Abstract
The flavin-adenine-dinucleotide-binding BLUF domain constitutes a new class of blue-light receptors, and the N-terminal domain of AppA is a representative of this family. The BLUF domain is of special interest because it uses a rigid flavin rather than an isomerizable chromophore, such as a rhodopsin or phytochrome, for its light-activation process. Crystal and solution structures of several BLUF domains were recently obtained, and their overall structures are consistent. However, there is a key ambiguity regarding the position of a conserved tryptophan (Trp-104 in AppA), in that this residue was found either close to flavin (Trpin conformation) or exposed to the solvent (Trpout conformation). The location of Trp-104 is a crucial factor in understanding the photocycle mechanism of BLUF domains, because this residue has been shown to play an essential role in the activation of AppA. In this study, we demonstrated a Trpin conformation for the BLUF domain of AppA through direct observation of the vibrational spectrum of Trp-104 by ultraviolet resonance Raman spectroscopy, and also observed light-induced conformational and environmental changes in Trp-104. This study provides a structural basis for future investigations of the photocycle mechanism of BLUF proteins.
Introduction
Understanding protein function at the atomic level is a major challenge in the field of biophysics and requires a combination of structural and functional methods. We use photoreceptor proteins as a model system to understand in atomic detail how a chromophore and a protein interact to sense light and send a biological signal. In many biological photoreceptors, such as rhodopsins, phytochromes, photoactive yellow protein, and phototropins, gross structural changes are photoinduced in the chromophore molecule, triggering a series of protein structural changes that ultimately lead to the formation of a signaling state (1). In contrast, flavin-containing BLUF (blue-light using flavin adenine dinucleotide (FAD)) (2) proteins show a different photoactivation mechanism that is not accompanied by prominent structural changes in the chromophore (3). In BLUF proteins, the signaling state is characterized by a light-induced ∼10 nm red shift of the ultraviolet-visible (UV-vis) absorption spectrum (4–10) and an ∼16 cm−1 downshift of the C4=O stretching mode of the flavin ring (11–13). These relatively minor spectroscopic changes are characteristic of BLUF proteins.
Recently, crystal and solution structures of several BLUF domains were obtained (7,14–20). These structures show that the BLUF domain consists of a five-stranded β-sheet with two α-helices packed on one side of the sheet, and the noncovalently bound isoalloxazine ring of FAD positioned between the two α-helices. AppA from Rhodobacter sphaeroides is representative of BLUF proteins and functions as a transcriptional antirepressor of photosynthetic genes (3). Fig. 1 shows the x-ray structures of the BLUF domain of WT AppA (panel A) (14) and the Cys-20→Ser (C20S) mutant (panel B) (17). FAD is involved in an extensive hydrogen-bond network with the side chains lining its binding pocket, including a highly conserved tyrosine (Tyr-21), glutamine (Gln-63), and tryptophan (Trp-104). A light-induced rearrangement of the hydrogen-bond network is believed to result in the formation of a signaling state. In fact, site-specific mutation of Tyr-21 and Gln-63 results in loss of the light-induced spectral changes (4,21,22). Furthermore, in vivo and in vitro site-directed analyses of AppA (23) have demonstrated a crucial role for Trp-104 in the light-dependent antirepressor activity of AppA, although the Trp-104 mutants show a nearly normal red shift in the absorption spectrum (24,25). Because these spectroscopic and biochemical studies indicate that the active-site residues play an essential role in blue-light sensing by BLUF domains, information about their structure is required to understand the photocycle mechanism of BLUF proteins. However, although an overall protein structure is well established, there still exists a key ambiguity regarding the position and orientation of Gln-63 and Trp-104 with respect to the flavin moiety. As demonstrated in Fig. 1 A, the crystal structure of the BLUF domain of WT AppA shows that Trp-104 is found in the vicinity of FAD and forms a hydrogen bond with a side chain of Gln-63 (14). We denote this structure as Trpin. The Trpin conformation in the dark state is supported by previous ultrafast experiments (26) as well as recent fluorescence (27) and NMR (28) spectroscopic studies on the BLUF domain of AppA. On the other hand, the x-ray crystal structure of the C20S mutant of AppA showed the Trpout conformation, where Trp-104 is exposed to the solvent and is located >10 Å from Gln-63 (17). More recently, spectroscopic studies of the AppA BLUF domains showed an influence of a N-terminal truncation on the conformation and/or environment of Trp-104 and suggested that the Trpout conformation is a physiologically relevant structure (29). The Trpout conformation has been also observed in several BLUF proteins, such as PixD from Thermosynechococcus elongatus BP-1 (Tll0078) (15) and BlrB from R. sphaeroides (7). Furthermore, the crystal structures of PixD from Synechocystis sp. PCC6803 (Slr1694) show the Trpout conformation, with the exception that one of 10 crystallographic subunits adopts the Trpin conformation (18). It is clear that, although detailed structural information has been accumulated, an important question remains regarding the location of Trp-104. Furthermore, contradictory observations regarding the structures of Trp-104 make the photocycle mechanism of the BLUF domain a matter of controversy.
Figure 1.
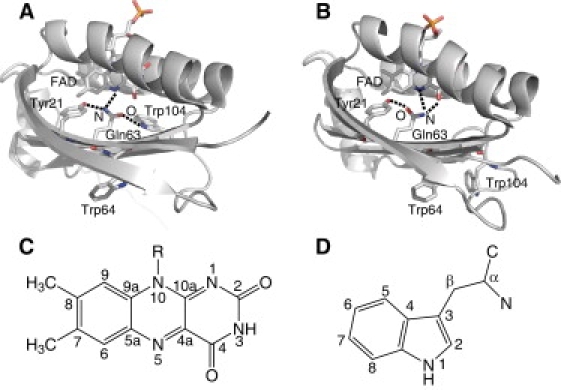
X-ray structures of the BLUF domain of AppA for the (A) WT and (B) C20S mutant. The vicinity of the FAD chromophore and two tryptophan residues is highlighted with the hydrogen-bond patterns lining the FAD binding pocket. The coordinates were taken from Protein Data Bank ID (A) 1YRX (14) and (B) 2IYG (17). Panels C and D show the structure and numbering of the FAD isoalloxazine ring and tryptophan residue, respectively.
