Abstract
Aims
Recent genetic studies identified the rs1333049 variant on chromosome 9p21 as a major susceptibility locus for coronary artery disease and myocardial infarction (MI). Here, we evaluated whether this variant also contributes to recurrent MI or cardiac death following an acute coronary syndrome (ACS).
Methods and results
A total of 3247 patients with ACS enrolled in the Global Registry of Acute Coronary Events (GRACE) in three distinct populations (UK, Belgium and Poland) were prospectively followed for 6 months and genotyped for rs1333049, in addition to 3004 and 2467 healthy controls from the UK and Belgium. After having confirmed that the at-risk C allele of rs1333049 was associated with index ACS in the UK and Belgian populations, we found that the rs1333049 at-risk C allele was significantly and independently associated with recurrent MI [age- and gender-adjusted hazard ratio (HR) 1.48, CI = 1.00–2.19, P = 0.048; and multivariable-adjusted HR 1.47, CI = 0.99–2.18; P = 0.053] and with recurrent MI or cardiac death (age- and gender-adjusted HR 1.58, CI = 1.00–2.48; P = 0.045; and multivariable adjusted HR 1.49, CI = 1.03–1.98; P = 0.028) within 6 months after an index ACS. Inclusion of rs1333049 into the GRACE risk score significantly improved classification for recurrent MI or cardiac death (P = 0.040), as calculated by the integrated discrimination improvement method.
Conclusion
In this large observational study, the 9p21 variant was independently associated with adverse cardiac outcome after ACS.
Keywords: Chromosome 9p21, Rs1333049, Genetics, Acute coronary syndrome, Myocardial infarction, Plaque rupture
See page 1038 for the editorial comment on this article (doi:10.1093/eurheartj/ehq054)
Introduction
Coronary artery disease (CAD) is a complex cardiovascular disorder caused by the interaction of environmental factors and hereditary predisposition.1,2 Recently, the unbiased identification of genetic variations underlying cardiovascular disease has become a fruitful enterprise, with at least four genome-wide association studies identifying chromosome 9p21 as a major locus for risk of CAD or myocardial infarction (MI).3–6 Strikingly, the pathophysiologic effects of the 9p21 locus are not restricted to the coronary vasculature, since genetic variants in this locus have also been linked to abdominal aortic aneurysm, peripheral artery disease, and stroke.7 In addition, the 9p21 locus has been associated with prevalence of atherosclerotic plaque in carotid arteries from healthy individuals, suggesting that the 9p21 locus promotes atherosclerosis.8
Unresolved questions regarding the role of chromosome 9p21 in CAD still remain. Little is known, for instance, about the effect of 9p21 in an unbiased population of acute coronary syndrome (ACS) patients presenting to hospital. Indeed, most CAD/MI studies so far were performed in patients with a familial predisposition (e.g. patients with at least one affected sibling) or in patients presenting at an early age.3,4,9,10 Furthermore, none of these studies focused on the full spectrum of ACS, including the ST-elevation MI (STEMI), non-STEMI, and unstable angina (UA) subtypes, which differ from each other in the degree of thrombus formation.11 Importantly, it is also unknown whether the 9p21 locus confers a risk for recurrent MI or cardiac death following an index ACS. If genetic factors predispose to adverse outcome, this may influence clinical management. Acute coronary syndrome patients are most vulnerable to recurrent and lethal cardiovascular complications during the first 6 months after the primary event, but identification of those most at risk is elusive, when based on clinical phenotypes and presentation characteristics. Improved risk prediction using a combination of phenotypic characteristics and genetic testing could therefore guide patient triage and improve acute and longer-term patient care.12
In the present study, we used the Global Registry of Acute Coronary Events (GRACE) program, which is a large, prospective, multinational observational study of patients hospitalized with ACS. The GRACE Genetics Study employs standardized criteria for inclusion and outcome events across the geographically distinct populations in the UK, Belgium, and Poland. After confirming that the 9p21 locus associated with primary MI in the GRACE Genetics Study, we tested whether this association was independent from the ACS subtype. We next assessed whether the 9p21 locus correlates with recurrent MI and the combined endpoint consisting of recurrent MI or cardiac death.
Methods
Study populations and phenotype descriptions
Grace genetics study
The GRACE registry is a large scale, multinational, prospective observational study of the spectrum of patients with an ACS hospitalized between July 1999 and December 2007. The GRACE Genetics Study consists of the 3473 patients from whom DNA was collected by the eight participating centres in three countries (UK, Belgium, and Poland) between May 2001 and June 2007. Collected blood samples were transferred to the core laboratory (The Queens Medical Research Institute, Edinburgh, UK), where DNA was extracted and stored at −80°C.
Full details of the methods used in the GRACE registry have been published previously.13 Briefly, patients were eligible if they were admitted to participating hospitals with a clinical diagnosis of ACS and at least one of the following features: electrocardiographic changes consistent with ACS, increases in serum biochemical markers of cardiac necrosis [positive troponins, creatinine phosphokinase (CPK), or creatine kinase MB (CK-MB) fraction above two times the upper limit of normal (ULN)], documented CAD [i.e. history of MI; congestive heart failure believed to be due to ischaemia or resuscitated sudden death; history of/or new positive stress test or angina, with or without imaging; prior or new cardiac catheterization documenting artery disease, percutaneous coronary intervention (PCI), or coronary artery bypass graft (CABG)], or both. The presentation of the patients with ACS was not the consequence of trauma, surgery, or other significant co-morbidity (for example anaemia). To ensure enrolment of an unselected ACS population, sites recruited the first 10–20 consecutive eligible patients each month. Regular audits were performed at all participating hospitals to confirm conformance to eligibility criteria and correct interpretation of data in the case report form. Trained study coordinators collected data using standardized case report forms. Demographic characteristics, medical history, presenting symptoms, duration of pre-hospital delay, biochemical and electrocardiographic findings, treatment practices, and outcomes were collected. All cases were assigned to one of the following categories using predefined criteria: (i) STEMI (including left bundle-branch block), (ii) non-STEMI, (iii) UA, and (iv) other cardiac or (v) non-cardiac diagnoses.13 All patients were prospectively followed-up by trained study coordinators at 6 months following admission. Methods of follow-up involved one or more of the following: phone call to patient, hospital visit, phone call to patient's general physician, and/or use of hospital records. All participating hospitals were subject to a rigorous on-site 3 years audit cycle, which involved source verification of randomly selected cases and validation of outcome events. Outcome events (including recurrent MI) were prospectively defined using standard definitions for the entire study, independent of local interpretations.
