Abstract
The control of cell division is critical to organogenesis, but how this control is achieved is not fully understood. We found that mutations in bed-3, encoding a BED Zn-finger domain transcription factor, confer a phenotype where a specific set of cell divisions during vulval organogenesis is lost. Unlike general cell cycle regulators in C. elegans, the function of bed-3 is restricted to specific lineages. Transcriptional reporters suggest that bed-3 is expressed in a limited number of cell types including vulval cells whose divisions are affected in bed-3 mutants. A bed-3 mutation also affects the expression pattern of the cdh-3 cadherin gene in the vulva. The phenotype of bed-3 mutants is similar to the phenotype caused by mutations in cog-1 (Nkx6), a component of a gene regulatory network controlling cell type specific gene expression in the vulval lineage. These results suggest that bed-3 is a key component linking the gene regulatory network controlling cell-type specification to control of cell division during vulval organogenesis.
Keywords: C. elegans, organogenesis, cell cycle regulation, cell division, cell proliferation, BED domain, zinc finger, transcription, gene regulatory network (GRN)
Introduction
The control of cell division is a critical part of organogenesis, and determines the size and the structure of the organ produced. This is especially apparent in C. elegans, where the pattern of cell division (the cell lineage) during development is nearly invariant from one animal to another, causing individual C. elegans worms to be identical at the cellular level (Sulston and Horvitz, 1977). Despite its importance, developmental regulation of cell division, and how it is tied to cell fate specification, is not fully understood.
To understand cell division control, we are investigating C. elegans vulval development, a well-studied model of organogenesis (reviewed in; Sternberg, 2006). During the initial phase of vulval development (third larval stage; L3), EGF, Wnt and Notch signaling induce three precursor cells (called P5.p, P6.p and P7.p, also vulval precursor cells; VPC) to adopt vulval fates. In the fourth larval (L4) stage, these three cells undergo three rounds of division in a stereotyped pattern to produce the specific number and arrangement of cells needed to form the adult vulva (Sulston and Horvitz, 1977) (Figure 1). The first two rounds of division occur along a longitudinal (anterior/posterior; a/p) axis and produce a row of twelve Pn.p granddaughter cells (Pn.pxx). In contrast, the third (final) round of division occurs in sublineage-specific orientations (Figure 1): The outermost four Pn.p granddaughters divide along a longitudinal (a/p) axis, whereas two Pn.p granddaughters, P5.ppp and P7.paa do not undergo the third round of division (see Figure 1 legend for cell nomenclature). The remaining six Pn.p granddaughters divide along a transverse (left/right; l/r) axis. Whereas the mechanism by which Pn.p cells are induced to adopt vulval fates is well understood, how division of each Pn.p granddaughter is specified is not known.
Figure 1. Vulval development of C. elegans and lineage defect of bed-3 mutants.
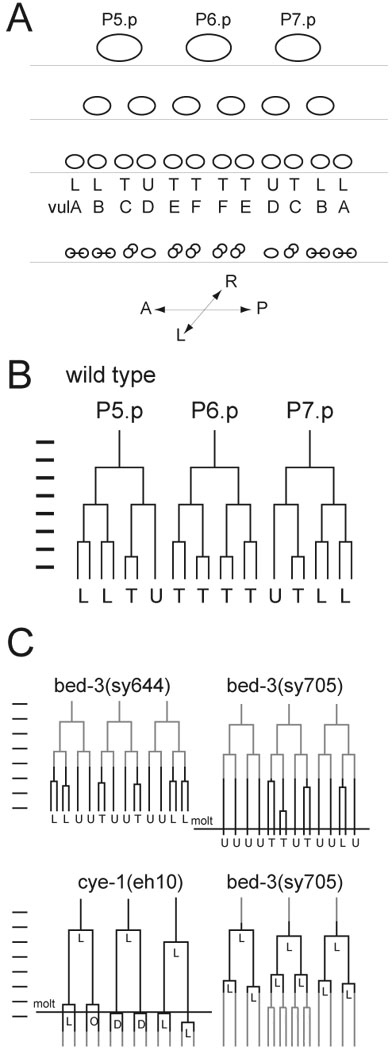
A. Arrangements of Pn.p descendant cells through three rounds of division. Ovals represent cell nuclei. First two divisions occur in anterior/posterior orientation and produce a linear array of twelve Pn.p granddaughters. The twelve cells then divide in sublineage-specific orientations (indicated by letters below: L = longitudinal (a/p), T = transverse (l/r), U = undivided). Letters A through F represent cell types produced by each Pn.p granddaughter, vulA through vulF. In general, the third round of cell division produces two cells of the same type, thus each of the twelve Pn.p granddaughters corresponds to a single cell type. The only exceptions are the cells marked by the letter "B". These divide to produce vulB1 and vulB2 cells, which exhibit distinct gene expression patterns. B. The wild-type vulval cell divisions represented as a lineage diagram. The time is plotted on the vertical axis, and markers on the left are at one hour intervals. Each branching of the inverted tree figure represents a cell division. The orientation of the terminal division is marked below each sublineage. Cells are referred to by their ancestry: The posterior daughter of P5.p is P5.pp. The anterior daughter of P5.pp is P5.ppa and so on. We use x(e.g. P5.ppx) to refer to an unspecified daughter cell, and Pn.p refers to P5.p, P6.p or P7.p. C. Top: Lineage diagrams of representative bed-3(sy644) and bed-3(sy705) mutant animals. Observations were initiated after the completion of the first two rounds of Pn.p divisions and continued until several hours after the last division. Bottom: Lineage diagrams of representative cye-1(eh10) and bed-3(sy705) animals. Observations were started before the first Pn.p division and stopped after the second round of division. Whether these cells had divided further was not determined. The observed portion of the lineage is marked by black lines. Grey lines represent portions of the lineage that were inferred based on cell size and position. The time of molting is indicated by a horizontal line across the lineage diagram. Molting was not scored in the bed-3(sy644) animal, and probably took place after all divisions were completed.
Examination of cell morphology and gene expression reveals that vulval lineages produce seven terminally differentiated cell types, vulA, vulB1, vulB2, vulC, vulD, vulE and vulF (Inoue et al., 2002; Sharma-Kishore et al., 1999). With the exception of the vulB1/B2 case, the terminal division leads to cells with identical fates, suggesting that cell-type specification could operate at the level of Pn.p granddaughters (Pn.pxx cells; Figure 1). A set of genes, including lin-11 (Freyd et al., 1990; Gupta et al., 2003), cog-1 (Palmer et al., 2002), lin-29 (Inoue et al., 2002; Inoue et al., 2005; Ririe et al., 2008), egl-38 (Chang et al., 1999; Rajakumar and Chamberlin, 2006) and nhr-67 (Fernandes and Sternberg, 2007) establish the cell-type specific pattern of gene expression in the seven vulval cell types. These genes encode sequence-specific transcription factors and are mutually interacting, suggesting that they form a gene regulatory network (GRN; (Davidson and Levine, 2008) that specifies the cell type through regulation of cell type specific gene expression (Inoue et al., 2005; Ririe et al., 2008).
Because division of each Pn.pxx cell (presence/absence of division and orientation) correlates with the cell type produced by the Pn.pxx cell, it is reasonable to hypothesize that division and cell fate are controlled by a single mechanism. Of genes in the vulval gene regulatory network, mutations in cog-1 and lin-11 alter the cell division pattern (Freyd et al., 1990; Palmer et al., 2002). In contrast, mutations in lin-29, egl-38 and nhr-67 do not visibly affect cell division. General regulators of cell cycle, such as cye-1 (cyclin E) (Fay and Han, 2000), cul-1 (cullin) (Kipreos et al., 1996), cdc-14 (Cdc14) (Saito et al., 2004) and cki-1 (cyclin dependent kinase inhibitor) (Hong et al., 1998) regulate cell division in multiple cell lineages, including the vulval lineage. How these genes contribute to the specification of cell division pattern, however, is unclear.
Here we describe bed-3, a gene required for the terminal division of Pn.p lineages. bed-3 encodes a protein containing a single BED Zinc-finger domain (Aravind, 2000), a sequence-specific DNA-binding domain (Hirose et al., 1993; Hirose et al., 1996; Zhao et al., 1995). Analyses of bed-3 mutant phenotypes and expression pattern suggest that bed-3 is not required for all cell divisions in C. elegans. Instead, bed-3 mutations are similar to, and interact with mutations in cog-1, a component of the vulval gene regulatory network, indicating that this network controls both gene expression and the terminal divisions in vulval lineages.
Materials and Methods
Worm keeping and informatics
C. elegans was cultured and manipulated as described (Brenner, 1974; Mello et al., 1991). All strains are in the N2 Bristol background unless noted otherwise. Mutations and markers used in this study include: LGII cog-1(sy275), cog-1(sy607); LGIII unc-119(ed4), cul-1(e1756), cye-1(eh10); LGIV unc-24(e138), egl-20(n578), dpy-20(e1282), egl-38(sy294), egl-38(s1775); LGV kuIs29[cog-2::gfp], syIs51[cdh-3::cfp]; LGX syIs55[ceh-2::yfp], syIs59[egl-17::cfp]; Translocations nT1[qIs51](LGIV/LGV), hT2[qIs48] (LGI/LGIII).
