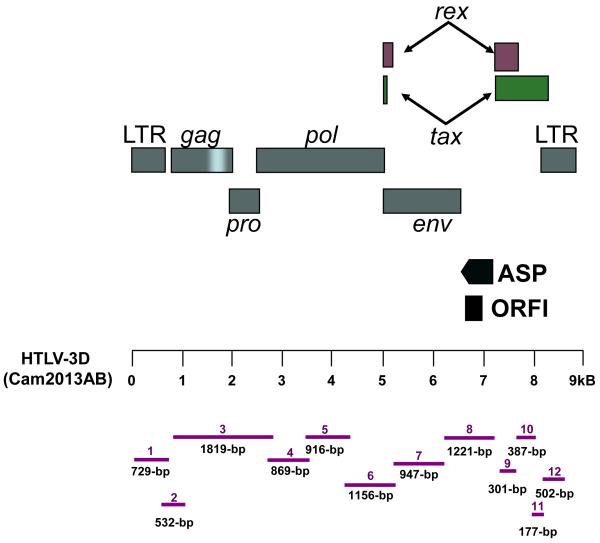
By using uncultured PBMC DNA and a series of 12 overlapping nested PCR amplifications (Fig. 5), as previously described (Sintasath et al., 2009b; Switzer et al., 2006b), the entire proviral genome of HTLV-3D(Cam2013AB) was obtained and was determined to be 8913-bp in length (GenBank accession number GQ463602). The primers used to obtain the full-length HTLV-3D genome are provided in Table 1. HTLV-3D(Cam2013AB) was about 23% divergent from prototypical PTLV-3 subtypes, with the exception of STLV-3D(Cmo8699AB) with which it shared 99.8% sequence identity across the genome (Table 2). Of the 34 nucleotide changes from STLV-3D(Cmo8699AB), 8 occurred in the LTR but did not affect important regulatory elements, 3 occurred in gag, 1 in protease, 11 in polymerase (pol), 11 in envelope (env), and 1 in tax. These mutations resulted in 13 synonymous and 8 nonsynonymous amino acid substitutions. The nonsynonymous changes were localized in pol (n=6) and env (n=2). The organization of the HTLV-3D(Cam2103AB) genome was similar to that of the PTLV-3 group and consisted of the regulatory and structural genes gag, pro, pol, env, tax, rex all flanked by LTRs (Fig. 5). The LTR contained only two 21-bp repeat elements typical of PTLV-3 (Sintasath et al., 2009b; Switzer et al., 2006b; Switzer et al., 2009).
Figure 5.
HTLV-3D(Cam2013AB) (a) genomic organization and (b) schematic representation of PCR-based genomic walking strategy. (a) Non-coding long terminal repeats (LTR), coding regions for all major proteins (gag, group specific antigen; pro, protease; pol, polymerase; env, envelope; rex, regulator of expression; tax, transactivator). (b) Using a previously described PCR-based genomic walking strategy (Sintasath et al., 2009b; Switzer et al., 2006b), the complete proviral sequence (8913-bp) was then obtained by using PTLV-3D-specific primers located within each major gene region in combination with generic PTLV primers (fragments 1 – 12). Amplicon sizes are approximated with the solid bars.
Table 1.
Primers used to generate the complete HTLV-3D(Cam2013AB) proviral genomea
Forward primer (5′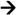 3′) 3′) |
Reverse primer (5′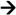 3′) 3′) |
|||||
|---|---|---|---|---|---|---|
| Regionb | Primer Set |
Name | Primer sequencea Name | Primer sequence | Product (bp) |
|
| 1. LTR | Outer | 8699LTRF1 | 1-TGACAGTGACAGCAAGCCCCAAGGCGA-27 | P3GR6 | 1225-AYTGGTGGCTRCCWGGGGGCGGAAG-1201 | 1225 |
| Inner | 8699LTRF2 | 12-GCAAGCCCCAAGGCGAGCCAC-32 | 8699LTRR1 | 740-TAGGAGGGTGATCAATCCCGGACGA-715 | 729 | |