The primary endpoint for this study was recurrent MI within the first 6 months following the day of hospital admission. A second endpoint consisted of recurrent MI or cardiac death. Non-cardiac death was deliberately excluded as an outcome event since there may be multiple different mechanisms of death without implicating a recurrent myocardial ischaemia or infarction. In-hospital recurrent infarction was confirmed by ECG changes or elevation of cardiac markers, as described previously.12 In patients with acute MI, biomarker criteria for in-hospital recurrent infarction were: (i) re-elevation of the troponin above ULN and increase by at least 20% over the previous value or CK-MB above the ULN and increased by at least 50% over the previous value. (ii) If CK-MB or troponin were not available, CPK had to be either >2 × ULN and increased by 25% over the previous value, or >1.5 × ULN and increased by at least 50% over the previous value. (iii) Following PCI or CABG, CK-MB (or CPK) elevation had to be >3× or >5 × ULN, respectively, and increased by at least 50% over the previous value. (iv) Following CABG, recurrent MI required biomarker and ECG criteria consistent with MI.13
Local hospital ethics committees approved the GRACE Genetics Study. All patients provided written informed consent for follow-up contact and DNA sampling at enrolment, in accordance with the declaration of Helsinki.
Asklepios study cohort
A total of 2524 participants (1301 women) of 35–55 years, free from overt cardiovascular disease at study initiation, were randomly sampled from the twinned Belgian communities of Erpe-Mere and Nieuwerkerken, Belgium, between October 2002 and October 2004. Exclusion criteria were: (i) clinical presence of atherosclerosis or atherothrombosis (symptomatic or haemodynamically significant stenosis, prior atherothrombotic event, and revascularization); (ii) major concomitant illness; (iii) diabetes mellitus type 1 and type 2 if proven macro-vasculopathy or end-stage renal disease; (iv) conditions precluding accurate haemodynamic assessment (atrial fibrillation, pregnancy); (v) inability to provide informed consent.14 Rs1333049 genotypes were available for 2467 participants (97.7%).
Wellcome Trust Case Control Consortium (WTCCC) Controls
A total of 3004 controls from the Wellcome Trust Case Control Consortium (WTCCC) (1504 individuals from the 1958 British Birth Cohort sampled between 2002 and 2004 and 1500 individuals from the UK Blood Service Group) were used as control population for the GRACE patients from the UK.3 Genotypic data for rs1333049 were obtained for 2936 participants (97.7%) from the European Genotype Archive (http://www.ebi.ac.uk/ega).
Genotyping of SNPs in the 9p21 locus
The 9p21 locus that is linked with CAD encompasses two linkage disequilibrium (LD) blocks. Based on r2 calculations between SNPs in both LD blocks, it was demonstrated that rs1333049 is the most representative SNP for the first block. The association signal in the second block can be most consistently captured by three SNPs, i.e. rs7044859, rs1292136, and rs7865618.4,9 We therefore genotyped these four SNPs on the 9p21 locus in a blinded manner using iPLEX technology on a MassARRAY Compact Analyser (Sequenom Inc., CA, USA), as described previously.15 However, since the first LD block (represented by rs1333049) is most prominently linked to CAD, we focused most of the analysis on rs1333049.9 The WTCCC controls were previously genotyped using the Affymetrix platform (Affymetrix Inc., CA, USA).3
Statistical analysis
Data are summarized as frequencies and percentages for categorical variables. Continuous variables are presented as medians with 25th and 75th percentiles. Hardy–Weinberg equilibrium of the genotypes was assessed using Pearson's χ2-test. For the case–control studies, the difference in genotype frequencies was calculated using Pearson's χ2-test. Meta-analysis was performed with the Mantel–Haenszel and the DerSimonian and Laird method.16 Heterogeneity across the study populations was calculated using the χ2-based Cochran Q statistic.17 Lambda statistics was used to calculate the genetic model of inheritance, as previously published.18
For most variables, less than 1% of the data was missing. However, for a number of variables, the percentage of missing values was higher, including serum total cholesterol (12.6%), creatinine (3.6%), systolic blood pressure (2.9%), pulse (3.3%), initial cardiac enzymes (4.0%), Killip class (2.3%), and the use of thrombolytics (2.3%). Missing values were sampled using a multiple imputation technique based on the Markov chain Monte Carlo method (the SAS procedure MI) with seven imputation data sets. Results were combined using the SAS procedure MIANALYSE. In 38.2% of the patients with a recurrent MI, the time of the recurrence was missing since it was not collected in initial case report forms used before July 2002. Missing event times were therefore imputed by a random draw from observed event times. Incident recurrent MI analysis was performed by Cox proportional hazard model using the SAS procedure PHREG, while considering clinical variables known to be associated with recurrent MI as covariates.12,19 These included: age, gender, history of angina, prior MI, diabetes, current smoking status, systolic blood pressure, heart rate, total cholesterol, initial serum creatinine level, elevated initial cardiac enzymes, ST segment depression, Killip class, PCI, CABG, thrombolytics as well as participating country. The cumulative incidence function (CIF) was calculated and plotted for recurrent MI, assuming competing risk of death. For recurrent MI or death, the Kaplan–Meier curve was calculated, using the SAS procedure LIFETEST. Consistency with non-imputed data was confirmed by repeating the analysis with non-imputed data.
Comparison of the GRACE score for improvement in reclassification with and without rs1333049 was done by the Integrated Discrimination Improvement method, which compares the average difference in correctly predicting the risk for patients who have a recurrent MI with those who do not.20
All P-values were two-tailed and <0.05 was considered statistically significant. This was a hypothesis-generating and exploratory study, in which the primary endpoint of interest was the association between rs1333049 and recurrent MI or cardiac death. Therefore, no adjustments were made to the significance level to account for multiple testing. Statistical analyses were performed using the SPSS software version 17.0 (SPSS Inc., IL, USA) and the SAS software version 9.1 (SAS Institute Inc., NC, USA).
