Abstract
Once a mutation in the gene tub was identified as the cause of obesity, retinal degeneration and hearing loss in tubby mice1–2, it became increasingly evident that the members of the tub gene family (tulps) influence maintenance and function of the neuronal cell lineage3–6. Suggested molecular functions of tubby-like proteins include roles in vesicular trafficking4,7, mediation of insulin signaling8 and gene transcription9,10. The mechanisms through which tub functions in neurons, however, have yet to be elucidated. Here we report the positional cloning of an auditory quantitative trait locus (QTL), the modifier of tubby hearing 1 gene (moth1)11, whose wildtype alleles from strains AKR/J, CAST/Ei and 129P2/OlaHsd protect tubby mice from hearing loss. Through a transgenic rescue experiment, we verified that sequence polymorphisms in the neuron-specific microtubule-associated protein 1a gene (Mtap1a) observed in the susceptible strain C57BL/6J (B6) are crucial for the hearing-loss phenotype. We also show that these polymorphisms change the binding efficiency of MTAP1A to postsynaptic density molecule 95 (PSD95), a core component in the cytoarchitecture of synapses. This indicates that at least some of the observed polymorphisms are functionally important and that the hearing loss in C57BL/6J-tub/tub (B6-tub/tub) mice may be caused by impaired protein interactions involving MTAP1A. We therefore propose that tub may be associated with synaptic function in neuronal cells.
The gene tub was first identified, through positional cloning efforts, as a gene with unknown function. A spontaneous mutation at the splice donor site in the last intron, which causes a replacement of the carboxy-terminal 44 amino acids with 24 amino acids that are not observed in the normal protein, leads to obesity associated with hyperinsulinemia and neurosensory deficits1–2. The fact that the targeted tub null allele12 has the same phenotype as the spontaneous mutant confirms that this splice-site mutation is also the molecular basis for the spontaneous loss-of-function mutation.
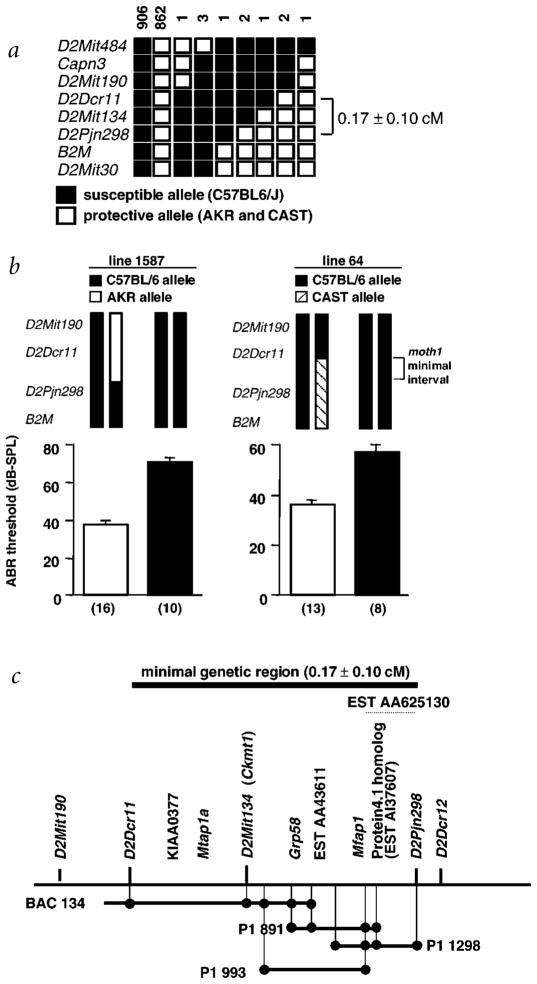
The moth1 locus is a major QTL that affects hearing in tubby mice. We previously showed that a single moth1 allele from strains AKR/J, CAST/EiJ or 129P2/OlaHsd was sufficient to protect B6-tub/tub mice from hearing loss11. The high level of significance for linkage of suppression of hearing loss (lod=33.4) observed on chromosome 2 suggested that fine-mapping by recombinant progeny testing13 could be applied to the moth1 locus. We collected a total of 1,780 meioses, of which 1,330 came from an F2 (B6-tub/tub × AKR) intercross and 450 from an F2 (B6-tub/tub × CAST.B6-tub/tub) intercross (Fig. 1a). All animals recombinant with respect to the critical region were progeny-tested to confirm the presence of the protective moth1 allele in the recombinant region (Fig. 1b). The minimal moth1 region is 0.17 ± 0.1 cM and flanked by markers D2Dcr11 and D2Pjn298. We assembled a physical contig of BACs and P1s spanning approximately 350 kb across the critical region (Fig. 1c). Using BAC/P1 end-sequencing, we identified two genes, mitochondrial creatine kinase (Ckmt1) and glucose-regulated protein, 58 kD (Grp58, also known as Erp57). We also carried out direct cDNA selection using eye and brain cDNA from AKR mice. Sequencing 50 cDNA clones from the screen yielded four additional genes—Protein 4.1–like, mouse KIAA0377 homolog, Mfap1 (mouse homolog of human microfibrillar associated protein 1) and Mtap1a—and three EST contigs (Fig. 1c). By Southern-blot analysis, we confirmed that the cDNA-selected genes and EST contigs were indeed from the critical region.