| 2. LTR-gag | Outer | P5LF7 | 540-TCGGTCTCCTTTCTTTGGCGGTCT-563 | PGGAGR1 | 1237-AIIGTCTGCATRAAYTGGGGGCT-1215 | 698 |
| Inner | P5LF8 | 650-CCAGGGGCTCAGAAAGTAAAGGCT-673 | PGGAGR2 | 1181-TGCYTGIARRTCTTTCATYTGCCA-1153 | 532 | |
| 3. gag-pol | Outer | P5LF6 | 391-GCACCTTCGCTTCTCCTGTCCTGG-414 | P3GR1 | 2565-GATAGGGTTATTGCCTGGTCCTTGATA-2539 | 2175 |
| Inner | 8699P19F1 | 747-CACCGGAAATTCATACAGCCGTGC-769) | P3GR1 | 2565-GATAGGGTTATTGCCTGGTCCTTGATA-2539) | 1819 | |
| 4. pol | Outer | 8699GF20 | 2093-ACCCCCCCAGTAAGCATCCAGGCG-2116 | PGPOLR2 | 3206-RYRGGIGTICCTTTIGAGACCCA-3184 | 1113 |
| Inner | 8699GF21 | 2338-AGATGTCCTCCAGCAATGCCAAAG-2361 | PGPOLR2 | 3206-RYRGGIGTICCTTTIGAGACCCA-3184 | 869 | |
| 5. pol | Outer | 2013F1 | 3104-CTTATGAAACCCTCCCTACC-3123 | 2013R1 | 5136-GTGCACCGACTGGGGTCGCC-5117 | 2033 |
| Inner | 2013F2 | 3117-CCCTACCATACATGTCAAGCC-3137 | 2013R3 | 4032-GTCACATTGGGTGGATGGACC-4022 | 916 | |
| 6. pol-env | Outer | 2013F1 | 3104-CTTATGAAACCCTCCCTACC-3123 | 2013R1 | 5136-GTGCACCGACTGGGGTCGCC-5117 | 2033 |
| Inner | 2013F3 | 3976-GGCCAGCTCCGGCGCCTGGCC-3996 | 2013R2 | 5131-CCGACTGGGGTCGCCAGGGAC-5111 | 1156 | |
| 7. env | Outer | PTLVENVF1 | 5040-AGACCAYCAACWCCATGGGTAA-5061 | PTLVENVR1 | 7256-CTYTGYCCRAAICCTGGRAARTGGGCTGA-7228 | 2217 |
| Inner | 8699GP46NF1 | 5057-CTACATTTCTCAAAATGCGGATCCTCC-5083 | 8699GP46CR1 | 6004-GACGGCTCGGCGCTGACGA-5984 | 947 | |
| 8. env | Outer | PTLVENVF1 | 5040-AGACCAYCAACWCCATGGGTAA-5061 | PTLVENVR1 | 7256-CTYTGYCCRAAICCTGGRAARTGGGCTGA-7228 | 2217 |
| Inner | 2013F4 | 5959-GTCCCCTGTCCCTGATCTCTCC-5980 | 2013R4 | 7179-CAGAGACCACAACTGCGGGGAC-7158 | 1221 | |
| 9. env-tax | Outer | OPH1F1 | 7080-AYCGGYGGTCCCASACTCC-7098) | PH2R | 7460-AAGGAGGGGAGTCGAGGGATAAGG-7437 | 381 |
| Inner | OPH1F2 | 7136-GCAGGAATAYACCACAGGCA-7155 | 2013VIR2 | 7437-GTATTGTAGAGGCGAGCTGA-7418 | 301 | |
| 10. tax | Outer | 2013F6 | 7391-CCTGGGACCCCATCGATGGAC-7411 | 8699TR5 | 7794-TTTGGTAGGGATTTTTGTTAGGAAGG-7769 | 404 |
| Inner | 2013F7 | 7408-TTTGGTAGGGATTTTTGTTAGGAAGG-7429 | 8699TR5 | 7794-TTTGGTAGGGATTTTTGTTAGGAAGG-7769 | 387 | |
| 11. tax | Outer | 2013F7 | 7408-GGACGCGTTGTCAGCTCGCCTC-7429 | 8699TF8R | 7899-TGGTGCGCGGGTGGGCTGAAACAGG-7875 | 492 |
| Inner | 8699TF5 | 7673-GCACCATCGTGTGCTGATACCTC-7695 | 8699TR6 | 7849-GGATAAGTATGGCCCCTGTAC-7829 | 177 | |
| 12. tax-LTR | Outer | 8699TF6 | 7750-CATCCGGACCAACTAGGGGCCTTC-7773 | 2013LF2R | 8298-CTGGGTGCGAGACGTCCCCTAGACAG-8273 | 547 |
| Inner | 8699TF7 | 7776-AACAAAAATCCCTACCAAACGCTT-7799 | 2013LF1R | 8277-GACAGATGATTCAACTGTATGCCCTTTGGC-8248 | 502 | |
Table 2.
