Abstract
The pathophysiologic pathways and clinical expression of mitochondrial DNA (mtDNA) mutations are not well understood. This is mainly the result of the heteroplasmic nature of most pathogenic mtDNA mutations and of the absence of clinically relevant animal models with mtDNA mutations. mtDNA mutations predisposing to hearing impairment in humans are generally homoplasmic, yet some individuals with these mutations have severe hearing loss, whereas their maternal relatives with the identical mtDNA mutation have normal hearing1,2. Epidemiologic, biochemical and genetic data indicate that nuclear genes are often the main determinants of these differences in phenotype3–5. To identify a mouse model for maternally inherited hearing loss, we screened reciprocal backcrosses of three inbred mouse strains, A/J, NOD/LtJ and SKH2/J, with age-related hearing loss (AHL). In the (A/J×CAST/Ei)×A/J backcross, mtDNA derived from the A/J strain exerted a significant detrimental effect on hearing when compared with mtDNA from the CAST/Ei strain. This effect was not seen in the (NOD/LtJ × CAST/Ei)×NOD/LtJ and (SKH2/J×CAST/Ei)×SKH2/J backcrosses. Genotyping revealed that this effect was seen only in mice homozygous for the A/J allele at the Ahl locus on mouse chromosome 10. Sequencing of the mitochondrial genome in the three inbred strains revealed a single nucleotide insertion in the tRNA-Arg gene (mt-Tr) as the probable mediator of the mitochondrial effect. This is the first mouse model with a naturally occurring mtDNA mutation affecting a clinical phenotype, and it provides an experimental model to dissect the pathophysiologic processes connecting mtDNA mutations to hearing loss.
Mouse models for mitochondrial disorders include nuclear gene mutants that lead to oxidative phosphorylation disorders6,7 or, in the case of the Tfam-deficient mouse, to mtDNA transcription-deficient mice8,9. These models are potentially useful, especially because the defects can be expressed in specific tissues through the use of the loxP-Cre system9, but they are unlikely to represent good models to elucidate the pathogenetic pathways caused by human mtDNA disease mutations. Efforts to create an mtDNA defect in mouse recently led to the introduction of a chloramphenicol-resistant mitochondrial chromosome into the embryonic stem cells of mice, with the creation of heteroplasmic mice10. These mice have cataracts and retinal changes, and when the mutation is passed through the germ line, it is lethal in utero and in the perinatal period11. Most recently, a similar approach led to a mouse with a mitochondrial deletion that, when transmitted maternally, causes multiple organ dysfunction and death before the age of 1 year, mostly because of renal failure12. Although the technical hurdles to create true models of pathogenic mtDNA mutations remain formidable, the establishment of such models remains a very important and rate-limiting step in mitochondrial research.
We previously reported on the hearing sensitivity of 80 inbred strains of mice as assessed by auditory-evoked brainstem response (ABR) threshold measurements13. To assess the potential mitochondrial effects on AHL, reciprocal matings were made between each of three hearing-impaired laboratory inbred strains—A/J, NOD/LtJ and SKH2/J—and the wild-derived inbred strain CAST/Ei. The choice of the three strains was fortuitous in that it was the first step of a systematic search to identify putative maternal effects. CAST/Ei retains good hearing beyond 12 months of age and is genetically distinct from the laboratory inbred strains. Female and male mice from each hearing-impaired strain were mated with, respectively, male and female CAST/Ei mice to produce reciprocal F1 hybrids. All F1 hybrids retained good hearing, similar to the parental CAST/Ei mice, at three and six months of age. Because of the apparent recessive nature of this hearing loss, F1 hybrids were backcrossed to the hearing-impaired strains (Fig. 1). We analyzed associations of maternal strain lineage with ABR thresholds in all three backcross populations at three and six months of age (Table 1). There was a statistically significant association of maternal origin with hearing loss in the A/J backcross with CAST/Ei, which was not seen in the NOD/LtJ and SKH2/J backcrosses (Table 1).
Fig. 1.
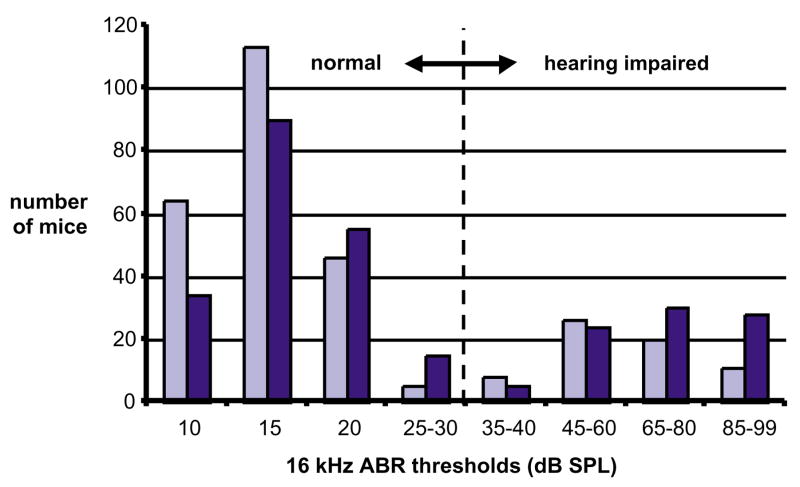
Frequency distribution of auditory-evoked brainstem response (ABR) thresholds in (A/J×CAST/Ei)×A/J backcross progeny. Hearing loss occurs earlier in NOD/LtJ and SKH2/J backcross mice than in A/J backcross mice, but otherwise their ABR threshold frequency distributions are similar. Mice were tested successively at ages 3 months (light bars) and 6 months (dark bars). The vertical dashed line indicates division between normal and elevated ABR thresholds. Backcross matings were set up using males and females from both types of reciprocal F1 hybrid. Maternal strain origins in N2 backcross mice were noted.