Fig. 1.
Generation of physical contig and gene expression map. a, Fine-structure map of the mothl locus compiled by examining 1,780 meioses from B6-tub/tub intercrosses with AKR/J and CAST/Ei. The genetic size of the critical region is 0.17 ± 0.10 cM. The number of chromosomes sharing the corresponding haplo-types are indicated above each column. b, Representative click ABR thresholds, of offspring from two recombinant individuals, that were key in determining the minimal genetic region of the moth1 locus. Genotypes of recombinant progeny are indicated (top) and corresponding ABR thresholds are shown below (mean ± s.e.m.). The number of progeny tested in each recombinant line is in parentheses. c, Schematic representation of the genetic and physical maps of four overlapping P1/BAC large-insert clones. Transcripts derived by direct cDNA selection and from end-sequencing of clones as well as STS markers are localized on the map. The approximate location of EST AA625130 is marked by a dotted line.
We compiled full-length mouse cDNA sequences for the genes Protein 4.1–like, KIAA0377 homolog, Mfap1 and Mtap1a and compared the sequences between B6 and AKR/J. No sequence differences were found in the cDNA of Protein 4.1–like, the KIAA0377 homolog and Mfap1. The Mtap1a cDNA derived from B6, however, contained 12 single-nucleotide differences which could lead to amino-acid alterations and a difference in the length of a GCTCCA (Ala-Pro) repeat in the open reading frame, when compared with AKR. Further sequencing of the 129P2/OlaHsd and CAST/Ei alleles of Mtap1a revealed ten amino-acid changes and a shorter Ala-Pro motif than that found in B6 mice (Table 1). We also observed two nucleotide differences among the alleles that did not segregate with the hearing-loss phenotype. A polymorphism at nt 2368, which potentially causes a Glu→Lys change, was seen only in the AKR allele. Moreover, whereas in the B6 and CAST alleles, nt 5509 was C (Pro), the corresponding AKR and 129P2/OlaHsd alleles contained a G residue (Ala).
Table 1.
Comparison of sequence alterations in Mtap1A, from protective alleles derived from AKR, CAST and 129/Ola, with the C57BL/6 allele
| Single-nucleotide alteration | ||
| Nucleotide alteration | Codon alteration | Amino-acid alteration |
| 2020A→G | ACA→GCA | Thr→Ala |
| 2795T→C | ATC→ACC | Ile→Thr |
| 2890T→G | TCC→GCC | Ser→Ala |
| 4093G→C | GTT→CTT | Val→Leu |
| 4302C→T | TCA→TTA | Ser→Leu |
| 4771A→G | AAT→GAT | Asn→Asp |
| 5772C→G | CAC→CAG | His→Gln |
| 5854T→C | TCT→CCT | Ser→Pro |
| 5971C→G | CCT→GCT | Pro→Ala |
| 5994A→G | CAA→CAC | Gln→His |
| Alteration in the alanine-proline repeat 6871–6976: GCTCCA..........GCTCCA (90 bp) to 6871–6900: GCTCCA…GCTCCA (30 bp) | AlaPro..........AlaPro (30 aa) to AlaPro…AlaPro (10 aa) | |
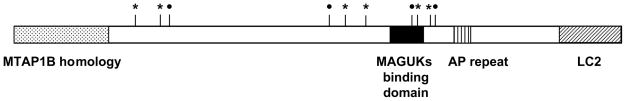
The AKR/J-derived mouse Mtap1a cDNA sequence encodes a polyprotein precursor of 2,796 amino acids, which is processed to a heavy chain (aa 1–2575) and a light chain (aa 2576–2796). The polymorphic Ala-Pro motif starting at aa 2291, and seven of the ten single–amino acid changes in B6 localized between aa 1365 and 1998 (Fig. 2), suggest ‘hot spots’ for sequence alterations. Although few QTL loci have been identified, multiple polymorphisms and hot spots for sequence polymorphisms may be a common feature of genes responsible for QTLs. For example, several sequence variations were identified in the gene Cd36 that lead to an insulin-resistance phenotype. In that case, most of the sequence alterations were localized in the same exon14.
