Abstract
Here we report the nearly complete base assignments and partial sugar assignments of the 35-residue terminator hairpin of the Bacillus subtilis xpt-pbuX-mRNA guanine sensing riboswitch.
Keywords: Gene regulation, mRNA, Resonance assignment, Riboswitch, Transcription termination
Biological context
The Bacillus subtilis xpt-pbuX-mRNA guanine riboswitch, which is representative of all known purine riboswitches, regulates transcription by binding guanine with high specificity (Mandal et al. 2003). mRNA transcription can be completed when no guanine is present, however mRNA transcription is prematurely aborted when guanine is present and binds to the aptamer part of the riboswitch, which is partially pre-arranged to facilitate ligand binding (Ottink et al. 2007). Guanine binding promotes the formation of a stable terminator hairpin, which interacts with the RNA polymerase, signalling the protein to abort the transcription (Mandal et al. 2003).
In contrast to the highly conserved binding pocket of the riboswitch’s aptamer part, variations in length and base pairing have been observed in the terminator hairpin, resulting in variations in stability. These differences in stability, and consequently differences in structure play an important role in the kinetic trapping mechanism employed by the guanine riboswitches, where during transcription a decision between termination and antitermination is made.
Methods and experiments
The RNA sequence of the terminator hairpin (Fig. 1) was prepared as previously described (Girard et al. 2007). The unlabelled NMR sample (0.44 mM) was prepared in 10 mM Na-phosphate buffer (pH 6.7) containing 0.1 mM EDTA. All NMR spectra were recorded at 15°C on an 800 MHz Varian Inova spectrometer equipped with a cryo-probe. NOESY spectra in water (93% H2O, 7% D2O) were recorded at 15°C with 100, 200, and 300 ms mixing times. The water signal was suppressed with a jump-return pulse (Plateau and Gueron 1982) in combination with a Watergate water suppression scheme (Piotto et al. 1992). In 100% D2O, NOESY spectra were recorded with 100, 300 and 500 ms mixing times. In addition, DQF-COSY spectra and proton decoupled natural abundance 1H-13C-HMQC spectra were recorded to further aid in the assignment process (Wijmenga and Van Buuren, 1998; Cromsigt et al., 2001). Imino protons and cytosine amino protons were assigned from the spectra measured in H2O, whereas the aromatic protons and carbons and the sugar C/H1′ and C/H2′ resonances were assigned using the spectra measured in D2O. All acquired data were processed with NMRPipe (Delaglio et al. 1995). Peak picking and assignment of the spectra was performed with the Sparky software (Kneller and Kuntz 1993).
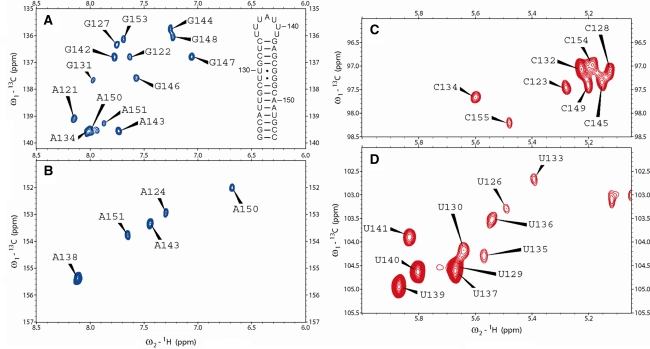
Fig. 1.
Natural abundance 1H-13C-HMQC spectra of the 35-nucleotide guanine riboswitch terminator hairpin. a C8H8 region of purine residues; the C8H8 signal of loop residue A138 is shifted just out of displayed spectral window (142.6 ppm, 8.264 ppm); the relatively high ppm value of H8 indicates de-stacking of this loop residue, which is also evident from the relatively high H2 ppm value seen in panel B. Also the sequence and secondary structure of the terminator hairpin is shown; numbering is according to Mandal et al. 2003 b C2H2 region of adenine residues c C5H5 region of cytosine residues d C5H5 region of uracil residues
Assignments and data deposition
The assignments have been deposited into the BMRB database under number 16479. The nearly complete base assignment (1H: 100%; 13C: 97%) and partial sugar assignment (1H: 21%; 13C: 18%) was achieved using high-quality 2-dimensional homonuclear and heteronuclear NMR spectra (Fig. 1a–d) recorded from one unlabelled RNA sample in H2O and one in D2O each of moderate concentration (0.44 mM).
From an imino-imino sequential walk in the NOESY spectra recorded in H2O, all imino protons could be assigned except the one from G121 (the closing GC base pair, which is partially opened due to end fraying and whose imino exchanges with H2O), and from the loop residues U136 and U140, which are only weakly base paired or not base paired at all. The iminos from loop residues U135 and U141 could be detected in the NOESY with 100 ms NOE mixing time, showing that they do form a base pair. From the H2O-NOESYs, the cytosine-amino protons were also assigned.
Aromatic (C/H2, C/H5, C/H6, C/H8) and sugar (C/H1′, C/H2′) chemical shifts were determined from the spectra recorded in D2O. Due to heavy overlap in the spectra of this relatively large RNA, the C/H2′ resonances could only partially be assigned and the other sugar shifts (C/H3′, C/H4′, C/H5′, C/H5″) could not be determined.
A full H1′–H6/8 sequential walk, continuing throughout the entire stem and loop could be obtained from the NOESY spectra recorded in D2O. Using these spectra, combined with a DQF-COSY spectrum and natural abundance 1H-13C-HMQC spectra (Fig. 1), nearly all aromatic and sugar-H1′ chemical shifts of stem-residues could be assigned and all resonances of the loop residues. The only exception is the H1′ of C134, which could not be assigned unambiguously due to heavy spectral overlap. The H2’s of C2′-endo puckered sugars could readily be determined from the observable H1′–H2′ coherences in the COSY spectrum (Girard et al. 2007). From the NOESY spectra, this assignment could be extended with 6 more H2′ resonances. Using the 1H-13C-HMQC spectra, nearly all C2, C5, C6, C8 and C1′ resonances could be assigned. Due to spectral overlap, the carbon assignments could not fully be completed. The H8 and H2 of A138 show relatively high chemical shift values, 8.26 ppm and 8.10 ppm, respectively; the experience little ring current, indicating, as expected, a lack of stacking interactions for this loop residue.
Acknowledgments
This research was supported by NWO, The Netherlands (SW).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Cromsigt J, Van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Meth Enzymol. 2001;338:371–399. doi: 10.1016/S0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe—a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Girard FC, Ottink OM, Ampt KAM, Tessari M, Wijmenga SS. Thermodynamics and NMR studies on duck, heron and human HBV encapsidation signals. Nucleic Acids Res. 2007;35:2800–2811. doi: 10.1093/nar/gkm131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller DG, Kuntz ID (1993) Ucsf sparky—an NMR display, annotation and assignment tool. J Cell Biochem 53:254–254
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/S0092-8674(03)00391-X. [DOI] [PubMed] [Google Scholar]
- Ottink OM, Rampersad SM, Tessari M, Zaman GJR, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Plateau P, Gueron M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J Am Chem Soc. 1982;104:7310–7311. doi: 10.1021/ja00389a067. [DOI] [Google Scholar]
- Wijmenga SS, Van Buuren BNM. The use of NMR for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc. 1998;32:287–387. doi: 10.1016/S0079-6565(97)00023-X. [DOI] [Google Scholar]