Results
Grace genetics study
Overall, 3473 patients were enrolled in the GRACE Genetics Study. Patients with no diagnosis, a non-cardiac diagnosis, or with no ACS at the time of hospital discharge (16, 111, and 102 patients, respectively) were excluded. For the remaining 3247 patients with confirmed ACS, follow-up status 24 h after initial presentation and at a median of 6 months was available for 2980 (91.8%) ACS patients. Of these 2980 patients, genotypes for the rs1333049 variation were available in 2942 patients (98.7%). The genotyping success rate for rs1333049 was ≥98.7%, and 95.2% for rs7044859, 98.9% for rs496892, and 97.3% for rs7865618. Fifty-five samples were genotyped twice with an average concordance of ≥99.8%. No deviation from Hardy–Weinberg equilibrium was seen in any of the cohorts (P > 0.05).
The median follow-up in these 2942 patients was 195 days [interquartile range (IQR) 185–212] following admission, with a recurrent MI occurring in 170 patients (5.8%) and a recurrent MI or cardiac death in 205 patients (7.0%). The median time to recurrence was 12 days (IQR 4–57) for recurrent MI and 29 days (IQR 4–121) for recurrent MI or cardiac death. In Table 1, demographic characteristics, medical history, and presenting clinical features are shown for all patients with a primary ACS. Approximately 28.7% of the primary ACS patients presented with an ST-segment elevation or a left bundle branch block, 70.5% had positive cardiac biomarkers of necrosis, 47.5% had a PCI, and 13.2% received thrombolytics. Consistent with previous studies,21,22 patients with recurrent MI were older, more likely to have prior history of vascular disease, exhibited a higher GRACE risk score for an endpoint predicting death or recurrent MI, and were more likely to have three-vessel disease, as calculated by the number of coronary vessels with >50% stenosis (Table 1). In addition, patients with a non-STEMI and those treated with thrombolytics were more prone to recurrent MI.
Table 1.
Baseline clinical variables predicting a recurrent myocardial infarction in the GRACE Genetics Study
| Variables | Total | No recurrent MI | Recurrent MI | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| No. of patients (%) | 2942 | 2772 (94.2%) | 170 (5.8%) | ||
| Demographics | |||||
| Men, n (%) | 1984 (67.9%) | 1869 (67.9%) | 115 (68.9%) | 1.05 (0.75–1.46) | 0.79 |
| Age, median (25th–75th percentiles), year | 65 (55–73) | 65 (55–73) | 70 (61–79) | 1.04 (1.03–1.06) | <0.001 |
| BMI, median (25th–75th percentiles), kg/m2 | 27.04 (24.37–30.10) | 27.04 (24.38–30.10) | 27.04 (24.22–29.91) | 1.00 (0.99–1.00) | 0.85 |
| Medical History, n (%) | |||||
| Angina | 1628 (55.5%) | 1515 (54.8%) | 113 (66.5%) | 1.64 (1.18–2.27) | 0.003 |
| Diabetes | 500 (17.0%) | 458 (16.6%) | 42 (24.7%) | 1.65 (1.15–2.38) | 0.007 |
| Prior MI | 799 (27.3%) | 729 (26.4%) | 70 (41.2%) | 1.95 (1.42–2.68) | <0.001 |
| Congestive heart failure | 200 (6.8%) | 179 (6.5%) | 21 (12.4%) | 2.05 (1.26–3.31) | 0.004 |
| Stroke or transient ischaemic attack | 173 (5.9%) | 164 (5.8%) | 12 (7.1%) | 1.23 (0.67–2.26) | 0.51 |
| Prior PCI | 408 (13.9%) | 381 (13.8%) | 27 (15.9%) | 1.18 (0.77–1.81) | 0.44 |
| Prior CABG | 240 (8.2%) | 223 (8.1%) | 17 (10%) | 1.27 (0.75–2.13) | 0.37 |
| Peripheral arterial disease | 217 (7.4%) | 190 (6.9%) | 27 (16.0%) | 2.57 (1.66–3.98) | <0.001 |
| Hypertension | 1587 (54.2%) | 1491 (54.0%) | 96 (56.5%) | 1.10 (0.81–1.51) | 0.53 |
| Hyperlipidaemia | 1192 (40.7%) | 1121 (40.6%) | 71 (41.8%) | 1.05 (0.76–1.43) | 0.77 |
| Combined history of atherosclerosis | 1853 (63.2%) | 1722 (62.3%) | 131 (77.1%) | 2.03 (1.41–2.93) | <0.001 |
| Family history of coronary artery disease | 581 (26.1%) | 559 (26.3%) | 22 (22.4%) | 0.81 (0.50–1.31) | 0.39 |
| Smoking status, n (%) | |||||
| Current | 874 (30.3%) | 831 (30.6%) | 43 (25.4%) | 0.77 (0.54–1.10) | 0.16 |
| Former | 786 (27.2%) | 739 (27.2%) | 47 (27.8%) | 1.03 (0.73–1.46) | 0.86 |
| Presenting characteristics | |||||
| Positive initial enzymes, n (%) | 1991 (70.5%) | 1860 (69.9%) | 131 (79.1%) | 1.71 (1.15–2.52) | 0.007 |
| ST elevation or LBBB, n (%) | 843 (28.7%) | 793 (28.6%) | 50 (29.4%) | 1.04 (0.74–1.46) | 0.82 |
| ST depression or T wave inversion, n (%) | 1032 (35.1%) | 949 (34.2%) | 83 (48.8%) | 1.83 (1.34–2.50) | <0.001 |
| Systolic blood pressure, median (25th–75th percentiles), mmHg | 140 (120–160) | 140 (120–159) | 140 (120–160) | 1.00 (0.99–1.01) | 0.21 |
| Diastolic blood pressure, median (25th–75th percentiles), mmHg | 80 (70–90) | 80 (70–90) | 80 (70–90) | 1.00 (0.99–1.01) | 0.66 |