| PTLV-3 (subtype A) | PTLV-3 (subtype B) | PTLV-3 (subtype D) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| STLV-3 (TGE-2117) |
STLV-3 (PH969) |
STLV-3 (CTO604) |
STLV-3 (NG409) |
STLV-3 (PPA-F3) |
HTLV-3 (Pyl43) |
HTLV-3 (Lok18) |
HTLV-3 (2026ND) |
STLV-3 (8699cmo) |
|
| Genome | 77.3 | 77.3 | 77.5 | 77.4 | 77.5 | 74.8 | 77.7 | 77.4 | 99.8 |
| LTR | 73.9 | 74.4 | 75.9 | 75.6 | 74.7 | 76.1 | 76.6 | 75.2 | 98.9 |
| gag | 79.8 (88.6) | 79.3 (87.9) | 79.7 (88.6) | 79.4 (87.7) | 80.2 (88.6) | 79.6 (88.4) | 79.6 (87.8) | 79.4 (87.4) | 99.8 (99.8) |
| p19 | 78.0 (81.3) | 79.6 (82.1) | 77.0 (82.1) | 76.6 (78.9) | 76.6 (81.3) | 76.8 (82.1) | 77.3 (82.1) | 75.3 (80.5) | 100.0 (100) |
| p24 | 82.6 (95.8) | 80.9 (93.5) | 82.1 (95.3) | 82.1 (96.3) | 83.5 (96.3) | 82.6 (95.8) | 81.8 (94.9) | 82.2 (93.9) | 99.8 (100) |
| p15 | 75.7 (81.4) | 73.4 (80.5) | 77.7 (81.4) | 76.8 (79.1 | 77.2 (80.2) | 76.5 (79.1) | 77.3 (80.2) | 78.4 (81.4) | 99.6 (100) |
| pro | 71.9 (74.7) | 72.9 (74.2) | 74.3 (75.3) | 73.4 (74.2) | 72.6 (74.7) | 73.5 (74.7) | 73.9 (74.7) | 73.8 (76.4) | 99.8 (100) |
| pol | 76.2 (81.5) | 76.1 (82.0) | 75.9 (81.1) | 75.7 (81.4) | 75.4 (81.80 | 75.8 (81.7) | 76.1 (80.1) | 75.8 (81.7) | 99.6 (99.3) |
| env c | 77.3 (84.4) | 77.3 (83.2) | 77.1 (83.4) | 78.4 84.0) | 78.3 (85.0) | 77.3 (83.8) | 77.4 (83.8) | 78.7 (84.8) | 99.5 (99.6) |
| SU | 76.6 (80.4) | 75.7 (78.5) | 75.5 (79.5) | 77.6 (80.1) | 76.9 (80.8) | 76.1 (79.8) | 76.1 (79.8) | 77.8 (80.8) | 99.3 (99.6) |
| TM | 78.5 (91.5) | 80.1 (91.5) | 79.5 (89.8) | 79.8 (90.9) | 80.6 (92.6) | 79.7 (90.9) | 79.8 (90.9) | 80.4 (92.1) | 99.8 (99.4) |
| rex | 88.2 (73.7) | 87.8 (72.1) | 86.7 (69.4) | 87.4 (72.7) | 87.1 (71.6) | 86.9 (69.9) | 86.7 (69.4) | 86.3 (71.0) | 100.0 (100) |
| tax | 84.6 (90.3) | 84.6 (90.3) | 83.5 (89.2) | 83.7 (89.2) | 83.7 (89.2) | 83.7 (89.7) | 83.9 (89.7) | 82.6 (87.7) | 99.9 (100) |
Amino acid similarities given in parentheses.
Full-length genomes are not available from PTLV-3 subtype C for comparison.
SU, surface; TM, transmembrane
Like STLV-3D, a putative antisense protein (ASP), reportedly involved in viral replication and persistence, is present on the negative RNA strand of HTLV-3D(Cam2013AB). In addition, all Tax regulatory and functional motifs were conserved, including a potential PDZ domain in the C-terminus, an important binding site for Tax in mediating signal transduction and interleukin-2-independent growth induction for T-cell transformation (Rousset et al., 1998; Tsubata et al., 2005). These results suggest Tax3D interactions with cellular regulatory pathways similar to those of both PTLV-1 and PTLV-3 (Chevalier et al., 2006; Sintasath et al., 2009b; Switzer et al., 2006b; Switzer et al., 2009). All major structural, enzymatic, and regulatory gene regions of HTLV-3d(Cam2013AB) are intact and suggest viral replication and a predicted pathogenic potential comparable to other PTLV-3s (Fig. 5) (Calattini et al., 2006; Chevalier et al., 2007; Sintasath et al., 2009b; Switzer et al., 2006b). Like STLV-3D(Cmo8699AB), a possible accessory protein (ORF-I) of unknown function and 131 aa in length is present in HTLV-3D between env and the 3′ LTR (Fig. 5, nucleotides 6559 – 6951).