Therefore, it is crucial to determine the position of Trp-104 in solution to elucidate the mechanism of the signaling state formation. To that end, we applied ultraviolet resonance Raman (UVRR) spectroscopy to BLUF proteins, for the first time to our knowledge. Raman spectroscopy can provide structural information via the sensitivity of vibrational bands to molecular conformation and environmental changes (30,31). If the excitation wavelength is out of resonance with the electronic transitions of a sample, both the chromophore and protein moieties will contribute to the Raman spectra. However, excitation at wavelengths below 250 nm results in strong enhancement of vibrational modes for aromatic residues such as tryptophan and tyrosine, with little interference from the chromophore and the remaining protein moiety. Thus, the UVRR spectra of the BLUF domain of AppA directly probe structural and/or environmental changes in Trp-104. As discussed below, the UVRR data provide evidence for the Trpin conformation and structural and/or environmental changes in Trp-104 during the light-activation process.
Materials and Methods
Sample preparations
The W64F and Q63L/W64F mutations were achieved by polymerase chain reaction (PCR) primer mutagenesis using Pyrobest DNA polymerase (TaKaRa, Shiga, Japan). The primers used were pET28-F (4), AppA126-R (24), AppA-W64F-F (5′-GGCGTCTTCTTCCAGTTCCTCGAAGGCCGCCCC-3′), AppA-W64F-R (5′-GGGGCGGCCTTCGAGGAACTGGAAGAAGACGCC-3′), AppA-Q63L-W64F-F (5′-CAGGGCGTCTTCTTCCTCTTCCTCGAAGGCCGCCC-3′), and AppA-Q63L-W64F-R (5′-GGGGCGGCCTTCGAGGAAGAGGAAGAAGACGCCCTG-3′). The AppA-W64F-F and AppA-W64F-R, and AppA-Q63L-W64F-F and AppA-Q63L-W64F-R were the complementary primer pairs to yield specific W64F and Q63L/W64F mutations, respectively. The first PCR was achieved with the primer pairs pET28-F/AppA-W64F-R (or AppA-Q63L-W64F-R) and AppA126-R/AppA-W64F-F (or AppA-Q63L-W64F-F), using the isolated plasmid pETAppA156 (4) as a template DNA. The two DNA fragments from the first PCR were mixed and then used as templates for the second PCR amplification using the primer pair pET28-F/AppA126-R. The second PCR products were digested with NdeI and EcoRI, and cloned into NdeI-EcoRI-cut pTYB12 vector (New England BioLabs, Ipswich, MA). The resulting plasmids pTYAppA126-W64F and pTYAppA126-Q63L-W64F were used to transform Escherichia coli strain BL21(DE3) (Novagen, San Diego, CA), and the resulting strains were used to express AppA126 W64F and Q63L/W64F mutant proteins, respectively. Previously published expression plasmids for AppA126 WT (11), Q63L (22), and W104A (24) mutant proteins were used, and the recombinant proteins were expressed and purified as described previously (11).
UVRR spectroscopy
The UVRR spectrometer is composed of a hollow cathode HeAg Laser (70-224SL; Photon Systems, Covina, CA), a 0.5 m single spectrometer (Spex 500M) equipped with a 3600 groove/mm holographic grating, and a liquid nitrogen-cooled, UV-coated charge coupled device (CCD) detector (Spec-10:400B; Roper Scientific, Trenton, NJ). The HeAg laser was run at 15 Hz and produced a 224.3 nm light of ∼100 μs pulse width. The system was operated to produce 0.3 μJ/pulse at the sample. A 90° scattering geometry was employed, and the scattered photons were collected and focused onto the entrance slit of a spectrometer by using two quartz plano convex lenses. A polarization scrambler was placed at the entrance slit to remove the effects of polarization from the spectrometer throughput. A Triax190 spectrometer (HORIBA Jobin Yvon, Edison, NJ) removed the excitation light, and the first order of the dispersed light by the 500 M spectrometer was imaged on the detector. An entrance slit width of 0.2 mm corresponds to a spectral resolution of ∼14 cm−1. All spectra were taken at room temperature (∼25°C), and an in-house-made software eliminated the noise spikes in the spectra caused by cosmic rays. All Raman spectra were calibrated by using neat cyclohexane as a standard. Sample volumes were 150 μL and were contained in a quartz spinning cell (10 mm in diameter). The cell was spun at 1600 rpm. The sample was dissolved in 5 mM Tris-HCl, 1 mM NaCl, pH 8.0, 0.1 M NaClO4. The protein concentration was estimated by measuring absorption spectra, and ∼50 μM of samples were used for the measurements. The extinction coefficients of the FAD chromophore are assumed to be the same in both the wild-type (WT) and mutants. The effect of sample degradation during the measurements was carefully checked by comparing the observed Raman spectra. If some spectral changes were recognized, the Raman spectrum was discarded.
For measurements of the signaling state, the sample solutions were irradiated by a 441.6 nm light from a helium-cadmium laser (IK5651R-G; Kimmon Electric, Tokyo, Japan) to produce the signaling state as a photostationary state. The pump beam was spatially displaced from the probe beam to avoid fluorescence generated by the pump beam, as well as possible contamination of short-lived intermediates. The laser power at the samples was 0.5 mW. The spectral intensities were normalized by adjusting the heights of the perchlorate bands at 934 cm−1 to be the same for both the dark and signaling states. Typically, a 1 mL of sample solutions was prepared and split into six solutions, and these sample solutions were used to measure the UVRR spectra for either the dark or the signaling state. This ensured that the concentrations of NaClO4 were exactly the same in both states.