| Pulse (25th–75th percentiles), b.p.m. | 75 (64–87) | 75 (64–86) | 80 (65–96) | 1.01 (1.00–1.02) | 0.005 |
| Killip class, n (%) | <0.001 | ||||
| I | 2271 (79.0%) | 2163 (79.9%) | 108 (65.1%) | Reference | |
| II | 524 (18.2%) | 476 (17.6%) | 48 (28.9%) | 2.02 (1.42–2.88) | <0.001 |
| III | 68 (2.4%) | 58 (2.1%) | 10 (6.0%) | 3.45 (1.72–6.94) | 0.001 |
| IV | 11 (0.4%) | 11 (0.4%) | 0 (0%) | N/A | N/A |
| Creatinine, median (25th–75th percentiles), mg/dL | 1.04 (0.90–1.22) | 1.04 (0.90–1.22) | 1.13 (0.96–1.39) | 1.22 (1.03–1.45) | 0.02 |
| Serum cholesterol, median (25th–75th percentiles), mg/dL | 194 (166–228) | 194 (166–229) | 184 (159–213) | 0.99 (0.99–1.00) | 0.03 |
| GRACE risk score | |||||
| Probability of in-hospital death or MI, median (25th–75th percentiles), % | 11 (8–17) | 11 (8–17) | 14 (9–19) | 1.04 (1.02–1.06) | <0.001 |
| Probability of death or MI 6 months after discharge, median (25th–75th percentiles), % | 4 (3–8) | 4 (3–8) | 7.5 (4–15) | 1.05 (1.03–1.07) | <0.001 |
| Diagnosis | 0.01 | ||||
| STEMI, n (%) | 768 (27.2%) | 724 (27.2%) | 44 (26.8%) | 0.99 (0.69–1.41) | |
| Non-STEMI, n (%) | 1223 (43.3%) | 1136 (42.7%) | 87 (53.0%) | 1.51 (1.11–2.06) | |
| UA, n (%) | 833 (29.5%) | 800 (30.1%) | 33 (20.1%) | 0.59 (0.40–0.88) | |
| Treatment | |||||
| Cardiac catheterization, n (%) | 1926 (65.8%) | 1817 (65.9%) | 109 (64.5%) | 0.94 (0.60–1.30) | 0.71 |
| Three-vessel disease (>50% stenosis), n (%) | 445 (23.1%) | 400 (22.0%) | 45 (41.3%) | 2.35 (1.58–3.51) | 0.001 |
| Left main (>50% stenosis), n (%) | 138 (7.2%) | 127 (7.0%) | 11 (10.1%) | 0.70 (0.35–1.28) | 0.22 |
| PCI, n (%) | 1397 (47.5%) | 1325 (47.9%) | 72 (42.4%) | 0.80 (0.58–1.09) | 0.16 |
| Thrombolytics, n (%) | 380 (13.2%) | 342 (12.6%) | 38 (22.8%) | 2.04 (1.39–2.98) | <0.001 |
| CABG, n (%) | 111 (3.8%) | 103 (3.7%) | 8 (4.7%) | 1.28 (0.61–2.67) | 0.51 |
Data were analysed by logistic regression. Combined history of atherosclerosis comprehends medical history of angina, prior MI, stroke or transient ischaemic attack, prior PCI, CABG, or peripheral arterial disease. A 50% stenosis cut-off was used to determine presence or absence of CAD, instead of a 70% stenosis cut-off, which is used to denominate clinically significant stenosis.
CABG, coronary artery bypass graft; CI, confidence interval; LBBB, left bundle branch block; MI, myocardial infarction; N/A, non-applicable; OR, odds ratio; PCI, percutaneous coronary intervention; STEMI, ST-segment elevated myocardial infarction; UA, unstable angina.
9p21 increases the risk for acute coronary syndrome in a hospital-based population
In order to validate the association of rs1333049 with CAD in the GRACE Genetics Study, we first assessed whether the rs1333049 variation was associated with ACS and therefore performed a case–control association study for the UK and Belgian GRACE patients. To this extent, we used population-based DNA cohorts from the two countries: 2936 participants from the WTCCC study3 and 2467 individuals from the Asklepios Study.14 GRACE patients from Poland were not used because no population-based control cohort with existing genotypic information for rs1333049 was available. In the UK and Belgian sub-studies, the at-risk C allele of rs1333049 was uniformly associated with ACS with respective odds ratios of 1.25 (CI = 1.13–1.39; P = 1.0 × 10−5) and 1.15 (CI = 1.02–1.29; P = 0.024), and a pooled OR of 1.21 (CI = 1.10–1.32; P = 2.4 × 10−6 under a fixed effects model, P = 4.6 × 10−5 under a random effects model; P for heterogeneity = 0.189) for both studies. Genotypic analysis showed that the pooled risk of carrying a single C allele was 1.20 and the risk of carrying two at-risk C alleles was 1.45 (see Supplementary material online, Table S1). Overall, the observed risk effects were in accordance with those observed in previous studies3,4,9 demonstrating that the rs1333049 variant is also a risk factor for unselected patients with ACS seen in daily practice.
Association of 9p21 with clinical subtypes of acute coronary syndrome
Having confirmed that rs1333049 is a risk factor for ACS also in the GRACE Genetics Study, we next assessed whether the rs1333049 acts independently from known clinical risk factors for cardiovascular disease to increase susceptibility for ACS. As shown in Table 2, no significant differences for any of the demographic or phenotypic risk factors were seen. Remarkably, rs1333049 genotypes were also similarly distributed among the different ACS subtypes (STEMI, non-STEMI, and UA), thus indicating that the full spectrum of ACS, rather than a particular subtype, was associated with rs1333049 (Table 2).
Table 2.