Fig. 2.
Domain structure of MTAP1A and positions of polymorphisms. The location of the light chain 2 (LC2), membrane-associated guanylate kinases (MAGUKs) binding domain and the Ala-Pro (AP) repeat are indicated. Filled circles indicate conservative replacement, and asterisks nonconservative replacement, of amino acids. Note that an AP repeat-length polymorphism is seen between susceptible and protective alleles.
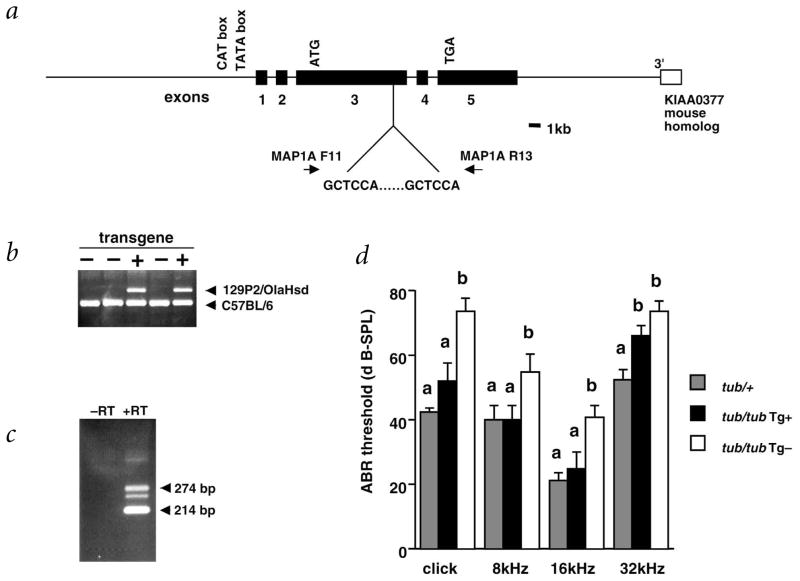
To verify that Mtap1a is indeed moth1, we tested whether an exogenous protective allele from 129P2/OlaHsd could prevent hearing loss in tubby mice. We generated transgenic mice using a construct containing approximately 20 kb each of 5′ regulatory and 3′ downstream genomic regions of Mtap1a (Fig. 3a). We confirmed the generation of transgene positive mice by allele-specific PCR (Fig. 3b) and determined expression of the trans-gene by RT–PCR analysis of mRNA from the cochlea (Fig. 3c).
Fig. 3.
Transgenic rescue of hearing impairment in B6-tub/tub by the 129P2/OlaHsd allele of Mtap1a. a, Transgenic rescue construct. Primers for amplification of the GCTCAA repeat used to distinguish between the B6 allele and the transgene derived from 129P2/OlaHsd alleles are indicated. b, Detection of transgene-positive (Tg+) mice by PCR. The size of the resultant PCR products are 274 bp for 129P2/OlaHsd and 214 bp for B6. c, Expression of the transgene-derived Mtap1a in the cochlea as detected by RT–PCR with the primers used for genotyping (the third band which appears under the transgene-derived 274 bp band represents the heteroduplex between the B6 and 129P2/OlaHsd alleles). d, ABR thresholds in C57BL/6-tub/tub-Tg+ mice. The mean ± s.e.m. of the sound pressure threshold is shown for each stimulus and genotype. Five or six mice at 5–6 wk were tested for each genotype. Threshold levels with different superscripts are significantly different (P<0.01).