All references to C. elegans genome resources are for the WormBase referential data freeze WS160. Accession numbers (RefSeq) of homologs discussed here are; hDREF (ZBED1), NP_004720; ZBED2, NP_078784; ZBED3, NP_115743; ZBED4, NP_055653; BED-1, NP_001024227; BED-2, NP_499474; BED-3, NP_501785; DREF, NP_523529; BEAF32, NP_523747. Locations of BED, hATC and BESS domains are based on NCBI annotations. The region of weak homology was detected by performing a PSI-BLAST search using a portion of BED-3 sequence not containing the BED domain (amino acid 200 to 599). The PSI-BLAST search was done using the NCBI website (http://www.ncbi.nlm.nih.gov/).
Isolation of bed-3 mutations
bed-3(sy644) was isolated in a screen for late vulval development defects using vulval cell fate markers cdh-3::cfp and ceh-2::yfp. The PS3528 syIs51[cdh-3::cfp]; syIs55[ceh-2::yfp] strain (Inoue et al., 2002) was mutagenized with EMS (ethylmethanesulfonate) and propagated for two generations. Candidate mutants were picked in the F2 generation based on the protruding vulva (Pvl) phenotype or based on gross alteration of marker expression as scored using a dissecting microscope equipped for epifluorescence. Lines generated from these animals were later examined under high magnification. Lines that exhibited Pn.p fate changes (i.e. 1° vs. 2° vs. 3° VPC fates; Sternberg and Horvitz, 1986) were discarded, because this type of mutant has been studied extensively. Only those lines with later lineage defects or those altering marker expression were retained. From approximately 10000 mutagenized genomes, we identified six mutations that met these criteria. Based on frequencies of similar mutations from other screens, it is unlikely that all such mutations present in the 10000 mutagenized genomes were recovered. Three mutations, sy633 I, sy635 I and sy636 III increased the level of ceh-2::yfp expression in cells that normally express ceh-2::yfp. One mutation, sy638, caused a low penetrance P-Rvl (posterior reversal) phenotype indicative of anterior-posterior reversal of the P7.p lineage (Green et al., 2008; Inoue et al., 2004). One mutation, sy634 I was found to be a mutant allele of lin-11 by a complementation test. bed-3(sy644) was initially identified as a mutation altering the numbers of cdh-3::cfp expressing cells.
To generate additional alleles of bed-3, we crossed EMS mutagenized wild-type males to unc-24 bed-3(sy644) dpy-20 IV hermaphrodites. Cross progeny were distinguishable from self-progeny as non-Unc non-Dpy animals, and among these, Egl animals were picked as candidate bed-3 alleles. Although some F1 animals had mated with siblings, it was possible to establish lines of new mutants by picking Egl animals. Where possible, the new allele was made homozygous by following the segregation of unc-24 and dpy-20, which were linked in cis to the original allele. sy700 was maintained originally balanced by the unc-24 bed-3(sy644) dpy-20 chromosome, and was later balanced by nT1[qIs51]. Mutations bed-3(sy704) and bed-3(sy710) were isolated in the same round of screening as bed-3(sy705). Although sy704, sy705 and sy710 were derived from separate F1 animals, subsequent analysis showed that sy704 and sy710 are the same mutation as sy705. These lines were not analyzed further.
Mapping and cloning of bed-3
bed-3(sy644) was mapped to the unc-24 to egl-38 interval on chromosome 4 using standard methods. Mutations in this interval, egl-38(sy294), egl-38(s1775) and egl-20(n578) complemented the Egl phenotype of bed-3(sy644). For fine-scale mapping, an unc-24 bed-3(sy644) dpy-20 IV chromosome was placed in trans to a chromosome from the strain CB4856, which is a wild-type isolate of C. elegans containing numerous single nucleotide polymorphisms (snp) in comparison to N2. From the heterozygous parent, Unc non-Dpy and Dpy non-Unc recombinant progeny were picked and lines established. These lines were tested for the presence of bed-3(sy644) and for N2/CB4856 polymorphisms in the region. The most informative polymorphic markers were pkP4081 (cosmid K07F5), snp_T13H10[3] (cosmid T13H10) and dbP3 (cosmid K08F4). Each of these polymorphisms alters a restriction enzyme recognition site, and was scored by PCR amplification of short DNA fragments followed by a restriction digest. Multiple recombinants placed sy644 between pkP4081 on the left and dbP3 on the right (288,836 bp interval). However, among 80 recombination events scored between unc-24 and dpy-20, none recombined between bed-3(sy644) and snp_T13H10[3], suggesting that these loci are very close.
Cosmid clones of genomic DNA from the interval between pkP4081 and egl-38 were tested for transformation rescue of the Egl phenotype. The following cosmids did not rescue the bed-3 mutant: W08D2, K07F5, F49C8, F32B6, F25H8, T13H10, C47E12, F44D12, T04B2, W01B6, C04G2, W01E11, W01E2. A transgenic line made by coinjecting a mixture of overlapping cosmids F26D3 and C13H3 rescued bed-3(sy702), whereas multiple lines made from injection of F26D3 failed to rescue, suggesting that bed-3 is on the part of cosmid C13H3 not overlapped by F26D3. Although exact ends of C13H3 and F26D3 clones are not known, examination of the physical map suggested that bed-3 is near F25H8.6. Multiple lines made with cosmid F25H6 failed to rescue bed-3(sy644) even though the gene should be fully contained in this cosmid, possibly because of cosmid rearrangement. Microinjection of a single-gene subclone of C13H3 which contains only F25H8.6 (pTI05.37) rescued mutant F1 animals. To determine if F25H8.6 is bed-3, mutant alleles were sequenced. Because four mutant alleles contain four different mutations in the coding region of this gene, F25H8.6 is bed-3.
Lineage analysis and scoring of cell numbers
Lineage analyses of bed-3 and cye-1 mutants were performed as previously described by keeping live animals on agar pads on sealed slides (Sulston and Horvitz, 1977). cye-1 and cul-1 mutants were maintained balanced by hT2[qIs48[myo-2::gfp]] and non-GFP progeny were scored. For Table 2, lineages were typically observed from Pn.p daughters or granddaughters. Worms were observed for at least one hour past the last division before undivided cells were scored as "U" (undivided). We did not find a strong effect of bed-3(sy644) on the timing of cell divisions; however, we cannot rule out the possibility that some of the "U" cells divided later.
Table 2.
Lineages of bed-3 mutants based on observations of cell divisions
| genotype | lineage (P5.p, P6.p, P7.p granddaughters) |
|---|---|
| Wild type | LLTU TTTT UTLL* |
| cog-1(sy275) | LLTU TUUT UTLL* |
| cog-1(sy607) | LLUU TTTT UULL* |
| bed-3(sy644) | LLUU TTTT UTLL |
| LLUU TTTT UOLL | |
| LLUU TUUT UULL | |
| LLOU TTTT UTLL | |
| LLUU TTTT UTLL | |
| LLOU TTTT UTLL | |
| LLTU TTTT UTLL | |
| LLUU TTTT UULL | |
| bed-3(sy705) | UUUU TTTU UULU |
| UUUU TTTU UUUU | |
| UUUU TTUT ULUU | |
| cog-1(sy275) bed-3(sy644) | UUUU TUUT UUUU |
| UUUU TUUT UUUU | |
| UUUU TUUT UUUU | |
| UUUU TUUT UUUU | |
| ULUU TUUT UODU | |
| UUUU TUUT UULL | |
| cog-1(sy607); bed-3(sy644) | UUUU TTUT UUUU |
| UUUU TUUU UUUU | |
| UUUU UTTT UUUU | |
| UUUU UTTT UUUU | |
Divisions of Pn.p granddaughters are represented by "T" = transverse, "L" = longitudinal, "O" = oblique, or "D" = divided with axis undetermined, based on the orientation of the division axis. The failure of a Pn.p granddaughter to divide is represented by "U" = undivided.
Idealized lineages. Penetrances of division defects in cog-1 mutants are less than 100%. See Palmer et al. for details. For bed-3 mutants, each line represents a single animal.
Vulval cell numbers were determined in the mid-L4 stage. In most animals, cell size, arrangement and morphology could be used to infer which cells were derived from which precursor. The organization of P6.p-derived cells was wild type in most bed-3 mutants. However, because the cell arrangement was disorganized in some animals, it is possible that scoring of cells as P5.p-, P6.p- or P7.p-derived was incorrect in some animals (Supplementary Figure S1).
For cul-1, we scored non-GFP progeny of cul-1/hT2[qIs48[myo-2::gfp]] parents. We counted 38 ± 2.9 (n = 4) vulval cells for cul-1(e1756) mutants and 33 ± 6.6 (n = 3) vulval cells for cul-1(e1756); bed-3(sy702) double mutants. While these results and the small size of cul-1; bed-3 vulval cells demonstrate that cul-1; bed-3 mutants undergo extra rounds of cell division, the numbers for cul-1 are smaller than previously reported by Kipreos et al. (1996) and are likely to be undercounts due to the difficulty in distinguishing vulval and non-vulval cells in highly disorganized cul-1 animals. Notably, Kipreos et al. (1996) used a ceh-24::gfp marker and videomicroscopy to obtain accurate cell counts, which was not done for this study. Because of these uncertainties, it is unclear whether cul-1; bed-3 double mutants have reduced cell numbers compared to cul-1 single mutants.