Instrumental resolution and spectral shifts
The instrumental resolution of the UVRR spectrometer (∼14 cm−1) describes our ability to distinguish adjacent Raman bands in a spectrum. Relative frequency shifts in the Raman bands of different samples or conditions, however, can be determined with much greater accuracy (32). Important factors that affect the observable spectral shifts are the signal/noise ratios and the number of data points. Fig. 2 considers two Gaussian bands centered at 49 and 50 cm−1, with a full-width at half maximum of 14 cm−1. The number of data points in the simulated spectra shown in Fig. 2, A–C, is 51, which corresponds to 1 cm−1 increments. When there is no noise (Fig. 2 A), the small shift of 1 cm−1 can be observed, and their difference spectrum (a lower trace) exhibits a clear, derivative-like feature. In Fig. 2, B and C, on the other hand, two Gaussian bands contain 4% and 10% levels of Gaussian noise, respectively, and the 1 cm−1 shift becomes unclear when the noise level increases (Fig. 2, A–C). Panels D–F and G–I in Fig. 2 display similar simulated Gaussian bands with 5 and 10 cm−1 increments, respectively (the number of data points is 11 and 5, respectively). As the increment increases (and the number of data points decreases), it is difficult to recognize a 1 cm−1 shift even when there is no noise (Fig. 2 G). In this study, the increments of the reported UVRR spectra are ∼2 cm−1. Thus, the signal/noise ratios of the spectra mainly determine whether shifts of a few wave numbers can be detected. Note that we repeated the same measurements several times and confirmed that the frequency of the perchlorate bands at 934 cm−1 was accurate, with an accuracy of ±0.2 cm−1. This verifies the integrity of the observed shifts reported here.
Figure 2.
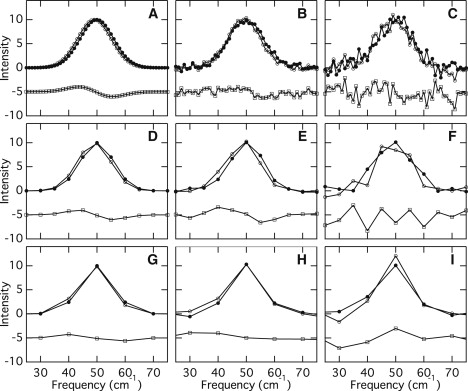
Simulated Gaussian bands centered at 49 cm−1 (open circle) and 50 cm−1 (closed circle) cm−1 with different noise and increments, and their difference spectrum (open square). In panels B, E, and H, a random value from a Gaussian distribution is added such that the standard deviation is 4% of the band intensity. In panels C, F, and I, a random value from a Gaussian distribution is added such that the standard deviation is 10% of the band intensity. Spectral increments: (A–C) 1 cm−1, (D– F) 5 cm−1, and (G– I) 10 cm−1.
Results
UVRR spectra for the dark state of AppA126
Fig. 3 shows the UVRR spectra for the dark state of WT AppA126, as well as aqueous solutions of tryptophan, tyrosine, and FAD at 224.3 nm excitation. A comparison of the spectra indicates that the UVRR spectrum for WT AppA126 (trace a) contains several Raman bands that arise mainly from tryptophan (trace b) and tyrosine residues (trace c); their mode labels (W for tryptophan, Y for tyrosine) (30) are indicated in the figure. The bands at 1551, 1011, and 761 cm−1 are assigned to W3 (C-C stretching of the pyrrole ring), W16 (benzene ring-breathing), and W18 (the indole ring-breathing), respectively (30,31). A doublet at 1360/1340 cm−1 is ascribed to W7, which arises from a Fermi resonance between a fundamental mode at ∼1340 cm−1 and a combination of two out-of-plane modes (30). The tryptophan bands dominate the UVRR spectra of AppA126 because 224.3 nm excitation occurs within the strong tryptophan π–π∗ electronic transition around 220 nm (denoted as Bb) (33,34). In addition to tryptophan, we observed Raman bands for a tyrosine residue. A main Raman band of tyrosine is observed at 1616 cm−1 and is assigned to the ring C-C stretching mode Y8a (30,31). Although this mode is overlapped with the W1 mode of tryptophan, the intensity ratio of W1 to W3 bands of tryptophan in an aqueous solution (trace b) indicates that Y8a is a dominant contributor to the band at 1616 cm−1. A remaining Raman band of tyrosine is observed at 1178 cm−1, which is due to Y9a (CH in-plane bending) (30,31). The 932 cm−1 band marked with an asterisk derives from the internal standard perchlorate band.
Figure 3.
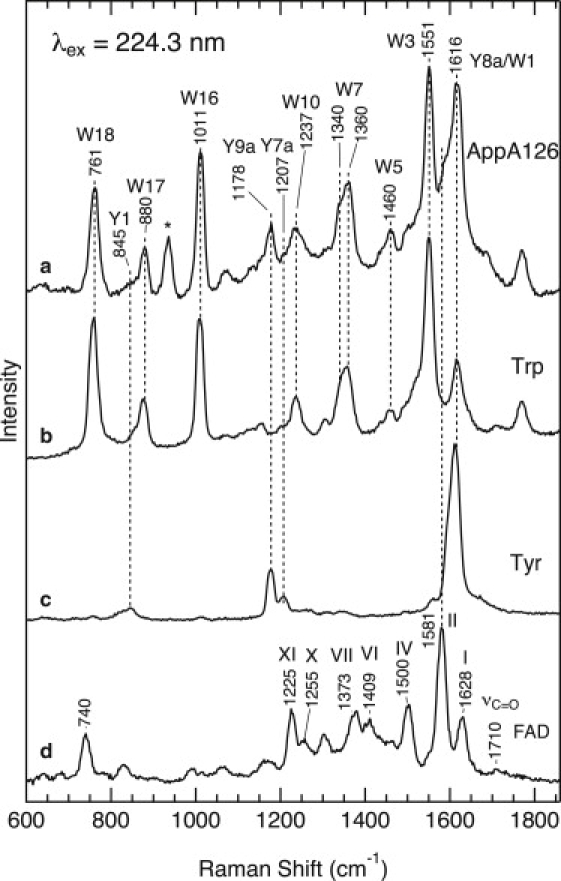
UVRR spectra of WT AppA126 (a), aqueous tryptophan (b), tyrosine (c), and FAD (d). The spectra were obtained at 224.3 nm excitation.