Phenotypic analysis of transgene-positive mice from Line 56 that were homozygous with respect to the tubby mutation showed significantly improved auditory brainstem response (ABR) thresholds, compared with B6-tub/tub littermates (Fig. 3d). The exception was the reduction of ABR thresholds for the 32-kHz tone, which did not quite reach statistical significance. RT–PCR results suggest that the transgene is not expressed at levels as high as the susceptible B6 allele. It is possible that higher levels of expression are necessary to effect total phenotypic rescue in the basal cochlear hair cells responsible for high frequency hearing, compared with the expression required for rescue in apical portions of the cochlea.
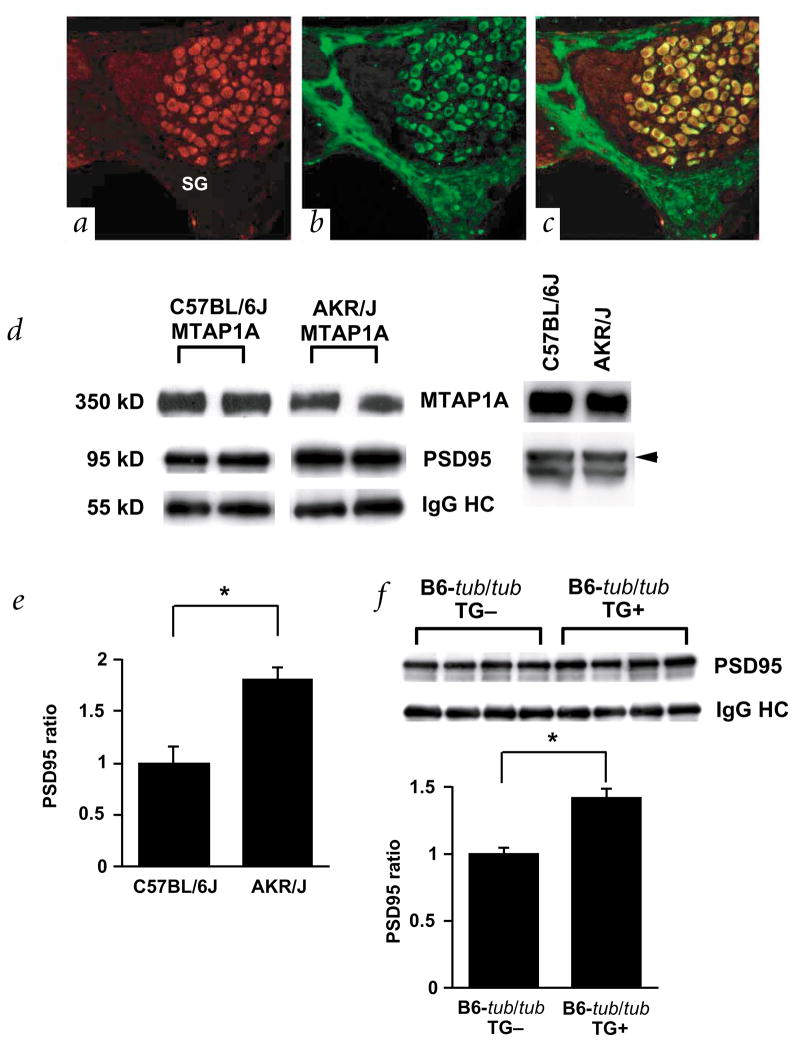
The nearly complete rescue of the hearing-loss phenotype by the transgene suggests that the polymorphisms observed between cDNA of the susceptible B6 and protective alleles in Mtap1a are key in determining the hearing phenotype in tubby mice. To further understand the interaction between tub and Mtap1a, we examined their expression in cochlea. As previously reported, TUB is localized in the hair cells, supporting cells and spiral ganglion cells of cochlea11. Notably, MTAP1A is prominently expressed in the spiral ganglion cells (Fig. 4b). Double-labeling showed that both TUB and MTAP1A were co-localized in the cytosol of these cells (Fig. 4c). Although both hair and spiral ganglion cell degeneration first occur in tubby mice15,16 by 20 weeks, functional defects assessed by ABR analysis are apparent at only 3 weeks11,15, suggesting that the degeneration of these cells might be a secondary event. As hearing loss in tubby mice was rescued by the 129P2/OlaHsd allele of Mtap1a, which is expressed specifically in the spiral ganglion cells, the tubby gene may significantly influence auditory signal transmission in these cells.