Examination of non-vulval lineages
The number of π cell descendants was scored in the mid-L4 stage using kuIs 29[cog-2::gfp] (Hanna-Rose and Han, 1999), which is expressed in daughters of π cells and the anchor cell. Ventral cord neurons were counted in the P5.p to P8.p interval in L2 animals using Nomarski optics. Lateral hypodermal nuclei were counted using the syIs51[cdh-3::cfp] marker (expressed in seam cells as well as vulval cells) in mid-L4 to young adult animals.
Molting phenotype of bed-3 mutants
Semi-synchronized populations of N2, bed-3(sy702) and bed-3(sy705) strains were generated by overnight egg lays and scored two days later as adults. 21 of 55 bed-3(sy705) mutants examined had not shed the L4 cuticle completely. In contrast, no molting defect was observed in N2 (n=20) and bed-3(sy702) (n=57) animals.
bed-3::gfp fusion
The genomic DNA from 1.4 kb (kilobase) upstream of the bed-3 gene was PCR amplified using primers TTAAGCATGCAGAAGCGTAGAAGGTAGGCAAGTGGG and GAGAGTCGACTCCAGGTAATGGGATACCTG. The resulting fragment was cloned into pCR-XL-TOPO (Invitrogen) using TOPO cloning. The promoter fragment was then excised from the vector using NsiI (site in vector multiple cloning site) and SalI (site in downstream primer) and cloned into 4xNLSgfp vector pPD121.83 cut with PstI and SalI to generate the promoter-gfp construct pTI06.15. To test for enhancers, intron-containing fragments were isolated from the genomic clone pTI05.37 cut with HindIII or NspI and cloned into pTI06.15 cut with HindIII and SphI respectively. This places each intron-containing fragment 5' to the promoter fragment. All reporter constructs were injected into unc-119(ed4) using unc-119(+) as a coinjection marker (Maduro and Pilgrim, 1995; Mello et al., 1991).
Results
Isolation and molecular characterization of bed-3 mutations
We isolated the bed-3(sy644) mutation in a screen for mutations that affect the expression of vulval cell type markers cdh-3::cfp and ceh-2::yfp (see Materials and Methods) (Inoue et al., 2002). cdh-3::cfp is expressed in vulC, vulD, vulE and vulF (Pettitt et al., 1996), and bed-3(sy644) affected the number of cdh-3::cfp expressing cells. In addition, bed-3(sy644) caused Egl (egg-laying defective) and weak Pvl (protruding vulva) phenotypes. Mapping and complementation tests found that bed-3 is a new genetic locus on chromosome 4. To isolate additional alleles, we carried out a genetic screen for mutations that fail to complement bed-3(sy644) for the Egl phenotype (see Materials and Methods). From a screen of approximately 4400 EMS mutagenized genomes, we identified three additional mutations that fail to complement sy644 (Table 1). This frequency is consistent with loss-of-function alleles (Greenwald and Horvitz, 1980).
Table 1.
bed-3 mutations and phenotypes
| allele name | nature of mutation | phenotypes |
|---|---|---|
| sy644 | nonsense, exon 4 | Egl, Pvl |
| sy702 | nonsense, exon 5 | Egl, Pvl |
| sy705 | nonsense, exon 3 | Egl, Pvl |
| sy700 | deletion, exon 4 to 5 | Egl, Maternal effect lethal |
Egl = egg laying defect, Pvl = protruding vulva
We cloned bed-3 using a combination of SNP mapping and germline transformation rescue (see Materials and Methods) and found that it corresponds to the predicted gene F25H8.6 (Figure 2). The mRNA structure was determined by RT-PCR, and was identical to the structure in WormBase (release WS160). bed-3 is predicted to encode a 599 amino acid protein (BED-3) with a single BED-type zinc finger domain (Figure 3, Supplementary Figure 2). BED Zn-finger proteins are sequence-specific DNA binding proteins (Aravind, 2000). In C. elegans, a BED domain protein DPY-20 regulates body shape (Clark et al., 1995). In Drosophila melanogaster, a BED domain protein DREF activates transcription of genes involved in cell proliferation (Hirose et al., 1993; Hirose et al., 1996; Matsukage et al., 1995), and a BED domain protein BEAF32 (boundary element associated factor) associates with elements that separate chromatin domains (Emberly et al., 2008; Zhao et al., 1995). The human ortholog of DREF (hDREF) also regulates cell proliferation in cell culture (Ohshima et al., 2003). The sequence conservation within this family is mostly in the BED domain, although weak similarity outside the BED domain exists between the C. elegans BED-3 and human proteins hDREF and ZBED4 (Figure 3; see Materials and Methods). Because the level of sequence conservation is low even within the BED domain, a convincing orthology relationship was not detected between BED-3 and any other protein outside of nematodes. However, BED-3 is architecturally similar to hDREF in containing only a single BED domain and a weakly conserved C-terminal region. In contrast, some proteins (e.g. human ZBED4, C. elegans BED-2) contain multiple BED domains. Also, some BED domain proteins (e.g. BEAF32, human ZBED2) show no similarity to BED-3 in the C-terminal region.
Figure 2. bed-3 gene structure.
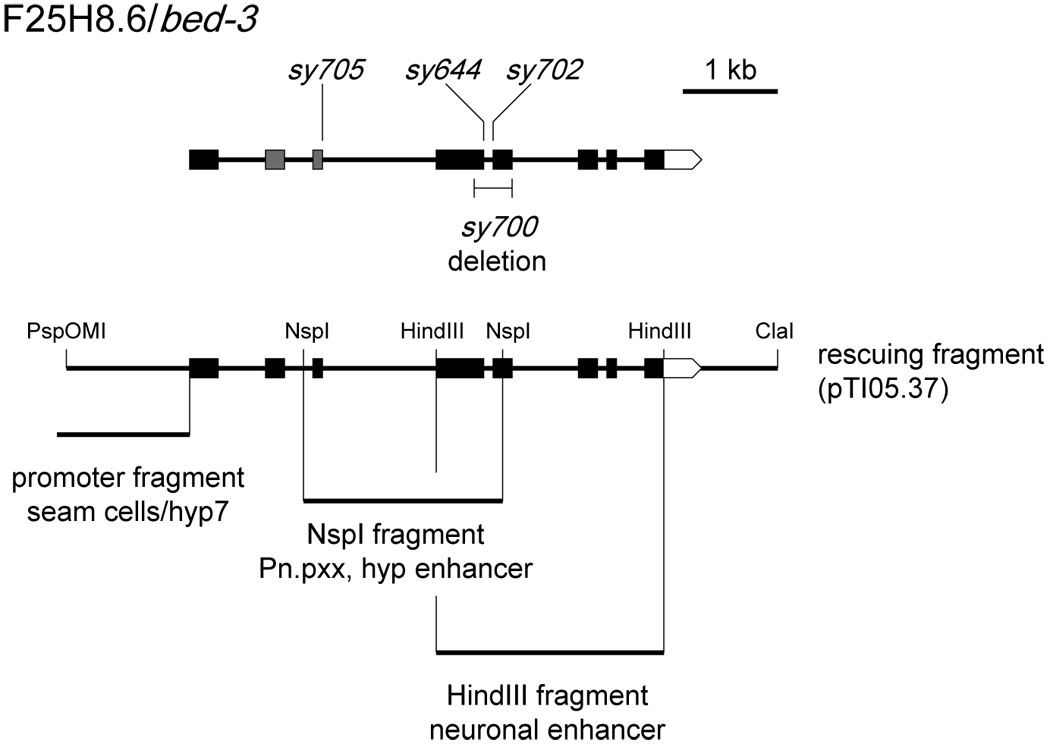
Top: the intron/exon structure of bed-3 and locations of mutations. The BED Zn-finger domain is encoded in second and third exons (gray). Bottom: genomic regions tested for promoter and enhancer activity. The promoter fragment, placed upstream of a gfp reporter, drives expression in seam and hyp7 cells. When placed upstream of the promoter fragment, NspI and HindIII fragments drive additional expression, demonstrating the presence of enhancer activity.
Figure 3. Domain structures of BED Zn-finger domain containing proteins.
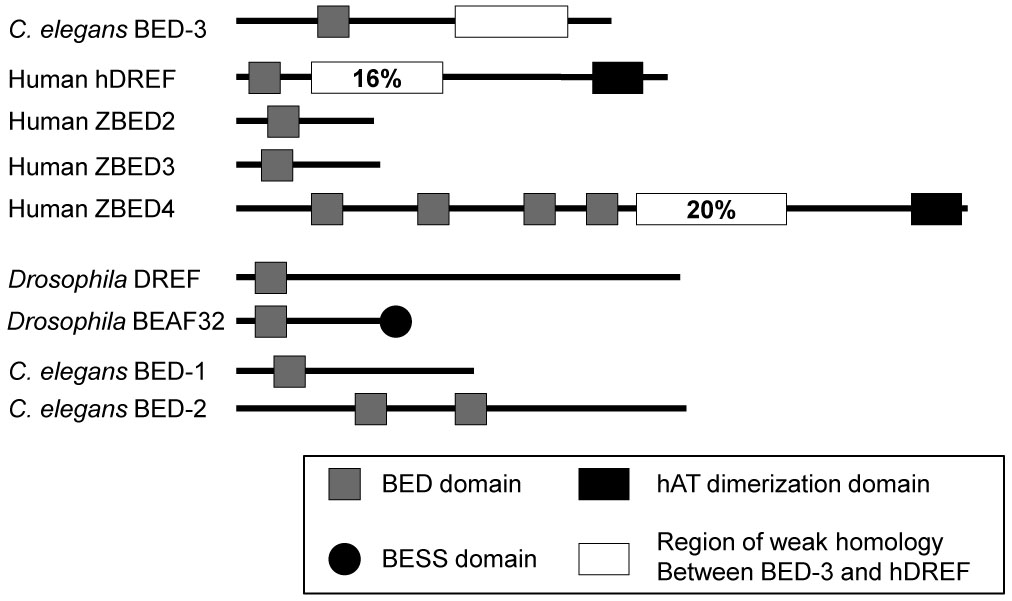
N-terminus is to the left. Percentages indicate amino acid identity with the homologous region of BED-3.