Discovery of a new HTLV-3 and Estimation of Divergence Dates for PTLV-3D
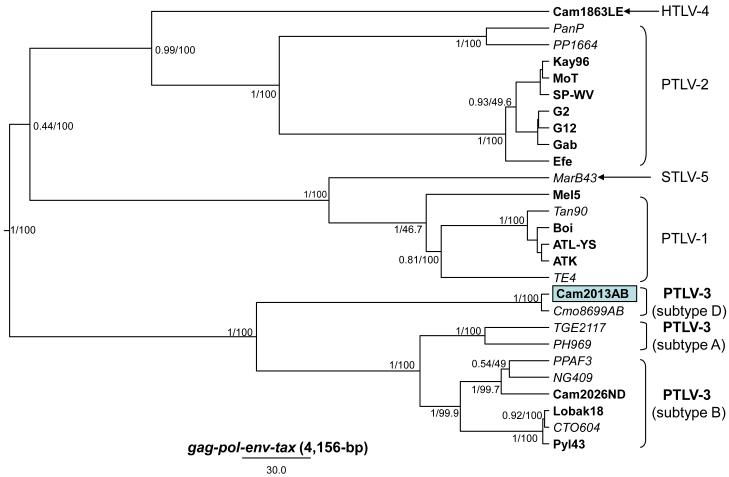
Robust phylogenetic analysis of first and second codon positions (12cdp) of concatenated 4156-bp gag-pol-env-tax nucleotide sequences using Bayesian and ML inference, as described in more detail elsewhere, (Sintasath et al., 2009b) confirmed with strong statistical support the high genetic similarity of STLV-3D(Cmo8699AB) and HTLV-3(Cam2013AB) (Fig 6). Both simian and human viruses clustered tightly in a distinct lineage independent of other PTLV-3 (Fig. 6). These results strongly support the discovery of a highly divergent HTLV-3 that we provisionally name HTLV-3D. Our taxonomic classification is based on guidelines used by the International Committee on Taxonomy of Viruses (ICTV, http://www.ictvonline.org) and those published elsewhere for deltaretroviruses using recommendations described for the nomenclature of new HTLVs (Sintasath et al., 2009b; Switzer et al., 2006b; Switzer et al., 2009). Specifically, the subtype classification is based upon highly supported phylogenetic division of the subtypes and genetic distances of at least 5% across the genome, as recently proposed (Sintasath et al., 2009b). This classification system is consistent with the phylogenetic relationships and genetic divergence seen between HTLV-1 and HTLV-2 subtypes using only LTR sequences (Liegeois et al., 2008; Mahieux et al., 1998; Mahieux et al., 2000a; Sintasath et al., 2009a; Switzer et al., 1995).
Figure 6.
Identification of a new HTLV-3 subtype by phylogenetic analysis of concatenated gag-pol-env-tax sequences. Bayesian and ML trees inferred from 1st and 2nd codon positions (cdp) of 4,156-bp concatamer alignments and 200 million MCMC chains and a relaxed molecular clock. Posterior probabilities over ML bootstrap values are provided at each node in the concatamer gene tree. HTLV sequences are in bold, STLV sequences are italicized.
To gain insight into the evolutionary history of this new virus, molecular dating of HTLV-3D was estimated using a Bayesian Markov Chain Monte Carlo (MCMC) approach and a relaxed molecular clock implemented in the BEAST software package (Drummond and Rambaut, 2007), as previously described (Lemey et al., 2005; Sintasath et al., 2009b; Switzer et al., 2006b; Switzer et al., 2009). Calibration of the relaxed molecular clock was done using the dates of 40,000 – 60,000 years ago (ya) for the Melanesian HTLV-1 lineage (HTLV-1mel) and 15,000-30,000 ya for the most recent common ancestor of HTLV-2a/HTLV-2b native American strains as strong priors in a Bayesian MCMC relaxed molecular clock method. These dates and the corresponding HTLV sequences represent best estimates of human colonization of Australo-Melanasia and the Americas, respectively. The use of two calibration points has previously been shown to provide more reliable estimates of PTLV substitution rates than a single calibration date (Lemey et al., 2005; Switzer et al., 2009). The upper and lower divergence times estimated from anthropological data were used to define the interval of a strong uniform prior distribution from which the MCMC sampler would sample possible divergence times for the corresponding node in the tree. Using these methods a more recent common ancestor for HTLV-3D and STLV-3D(Cmo8699AB) was inferred about 3,600 ya. The most recent common ancestor (MRCA) of the PTLV-3 clade was inferred to have occurred 115,688 ya which is consistent with dates reported previously (Sintasath et al., 2009b; Switzer et al., 2006b; Switzer et al., 2009). Similar divergence dates were obtained using an alignment of 628-bp 12cdp tax sequences that included all PTLV-3 subtypes, except the mean substitution rates were slightly higher for the partial tax alignment (5.92 × 10−7 vs 5.41 × 10−7 substitutions/site/year).
Cameroon continues to be an epicenter for the emergence of a broad range of divergent retroviruses (Calattini et al., 2009; Calattini et al., 2005; Wolfe et al., 2005b). All HIV-1 groups (M, N, and O) circulate in Cameroon (Hahn et al., 2000), including the newly identified HIV-1 group P that is genetically similar to a simian immunodeficiency virus (SIV) found in wild gorillas (Plantier et al., 2009). We describe here the identification of a new HTLV-3 in a Cameroonian primate hunter that we call subtype D based on its high genetic relatedness to STLV-3D discovered recently in two NHPs located within 20 kilometers of the hunter’s village documented by GPS mapping performed at the time of both human and animal specimen collections (Fig. 1). These results strongly support the phylogeographic clustering of STLV-3 and HTLV-3, a finding consistent with those of STLV-1 and HTLV-1, re-affirming further the ease of transmission of STLVs to humans who live in proximity to and are exposed to NHPs via hunting, butchering, or keeping NHP pets (Calattini et al., 2009; Calattini et al., 2005; Wolfe et al., 2005b). Interestingly, none of the participants reported bite wounds following NHP exposure suggesting that cross-species transmission occured by contact with infected body fluids possibly following a mucocutaneous or other exposure. Our findings also demonstrate that HTLV-3 genetic variation is driven by the diversity of STLV-3 existing in NHPs and crossing into humans and that these zoonotic transmissions have occurred on at least two separate occasions. Given the wide genetic diversity and distribution of STLV-3 over the African biomes, the historical exposure of Africans to NHPs, and the inferred ancient origin of STLV-3, it is possible that more HTLV-3 variants will likely be found.