The upper part of Fig. 4 (traces a and b) compares the UVRR spectra for the dark state of WT AppA126 and Trp-64→Phe (W64F) and Trp-104→Ala (W104A) mutants. We selected these mutants for this study because their spectroscopic and/or biochemical characterizations have already been reported (23,24,27,29). In Fig. 4, the UVRR spectra are normalized by adjusting the heights of the tyrosine Y9a bands at 1178 cm−1 to be the same, because the number of tyrosine residues remains unchanged in the mutants. On the other hand, since the mutant proteins lack one of the tryptophan residues, the intensities of the tryptophan Raman bands, such as W3, W16, and W18, are significantly reduced. The difference Raman spectrum of trace c in Fig. 4, obtained by subtracting the W104A spectrum from the WT spectrum, displays the spectrum of the Trp-104 residue. Similarly, by subtracting the W64F spectrum from spectrum of WT, we can obtain the Raman spectrum of Trp-64 (trace d). For comparison, Fig. 4 also displays the UVRR spectrum of tryptophan in an aqueous solution (trace e). As can be seen in the difference spectrum (trace f), the UVRR spectra of Trp-104 and Trp-64 are distinctly different. For instance, the frequency of the W3 band for Trp-104 (1552 cm−1) is slightly higher than that for Trp-64 (1549 cm−1), and the band shape of the W7 doublet also differs between the two residues. In addition, we observed a larger intensity of the W3, W7, and W16 bands for Trp-104 compared to Trp-64. These spectral differences between the two residues are due to their different conformations and/or environments, as discussed in detail in the Discussion section.
Figure 4.
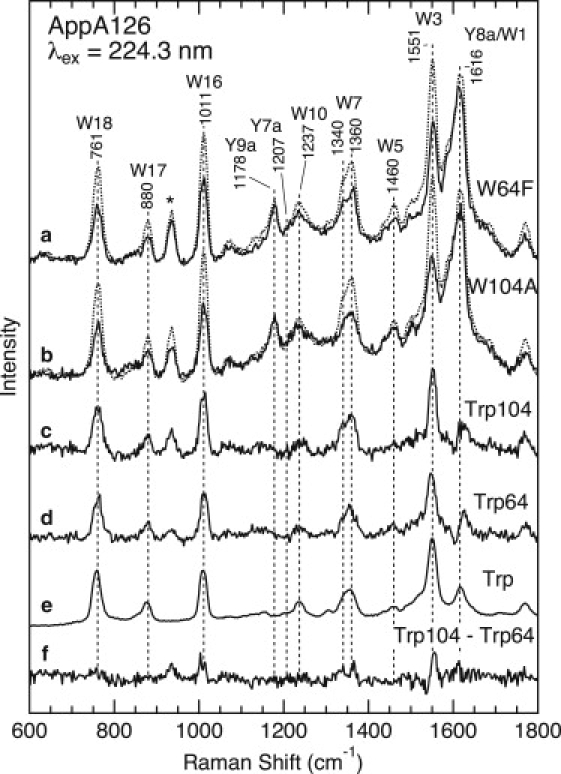
UVRR spectra of WT AppA126 and the W64F and W104A mutants in the dark state with 224.3 nm excitation. (a) WT AppA126 (dotted line) and the W64F mutant (solid line). (b) WT AppA126 (dotted line) and the W104A mutant (solid line). Estimated UVRR spectra for Trp-104 (c) and Trp-64,(d) as well as a spectrum of aqueous tryptophan (e), are also shown. The protein concentration was ∼50 μM and the buffer composition was 5 mM Tris-HCl, 1 mM NaCl, pH 8.0, 0.1 M NaClO4. The concentration of tryptophan was 1 mM, and the sample was dissolved in 10 mM Tris-HCl, pH 7.4. Trace f is the Trp-104 (c) minus Trp-64 (d) difference spectrum. The asterisk indicates a Raman band of ClO4−.
UVRR spectra for the signaling state of AppA126
As a next step, we examine the protein structural changes associated with the formation of the signaling state. The signaling state was produced as a photostationary state, and Fig. 5 shows the light-induced spectral changes in the UVRR spectra. The upper part of Fig. 5 compares the UVRR spectra of the dark (solid line in trace a) and signaling (dotted line) states of WT AppA126, and trace b is the light-induced difference spectrum. The samples contain 0.1 M ClO4− as an internal intensity standard, and its 934 cm−1 band was used to normalize the spectra for subtraction as described in Materials and Methods. As can be seen in the difference spectrum, some of the Raman bands give rise to difference features that reflect intensity changes and/or frequency shifts. A comparison with its parent spectra (trace a) indicates that most of the features in the difference spectrum arise from tryptophan residues. Fig. 5 also illustrates the light − dark difference spectra for the W64F (trace c) and W104A (trace d) mutants. As shown in the figure, the difference spectrum for the W64F mutant is very similar to that for WT, especially for features observed at ∼1550, 1350, and 1010 cm−1. On the other hand, there is no detectable feature for the W104A mutant. These observations demonstrate spectral changes in the UVRR spectrum of Trp-104, implying that Trp-104 experiences structural and/or environmental changes during the formation of the signaling state.