To determine the nature of the mutations, we sequenced mutant alleles of bed-3 and found sy644, sy702 and sy705 to be nonsense mutations in exons 4, 5, and 3, respectively (Figure 2, Supplementary Figure 2, Table 1). None of these completely disrupt the BED domain, and they are unlikely to be null mutations. The bed-3(sy700) mutation is a frame-shifting deletion. bed-3(sy700), unlike other alleles, cannot be maintained in a homozygous state because of an apparent maternal effect embryonic lethality. However, the deletion occurs downstream of the nonsense mutation in homozygous-viable bed-3(sy705), and sy700 exhibits a weaker vulval lineage phenotype (see below). Based on these results, the lethal phenotype in the sy700 strain is likely caused by a linked background mutation rather than by sy700 itself.
We tested the effect of RNAi against bed-3 by using a clone from a bacteria-feeding library (Open Biosystems) (Rual et al., 2004). Wild-type animals that grew on E. coli expressing bed-3 dsRNA exhibited both the Egl defect and the vulval cell-lineage defect (Supplementary Figure S1), demonstrating that these phenotypes are caused by a reduction or loss of function in the bed-3 gene. Previously, RNAi against bed-3 (F25H8.6) was reported to cause molting defects (Frand et al., 2005), and we found that a strong bed-3 mutation also caused molting defects (Figure 4C, Materials and Methods).
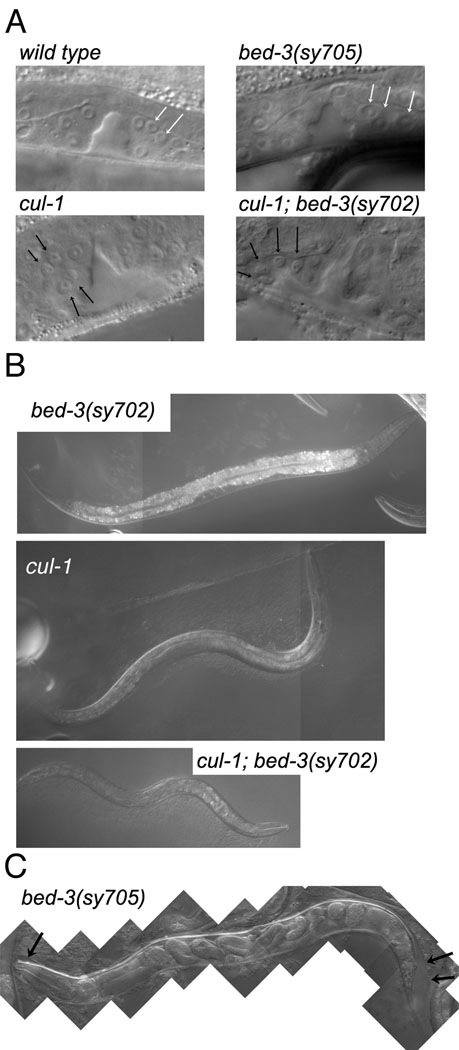
Figure 4. Phenotypes of bed-3 mutants and interaction with cul-1.
A. Mid-L4 stage vulva, after cell divisions had been completed. Arrows point to vulval nuclei. Not all nuclei are in the plane of focus. However, note that bed-3 mutant nuclei are larger than in the wild type due to the lack of terminal division. In contrast, cul-1 nuclei are smaller due to extra divisions. The cul-1; bed-3 double mutant is similar to cul-1. B. Body size of bed-3(sy702), cul-1 and cul-1; bed-3(sy702) mutants. All animals are in the mid-L4 stage and shown at the same scale. bed-3(sy702) mutants are about the same size as the wild-type (not shown). bed-3(sy705) mutants are slightly smaller (not shown). C. The molting defect of bed-3(sy705). This animal is an old adult, as indicated by the presence of late stage embryos in the uterus. However, because of the molting defect, the L4 cuticle has not been shed (black arrows).
bed-3 regulates cell division in the vulval lineages
To determine the function of bed-3 in vulval development, we observed the pattern of cell divisions (the lineage) in bed-3(sy644) and bed-3(sy705) mutants (Figure 1C, Figure 4A, Table 2). In both mutants, granddaughters of P5.p, P6.p and P7.p often failed to divide. Cells most affected by bed-3(sy644) were P5.ppa and P7.pap, the parents of vulC cells (failed to divide in 7/16 or 45% of cases). Parents of vulA and vulB cells divided normally. bed-3(sy705) caused a stronger defect in which most P5.p and P7.p granddaughters failed to divide (parents of vulA, vulB and vulC failed to divide in 16/18 or 89% of cases). In addition, in some bed-3(sy705) animals, divisions of P6.p granddaughters (3/12 P6.pxx cells) and P5.p and P7.p daughters were defective (data not shown). The penetrance of the cell division defect observed in bed-3(sy644) is less than the penetrance of the Egl phenotype (egg-laying defective; almost 100%), therefore the Egl phenotype is not caused solely by the cell division defect.
Since loss of cell division reduces the final number of vulval cells, we compared other bed-3 mutations by the cell number (Figure 5; Supplementary Figure S1). We found that all mutations caused a qualitatively similar phenotype, with sy705 exhibiting the strongest defect. The cell counts place the mutants in an allelic series with respect to the vulval cell division phenotype; sy700 < sy644 = sy702 < sy705.
Figure 5. Cell numbers in various lineages.
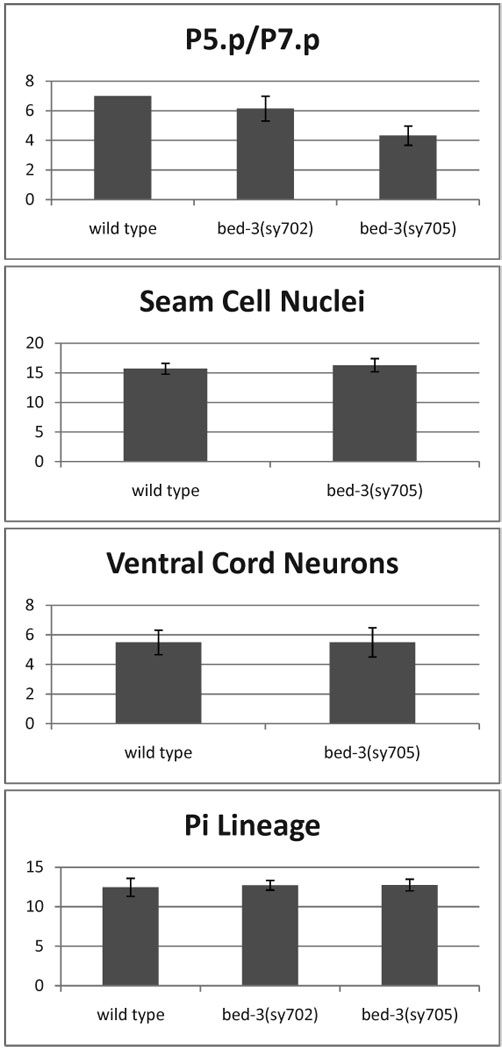
Numbers of cells produced by diverse lineages were assayed in the wild-type and bed-3 mutant backgrounds. P5.p/P7.p: Descendants of the P5.p or the P7.p lineage. Seam Cell Nuclei: The total number of seam cell nuclei (per side) in late L4 or young adult stage animals. Ventral Cord Neurons: The number of neuronal cell bodies in the ventral cord, per Pn.p-to-Pn.p interval. Pi Lineage: The number of π cell descendants in the mid-L4 stage. The counts include +1 for the anchor cell, which is also labeled by the cog-2::gfp marker used to identify π cell descendants. Error bars indicate standard deviations.
bed-3::gfp reporter is expressed in Pn.p granddaughters
To study where the bed-3 gene could be expressed, we assayed genomic sequences surrounding the bed-3 coding region for promoter and enhancer activities (Figure 2). A 6.5kb PspOMI to ClaI genomic clone rescues the bed-3 mutant (pTI05.37). This clone contains 1.3 kb of sequence upstream of the bed-3 coding region, all exons and introns, and 3' sequence to the middle of the next gene in the genome, dur-1.
We first tested the construct in which 1.4 kb of the upstream sequence directed the expression of a nuclear localized GFP variant (Figure 2; pTI06.15; see Materials and Methods). This bed-3::gfp reporter was most prominently expressed in lateral hypodermal (seam) cells, and occasionally in the hyp7 hypodermal cell. This expression was dynamic, with a notable increase in GFP expression around the time of the L3-to-L4 molt. Although occasional animals expressed GFP in vulval cells, this expression was faint and inconsistent. Genes required for molting are expressed in seam and hyp7 cells in a dynamic pattern with expression peaks near the time of the molt (Frand et al., 2005; Kostrouchova et al., 1998; Kostrouchova et al., 2001). Thus, the expression pattern of bed-3 is consistent with its role in molting, and suggests that bed-3 drives expression of other molting-related genes in hypodermal cells.