Baseline clinical variables according to rs1333049 genotype in the GRACE Genetics Study
| Variables | Total | rs1333049 Genotype |
P-value | ||
|---|---|---|---|---|---|
| GG | GC | CC | |||
| No. of patients (%) | 2942 | 726 (24.7%) | 1460 (49.6%) | 756 (25.7%) | |
| Demographics | |||||
| Men, n (%) | 1984 (67.9%) | 485 (67.4%) | 992 (68.4%) | 507 (67.6%) | 0.86 |
| Age, median (25th–75th percentiles), year | 65 (55–73) | 65 (56–73) | 65 (55–73) | 66 (56–74) | 0.18 |
| BMI, median (25th–75th percentiles), kg/m2 | 27.0 (24.4–30.1) | 26.8 (24.2–30.1) | 27.1 (24.5–30.1) | 27.1 (24.3–30.1) | 0.60 |
| Medical history, n (%) | |||||
| Angina | 1628 (55.5%) | 390 (53.9%) | 797 (54.7%) | 441 (58.5%) | 0.15 |
| Diabetes | 500 (17.0%) | 122 (16.8%) | 248 (17.0%) | 130 (17.3%) | 0.97 |
| Prior MI | 799 (27.3%) | 195 (26.2%) | 381 (26.2%) | 223 (29.5%) | 0.25 |
| Congestive heart failure | 200 (6.8%) | 53 (7.3%) | 101 (7.0%) | 46 (6.1%) | 0.62 |
| Stroke or transient ischaemic attack | 173 (5.9%) | 43 (5.9%) | 82 (5.6%) | 48 (6.4%) | 0.78 |
| Prior PCI | 408 (13.9%) | 99 (13.7%) | 204 (14.0%) | 105 (13.9%) | 0.98 |
| Prior CABG | 240 (8.2%) | 46 (6.4%) | 130 (8.9%) | 64 (8.5%) | 0.11 |
| Peripheral arterial disease | 217 (7.4%) | 48 (6.6%) | 113 (7.8%) | 56 (7.5%) | 0.63 |
| Hypertension | 1587 (54.2%) | 404 (55.9%) | 764 (52.6%) | 419 (55.6%) | 0.23 |
| Hyperlipidaemia | 1192 (40.7%) | 308 (42.7%) | 573 (38.4%) | 311 (41.4%) | 0.30 |
| Combined history of atherosclerosis | 1853 (63.2%) | 449 (62.1%) | 907 (62.3%) | 497 (65.9%) | 0.19 |
| Family history of coronary artery disease | 581 (26.1%) | 144 (26.1%) | 282 (25.4%) | 155 (27.6%) | 0.62 |
| Smoking status, n (%) | |||||
| Current | 874 (30.3%) | 214 (30.2%) | 445 (31.1%) | 215 (29.0%) | 0.62 |
| Former | 785 (27.2%) | 180 (25.4%) | 395 (27.6%) | 210 (28.3%) | 0.43 |
| Presenting characteristics | |||||
| Positive initial enzymes, n (%) | 1991 (70.5%) | 484 (69.4%) | 991 (70.2%) | 516 (72.1%) | 0.53 |
| ST elevation or LBBB, n (%) | 843 (28.7%) | 201 (27.7%) | 430 (29.5%) | 212 (28.7%) | 0.63 |
| ST depression or T wave inversion, n (%) | 1032 (35.1%) | 252 (34.7%) | 506 (34.7%) | 274 (36.2%) | 0.74 |
| Systolic blood pressure, median (25th–75th percentiles), mmHg | 140 (120–160) | 140 (120–160) | 138 (120–156) | 140 (120–160) | 0.072 |
| Diastolic blood pressure, median (25th–75th percentiles), mmHg | 80 (70–90) | 80 (70–90) | 80 (70–90) | 80 (70–90) | 0.33 |
| Pulse (25th–75th percentiles), b.p.m. | 75 (64–87) | 75 (64–88) | 75 (64–86) | 75 (65–90) | 0.59 |
| Killip class, n (%) | 0.86 | ||||
| I | 2271 (79.0%) | 558 (79.3%) | 1118 (78.6%) | 595 (79.5%) | |
| II | 524 (18.2%) | 127 (18.0%) | 267 (18.8%) | 130 (17.4%) | |
| III | 68 (2.4%) | 18 (2.6%) | 31 (2.2%) | 19 (2.5%) | |
| IV | 11 (0.4%) | 1 (0.1%) | 6 (0.4%) | 4 (0.4%) | |
| Creatinine, median (25th–75th percentiles), mg/dL | 1.04 (0.90–1.22) | 1.06 (0.90–1.23) | 1.04 (0.90–1.21) | 1.03 (0.90–1.23) | 0.57 |
| Serum cholesterol, median (25th–75th percentiles), mg/dL | 194 (166–228) | 194 (164–228) | 195 (166–228) | 193 (166–229) | 0.99 |
| GRACE risk score | |||||
| Probability of in-hospital death or MI, median (25th–75th percentiles), % | 11 (8–17) | 11 (8–16) | 11 (7–17) | 11 (8–17) | 0.63 |
| Probability of death or MI 6 months after discharge, median (25th–75th percentiles), % | 4 (3–8) | 4 (3–8) | 4 (3–8) | 5 (3–8) | 0.28 |
| Diagnosis | 0.53 | ||||
| STEMI, n (%) | 768 (26.1%) | 178 (24.5%) | 397 (27.2%) | 193 (25.5%) | |
| Non-STEMI, n (%) | 1223 (41.6%) | 306 (42.1%) | 594 (40.7%) | 323 (42.7%) | |
| UA, n (%) | 833 (28.3%) | 213 (29.3%) | 420 (28.8%) | 200 (26.5%) | |
| Treatment | |||||
| Cardiac catheterization, n (%) | 1926 (65.8%) | 469 (64.7%) | 980 (67.4%) | 577 (63.7%) | 0.16 |
| Three-vessel disease (>50% stenosis), n (%) | 445 (27.4%) | 108 (23.9%) | 231 (25.6%) | 106 (23.5%) | 0.64 |
| Left main (>50% stenosis), n (%) | 138 (7.2%) | 24 (5.1%) | 76 (7.8%) | 38 (8.0%) | 0.14 |
| PCI, n (%) | 1397 (47.5%) | 343 (47.2%) | 709 (48.6%) | 345 (45.8%) | 0.44 |
| Thrombolytics, n (%) | 380 (13.2%) | 94 (13.2%) | 193 (13.5%) | 93 (12.6%) | 0.83 |
| CABG, n (%) | 111 (3.8%) | 23 (3.2%) | 62 (4.3%) | 26 (3.5%) | 0.39 |
Data were analysed by Kruskal–Wallis or χ2 test. Abbreviations are the same as in Table 1.
Although previous studies reported an association between rs1333049 and prevalent atherosclerosis in carotid vessels from healthy individuals,23 we only observed a weak trend towards association of rs1333049 and prior history of atherosclerosis, with carriers of two at-risk C alleles being slightly more common (26.8 vs. 24.9%; P = 0.19, Table 2). In addition, there was also no significant association between rs1333049 and three-vessel disease (P = 0.64).