Figure 5.
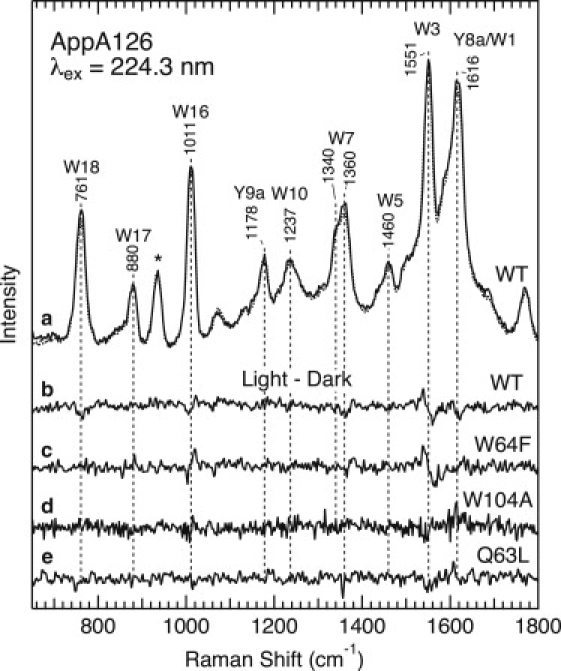
Light-induced changes in the UVRR spectra of WT AppA126 and the W64F, W104A, and Q63L mutants. (a) WT AppA126 in the dark (solid line) and light (dotted line) states. Light − dark difference spectra are shown for (b) WT AppA126 and the (c) W64F, (d) W104A, and (e) Q63L mutants.
The most characteristic feature in the difference spectrum is observed in the W3 band. A derivative-like difference signal indicates a downshift of the W3 band, and the shift for WT and the W64F mutant is also clearly seen in an expanded view of the UVRR spectra shown in Fig. S1 in the Supporting Material. The observed shifts are estimated as ∼−1 and −2 cm−1 for WT and W64F, respectively. A derivative-like feature at ∼1011 cm−1 indicates an upshift of the W16 mode in the signaling state. The small shifts for WT and the W64F mutant can be also discerned in an expanded view of the UVRR spectra shown in Fig. S1. Another feature in the difference spectrum is due to the W7 doublet of tryptophan. Fig. S1 displays the UVRR spectra in the region between 1270 and 1420 cm−1, and it can be seen that a higher-frequency component of the W7 doublet at 1359 cm−1 decreases in intensity upon formation of the signaling state.
Effects of Gln-63→Leu mutation on the UVRR spectra
We next examined the effects of the Q63L mutations and further explored the location of Trp-104. The upper part of Fig. 6 displays the UVRR spectra of the dark state for WT AppA126 (dotted line in trace a) and the Q63L mutant (solid line). In contrast to WT AppA126, this mutant did not show the characteristic light-induced spectral changes observed for WT AppA126 and the W64F mutant (see Fig. 5), which is consistent with a previous UV-vis absorption study (22). To visualize the effects of the mutation more clearly, Fig. 6 displays the Q63L − WT difference spectrum (trace b). It is clear that this mutation significantly affects the Raman bands of tryptophan residues. One of the remarkable spectral changes is the reduced intensity of the bands for W3, W7, W16, and W18. The mutation also causes ∼+2 and −2 cm−1 shifts for the W5 and W17 bands, respectively. In addition, the flavin moiety also contributes to the observed spectral changes. We assign the difference features near 1580 and 1225 cm−1 to the vibrational modes of the flavin ring, since the UVRR spectrum of FAD shows clear bands at the same frequencies (Fig. 3). The influence of the Q63L mutation on the Raman bands for the FAD chromophore is reasonable, since Gln-63 directly interacts with the isoalloxazine ring (14,16,17). Tyrosine Raman bands are also expected to show a sensitivity to the Q63L mutations because of a hydrogen bond between Tyr-21 and Gln-63 (see Fig. 1). A difference feature around 1250 cm−1 may be due to the Tyr Y7a band, but its origin is not clear at present. In Fig. 6, we also show the difference spectrum between the Q63L mutant and the signaling state of WT AppsA126 (trace c), and the difference spectra for the dark (trace b) and signaling states of WT AppA126 are very similar.
Figure 6.
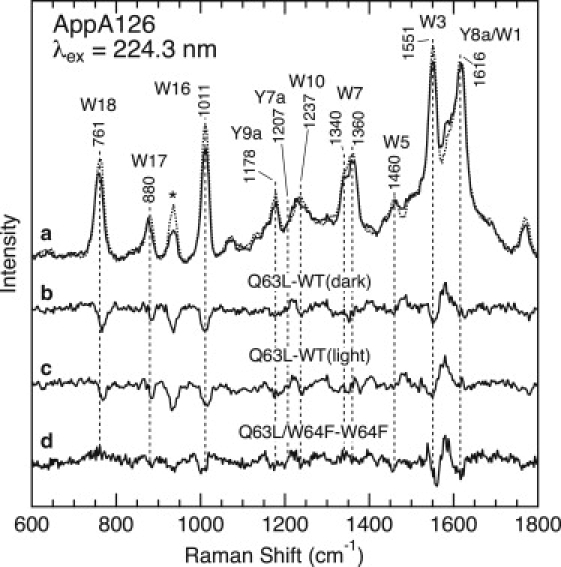
UVRR and difference spectra for WT AppA126 and the Q63L, W64F, and Q63L/W64F mutants. (a) UVRR spectra of WT AppA126 in the dark state (dotted line) and the Q63L mutant (solid line). (b) The Q63L mutant − WT AppA126 in the dark state. (c) The Q63L mutant −WT AppA126 in the light state. (d) The Q63L/W64F − W64F mutants in the dark state.