While HTLV-1-based screening tools such as EIA have been successful in finding HTLV-3 and HTLV-4, the sensitivity of the current screening assays to detect these viruses is currently unknown. Likewise, HTLV-3 and HTLV-4 exhibit a broad range of reactivity in HTLV-1-based WB tests, thus requiring PCR and sequence analysis for confirmation of infection (Calattini et al., 2009; Calattini et al., 2005; Switzer et al., 2006a; Wolfe et al., 2005b). Therefore, better diagnostic tools incorporating HTLV-3 and HTLV-4 antigens are required to improve serologic testing for these novel HTLVs and to determine whether these infections are currently misdiagnosed as HTLV-1 or HTLV-2. If so, the prevalence of HTLV-3 infections may be greater than currently appreciated. The absence of HTLV testing of blood donors in many African countries, including Cameroon, provides opportunities for occult dissemination of both HTLV-3 and HTLV-4. Thus, to better understand the public health significance of these new HTLVs, studies are needed to define their prevalence in the general population.
Our finding of a comparatively recent ancestor for STLV-3D and HTLV-3D suggests a contemporary zoonotic transmission event occurring in close geographic proximity in the forests of Cameroon, most likely via exposures occuring at the hunter/primate interface. This hypothesis is supported by both viruses having nearly identical LTR sequences, which is the most divergent region of the PTLV genome, suggesting a relatively short evolutionary history of HTLV-3D. This high genetic similarity is consistent with that observed in PTLV transmission pairs (Lal et al., 1993; Van Dooren et al., 2004a; Van Dooren et al., 2004b). Even less divergence is seen in LTR sequences of both HTLV-3(Pyl43) and HTLV-3(Lobak18) and STLV-3(CTO604) implying a more recent primate-to-human transmission for these viruses than that for HTLV-3D (Calattini et al., 2009; Calattini et al., 2007; Meertens et al., 2002). The relatively recent emergence of HTLV-3 and novel STLV-1-like viruses may be the result of further encroachment into the forests of Cameroon by hunters and lumber companies that employ hunters to provide food for their workers and the increased demand for bushmeat (Wolfe et al., 2005a). Additional studies are required to determine the prevalence of HTLV-3, to investigate further the evolutionary history of HTLV-3, and to evaluate its pathogenic potential and person-to-person transmissibility, all of which will help to define the public health significance of this new human retrovirus. The use of GPS defined locations of infected animals and humans, as described here, will be instrumental in understanding further the zoonotic transmission and evolutionary history of PTLVs and other pathogens.
Acknowledgements
NDW was supported by awards from the National Institutes of Health Director’s Pioneer Award (Grant DP1-OD000370), the WW Smith Charitable Trust, the US Military HIV Research Program, and grants from the NIH Fogarty International Center (International Research Scientist Development Award Grant 5 K01 TW000003-05), AIDS International Training and Research Program (Grant 2 D 43 TW000010-17-AITRP), and the National Geographic Society Committee for Research and Exploration (Grant #7762-04). DMS was funded through a National Science Foundation Graduate Research Fellowship and the Edward and Kathy Ludwig Scholarship. This research was supported in part by the Global Viral Forecasting Initiative, Google.org, and The Skoll Foundation. We thank the entire staff of GVFI-Cameroon for their support and assistance. The collaboration of numerous hunters participating voluntarily in the GVFI surveillance program is also appreciated. The Cameroon Ministry of Defense, Ministry of Scientific Research and Innovation, Ministry of Forestry and Fauna and Ministry of Public Health provided authorizations and support for this work. We also thank Dr. Donald Burke for helping to establish these study sites. Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blattner WA, Clark JW, Gibbs WN, Williams CK, Nomura A, Mann D, Saxinger C, Robert-Guroff M, Gallo RC. HTLV: epidemiology and relationship to disease. Princess Takamatsu Symp. 1984;15:93–108. [PubMed] [Google Scholar]