Fig. 4.
Localization and characterization of MTAP1A. a–c, Immunofluorescence micrographs of the spiral ganglion cells in the organ of corti after double-labeling with antibodies against TUB (red, a) and MTAP1A (green, b). Panel c shows a merged version of a and b. Note that TUB is expressed in both the cytosol and nucleus and that TUB and MTAP1A are co-localized in the cytosol of the spiral ganglion cells (yellow). d, Association of MTAP1A derived from B6 (left) and AKR/J (right) with PSD95 in the cerebellum. Immunoprecipitation with anti-MTAP1A followed by immunoblotting with anti-MTAP1A and anti-PSD95 reveals that the amount of PSD95 precipitated by equal amounts of anti-MTAP1A antibody is higher in AKR/J than B6 (left panel). Immunoblot analysis (right panel) shows that the level of protein expression of both MTAP1A and PSD95 is the same in the cerebellum of B6 and AKR/J mice. Levels of immunoprecipitates were determined for each strain using the same membrane. e, Measurement of the relative amount of immunoprecipitated PSD95 in four individual animals each of strains B6 (n=4) and AKR/J (n=4). Levels were normalized to the level of IgG heavy chain, and the B6 levels were arbitrarily set to 1. The mean ± s.e.m. is represented by a bar (asterisk indicates P<0.05). f, Association of MTAP1A with PSD95 in B6-tub/tub and B6-tub/tub-Tg+ mice. An immunoblot for PSD95 co-precipitated with MTAP1A in each animal (n=4 for each strain) is shown (top panel). Measurement of the relative amount of immunoprecipitated PSD95 in four individual animals of each strain (bottom panel). Levels were normalized to the level of IgG heavy chain, and the B6-tub/tub mice levels were arbitrarily set to 1. The mean ± s.e.m. is represented by a bar (asterisk indicates P<0.05).
The MAP proteins were originally shown to stabilize micro-tubules. Recent studies revealed other functions for this protein family, such as trafficking of vesicles and organelles17,18. MTAP1A is characterized by the presence of a binding site for a guanylate kinase (GUK) domain that is thought to act as a protein-interaction domain. GUK domains are present in members of the membrane-associated guanylate kinase (MAGUK) family of proteins, which are crucial in establishing the post synaptic cytoarchitecture17,18. One member of this protein family, PSD95, interacts with various neurotransmitter receptors, cell-adhesion molecules and signaling molecules, thus functioning as a ‘scaffolding’ protein and coordinating the function of the synapse19. Because PSD95 physically interacts with MTAP1A17, and the position of most sequence polymorphisms in the susceptible strain B6 are in the vicinity of the GUK binding domain, we examined the MTAP1A/PSD95 complex in cerebellar protein extracts from B6 and AKR mice (Fig. 4d). We found that the amount of PSD95 immunoprecipitated by excess MTAP1A antibodies was higher in the complex from AKR than in that precipitated from B6 mice (Fig. 4e). As we did not find any sequence polymorphisms in PSD95 between the B6 and AKR alleles, the immunoprecipitation results indicate that the sequence polymorphisms in the B6 MTAP1A allele may affect the binding between MTAP1A and PSD95. Similarly, in the transgenic mice, we saw an increase in binding of MTAP1A to PSD95, although less than that observed in AKR mice (Fig. 4f). Although it is possible that this difference in binding could be responsible for rescuing the hearing phenotype, we cannot exclude other mechanisms of rescue. For example, MTAP1A may be involved in additional interactions that could be responsible for the phenotypic difference. There are several molecules that have MAGUK domains, including PSD93, which is also known to bind MTAP1A17. In addition, binding of MTAP1A to PSD95 is influenced by a variety of interacting proteins, including NMDA receptor, APC and CRIPT17. Binding to proteins in regions other than the GUK binding domain of MTAP1A may also be affected, as the Ala-Pro motif from the B6 allele is significantly shorter in the MTAP1A protein than that of wildtype. Proline-rich regions generally form motifs that are known to interact with other proteins20. Thus, binding proteins for MTAP1A in addition to PSD95 may be important in linking MTAP1A with TUB.