In Fig. 6, we compare the UVRR spectra of the W64F mutant and the Q63L/W64F double mutants in the dark state. Since these mutants contain only one tryptophan residue at position 104, we can see whether the Q63L mutation influences the Raman bands of Trp-104. Trace d is the Q63L/W64F − W64F difference spectrum, and a comparison of the difference spectra shown as traces b and d indicates that most of the difference features for the Q63L − WT difference spectrum are preserved for the Q63L/W64F − W64F difference spectrum, with the exception of a negative feature for the W18 band. This result demonstrates an influence of the Q63L mutation on the Raman bands for Trp-104, implying a direct interaction between Trp-104 and Gln-63.
Discussion
Structure of the BLUF domain of AppA
In this study, we found clear distinctions in the UVRR spectra between Trp-104 and Trp-64 (Fig. 4). One remarkable difference is the higher intensity of the W3, W7, and W16 bands for Trp-104 compared to those for Trp-64. UVRR studies on tryptophan and its models have shown that the Raman intensities of tryptophan reflect its environmental hydrophobicity (35,36). For example, Chi and Asher (36) measured the UVRR spectra of a tryptophan derivative in water-propanol solvent mixtures and found that the intensities of tryptophan Raman bands with 229 nm excitation increase with decreasing water composition due to a red shift of the Bb absorption band. A similar change in intensity was also observed for the UVRR spectra of tryptophan in water-ethylene glycol mixtures with 223 nm excitation (34). Thus, the larger intensity of the bands for Trp-104 relative to those for Trp-64 suggests a more hydrophobic environment around Trp-104, in agreement with recent fluorescence studies on AppA (27,29). This observation is also consistent with the Trpin conformation as found in WT AppA structures in which Trp-104 is buried within the interior of protein and Trp-64 is located on the surface of protein (14,16). However, we cannot completely exclude the Trpout conformation, because Trp-64 may be more buried as a consequence of dimerization of the protein molecule (37). To obtain further evidence concerning the location of Trp-104, we next consider the results for the Q63L mutants as described below.
As shown in Fig. 6, we found significant effects of the Q63L mutation on the Raman spectrum of Trp-104. This suggests a direct interaction, such as a hydrogen bond, between Gln-63 and Trp-104, and implies a Trpin conformation in the dark state of AppA126. The direct interaction between the two residues is consistent with x-ray crystal (14) and NMR solution (16) structures for the BLUF domain of WT AppA. As illustrated in Fig. 1 A, these structures show a hydrogen bond between Gln-63 and Trp-104. This accounts for the significant effects of the Q63L mutation, since the Gln-63/Trp-104 hydrogen bond will be broken in the mutant. In addition, the Trpin conformation is also consistent with the tryptophan fluorescence data of the W64F mutants of AppA (27,29), which exhibit a fluorescence maximum (∼330 nm) that is typical for a buried environment. On the other hand, our finding provides evidence against the Trpout conformation as found in the crystal structure of the AppA C20S mutant (17). The reason for this discrepancy is unclear, but the C20S mutation and/or a binding of a DTT molecule, which is present in the crystalline buffer, may affect the protein structure of the mutant. We also note that the Trpin conformation for AppA126 is also in conflict with a proposal based on a recent spectroscopic study of AppA in solution. Dragnea et al. (29) made an interesting finding that the presence or absence of the N-terminal amino acids affects the environment of Trp-104, and suggested that the N-terminal truncation in AppA17-133 (a BLUF domain of AppA containing amino acids 17–133) may be responsible for the observation of the Trpin conformation in the crystal structure of WT AppA (14). However, because their results do not provide direct evidence for the location of Trp-104, we believe the data presented here for the Q63L mutant favor the Trpin conformation for the dark state of AppA126.
It should be noted that the above conclusion contradicts the crystal structures of Tll0078 (15), BlrB (7), and nine of 10 crystallographic subunits of Slr1694 (18). Because our UVRR study does not address the location of the tryptophan residues in these BLUF proteins, this remains to be resolved in a future study. However, a recent site-directed mutagenesis study on Slr1694 showed that although Trp-91 (Trp-104 of AppA) is not functionally important, its neighboring residue, Met-93, is crucial (38). This result may imply that the location of the tryptophan residue does indeed differ between AppA and other BLUF proteins, such as Tll0078, BlrB, and Slr1694. Further UVRR studies on the other BLUF proteins are necessary to clarify this issue.
Another controversial issue regarding the structure of the BLUF domain is the orientation or structure of a conserved Gln-63 residue. In various x-ray crystal and NMR solution structures (14–18,28), as well as spectroscopic (13,22,39–41) and theoretical (42–44) studies, the dark-state orientation of Gln-63 has remained ambiguous. As illustrated in Fig. 1 A, the crystal structure for WT AppA shows that the amino group of Gln-63 is hydrogen-bonded to Tyr-21 and the FAD N5 atom. No hydrogen bonding to the flavin C4=O moiety occurs. In the crystal structure for the C20S mutant (Fig. 1 B) (17), on the other hand, Gln-63 is rotated by ∼180° compared to WT AppA, Tyr-21 is a hydrogen-bond donor to the Gln-63 carbonyl, and the Gln-63 amino group is hydrogen-bonded to both the N5 and C4=O moieties of FAD. Although our UVRR study does not provide direct information concerning the orientation of Gln-63, the Trpin conformation proposed here appears to favor the former orientation, where the carbonyl group of Gln-63 forms a hydrogen bond with the NH moiety of the indole ring in Trp-104.