- Calattini S, Betsem E, Bassot S, Chevalier SA, Mahieux R, Froment A, Gessain A. New strain of human T lymphotropic virus (HTLV) type 3 in a Pygmy from Cameroon with peculiar HTLV serologic results. J Infect Dis. 2009;199(4):561–4. doi: 10.1086/596206. [DOI] [PubMed] [Google Scholar]
- Calattini S, Betsem E, Froment A, Bassot S, Chevalier SA, Mahieux R, Gessain A. Identification and Complete Sequence Analysis of a New HTLV-3 Strain from South Cameroon; 13th International Conference on Human Retrovirology: HTLV and Related Viruses; Hakone, Japan. 2007. [Google Scholar]
- Calattini S, Chevalier SA, Duprez R, Afonso P, Froment A, Gessain A, Mahieux R. Human T-cell lymphotropic virus type 3: complete nucleotide sequence and characterization of the human tax3 protein. J Virol. 2006;80(19):9876–88. doi: 10.1128/JVI.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S, Chevalier SA, Duprez R, Bassot S, Froment A, Mahieux R, Gessain A. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology. 2005;2(1):30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier SA, Meertens L, Pise-Masison C, Calattini S, Park H, Alhaj AA, Zhou M, Gessain A, Kashanchi F, Brady JN, Mahieux R. The tax protein from the primate T-cell lymphotropic virus type 3 is expressed in vivo and is functionally related to HTLV-1 Tax rather than HTLV-2 Tax. Oncogene. 2006;25(32):4470–82. doi: 10.1038/sj.onc.1209472. [DOI] [PubMed] [Google Scholar]
- Chevalier SA, Walic M, Calattini S, Mallet A, Prevost MC, Gessain A, Mahieux R. Construction and characterization of a full-length infectious simian T-cell lymphotropic virus type 3 molecular clone. J Virol. 2007;81(12):6276–85. doi: 10.1128/JVI.02538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Van Dooren S, Liegeois F, Pourrut X, Abela B, Loul S, Mpoudi-Ngole E, Vandamme A, Delaporte E, Peeters M. Simian T-cell leukemia virus (STLV) infection in wild primate populations in Cameroon: evidence for dual STLV type 1 and type 3 infection in agile mangabeys (Cercocebus agilis) J Virol. 2004;78(9):4700–9. doi: 10.1128/JVI.78.9.4700-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessain A, de The G. Geographic and molecular epidemiology of primate T lymphotropic retroviruses: HTLV-I, HTLV-II, STLV-I, STLV-PP, and PTLV-L. Adv Virus Res. 1996;47:377–426. doi: 10.1016/s0065-3527(08)60740-x. [DOI] [PubMed] [Google Scholar]
- Gessain A, Mahieux R. Epidemiology, origin and genetic diversity of HTLV-1 retrovirus and STLV-1 simian affiliated retrovirus. Bull Soc Pathol Exot. 2000;93(3):163–71. [PubMed] [Google Scholar]
- Gessain A, Mahieux R, de The G. Genetic variability and molecular epidemiology of human and simian T cell leukemia/lymphoma virus type I. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl 1):S132–45. doi: 10.1097/00042560-199600001-00022. [DOI] [PubMed] [Google Scholar]
- Gessain A, Meertens L, Mahieux R. Molecular epidemiology of human T cell leukemia/lymphoma viruses type 1 and type 2 (HTLV-1/2) and related simian retroviruses (STLV-1, STLV-2, and STLV-L/3) In: Leitner T, editor. The molecular epidemiology of human viruses. 1st ed. Kluwer Academic Publishers; Boston: 2002. [Google Scholar]
- Goubau P, Van Brussel M, Vandamme AM, Liu HF, Desmyter J. A primate T-lymphotropic virus, PTLV-L, different from human T-lymphotropic viruses types I and II, in a wild-caught baboon (Papio hamadryas) Proc Natl Acad Sci U S A. 1994;91(7):2848–52. doi: 10.1073/pnas.91.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287(5453):607–14. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Khabbaz RF, Fukuda K, Kaplan JE. Guidelines for counseling human T-lymphotropic virus type I (HTLV-I)- and HTLV type II-infected persons. Transfusion. 1993;33(8):694. doi: 10.1046/j.1537-2995.1993.33893342756.x. [DOI] [PubMed] [Google Scholar]
- Lal RB, Gongora-Biachi RA, Pardi D, Switzer WM, Goldman I, Lal AF. Evidence for mother-to-child transmission of human T lymphotropic virus type II. J Infect Dis. 1993;168(3):586–91. doi: 10.1093/infdis/168.3.586. [DOI] [PubMed] [Google Scholar]
- Lemey P, Pybus OG, Van Dooren S, Vandamme AM. A Bayesian statistical analysis of human T-cell lymphotropic virus evolutionary rates. Infect Genet Evol. 2005;5(3):291–8. doi: 10.1016/j.meegid.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Liegeois F, Lafay B, Switzer WM, Locatelli S, Mpoudi-Ngole E, Loul S, Heneine W, Delaporte E, Peeters M. Identification and molecular characterization of new STLV-1 and STLV-3 strains in wild-caught nonhuman primates in Cameroon. Virology. 2008;371(2):405–17. doi: 10.1016/j.virol.2007.09.037. [DOI] [PubMed] [Google Scholar]