We next tested for the presence of enhancers in introns. Of seven introns in the bed-3 gene, introns 3 and 5 are exceptionally large (>500 bp) and contain sequences that are conserved between C. elegans and C. briggsae (WABA alignment, WormBase WS160). To test whether these introns contained transcriptional enhancers, we placed intron-containing fragments upstream of the 1.4 kb bed-3 promoter in the bed-3::gfp (pTI06.15) construct. Two intron-containing fragments enhanced transcription from the bed-3 promoter in two distinct patterns (Figure 2). The construct containing introns 5, 6 and 7 expressed GFP in a subset of neuronal cell bodies in the ventral cord and in HSN neurons required for egg laying (HindIII fragment; pTI06.28; see Materials and Methods). The fragment containing introns 3 and 4 increased the level of expression in the lateral hypodermis and hyp7 cells (NspI fragment; pTI06.29; see Materials and Methods). Furthermore, this enhancer fragment caused expression in P5.p, P6.p and P7.p granddaughters (Figure 6). Although this expression was variable in that not all animals expressed GFP in all Pn.p granddaughters, it was consistently observed in multiple independently generated lines. These results suggest that bed-3 acts autonomously in cells whose divisions are affected in bed-3 mutants, however, the possibility that bed-3 acts in seam or hyp7 hypodermal cells to control vulval cell division has not been ruled out.
Figure 6. bed3::gfp expression in vulval cells.
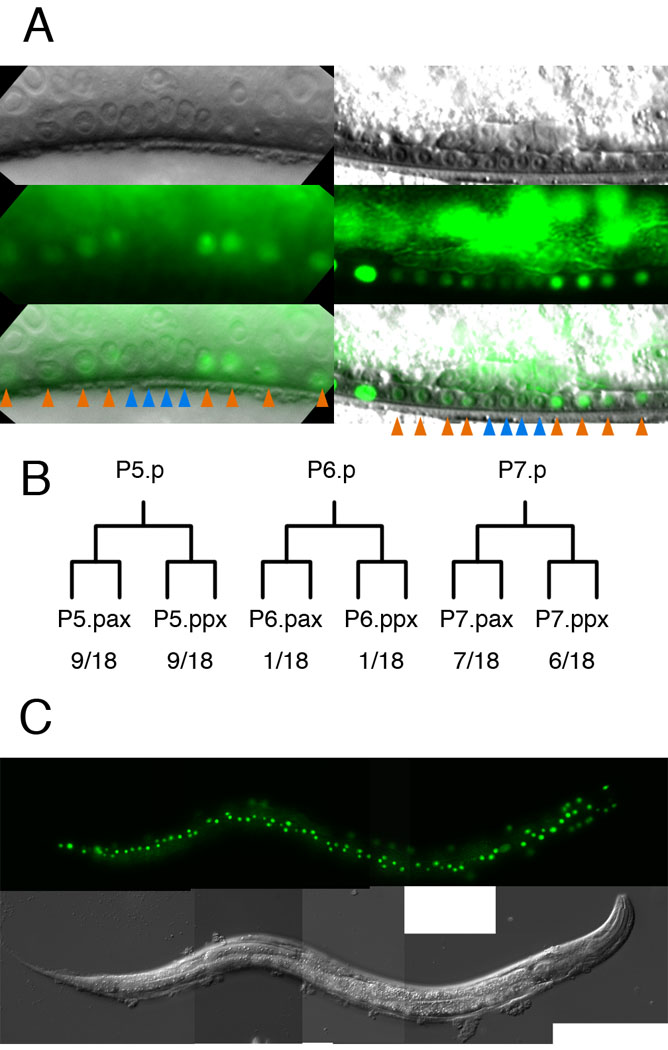
Expression of bed-3::gfp enhanced by the NspI fragment. The reporter shown (pTI05.29) expresses a nuclear localized variant of GFP. A. Nomarski, epifluorescence and combined images of two animals expressing bed-3::gfp in Pn.p granddaughters. Anterior is to the left. In the animal shown to the left, P5.p and P7.p granddaughters (orange arrows) express GFP. In the animal shown to the right, P6.p granddaughters (blue arrows) also express GFP. B. Frequencies of bed-3::gfp expression in individual Pn.p granddaughters. Because sister cells (e.g. P5.paa and P5.pap) always expressed similar levels of GFP, they were scored together. P5.pxx and P7.pxx often express bed-3::gfp, whereas expression in P6.pxx is observed less often. C. Expression of bed-3::gfp in hyp7 and seam cells.
Comparison of bed-3 with general cell-cycle regulators
Disruption of general cell cycle regulators can cause loss of cell division in vulval lineages. For example, a mutation in cye-1 (encoding cyclin E) causes vulval precursor cells to undergo only two rounds of division (Fay and Han, 2000). Because this phenotype is superficially similar to that of bed-3 mutants, we performed a detailed comparison (Figure 1C). During wild-type vulval development, the first two divisions occur along an anterior-posterior axis and the third (terminal) division occurs along sublineage-specific axes. In cye-1(eh10) mutants, the second (terminal) division occurs along sublineage-specific axes. Furthermore, the time delay between the first and the second division is abnormally long, so that the second (terminal) division of cye-1(eh10) occurs at about the same time as the third (terminal) division of the wild type. These results suggest that the cye-1(eh10) causes a lengthening of the interval between cell divisions, and loss of the second round of cell division (Fay and Han, 2000).
In contrast, we found that bed-3(sy705) caused a specific loss of the third (terminal) division (Figure 1C). Direct observation of division (n=5) and observation of cell arrangement after the second division (many) showed that the second division occurs in the anterior/posterior orientation. Furthermore, the time between cell divisions in bed-3(sy705) was much shorter than for cye-1(eh10). The interval between the first and second division in cye-1(eh10) was 320 minutes (standard deviation 20 min.; n=6). The same interval was 178 minutes (s.d. 31 min.; n=12) in bed-3(sy705). The second division of bed-3 mutants also occurs earlier than the third (terminal) division of the wild type.
To further investigate the relationship of bed-3 to general cell cycle regulators, we examined the interaction between bed-3 and cul-1. cul-1 encodes a cullin, a component of an E3 ubiquitin ligase complex (Kipreos et al., 1996). In cul-1 mutants, many postembryonic cell lineages undergo extra rounds of division, demonstrating that cul-1 negatively regulates cell division. Vulval lineages also undergo extra rounds of division, and consequently, the adult vulva of cul-1 mutants contains more cells than the wild type. To determine how bed-3 interacts genetically with cul-1, we examined bed-3(sy702); cul-1(e1756) double mutants. The vulva of bed-3; cul-1 double mutants contained more cells than the wild type; i.e., bed-3 does not fully suppress the cul-1 mutant phenotype (Figure 4A, Materials and Methods). This result is consistent with the hypothesis that bed-3 functions upstream of general cell cycle regulators like cul-1. The double mutant also had a smaller body size than either bed-3(sy702) or cul-1(e1756) single mutants (Figure 4B) and appeared to be slow-growing.
bed-3 does not affect all postembryonic cell divisions
Mutations in general regulators of cell division like cye-1 and cul-1 affect multiple lineages (Fay and Han, 2000; Kipreos et al., 1996; van den Heuvel, 2006). To determine whether bed-3 is a general regulator of cell division, we looked for bed-3 function in additional lineages. In particular, we analyzed cells that express bed-3::gfp, cells that undergo division at the same time as Pn.pxx cells, and cells closely related to vulval cells by lineage (Figure 5).
bed-3::gfp is expressed in the lateral hypodermal (seam) cells. In each larval stage, each seam cell undergoes one (L1, L3, L4 stages) or two (L2) rounds of division (Sulston and Horvitz, 1977). In a stem-cell-like pattern, one of two (or two of four in L2) cells produced retains the parental seam cell identity, while the other cell fuses with the terminally differentiated hyp7 hypodermal cell. Mutations in cye-1 and cul-1 respectively decrease and increase the number of seam cell nuclei in the adult (Fay and Han, 2000; Fujita et al., 2008; Kipreos et al., 1996). To determine whether bed-3 regulates seam cell lineages, we examined the number of seam cell nuclei in bed-3(sy705) mutants (see Materials and Methods). We found the same number of seam cell nuclei in bed-3(sy705) mutants as in the wild type, suggesting that bed-3 is not required in the seam cell lineage to regulate cell division.
We also looked at the function of bed-3 in uterine lineages (including π cell lineages), which undergo final rounds of cell division near the L3 to L4 molt, contemporaneous with terminal vulval cell divisions (Kimble and Hirsh, 1979; Newman et al., 1996; Sulston and Horvitz, 1977). cye-1 and cul-1 mutations affect uterine lineages and cause gross morphological defects in the uterus (Fay and Han, 2000; Fujita et al., 2008; Kipreos et al., 1996). In contrast, we found that the gross morphology of the uterus was normal in the strongest bed-3 mutant, bed-3(sy705). We also examined the uterine π cell lineage using the marker cog-2::gfp, which is expressed in twelve π cell daughters and the anchor cell (Hanna-Rose and Han, 1999) (see Materials and Methods). We found no difference in the number of cog-2::gfp expressing cells in the wild type, bed-3(sy702) and bed-3(sy705) mutants, suggesting that π cell divisions are not affected by bed-3 mutations.