Photocycle mechanism
The light-induced changes in the UVRR spectra of AppA126 shown in Fig. 5 provide spectroscopic evidence for structural and/or environmental changes in Trp-104 associated with the formation of a signaling state. A characteristic feature of the light-induced difference spectra is a downshift of the W3 mode. The frequency of the W3 band for tryptophan is a sensitive probe of the side-chain conformation, i.e., the frequency depends on the χ2,1 dihedral angle about the Cα-Cβ-C3=C2 linkage of the indole side chain (30,31). The observed downshifts of 1–2 cm−1 imply a decrease in the dihedral ∼5° in the signaling state based on a reported relationship between χ2,1 and the W3 frequency (30,31). The light-induced difference spectra also exhibit features for the W7 and W16 bands. For example, the band shape of the W7 doublet sensitively reflects environmental changes in a tryptophan residue, such as hydrophobic interactions and steric repulsion (30,31). Thus, the data shown in Fig. 5 unequivocally indicate light-induced environmental changes in Trp-104.
Previous crystallographic studies of BLUF domains (14,17,18) suggested that Trp-104 plays a central role in the formation of a signaling state. The importance of Trp-104 was also supported by a biochemical and genetic study showing that the Q63L and W104A mutants of AppA are insensitive to blue light and functionally locked in the signaling state (23). This observation suggests the importance of a hydrogen bond between Gln-63 and Trp-104, and that the protein structure of the Q63L mutant is similar to that of the signaling state of WT AppA126. However, our investigation demonstrates that the UVRR spectra of the Q63L mutant and the signaling state of WT AppA126 are clearly different (see Figs. 5 and 6). For instance, the formation of a signaling state causes an ∼2 cm−1 downshift of W3, whereas the Q63L mutation does not show this frequency shift. Another noticeable difference is that the characteristic reduction in the intensities of the tryptophan Raman bands caused by the Q63L mutation is not accompanied by the formation of a signaling state. Because the reduced intensities imply a decrease in hydrophobicity around a tryptophan residue, as discussed above, the Q63L mutation may lead to an altered protein conformation in which Trp-104 becomes exposed to solvent. On the other hand, Trp-104 of WT AppA126 could be buried in both the dark and signaling states, albeit with different conformations and environments. The buried nature of Trp-104 during a photocycle is also supported by recent fluorescence experiments on tryptophan in WT and mutants of AppA (27,29).
In conclusion, the UVRR data reported here strongly suggest that AppA126 in the dark state adopts a Trpin conformation in which the highly conserved tryptophan, Trp-104, is buried inside the protein. This result resolves a controversy concerning the location of the Trp-104 residue. Furthermore, we observed conformational changes in Trp-104 associated with the formation of the signaling state. This result is consistent with the proposal that Trp-104 mediates the light signal from the FAD cofactor to a protein surface, which ultimately generates a biological signal.
Supporting Material
One figure is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00102-5.
Supporting Material
Acknowledgments
This work was supported by a grant from the Mitsubishi Foundation to M.U.
References
- 1.van der Horst M.A., Hellingwerf K.J. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 2.Gomelsky M., Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 2005;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 3.Masuda S., Bauer C.E. The antirepressor AppA uses the novel flavin-binding BLUF domain as a blue-light-absorbing photoreceptor to control photosystem synthesis. In: Briggs W.R., Spudich J., editors. Handbook of Photosensory Receptors. Wiley-VCH Verlag GmbH; Weinheim: 2005. pp. 433–445. [Google Scholar]
- 4.Kraft B.J., Masuda S., Bauer C.E. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42:6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 5.Masuda S., Hasegawa K., Ono T.A. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of synechocystis sp. PCC6803. Biochemistry. 2004;43:5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 6.Masuda S., Bauer C.E. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 7.Jung A., Domratcheva T., Schlichting I. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 2005;102:12350–12355. doi: 10.1073/pnas.0500722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopal S., Key J.M., Moffat K. Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochem. Photobiol. 2005;80:542–547. doi: 10.1562/2004-06-16-RA-203. [DOI] [PubMed] [Google Scholar]
- 9.Fukushima Y., Okajima K., Itoh S. Primary intermediate in the photocycle of a blue-light sensory BLUF FAD-protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry. 2005;44:5149–5158. doi: 10.1021/bi048044y. [DOI] [PubMed] [Google Scholar]
- 10.Gauden M., Yeremenko S., Kennis J.T. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 11.Masuda S., Hasegawa K., Ono T.A. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 2005;44:1215–1224. doi: 10.1021/bi047876t. [DOI] [PubMed] [Google Scholar]
- 12.Unno M., Sano R., Yamauchi S. Light-induced structural changes in the active site of the BLUF domain in AppA by Raman spectroscopy. J. Phys. Chem. B. 2005;109:12620–12626. doi: 10.1021/jp0522664. [DOI] [PubMed] [Google Scholar]
- 13.Okajima K., Fukushima Y., Ikeuchi M. Fate determination of the flavin photoreceptions in the cyanobacterial blue light receptor TePixD (Tll0078) J. Mol. Biol. 2006;363:10–18. doi: 10.1016/j.jmb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Anderson S., Dragnea V., Bauer C. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kita A., Okajima K., Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J. Mol. Biol. 2005;349:1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 16.Grinstead J.S., Hsu S.-T.D., Kaptein R. The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling. ChemBioChem. 2006;7:187–193. doi: 10.1002/cbic.200500270. [DOI] [PubMed] [Google Scholar]