As MTAP1A may influence trafficking of synaptic components from the cytosol to the synaptic junction18, the difference in binding efficiency of MTAP1A to PSD95 observed between the B6 and AKR alleles suggests a potential alteration in the capacity to transport synaptic components. This may in turn affect synaptic functions such as synaptic plasticity, neurotransmitter signaling and maintenance of cell polarity. We therefore hypothesize that the function of tubby is, at least in part, associated with synaptic function. This idea is supported by two additional observations. TUB has been suggested to be downstream of G-protein receptor signaling10, which, in neuronal cells, occurs at synaptic junctions. In addition, TUB is highly expressed in at least one region where synaptic junctions are formed (outer plexiform layer of the retina)3. As we have not yet found evidence that TUB is a component of a protein complex with MTAP1A and PSD95, further studies are necessary to elucidate the genetic interaction between tub and Mtap1a. Nevertheless, identification of the neuronal-specific Mtap1a as the moth1 gene reveals an essential component of a neuronal-specific pathway.
Methods
BAC/P1 contig construction and cDNA selection
We identified BAC and P1 clones by PCR-based screening of mouse BAC (RPCI-21) and P1 (ref. 6) libraries with the mouse DNA marker, D2Mit134, and markers generated from end sequences of BACs and P1s (PCR primers are available upon request). We carried out direct selection on mouse brain and eye cDNA libraries that we generated. We used fragments derived from one randomly digested BAC (BAC 134) and two P1 (P1 891 and P1 1298) clones to generate a probe mixture.
ABR analysis
We used a computer-aided evoked potential system (Intelligent Hearing System, IHS) to test mice for ABR thresholds as previously described11. Mice were anesthetized with tribro-moethanol (5.3 mg 10 g−1 body weight, i.p.). We inserted subdermal needle electrodes at the vertex (active) and ventrolaterally to the right ear (reference) and to the left ear (ground). Specific acoustic stimuli were delivered binaurally through 1-cm plastic tubes channeled from high-frequency transducers. We tested mice with click stimuli and also with 8-, 16- and 32-kHz tone pips at varying intensity, from low to high (10–90 dB SPL). We determined an ABR threshold for each stimulus frequency by identifying the lowest intensity that produced a recognizable ABR pattern (at least two consistent peaks)11.
Immunofluorescence microscopy
B6 (n=4) and AKR/J (n=4) mice at 4–6 wk were deeply anesthetized and transcardially perfused with PBS, followed by 4% paraformaldehyde (PFA) in PBS. The cochlea was removed and stored in 4% PFA for 4 h. For decalcification, we incubated the cochlea in 7% EDTA-PBS for 4 d before embedding it in paraffin. We stained 7-μm serial sections of the cochlea with Mayer’s hematoxylin using standard procedures. Adjacent sections were subjected to immunohistochemical analysis. Briefly, after blocking with 2% goat serum in PBS, sections were incubated with rabbit polyclonal antiserum against the amino-terminal half of TUB (Tub-N2; 1:200)3 and then with mouse monoclonal antibody against MTAP1A (HM-1; 1:200; Sigma). Binding was detected using biotinylated goat anti-mouse IgG (1:200; Vector), followed by FITC-AvidinD (1:200; Vector) or Cy3-conjugated donkey anti-rabbit IgG (1:150; Jackson Immunoresearch). We carried out a nuclear counterstain with DAPI at a final concentration of 5 μg ml−1. We collected images on a Leica DMRXE fluorescent microscope equipped with a SPOT CCD camera, using appropriate bandpass filters for each fluorochrome.
Generation of transgenic mice
We identified two 129P2/OlaHsd-derived P1 clones that contained Mtap1a by PCR screening of a 129P2/OlaHsd library using Mtap1a-specific primers (MAP1A F11 and MAP1A R13). These P1 clones contained at least three genes, including Mtap1a. We partially digested a P1 clone by Sau3A and subcloned the fragments into a cos-mid vector (Stratagene). We identified Mtap1a-positive clones by Southern-blot hybridization using probes specific to Mtap1a. We selected a cos-mid clone encompassing approximately 20 kb of 5′ and 3′ flanking regions by a combination of sequencing and RFLP analyses, and used it for the transgene construct for the rescue experiment after linearization with ClaI. The construct was micro-injected into pronuclei of C57BL/6 embryos to generate transgenic mice. We obatined four transgene-positive founders and crossed them to B6-tub/tub mice, and then intercrossed to obtain transgene-positive tub/tub mice.