Finally, we examined cells derived from Pn.a neuroblasts, which are sister cells of Pn.p cells (Sulston and Horvitz, 1977). These cells divide two to three rounds in the late L1 stage to generate neurons whose cell bodies lie in the ventral cord region. This region contains 15 neuron cell bodies at hatching, to which Pn.a lineages add 52 additional neurons during the L1 stage. To assay for defects in Pn.a lineages, we counted the number of neuron cell bodies in a specific interval in L2 animals (between P5.p to P8.p; see Materials and Methods). We found no difference in the number of neurons in this interval between N2 and the bed-3(sy705) mutant, suggesting that bed-3 mutations do not affect Pn.a lineages.
bed-3 interacts genetically with cog-1
The observation that bed-3 mutations do not affect many lineages suggested that bed-3 function is specific to terminal division of the vulval lineage. Previously, mutations in only one other gene, cog-1, was known to affect specific terminal cell divisions during vulval development. cog-1 encodes a Nkx6 homeobox transcription factor regulating gene expression in terminal vulval cell types (Inoue et al., 2005; Palmer et al., 2002). cog-1(sy275) is a missense mutation in the DNA binding domain, and cog-1(sy607) is a deletion eliminating one of two cog-1 transcripts. Neither allele is likely to cause a complete loss of function. In cog-1(sy275) mutants, granddaughters of P6.p often fail to divide. In cog-1(sy607)mutants, granddaughters of P5.p and P7.p often fail to divide (Table 2). Interestingly, cell divisions which are most commonly affected in cog-1 mutants (P5.ppa, P6.pap, P6.ppa, P7.pap) are also most commonly affected in the weak bed-3 mutation bed-3(sy644) (Table 2). These results suggested that cog-1 and bed-3 function in the same process and may interact genetically.
To look for genetic interaction between cog-1 and bed-3, we examined cog-1(sy275); bed-3(sy644) and cog-1(sy607); bed-3(sy644) double mutants, and found that the cell division defect was strongly enhanced (Table 2). For example, P5.paa and P7.ppp (parents of vulA in wild-type) always divided in bed-3(sy644), cog-1(sy275) and cog-1(sy607) single mutants (Table 2) (Palmer et al., 2002). However, the same cells failed to divide in cog-1(sy275); bed-3(sy644) (11/12 cells) and in cog-1(sy607); bed-3(sy644) double mutants (8/8 cells). The phenotype of double mutants (Table 2) suggest that both cog-1 and bed-3 function in all divisions of Pn.p granddaughters, but are not required for earlier rounds of Pn.p divisions. Because none of the mutations analyzed are null, these results are consistent with both parallel and serial function of cog-1 and bed-3.
bed-3 affects cell type specific gene expression in vulval cells
Mutations in cog-1 affect cell type specific pattern of gene expression as well as cell division. The pattern of alteration in gene expression is not fully understood, and is not consistent with a simple transformation of cells to the non-dividing vulD fate. Nevertheless, the co-occurrence of both cell division and cell type specific gene expression defects in cog-1 mutants suggest that a common underlying mechanism controls both processes. We tested whether bed-3 acts strictly in controlling cell division or like cog-1, bed-3 also controls cell type specific pattern of gene expression. In particular we analyzed cdh-3 and egl-17 expression, which are affected by cog-1 mutations (Inoue et al., 2005) (Table 3).
Table 3.
Alteration of cell type specific expression in bed-3 mutants
| Genotype | Expression pattern in P5.p and P7.p* |
Number of Pn.p lineages |
|---|---|---|
| cdh-3::cfp | vulC and vulD (2/4 Pn.pxx sublineages) |
30 |
| bed-3(sy705); cdh-3::cfp† | 1/4 Pn.pxx | 8 |
| 2/4 Pn.pxx | 7 | |
| 3/4 Pn.pxx | 2 | |
| egl-17::cfp | vulC and vulD (2/4 Pn.pxx sublineages) |
16 |
| bed-3(sy705); egl-17::cfp† | 1/4 Pn.pxx | 0 |
| 2/4 Pn.pxx | 12 | |
| 3/4 Pn.pxx | 0 | |
No obvious difference was observed between P5.p and P7.p lineages. The numbers are combined tally for P5.p and P7.p.
Only lineages in which P5.p or P7.p produced four descendants were analyzed.
In a wild-type background, egl-17::cfp is expressed in vulC and vulD cells, but not in vulA and vulB cells (Inoue et al., 2002). (Expression in vulE and vulF cells was not scored in this experiment.) To determine whether this pattern is affected by the strongest bed-3 allele sy705, we focused our analysis on P5.p and P7.p lineages which produced four descendant cells of similar size, since each of these cells is likely to be a Pn.p granddaughter. (In other cases, it can be ambiguous which cell belongs to which Pn.pxx sublineage.) If the correspondence between Pn.p granddaughters and gene expression is maintained, we expect two granddaughters of each Pn.p (P5.ppa/P5.ppp, or P7.paa/P7.pap) to express egl-17::cfp. Indeed, we found that in bed-3(sy705) mutants, two out of four cells expressed egl-17::cfp in 12 out of 12 cases (Table 3). In 11 of these, the innermost two Pn.p granddaughters expressed egl-17::cfp (data not shown; the one exceptional pattern could be due to rearrangement of cells).
In contrast to egl-17::cfp, we found that bed-3(sy705) affected the expression pattern of cdh-3::cfp. In a wild-type background, cdh-3::cfp is expressed in vulC, vulD, vulE and vulF (Inoue et al., 2002; Pettitt et al., 1996). If the correspondence between Pn.p granddaughters and cell type specific gene expression is maintained, we expect the inner two granddaughters of each Pn.p (P5.ppa/P5.ppp, or P7.paa/P7.pap) to express cdh-3::cfp. Instead, we found that eight of 17 such Pn.p lineages expressed cdh-3::cfp in only one cell, always the innermost granddaughter (P5.ppp or P7.paa) (Figure 7, Table 3). Seven of 17 Pn.p lineages expressed cdh-3::cfp in two cells as expected, and two lineages expressed cdh-3::cfp in three out of four cells. In all animals examined, expression in vulD, vulE and vulF cells was intact. This phenotype is reminiscent of cog-1(sy607), which causes a loss of cdh-3::gfp expression in vulC, vulD and vulE cells (Inoue et al., 2005). The phenotype in bed-3(sy705) is weaker than is observed for cog-1(sy607), however, this is not simply due to the strength of the mutation, because the cell division defect is stronger in bed-3(sy705) than in cog-1(sy607).
Figure 7. Expression of vulval cell type marker cdh-3::cfp in bed-3(sy705) background.
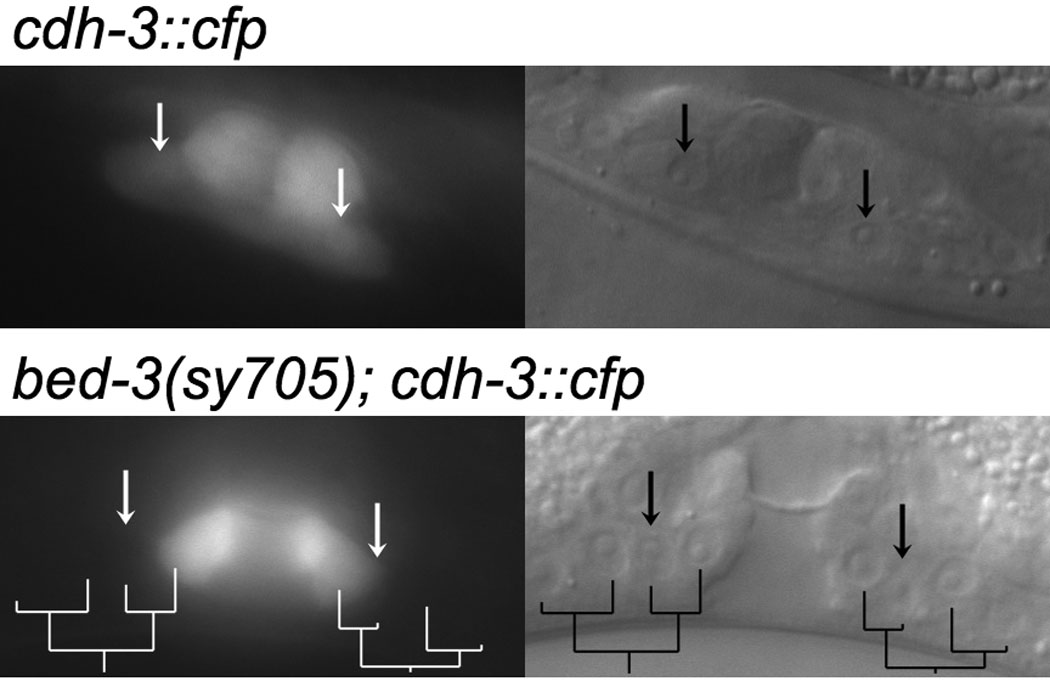
Corresponding Nomarski (right) and epifluorescence (left) images are shown. Top: Wild type. vulC cells are indicated by arrows. Bottom: bed-3(sy705). In this animal, P5.p and P7.p lineages divided only two rounds as shown on the lineage tree below. P5.ppa and P7.pap, which give rise to vulC in the wild type, are marked by arrows. vulC expression is lost in bed-3(sy705).