Association of 9p21 with recurrent myocardial infarction or cardiac death
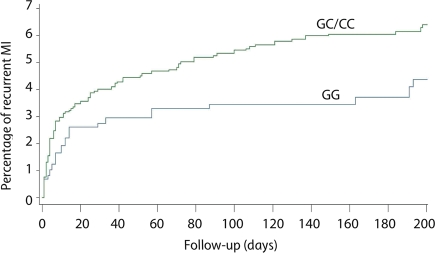
We then assessed whether 9p21 was also associated with recurrent atherothrombotic events. Survival analysis in the GRACE Genetics Study (n = 2942) revealed that rs1333049 was significantly associated with recurrent MI (n = 170) within the first 6 months after the primary event. This risk effect followed a dominant genetic model (see Supplementary material online, Table S2), in which the presence of any at-risk C allele predicted recurrent MI with an age- and gender-adjusted hazard ratio (HR) of 1.48 (CI = 1.00–2.19, P = 0.048) and a similar effect after multivariable adjustment (HR 1.47; CI = 0.99–2.18; P = 0.053; Table 3). The CIF illustrating that a recurrent MI is more common in C allele carriers vs. non-carriers is shown in Figure 1. Consistency with non-imputed data was confirmed by repeating the analysis without imputation and revealed comparable results (multivariable adjusted HR 2.30, CI = 1.27–4.09, P = 0.006). To further dissect this association with recurrent MI, genotype distributions of three additional SNPs in the 9p21 locus (rs7044859, rs1292136, and rs7865618), as well as their haplotype distributions, were studied. However, no additional relevant associations were detected (Supplementary material online, Tables S3 and S4).
Table 3.
Association of rs1333049 with recurrent myocardial infarction in the GRACE Genetics Study
| rs1333049 | No recurrent MI | Recurrent MI | Age- and gender-adjusted |
Multivariable-adjusteda |
||
|---|---|---|---|---|---|---|
| 2772 (94.2%) | 170 (5.8%) | HR (95% CI) | P-Value | HR (95% CI) | P-Value | |
| Genotypic, n (%) | ||||||
| CC | 710 (25.6%) | 46 (27.1%) | 1.41 (0.89–2.23) | 0.14 | 1.34 (0.84–2.12) | 0.21 |
| GC | 1367 (49.3%) | 93 (54.7%) | 1.52 (1.01–2.28) | 0.044 | 1.55 (1.03–2.33) | 0.037 |
| GG | 695 (25.1%) | 31 (18.2%) | Reference | Reference | ||
| Dominant, n (%) | ||||||
| CC + GC | 2077 (74.9%) | 139 (81.8%) | 1.48 (1.00–2.19) | 0.048 | 1.47 (0.99–2.18) | 0.053 |
| GG | 695 (25.1%) | 31 (18.2%) | Reference | Reference | ||
Data are analysed by a Cox proportional hazard model.
CI, confidence interval; HR, hazard ratio.
aAdjustments for: age, gender, history of angina, prior myocardial infarction, diabetes, current smoking status, systolic blood pressure, heart rate, total cholesterol, initial serum creatinine level, elevated initial cardiac enzymes, ST segment depression, Killip class, percutaneous coronary intervention (PCI), coronary artery bypass graft (CABG), thrombolytics, and participating country.
Figure 1.
Cumulative incidence function for recurrent myocardial infarction (MI) by rs1333049 genotype in the GRACE Genetics Study. The cumulative incidence function for recurrent MI by rs1333049 genotype is shown, comparing individuals carrying 1 or 2 at-risk C alleles (GC and CC) vs. individuals carrying no risk alleles (GG).
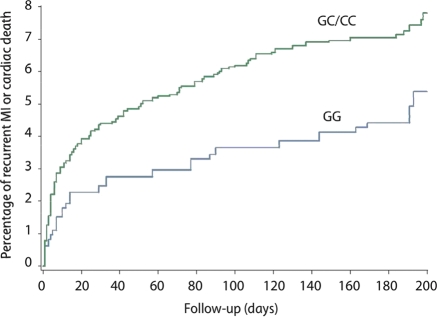
Since cardiac death is often precipitated by ischaemic heart disease, we used a second combined endpoint consisting of recurrent MI or cardiac death. In agreement with the primary endpoint analysis, rs1333049 was significantly associated with recurrent MI or cardiac death (n = 205) within the first 6 months after the primary event (P = 0.045 and P = 0.028, respectively, after age- and gender-adjustment and multivariable adjustment; Table 4 and Figure 2). Consistency with non-imputed data was confirmed by repeating the analysis without imputation (multivariable adjusted HR 2.28, CI = 1.27–4.09, P = 0.0057).
Table 4.
Association of rs1333049 with recurrent myocardial infarction or cardiac death in the GRACE Genetics Study
| rs1333049 | No recurrent MI or cardiac death | Recurrent MI or cardiac death | Age- and gender-adjusted |
Multivariable-adjusteda |
||
|---|---|---|---|---|---|---|
| 2737 (93.0%) | 205 (7.0%) | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Genotypic, n (%) | ||||||
| CC | 701 (25.6%) | 55 (26.8%) | 1.39 (0.82–2.37) | 0.22 | 1.33 (0.88–2.03) | 0.18 |
| GC | 1347 (49.2%) | 113 (55.1%) | 1.69 (1.06–2.69) | 0.028 | 1.58 (1.09–2.30) | 0.016 |
| GG | 689 (25.2%) | 37 (18.0%) | Reference | Reference | ||
| Dominant, n (%) | ||||||
| CC + GC | 2048 (74.8%) | 168 (82.0%) | 1.58 (1.00–2.48) | 0.045 | 1.49 (1.03–1.98) | 0.028 |
| GG | 689 (25.2%) | 37 (18.0%) | Reference | Reference | ||
Data are analysed by a Cox proportional hazard model.
CI, confidence interval; HR, hazard ratio.
aAdjustments as in Table 4.
Figure 2.
Cumulative incidence of recurrent MI or cardiac death by rs1333049 genotype in the GRACE Genetics Study. A Kaplan–Meier curve analysis for recurrent MI or cardiac death by rs1333049 genotype is shown, comparing individuals carrying 1 or 2 at-risk C alleles (GC and CC) vs. individuals carrying no risk alleles (GG).
We also assessed whether the 9p21 genetic risk locus improved the prediction of a recurrent MI or cardiac death by quantifying the improvement in risk classification after adding rs1333049 into the GRACE risk score for recurrent MI.12 To this extend, the integrated discrimination improvement (IDI) index was used.20 Inclusion of rs1333049 into the prediction model showed a trend towards improved classification of recurrent MI at 6 months; this effect failed, however, to reach significance (P = 0.073). However, risk classification for the combined endpoint consisting of recurrent MI or cardiac death was significantly improved (P = 0.040): an additional 5.93% of the patients had an increased probability of being correctly classified after inclusion of rs1333049 into the model.