- Mahieux R, Chappey C, Georges-Courbot MC, Dubreuil G, Mauclere P, Georges A, Gessain A. Simian T-cell lymphotropic virus type 1 from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type 1 subtype D. J Virol. 1998;72(12):10316–22. doi: 10.1128/jvi.72.12.10316-10322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieux R, Chappey C, Meertens L, Mauclere P, Lewis J, Gessain A. Molecular characterization and phylogenetic analyses of a new simian T cell lymphotropic virus type 1 in a wild-caught african baboon (Papio anubis) with an indeterminate STLV type 2-like serology. AIDS Res Hum Retroviruses. 2000a;16(18):2043–8. doi: 10.1089/088922200750054774. [DOI] [PubMed] [Google Scholar]
- Mahieux R, Gessain A. HTLV-1 and associated adult T-cell leukemia/lymphoma. Rev Clin Exp Hematol. 2003;7(4):336–61. [PubMed] [Google Scholar]
- Mahieux R, Horal P, Mauclere P, Mercereau-Puijalon O, Guillotte M, Meertens L, Murphy E, Gessain A. Human T-cell lymphotropic virus type 1 gag indeterminate western blot patterns in Central Africa: relationship to Plasmodium falciparum infection. J Clin Microbiol. 2000b;38(11):4049–57. doi: 10.1128/jcm.38.11.4049-4057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns A, Blattner WA. The epidemiology of the human T-cell lymphotrophic virus type I and type II: etiologic role in human disease. Transfusion. 1991;31(1):67–75. doi: 10.1046/j.1537-2995.1991.31191096189.x. [DOI] [PubMed] [Google Scholar]
- Manns A, Hisada M, La Grenade L. Human T-lymphotropic virus type I infection. Lancet. 1999;353(9168):1951–8. doi: 10.1016/s0140-6736(98)09460-4. [DOI] [PubMed] [Google Scholar]
- Meertens L, Gessain A. Divergent simian T-cell lymphotropic virus type 3 (STLV-3) in wild-caught Papio hamadryas papio from Senegal: widespread distribution of STLV-3 in Africa. J Virol. 2003;77(1):782–9. doi: 10.1128/JVI.77.1.782-789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Mahieux R, Mauclere P, Lewis J, Gessain A. Complete sequence of a novel highly divergent simian T-cell lymphotropic virus from wild-caught red-capped mangabeys (Cercocebus torquatus) from Cameroon: a new primate T-lymphotropic virus type 3 subtype. J Virol. 2002;76(1):259–68. doi: 10.1128/JVI.76.1.259-268.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Rigoulet J, Mauclere P, Van Beveren M, Chen GM, Diop O, Dubreuil G, Georges-Goubot MC, Berthier JL, Lewis J, Gessain A. Molecular and phylogenetic analyses of 16 novel simian T cell leukemia virus type 1 from Africa: close relationship of STLV-1 from Allenopithecus nigroviridis to HTLV-1 subtype B strains. Virology. 2001;287(2):275–85. doi: 10.1006/viro.2001.1018. [DOI] [PubMed] [Google Scholar]
- Meertens L, Shanmugam V, Gessain A, Beer BE, Tooze Z, Heneine W, Switzer WM. A novel, divergent simian T-cell lymphotropic virus type 3 in a wild-caught red-capped mangabey (Cercocebus torquatus torquatus) from Nigeria. J Gen Virol. 2003;84(Pt 10):2723–7. doi: 10.1099/vir.0.19253-0. [DOI] [PubMed] [Google Scholar]
- Nerrienet E, Meertens L, Kfutwah A, Foupouapouognigni Y, Gessain A. Molecular epidemiology of simian T-lymphotropic virus (STLV) in wild-caught monkeys and apes from Cameroon: a new STLV-1, related to human T-lymphotropic virus subtype F, in a Cercocebus agilis. J Gen Virol. 2001;82(Pt 12):2973–7. doi: 10.1099/0022-1317-82-12-2973. [DOI] [PubMed] [Google Scholar]
- Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemée V, Damond F, Robertson DL, Simon F. A new human immunodeficiency virus derived from gorillas. Nature Medicine. 2009;15(8):871–2. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- Roucoux DF, Murphy EL. The epidemiology and disease outcomes of human T-lymphotropic virus type II. AIDS Rev. 2004;6(3):144–54. [PubMed] [Google Scholar]
- Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16(5):643–54. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- Sintasath DM, Wolfe ND, Lebreton M, Jia H, Garcia AD, Le Doux-Diffo J, Tamoufe U, Carr JK, Folks TM, Mpoudi-Ngole E, Burke DS, Heneine W, Switzer WM. Simian T-lymphotropic virus diversity among nonhuman primates, Cameroon. Emerg Infect Dis. 2009a;15(2):175–84. doi: 10.3201/eid1502.080584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintasath DM, Wolfe ND, Zheng HQ, LeBreton M, Peeters M, Tamoufe U, Djoko CF, Diffo JL, Mpoudi-Ngole E, Heneine W, Switzer WM. Genetic characterization of the complete genome of a highly divergent simian T-lymphotropic virus (STLV) type 3 from a wild Cercopithecus mona monkey. Retrovirology. 2009b;6:97. doi: 10.1186/1742-4690-6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer WM, Hewlett I, Aaron L, Wolfe ND, Burke DS, Heneine W. Serologic testing for human T-lymphotropic virus-3 and -4. Transfusion. 2006a;46(9):1647–8. doi: 10.1111/j.1537-2995.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- Switzer WM, Pieniazek D, Swanson P, Samdal HH, Soriano V, Khabbaz RF, Kaplan JE, Lal RB, Heneine W. Phylogenetic relationship and geographic distribution of multiple human T-cell lymphotropic virus type II subtypes. J Virol. 1995;69(2):621–32. doi: 10.1128/jvi.69.2.621-632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer WM, Qari SH, Wolfe ND, Burke DS, Folks TM, Heneine W. Ancient origin and molecular features of the novel human T-lymphotropic virus type 3 revealed by complete genome analysis. J Virol. 2006b;80(15):7427–38. doi: 10.1128/JVI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer WM, Salemi M, Qari SH, Jia H, Gray RR, Katzourakis A, Marriott SJ, Pryor KN, Wolfe ND, Burke DS, Folks TM, Heneine W. Ancient, independent evolution and distinct molecular features of the novel human T-lymphotropic virus type 4. Retrovirology. 2009;6:9. doi: 10.1186/1742-4690-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubata C, Higuchi M, Takahashi M, Oie M, Tanaka Y, Gejyo F, Fujii M. PDZ domain-binding motif of human T-cell leukemia virus type 1 Tax oncoprotein is essential for the interleukin 2 independent growth induction of a T-cell line. Retrovirology. 2005;2:46. doi: 10.1186/1742-4690-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Brussel M, Salemi M, Liu HF, Gabriels J, Goubau P, Desmyter J, Vandamme AM. The simian T-lymphotropic virus STLV-PP1664 from Pan paniscus is distinctly related to HTLV-2 but differs in genomic organization. Virology. 1998;243(2):366–79. doi: 10.1006/viro.1998.9075. [DOI] [PubMed] [Google Scholar]
- Van Dooren S, Meertens L, Lemey P, Gessain A, Vandamme AM. Full-genome analysis of a highly divergent simian T-cell lymphotropic virus type 1 strain in Macaca arctoides. J Gen Virol. 2005;86(Pt 7):1953–9. doi: 10.1099/vir.0.80520-0. [DOI] [PubMed] [Google Scholar]
- Van Dooren S, Pybus OG, Salemi M, Liu HF, Goubau P, Remondegui C, Talarmin A, Gotuzzo E, Alcantara LC, Galvao-Castro B, Vandamme AM. The low evolutionary rate of human T-cell lymphotropic virus type-1 confirmed by analysis of vertical transmission chains. Mol Biol Evol. 2004a;21(3):603–11. doi: 10.1093/molbev/msh053. [DOI] [PubMed] [Google Scholar]
- Van Dooren S, Salemi M, Pourrut X, Peeters M, Delaporte E, Van Ranst M, Vandamme AM. Evidence for a second simian T-cell lymphotropic virus type 3 in Cercopithecus nictitans from Cameroon. J Virol. 2001;75(23):11939–41. doi: 10.1128/JVI.75.23.11939-11941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dooren S, Shanmugam V, Bhullar V, Parekh B, Vandamme AM, Heneine W, Switzer WM. Identification in gelada baboons (Theropithecus gelada) of a distinct simian T-cell lymphotropic virus type 3 with a broad range of Western blot reactivity. J Gen Virol. 2004b;85(Pt 2):507–19. doi: 10.1099/vir.0.19630-0. [DOI] [PubMed] [Google Scholar]
- Vandamme AM, Salemi M, Desmyter J. The simian origins of the pathogenic human T-cell lymphotropic virus type I. Trends Microbiol. 1998;6(12):477–83. doi: 10.1016/s0966-842x(98)01406-1. [DOI] [PubMed] [Google Scholar]
- Vandamme AM, Van Laethem K, Liu HF, Van Brussel M, Delaporte E, de Castro Costa CM, Fleischer C, Taylor G, Bertazzoni U, Desmyter J, Goubau P. Use of a generic polymerase chain reaction assay detecting human T-lymphotropic virus (HTLV) types I, II and divergent simian strains in the evaluation of individuals with indeterminate HTLV serology. J Med Virol. 1997;52(1):1–7. [PubMed] [Google Scholar]
- Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg Infect Dis. 2005a;11(12):1822–7. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, Torimiro JN, Prosser AT, Lebreton M, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Switzer WM. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A. 2005b;102(22):7994–9. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]