Discussion
During C. elegans vulval development, each vulval precursor cell (VPC; P3.p ~ P8.p) adopts one of three fates; 1°, 2° or 3°. Although VPC fate specification has been analyzed extensively over the last two decades, the mechanism by which VPC fates are 'executed' (i.e. how VPC descendants undergo fate-specific pattern of subsequent development) has received less attention. We found that the BED Zn-finger transcription factor BED-3 is involved in this phase of vulval development, specifically in the regulation of several cell division events in vulval cell lineages. Our analyses suggest that BED-3 is not a general regulator of cell division. Instead, similarities of bed-3 mutant phenotypes with those of cog-1 suggest that bed-3 interacts with, or is an integral component of the gene regulatory network (GRN) specifying cell type specific pattern of gene expression in vulval cells. The sequence similarity of BED-3 to cell proliferation regulators DREF and hDREF is intriguing and suggests possible conservation of function, however, it is unclear whether cell division control is the primary function of BED-3. Here we discuss these possibilities.
BED-3 is not likely to be a general regulator of cell division
Loss or gain of cell division in the vulval lineage can be caused by mutations in general cell cycle regulators (van den Heuvel, 2006). In C. elegans, the disruption of some cell cycle regulators (e.g. cul-1, cye-1 mutations) causes a relatively mild phenotype in which many lineages undergo hyper- or hypo-proliferation, but the overall pattern of development is not severely affected (i.e. mutants are viable) (Fay and Han, 2000; Kipreos et al., 1996). Some mutations such as cye-1(eh10) cause loss of vulval cell divisions, as do bed-3 mutations. Because bed-3 mutations we examined do not cause complete loss of function, we cannot rule out the possibility that bed-3 has a widespread function as a cell division regulator. Although we found no effect of bed-3 mutations on cell division outside of the vulva, this apparent specificity could be due to sensitivity of vulval lineages to mild reduction in bed-3 function. Nevertheless, several lines of evidence argue against this possibility. Uterine and seam cell lineages commonly affected in cell proliferation mutants (e.g. cul-1, cye-1) are normal in bed-3 mutants, and analysis of bed-3::gfp expression suggests that this gene functions in a rather limited number of tissues.
bed-3 and the vulval gene regulatory network
Our results indicate that the function of bed-3 is closely related to that of cog-1, which encodes an Nkx6 homeobox transcription factor. First, cog-1 and bed-3 mutations affect terminal cell division in vulval lineages. Second, cog-1 and bed-3 mutations affect the lineage-specific expression pattern of the cdh-3::cfp reporter. Finally, cog-1 and bed-3 mutations show synergistic interaction in the double mutant. Since cog-1 is a component of the vulval gene regulatory network, these results suggest that bed-3 may also function in this network.
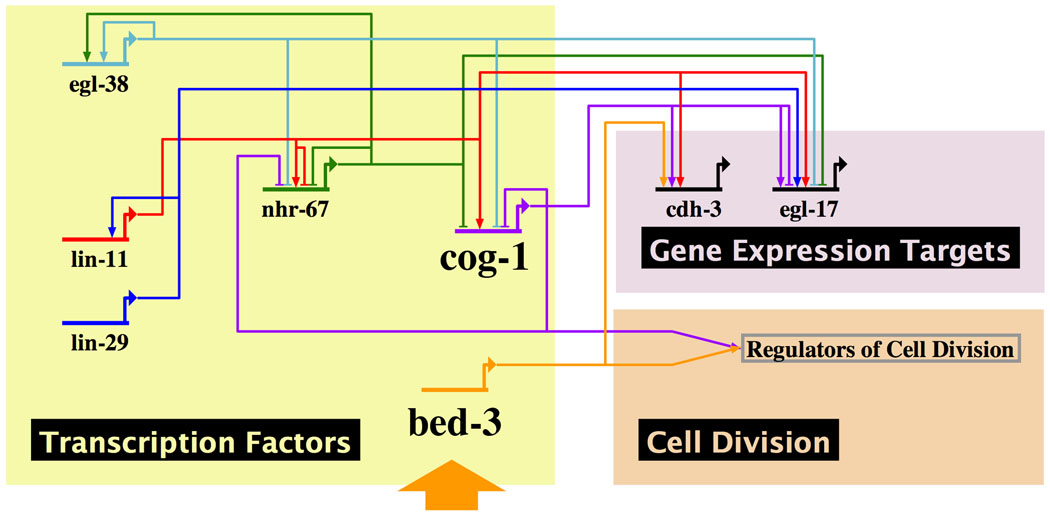
The VPC fate execution phase of vulval development is an excellent example of complex patterning events in which multiple cell types are generated in a specific spatial pattern. The underlying mechanism that enables the organism to perform this type of task is a network of interacting genes (gene regulatory network; GRN) that functions as a complex information-processing unit (Alon, 2007; Davidson and Levine, 2008; Davidson et al., 2002). Cell-type specification occurs when such a network responds to developmental signals and adopts a particular state in which genes appropriate for that cell type are activated. During VPC fate execution, transcription factor genes lin-11, lin-29, cog-1, egl-38 and nhr-67 constitute a part of the vulval GRN, as evidenced by the fact that mutations in these genes alter the pattern of vulval cell type specific gene expression (Figure 8) (Fernandes and Sternberg, 2007; Inoue et al., 2005; Ririe et al., 2008). Our current understanding of this network is incomplete, and the model is not yet predictive. Nevertheless, we can identify network motifs commonly found in GRNs (Alon, 2007), including negative auto-regulation (nhr-67), positive auto-regulation (egl-38), a double-negative feedback loop (cog-1 and nhr-67), and a feed-forward loop (lin-29, lin-11 and egl-17), indicating that the function of this network is likely similar to other described GRNs.
Figure 8. Vulval gene regulatory network regulates gene expression and cell division.
Only selected genes in the network are shown. The current knowledge of the vulval gene regulatory network is such that the model is not predictive. However, it can be observed that cell division and gene expression are regulated by related, but not fully linked mechanisms. (Note, since bed-3 and cog-1 encode transcription factors, regulation of cell division is also likely to involve expression of cell-division promoting genes.) Because mutations we analyzed are not nulls, our results are also consistent with cog-1 and bed-3 (orange arrow) acting in series to regulate cell division. The network was drawn using BioTapestry Editor (http://www.biotapestry.org) (Longabaugh et al., 2009).
Given that the cell division of a Pn.p granddaughter is correlated with the cell type, it is likely that the same gene regulatory network regulates cell division. Indeed, mutations in cog-1 and lin-11 alter the pattern of cell division as well as gene expression. However, although regulated by overlapping sets of genes, cell division and gene expression are not fully correlated. For example, the Pn.p granddaughter fated to become the vulD cell does not undergo the third round of VPC division. However, non-dividing Pn.p granddaughters in cog-1 mutants are not transformed to vulD based on gene expression. Similar loss of correlation between vulval lineage cell division and gene expression is apparent in Wnt pathway mutants (see Figure 5 in Deshpande et al., 2005). From these results, we may infer that there is no single master regulator for each vulval sublineage that controls both cell division and gene expression. Instead, cell division and gene expression can be considered separate outputs of a single gene regulatory network.
Where does bed-3 fit within this scheme? One possibility is that bed-3 is a component of the core decision making circuitry of the vulval gene regulatory network. Alternatively, bed-3 may be a peripheral component which connects the core decision making circuitry to specific outputs, which includes control of cell proliferation. The bed-3(sy705) mutation alters the pattern of cdh-3::cfp expression. This is perhaps suggestive of a more central function, although the latter possibility is not ruled out. Cell ablation studies indicate that regulation of cell type specific gene expression is mostly cell-autonomous after the second Pn.p division (Gupta et al., 2003). However, the possibility remains that the cdh-3::cfp expression defect is a secondary consequence of the cell division defect.
Are BED domain proteins conserved regulators of cell proliferation?
In Drosophila and human cells, BED Zn-finger transcription factors DREF (DNA replication element factor) and hDREF (human DREF) regulate cell proliferation (Matsukage et al., 2008). Promoters of many Drosophila genes required for cell proliferation (e.g. DNA polymerase alpha and proliferating cell nuclear antigen) contain a motif called the DNA replication-related element (DRE; TATCGATA) (Hirose et al., 1993; Matsukage et al., 1995). The DREF protein, in complex with TRF2 (TBP-related factor 2), binds to DREs and activates transcription (Hirose et al., 1993; Hirose et al., 1996; Hochheimer et al., 2002). DREF and TRF2 also interact with proteins regulating chromatin, including another BED Zn-finger domain protein BEAF-32 (Emberly et al., 2008; Hirose et al., 2001; Hochheimer et al., 2002; Nakamura et al., 2008; Zhao et al., 1995). Functional studies demonstrate that manipulation of DREF can have direct effect on cell proliferation during organogenesis (Hirose et al., 2001; Hirose et al., 1999; Hyun et al., 2005; Yoshida et al., 2001). In general, overexpression of DREF promotes cell proliferation whereas disruption of DREF inhibits cell proliferation. Combined with the observation that promoters of genes upregulated during cell proliferation in organ primordia are highly enriched for DRE (Jasper et al., 2002), these results suggest that DREF may play a central role in regulating cell proliferation in Drosophila organogenesis (Matsukage et al., 2008). The human homolog of DREF (hDREF) also promotes cell proliferation in cell culture (Ohshima et al., 2003).