Discussion
The GRACE Genetics Study was used to better characterize the 9p21 genetic risk locus for CAD. Our findings are relevant in several respects.
The most important finding is that the 9p21 locus predisposes to recurrent MI or cardiac death. Indeed, in ACS patients receiving optimal secondary prevention treatments, we found that carriers of the at-risk C allele exhibited an increased risk to develop a recurrent MI or cardiac death within 6 months after the primary event. Interestingly, the HR for developing a recurrent event in these carriers was higher than the risk of having a primary event (HR = 1.49 for recurrent MI or cardiac death vs. OR = 1.26 for primary ACS). This is remarkable, since patient populations with a primary ACS are already enriched for the rs1333049 at-risk C allele. In agreement herewith, the percentage of individuals carrying a C allele in the various patient groups increased gradually with the number of coronary events, i.e. 72.5% in healthy individuals compared with 74.9% in all primary ACS patients and as much as 81.8% in patients with a recurrent MI or cardiac death. The significant association of rs1333049 with recurrent MI or cardiac death could also generate intriguing insights into the biological role of 9p21 in CAD. Indeed, previous genetic studies investigating the 9p21 locus have used heterogeneous patient groups, consisting of stable CAD, coronary revascularization, or MI patients. Consequently, it has been impossible to conclude whether 9p21 drives atherosclerotic plaque formation, promotes plaque rupture, or triggers both. A study by Ye et al.,23 which demonstrates that 9p21 is associated with prevalence and 5 years progression of pre-clinical atherosclerosis in carotid vessels, suggests that 9p21 promotes plaque formation. It is therefore possible that GRACE patients carrying the rs1333049 allele develop more extensive atherosclerosis, and are more prone to a recurrent infarction. However, since we limited the follow-up period between the primary and recurrent event to 6 months, it is also possible −if not more likely− that recurrent events were due to subsequent rupture of pre-existing plaques. In this regard, our data also suggest a role for 9p21 in plaque rupture. Nevertheless, as long as the functional effect of rs1333049 on neighbouring genes is not known, it might be difficult to draw definite conclusions about the mechanisms by which rs1333049 exerts its effect on recurrent MI.
Our data may seem surprising since Horne et al.24 could not establish an association between the 9p21 locus and a combined endpoint of non-fatal MI and death, suggesting that 9p21 was not associated with MI. However, the differences in patient selection and outcome criteria between Horne's study and GRACE might explain why we did observe such association in the present study. Indeed, Horne et al. retrospectively selected patients from an angiographic database, which included stable and UA, and MI patients, while GRACE prospectively selected consecutive ACS patients only. As endpoint, Horne et al. used primary MI, recurrent MI, as well as all-cause moralities, whereas GRACE only considered recurrent MI and/or cardiac death.
Remarkably, the association of rs1333049 with recurrent MI or cardiac death was independent of all other risk factors, suggesting that rs1333049 may be used as an additional variable in clinical risk prediction models for recurrent MI. We tested this hypothesis using the GRACE risk score, which has been developed and validated for the robust prediction of cumulative 6 months risk of death or MI.12 We found that inclusion of rs1333049 into the GRACE risk score did not significantly improve risk classification for the prediction of recurrent MI (P = 0.073), but significantly improved risk classification of the combined endpoint of recurrent MI or cardiac death (P = 0.040) at 6 months using the integrated discrimination improvement method. It can therefore be anticipated that inclusion of additional genetic factors, which are likely to be identified in the near future, in combination with rs1333049 could enhance risk prediction models for adverse outcome following ACS.
Another finding of the study is that rs1333049 is a risk factor for primary ACS in two geographically different populations from the UK and Belgium. Although the primary reason for performing this analysis was to validate the GRACE Genetics Study, the findings from our case–control association study are of additional interest. Indeed, since the large majority of patients studied previously were a mixture of CAD or MI patients, which were either genetically enriched or exhibited early disease onset,3,4,9,10 risk effects in the latter cohorts may have been overestimated. However, in our prospective and consecutively collected GRACE Genetics Study, the risk associated with rs1333049 was largely comparable to that of previous reports: the odds ratio per C allele in the GRACE Genetics Study was 1.21 compared with 1.24 in the meta-analysis from Schunkert et al.9 Our study thus confirms that the risk associated with rs1333049 is applicable to patient populations as seen in clinical practice.
The strength of the GRACE Genetics Study is its specific design to capture an unselected and representative population of ACS patients with carefully characterized baseline and prospective follow-up data. In particular, standardized inclusion criteria and definitions of outcome events, which were independent of local interpretations in each of the participating clinical centres, were used. Another strength of the study is the inclusion of three geographically distinct populations, with variations in environmental risk factors and different incidence and prevalence rates for CAD. This suggests that the findings were not specific to a particular population or to a specific collection of risk factors in one country. It should be noted, however, that this study has also a number of limitations. First, we have used in the case–control study for primary ACS control populations that were not age- or gender-matched to the GRACE patients. However, since these control populations consisted of younger participants, which are yet to develop cardiovascular events, relatively more at-risk C allele carriers would be expected in these control populations compared with older cohorts, suggesting that our observed odds ratios would be an under-estimation, rather than over-estimation of the true risk effect. Second, since we do not have genome-wide SNP data for GRACE, we could not assess population substructure by multidimensional scaling or principal component analysis.25 A third limitation is the small effective sample size of the recurrent MI or cardiac death population. Indeed, only 5.8 (n = 170) and 7.0% (n = 205) of all primary ACS patients developed a recurrent MI or recurrent MI/cardiac death event. Additional replication studies are therefore needed to confirm our findings. Finally, periprocedural MI was not part of the initial case record form in GRACE, thereby precluding a sub-analysis excluding periprocedural MI from the endpoints. Because we only included a recurrent MI occurring at 24 h or later after the primary event and because periprocedural MI occurs in the majority of the patients within the first 24 h, we expect that most periprocedural MIs will automatically have been excluded by this definition.