For expression analysis of the transgene, we amplified cDNA sequences with primers MAP1A F11 and MAP1A R13. We isolated total RNA from the cochlea using the Qiagen RNA isolation kit (Qiagen) and synthesized first-strand cDNA with the SuperScript Choice System (Life Technology).
Immunoprecipitation and immunoblot analysis
We carried out immunoprecipitation and subsequent immunoblot analysis of cerebellar extracts as described17. Homogenates from each cerebellum were incubated with anti-MTAP1A at 4 °C overnight, followed by an overnight incubation with BioMag protein G (Polysciences). After washing, pellets were resuspended in 40 μl of 1× electrophoresis buffer. We loaded 6 μl from each sample onto a 10% SDS-polyacrylamide gel (Biorad). Proteins were electrophoretically transferred to PVDF membranes for immunoblot analyses. We used the following antibodies: anti-MTAP1A (Sigma) and anti-PSD95 (Upstate Biotechnology). We captured photo images of the immunoblots with an image scanner and measured band intensity with NIH image software (v. 1.62).
Acknowledgments
We are grateful to S. Ackerman, G. Cox and B.K. Knowles for careful review of the manuscript. This work was supported by grants from the Foundation for Fighting Blindness, the National Institute of Diabetes & Digestive & Kidney Diseases, the National Institute on Deafness and Other Communicative Disorders and AXYS Pharmaceuticals. Institutional shared services are supported by a National Cancer Institute Support grant. A.I. is a recipient of an American Heart Association postdoctoral fellowship.
References
- 1.Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 2.Kleyn PW, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda S, et al. Cell specific expression of tubby gene family members in the retina. Invest Ophthalmol Vis Sci. 1999;40:2706–2712. [PubMed] [Google Scholar]
- 4.Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 5.Ikeda S, et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet. 2000;22:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum Mol Genet. 2001;10:1325–1334. doi: 10.1093/hmg/10.12.1325. [DOI] [PubMed] [Google Scholar]
- 7.Hagstrom SA, et al. A role for the Tubby-like protein 1 in rhodopsin transport. Invest Ophthalmol Vis Sci. 2001;42:1955–1962. [PubMed] [Google Scholar]
- 8.Kapeller R, et al. Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J Biol Chem. 1999;274:24980–24986. doi: 10.1074/jbc.274.35.24980. [DOI] [PubMed] [Google Scholar]
- 9.Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 10.Santagata S, et al. G-protein signaling through Tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda A, et al. Genetic modification of hearing in tubby mice: evidence for the existence of a major gene (moth1) which protects tubby mice from hearing loss. Hum Mol Genet. 1999;8:1761–1767. doi: 10.1093/hmg/8.9.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubdal H, et al. Targeted deletion of the tub mouse obesity gene reveals that tubby is a loss-of-function mutation. Mol Cell Biol. 2000;20:878–882. doi: 10.1128/mcb.20.3.878-882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nature Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 14.Aitman TJ, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nature Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 15.Heckenlively JR, et al. Mouse model for Usher syndrome: linkage mapping suggests homology to Usher type I reported at human chromosome 11p15. Proc Natl Acad Sci USA. 1995;92:11100–11104. doi: 10.1073/pnas.92.24.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohlemiller KK, et al. Progression of cochlear and retinal degeneration in the tubby (rd5) mouse. Audiol & Neuro-Otol. 1997;2:175–185. doi: 10.1159/000259242. [DOI] [PubMed] [Google Scholar]
- 17.Brenman JE, et al. Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J Neurosci. 1998;18:8805–8813. doi: 10.1523/JNEUROSCI.18-21-08805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas U, et al. Synaptic targeting and localization of discs-large is a stepwise process controlled by different domains of the protein. Curr Biol. 2000;10:1108–1117. doi: 10.1016/s0960-9822(00)00696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata Y, Takai Y. Roles of postsynaptic density-95/synapse-associated protein 90 and its interacting proteins in the organization of synapses. Cell Mol Life Sci. 1999;56:461–472. doi: 10.1007/s000180050445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]