These results suggest that BED-3's role in regulation of cell division may be conserved. Other BED domain proteins, such as BEAF32 (Emberly et al., 2008; Zhao et al., 1995) and ZBED3 (Chen et al., 2009) are not direct regulators of cell proliferation. However, the overall domain structure of the BED-3 protein (a single N-terminal BED domain followed by a weakly conserved C-terminal region) is similar to hDREF and differs from BEAF32 or ZBED3. If the cell proliferation control is a conserved function, like hDREF, BED-3 may regulate cell division directly by activating transcription of genes that are required for cell cycle progression. Alternatively, the regulation may be indirect, and could involve transcriptional regulation of general cell cycle regulators such as cul-1 and cye-1.
Supplementary Material
P5.p and P7.p derived cells were counted in the mid-L4 stage, several hours after cells had finished dividing (see Materials and Methods). The numbers were tallied for each Pn.p cell. Data shown include both P5.p and P7.p since there was no obvious difference between these two symmetric lineages. Numbers of animals observed are: N2 (wild type), 15; sy644, 16; sy702, 26; sy705, 6; sy700, 16; sy700/sy644, 7; RNAi, 32. sy700 animals were homozygous progeny of sy700/nT1[qIs51]. sy700/sy644 were non-Unc progeny of unc-24 sy644 dpy-20/sy700 and also includes homozygous sy700 animals.
Sequence of bed-3 cDNA is shown with numbering in nucleotides starting from ATG. The BED zinc-finger domain is boxed. Nucleotide changes in missense mutations and the region deleted in bed-3(sy700) are shown above the sequence. The sequence of sy700 across the junction is (in the predicted reading frame) CTT GAA // CAG CAT TCC TGA.
Acknowledgment
We thank J. Gruber, Ng L.F., B. Halliwell, Shen Y.Q. and Fu X.Y. for assistance and resources at NUS. We thank J. Fernandes and T. Ririe for comments on the manuscript, and members of the Sternberg laboratory for helpful discussions. T.I. was supported by fellowship DRG-1646 from the Damon Runyon Cancer Research Foundation. This work was supported by the HHMI, with which P.W.S. is an investigator. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. An introduction to systems biology, Design principles of biological circuits. London: CRC Press, Taylor & Francis Group; 2007. [Google Scholar]
- Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–423. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Newman AP, Sternberg PW. Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Current Biology. 1999;9:237–246. doi: 10.1016/s0960-9822(99)80112-2. [DOI] [PubMed] [Google Scholar]
- Chen T, Li M, Ding Y, Zhang LS, Xi Y, Pan WJ, Tao DL, Wang JY, Li L. Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel Axin-interacting protein that activates Wnt/beta-catenin signaling. J Biol Chem. 2009;284:6683–6689. doi: 10.1074/jbc.M807753200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DV, Suleman DS, Beckenbach KA, Gilchrist EJ, Baillie DL. Molecular cloning and characterization of the dpy-20 gene of Caenorhabditis elegans. Mol. Gen. Genet. 1995;247:367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Levine MS. Properties of developmental gene regulatory networks. PNAS. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJC, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Deshpande R, Inoue T, Priess JR, Hill RJ. lin-17/Frizzled and lin-18 regulate POP-1/TCF-1 localization and cell type specification during C. elegans vulval development. Dev. Biol. 2005;278:118–129. doi: 10.1016/j.ydbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Shuettengruber B, Hennion M, Jiang N, Hart CM, Käs E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–2910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development. 2000;127:4049–4060. doi: 10.1242/dev.127.18.4049. [DOI] [PubMed] [Google Scholar]
- Fernandes JS, Sternberg PW. The tailless ortholog nhr-67 regulates patterning of gene expression and morphogenesis in the C. elegans vulva. PLoS Genetics. 2007;3:e69. doi: 10.1371/journal.pgen.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G, Kim S, Horvitz H. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Fujita M, Takeshita H, Sawa H. Cyclin E and CDK2 repress the terminal differentiation of quiescent cells after asymmetric division in C.elegans. 2008 doi: 10.1371/journal.pone.0000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008;134:646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BP, Wang M, Sternberg PW. The C. elegans LIM homeobox gene lin-11 specifies multiple cell fates during vulval development. Development. 2003;130:2589–2601. doi: 10.1242/dev.00500. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose W, Han M. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development. 1999;126:169–179. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- Hirose F, Ohshima N, Shiraki M, Inoue YH, Taguchi O, Nishi Y, Matsukage A, Yamaguchi M. Ectopic expression of DREF induces DNA synthesis, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: possible interaction with Polycomb and trithorax group proteins. Mol. Cell Biol. 2001;21:7231–7242. doi: 10.1128/MCB.21.21.7231-7242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J. Biol. Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Matsukage A. Targeted expression of the DNA binding domain of DRE-Binding Factor, a Drosophila transcription factor, attenuates DNA replication of the salivary gland and eye imaginal disc. Mol Cell Biol. 1999;19:6020–6028. doi: 10.1128/mcb.19.9.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- Hyun J, Jasper H, Bohmann D. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol. Cell Biol. 2005;25:5590–5598. doi: 10.1128/MCB.25.13.5590-5598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Inoue T, Sherwood DR, Aspock G, Butler JA, Gupta BP, Kirouac M, Wang M, Lee PY, Kramer JM, Hope I, Burglin TR, Sternberg PW. Gene expression markers for Caenorhabditis elegans vulval cells. Mech. Dev. 2002;119 Suppl 1:S203–S209. doi: 10.1016/s0925-4773(03)00117-5. [DOI] [PubMed] [Google Scholar]
- Inoue T, Wang M, Ririe TO, Fernandes JS, Sternberg PW. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H, Benes V, Atzberger A, Sauer S, Ansorge W, Bohmann D. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev. Cell. 2002;3:511–521. doi: 10.1016/s1534-5807(02)00297-6. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. PNAS. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta. 2009;1789:363–374. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukage A, Hirose F, Hayashi Y, Hamada K, Yamaguchi M. The DRE sequence TATCGATA, a putative promoter-activating element for Drosophila melanogaster cell-proliferation-related genes. Gene. 1995;166:233–236. doi: 10.1016/0378-1119(95)00586-2. [DOI] [PubMed] [Google Scholar]
- Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta. 2008;1779:81–89. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Ida H, Yamaguchi M. Transcriptional regulation of the Drosophila moira and osa genes by the DREF pathway. Nucleic Acids Res. 2008;36:3905–3915. doi: 10.1093/nar/gkn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, White JG, Sternberg PW. Morphogenesis of the C. elegans hermaphrodite uterus. Development. 1996;122:3617–3626. doi: 10.1242/dev.122.11.3617. [DOI] [PubMed] [Google Scholar]
- Ohshima N, Takahashi M, Hirose F. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J. Biol. Chem. 2003;278:22928–22938. doi: 10.1074/jbc.M303109200. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 2002;252:202–213. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Wood WB, Plasterk RH. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122:4149–4157. doi: 10.1242/dev.122.12.4149. [DOI] [PubMed] [Google Scholar]
- Rajakumar V, Chamberlin HM. The Pax2/5/8 gene egl-38 coordinates organogenesis of the C. elegans egg-laying system. Dev. Biol. 2006 doi: 10.1016/j.ydbio.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Ririe TO, Fernandes JS, Sternberg PW. The Caenorhabditis elegans vulva: A post-embryonic gene regulatory network controlling organogenesis. PNAS. 2008;105:20095–20099. doi: 10.1073/pnas.0806377105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito RM, Perreault A, Peach B, Satterlee JS, van den Heuvel S. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol. 2004;6:777–783. doi: 10.1038/ncb1154. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R, White JG, Southgate E, Podbilewicz B. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development. 1999;126:691–699. doi: 10.1242/dev.126.4.691. [DOI] [PubMed] [Google Scholar]
- Sternberg PW The C. elegans Research Community. WormBook. 2006. Vulval development. [Google Scholar]
- Sternberg PW, Horvitz HR. Pattern formation during vulval development in C. elegans. Cell. 1986;44:761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S. Cell-cycle regulation. In: The C. elegans Research Community, editor. WormBook. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Inoue YH, Hirose F, Sakaguchi K, Matsukage A, Yamaguchi M. Over-expression of DREF in the Drosophila wing imaginal disc induces apoptosis and a notching wing phenotype. Genes Cells. 2001;6:877–886. doi: 10.1046/j.1365-2443.2001.00473.x. [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P5.p and P7.p derived cells were counted in the mid-L4 stage, several hours after cells had finished dividing (see Materials and Methods). The numbers were tallied for each Pn.p cell. Data shown include both P5.p and P7.p since there was no obvious difference between these two symmetric lineages. Numbers of animals observed are: N2 (wild type), 15; sy644, 16; sy702, 26; sy705, 6; sy700, 16; sy700/sy644, 7; RNAi, 32. sy700 animals were homozygous progeny of sy700/nT1[qIs51]. sy700/sy644 were non-Unc progeny of unc-24 sy644 dpy-20/sy700 and also includes homozygous sy700 animals.
Sequence of bed-3 cDNA is shown with numbering in nucleotides starting from ATG. The BED zinc-finger domain is boxed. Nucleotide changes in missense mutations and the region deleted in bed-3(sy700) are shown above the sequence. The sequence of sy700 across the junction is (in the predicted reading frame) CTT GAA // CAG CAT TCC TGA.