In conclusion, our study confirms that the 9p21 locus confers a risk for primary ACS in an unselected patient population. Importantly, we also provide unprecedented evidence that the 9p21 locus is independently associated with recurrent MI or cardiac death following an acute coronary event. Since the follow-up period between the primary and recurrent event was restricted to 6 months, our data suggest that the 9p21 locus—in addition to promoting plaque formation—might also be involved in stimulating plaque instability and rupture. Finally, we provide suggestive evidence that prediction models for recurrent events might be complemented by genetic risk factors such as the 9p21 locus.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
Funding for the project was provided by the Wellcome Trust under award 076113.3 GRACE is supported by an unrestricted educational grant from Sanofi-Aventis to the Center for Outcomes Research, University of Massachusetts Medical School. Phenotypic characterization and DNA extraction were funded by an award from the British Heart Foundation (CH/92010) to K.A.A.F. Genotyping was funded by a ‘Fonds voor Wetenschappelijk onderzoek’ (FWO) ‘Krediet aan Navorsers’ grant (1516009N) to D.L. The Asklepios Study was funded by a FWO research grant (G042703) to E.R. D.R.D. was funded by the Wellcome Trust Cardiovascular Research and Cardiovascular Functional Genomics Initiatives, and by a British Heart Foundation CoRE award. D.L. was supported by the FWO. We are grateful to Désiré Collen and the Life Sciences Research Partners VZW (LSRP) for providing access to the MassARRAY Compact Analyser (Sequenom). None of the funding organizations had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Conflict of interest: none declared.
Acknowledgement
We thank the patients, physicians, and nurses participating in the GRACE, Asklepios, and WTCCC studies. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. We thank Laura De Velder for her excellent assistance to this study.
References
- 1.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, Lisheng L. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case–control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. doi:10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 2.Fischer M, Mayer B, Baessler A, Riegger G, Erdmann J, Hengstenberg C, Schunkert H. Familial aggregation of left main coronary artery disease and future risk of coronary events in asymptomatic siblings of affected patients. Eur Heart J. 2007;28:2432–2437. doi: 10.1093/eurheartj/ehm377. doi:10.1093/eurheartj/ehm377. [DOI] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. doi:10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. doi:10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. doi:10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 7.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. doi:10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 8.Yamagishi K, Folsom AR, Rosamond WD, Boerwinkle E. A genetic variant on chromosome 9p21 and incident heart failure in the ARIC study. Eur Heart J. 2009;30:1222–1228. doi: 10.1093/eurheartj/ehp087. doi:10.1093/eurheartj/ehp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, Blankenberg S, Tiret L, Ducimetiere P, Keniry A, Ghori MJ, Schreiber S, El Mokhtari NE, Hall AS, Dixon RJ, Goodall AH, Liptau H, Pollard H, Schwarz DF, Hothorn LA, Wichmann HE, Konig IR, Fischer M, Meisinger C, Ouwehand W, Deloukas P, Thompson JR, Erdmann J, Ziegler A, Samani NJ. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. doi:10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, Seedorf U, Rust S, Eriksson P, Hamsten A, Farrall M, Watkins H. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. doi:10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 11.Mizuno K, Satomura K, Miyamoto A, Arakawa K, Shibuya T, Arai T, Kurita A, Nakamura H, Ambrose JA. Angioscopic evaluation of coronary-artery thrombi in acute coronary syndromes. N Engl J Med. 1992;326:287–291. doi: 10.1056/NEJM199201303260502. [DOI] [PubMed] [Google Scholar]
- 12.Fox KA, Dabbous OH, Goldberg RJ, Pieper KS, Eagle KA, Van de Werf F, Avezum A, Goodman SG, Flather MD, Anderson FA, Jr, Granger CB. Prediction of risk of death and myocardial infarction in the six months after presentation with acute coronary syndrome: prospective multinational observational study (GRACE) BMJ. 2006;333:1091. doi: 10.1136/bmj.38985.646481.55. doi:10.1136/bmj.38985.646481.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The GRACE Investigators. Rationale and design of the GRACE (Global Registry of Acute Coronary Events) Project: a multinational registry of patients hospitalized with acute coronary syndromes. Am Heart J. 2001;141:190–199. doi: 10.1067/mhj.2001.112404. doi:10.1067/mhj.2001.112404. [DOI] [PubMed] [Google Scholar]
- 14.Rietzschel ER, De Buyzere ML, Bekaert S, Segers P, De Bacquer D, Cooman L, Van Damme P, Cassiman P, Langlois M, van Oostveldt P, Verdonck P, De Backer G, Gillebert TC. Rationale, design, methods and baseline characteristics of the Asklepios Study. Eur J Cardiovasc Prev Rehabil. 2007;14:179–191. doi: 10.1097/HJR.0b013e328012c380. doi:10.1097/HJR.0b013e328012c380. [DOI] [PubMed] [Google Scholar]
- 15.Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. doi:10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. doi:10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. doi:10.2307/3001666. [Google Scholar]
- 18.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34:1319–1328. doi: 10.1093/ije/dyi169. doi:10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 19.Newby LK, Bhapkar MV, White HD, Topol EJ, Dougherty FC, Harrington RA, Smith MC, Asarch LF, Califf RM. Predictors of 90-day outcome in patients stabilized after acute coronary syndromes. Eur Heart J. 2003;24:172–181. doi: 10.1016/s0195-668x(02)00325-1. doi:10.1016/S0195-668X(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 20.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. doi:10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 21.Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D, Braunwald E. The TIMI risk score for unstable angina/non-ST elevation MI: a method for prognostication and therapeutic decision making. JAMA. 2000;284:835–842. doi: 10.1001/jama.284.7.835. doi:10.1001/jama.284.7.835. [DOI] [PubMed] [Google Scholar]
- 22.Boersma E, Pieper KS, Steyerberg EW, Wilcox RG, Chang WC, Lee KL, Akkerhuis KM, Harrington RA, Deckers JW, Armstrong PW, Lincoff AM, Califf RM, Topol EJ, Simoons ML. Predictors of outcome in patients with acute coronary syndromes without persistent ST-segment elevation. Results from an international trial of 9461 patients. The PURSUIT Investigators. Circulation. 2000;101:2557–2567. doi: 10.1161/01.cir.101.22.2557. [DOI] [PubMed] [Google Scholar]
- 23.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008;52:378–384. doi: 10.1016/j.jacc.2007.11.087. doi:10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 24.Horne B, Carlquist JF, Muhlestein B, Blair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. doi:10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miclaus K, Wolfinger R, Czika W. SNP selection and multidimensional scaling to quantify population structure. Genet Epidemiol. 2009;33:488–496. doi: 10.1002/gepi.20401. doi:10.1002/gepi.20401. [DOI] [PubMed] [Google Scholar]