- 17.Jung A., Reinstein J., Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J. Mol. Biol. 2006;362:717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Yuan H., Anderson S., Bauer C. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 2006;45:12687–12694. doi: 10.1021/bi061435n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Q., Gardner K.H. Structure and insight into blue light-induced changes in the BlrP1 BLUF domain. Biochemistry. 2009;48:2620–2629. doi: 10.1021/bi802237r. [DOI] [PubMed] [Google Scholar]
- 20.Barends T.R.M., Hartmann E., Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 21.Laan W., van der Horst M.A., Hellingwerf K.J. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem. Photobiol. 2003;78:290–297. doi: 10.1562/0031-8655(2003)078<0290:icotpp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Unno M., Masuda S., Yamauchi S. Orientation of a key glutamine residue in the BLUF domain from AppA revealed by mutagenesis, spectroscopy, and quantum chemical calculations. J. Am. Chem. Soc. 2006;128:5638–5639. doi: 10.1021/ja060633z. [DOI] [PubMed] [Google Scholar]
- 23.Masuda S., Tomida Y., Takamiya K. The critical role of a hydrogen bond between Gln63 and Trp104 in the blue-light sensing BLUF domain that controls AppA activity. J. Mol. Biol. 2007;368:1223–1230. doi: 10.1016/j.jmb.2007.02.087. [DOI] [PubMed] [Google Scholar]
- 24.Masuda S., Hasegawa K., Ono T.A. Tryptophan at position 104 is involved in transforming light signal into changes of β-sheet structure for the signaling state in the BLUF domain of AppA. Plant Cell Physiol. 2005;46:1894–1901. doi: 10.1093/pcp/pci208. [DOI] [PubMed] [Google Scholar]
- 25.Laan W., Gauden M., Hellingwerf K.J. On the mechanism of activation of the BLUF domain of AppA. Biochemistry. 2006;45:51–60. doi: 10.1021/bi051367p. [DOI] [PubMed] [Google Scholar]
- 26.Gauden M., Grinstead J.S., Kennis J.T. On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry. 2007;46:7405–7415. doi: 10.1021/bi7006433. [DOI] [PubMed] [Google Scholar]
- 27.Toh K.C., van Stokkum I.H., Kennis J.T. On the signaling mechanism and the absence of photoreversibility in the AppA BLUF domain. Biophys. J. 2008;95:312–321. doi: 10.1529/biophysj.107.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinstead J.S., Avila-Perez M., Kaptein R. Light-induced flipping of a conserved glutamine sidechain and its orientation in the AppA BLUF domain. J. Am. Chem. Soc. 2006;128:15066–15067. doi: 10.1021/ja0660103. [DOI] [PubMed] [Google Scholar]
- 29.Dragnea V., Arunkumar A.I., Bauer C.E. Spectroscopic studies of the AppA BLUF domain from Rhodobacter sphaeroides: addressing movement of tryptophan 104 in the signaling state. Biochemistry. 2009;48:9969–9979. doi: 10.1021/bi9009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada I., Takeuchi H. Raman and ultraviolet resonance Raman spectra of proteins and related compounds. In: Clark R.J.H., Hester R.E., editors. Spectroscopy of Biological Systems, Advances in Spectroscopy. John Wiley & Sons; Chichester, UK: 1986. pp. 113–175. [Google Scholar]
- 31.Austin J.C., Jordan T., Spiro T.G. Ultraviolet resonance Raman studies of protein and related model compounds. In: Clark R.J.H., Hester R.E., editors. Biomolecular Spectroscopy, Part A, Advances in Spectroscopy. John Wiley & Sons; Chichester, UK: 1993. pp. 55–127. [Google Scholar]
- 32.Christian J.F., Unno M., Sligar S.G. Spectroscopic effects of polarity and hydration in the distal heme pocket of deoxymyoglobin. Biochemistry. 1997;36:11198–11204. doi: 10.1021/bi9710075. [DOI] [PubMed] [Google Scholar]
- 33.Rava R.P., Spiro T.G. Resonance enhancement in the ultraviolet Raman spectra of aromatic amino acids. J. Phys. Chem. 1985;89:1856–1861. [Google Scholar]
- 34.Asher S.A., Ludwig M., Johnson C.R. UV resonance Raman excitation profiles of the aromatic amino acids. J. Am. Chem. Soc. 1986;108:3186–3197. [Google Scholar]
- 35.Rodger K.R., Su C., Spiro T.G. Hemoglobin R→T structural dynamics from simultaneous monitoring of tyrosine and tryptophan time-resolved UV resonance Raman signals. J. Am. Chem. Soc. 1992;114:3697–3709. [Google Scholar]
- 36.Chi Z., Asher S.A. UV Raman determination of the environment and solvent exposure of Tyr and Trp residues. J. Phys. Chem. B. 1998;102:9595–9602. [Google Scholar]
- 37.Hazra P., Inoue K., Terazima M. Tetramer formation kinetics in the signaling state of AppA monitored by time-resolved diffusion. Biophys. J. 2006;91:654–661. doi: 10.1529/biophysj.106.083915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda S., Hasegawa K., Ono T.A. Crucial role in light signal transduction for the conserved Met93 of the BLUF protein PixD/Slr1694. Plant Cell Physiol. 2008;49:1600–1606. doi: 10.1093/pcp/pcn132. [DOI] [PubMed] [Google Scholar]
- 39.Stelling A.L., Ronayne K.L., Meech S.R. Ultrafast structural dynamics in BLUF domains: transient infrared spectroscopy of AppA and its mutants. J. Am. Chem. Soc. 2007;129:15556–15564. doi: 10.1021/ja074074n. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi R., Okajima K., Noguchi T. FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry. 2007;46:6459–6467. doi: 10.1021/bi7004653. [DOI] [PubMed] [Google Scholar]
- 41.Bonetti C., Mathes T., Kennis J.T. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys. J. 2008;95:4790–4802. doi: 10.1529/biophysj.108.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obanayama K., Kobayashi H., Sakurai M. Structures of the chromophore binding sites in BLUF domains as studied by molecular dynamics and quantum chemical calculations. Photochem. Photobiol. 2008;84:1003–1010. doi: 10.1111/j.1751-1097.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- 43.Ishikita H. Light-induced hydrogen bonding pattern and driving force of electron transfer in AppA BLUF domain photoreceptor. J. Biol. Chem. 2008;283:30618–30623. doi: 10.1074/jbc.M803864200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadeghian K., Bocola M., Schütz M. A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J. Am. Chem. Soc. 2008;130:12501–12513. doi: 10.1021/ja803726a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