Table 1.
Effects of mitochondrial origin on ABR thresholds of 3- and 6-month-old N2 mice from three backcrosses
| Mitochondrial origin | Number of N2 mice | ABR thresholds | Probability values Mann–Whitney | |
|---|---|---|---|---|
| Average | s.d. | |||
| (A/J×CAST/Ei) F1×A/J backcross | ||||
| 3-month test age | ||||
| A/J | 135 | 30.7 | 25.4 | 0.0043* |
| CAST | 157 | 21.1 | 17.2 | |
| 6-month test age | ||||
| A/J | 130 | 38.9 | 31.5 | 0.012* |
| CAST | 151 | 29.2 | 23.4 | |
| (NOD/LtJ×CAST/Ei) F1×NOD/LtJ backcross | ||||
| 3-month test age | ||||
| NOD | 144 | 37.8 | 34.4 | 0.27 |
| CAST | 156 | 40.9 | 34.6 | |
| 6-month test age | ||||
| NOD | 133 | 44.4 | 37.4 | 0.31 |
| CAST | 141 | 47.5 | 36.9 | |
| (SKH2/J×CAST/Ei) F1×SKH2/J backcross | ||||
| 3-month test age | ||||
| SKH | 141 | 33.3 | 30.2 | 0.30 |
| CAST | 74 | 35.6 | 29.6 | |
| 6-month test age | ||||
| SKH | 141 | 45.3 | 34.9 | 0.80 |
| CAST | 74 | 45.9 | 34.1 | |
Denotes significant results.
We have shown that all 10 inbred strains with an elevated ABR threshold so far chosen for genetic analysis, including the three backcrosses described above, share at least one common gene (designated Ahl) on mouse chromosome 10 that has a major effect on AHL (ref. 13). We examined the combined effects on hearing impairment of this nuclear gene with maternal origin in A/J back-cross mice (Table 2). Ahl genotypes exhibit an extremely high association with ABR threshold variation, which increases with the age of the mice. The maternal effect on AHL in this backcross, albeit less than that exerted by Ahl, is statistically highly significant. Maternal effects on average ABR thresholds were highly significant in backcross mice that inherited two copies of the A/J Ahl allele, whereas a single copy of the CAST/Ei Ahl allele prevented AHL regardless of maternal origin (Table 2). These results show an interaction between the effects of the nuclear Ahl gene and maternal origin in determining hearing impairment in (A/J×CAST/Ei)×A/J backcross mice.
Table 2.
Effects of the Chr 10 Ahl gene (as marked by D10Mit138) and mitochondrial origin on ABR thresholds of 3- and 6-month old N2 mice from the (A/J×CAST/Ei) F1×A/J backcross
| D10Mit138 Chr 10 Ahl | Mitochondrial origin | Number of N2 mice | ABR thresholds | Probability values Mann–Whitney | |
|---|---|---|---|---|---|
| Average | s.d. | ||||
| 3-month test age | |||||
| AA | – | 125 | 40.0 | 26.7 | <0.0001* |
| AC | – | 167 | 14.7 | 5.8 | |
| – | A/J | 135 | 30.7 | 25.4 | 0.0043* |
| – | CAST | 157 | 21.1 | 17.2 | |
| AA | A/J | 68 | 47.3 | 26.8 | 0.0002* |
| AA | CAST | 57 | 31.3 | 24.0 | |
| AC | A/J | 67 | 13.8 | 3.4 | 0.55 |
| AC | CAST | 100 | 15.7 | 6.9 | |
| 6-month test age | |||||
| AA | – | 120 | 56.1 | 28.9 | <0.0001* |
| AC | – | 161 | 17.0 | 8.7 | |
| – | A/J | 130 | 38.9 | 31.5 | 0.012* |
| – | CAST | 151 | 29.2 | 23.4 | |
| AA | A/J | 64 | 63.1 | 28.7 | 0.0005* |
| AA | CAST | 56 | 48.1 | 27.4 | |
| AC | A/J | 66 | 15.4 | 6.3 | 0.14 |
| AC | CAST | 95 | 18.1 | 9.9 | |
Denotes significant results.
Although this maternal effect could be due to imprinting or to the placental transmission of an ototoxic teratogenic agent in A/J mice, mtDNA is by far the most probable cause of this effect. In humans, hearing loss is associated with a number of different mtDNA mutations2, but not with imprinting or an ototoxic-specific congenital agent. Imprinting can also be directly excluded because back-cross mice derived from F1 females with A/J mothers (77 mice) had significantly higher hearing thresholds than back-cross mice derived from F1 females with CAST/Ei mothers (157 mice). At 3 months of age, the Mann–Whitney P value was 0.0023, and at 6 months of age, it was 0.01. These results are not consistent with imprinting because, for imprinting, all female F1 hybrids are equivalent (as all alleles should imprint as females regardless of the sex of the parents).
As we observed the maternal effect only for the A/J backcross and not for the NOD/LtJ and SKH2/J backcrosses (Table 1), it seems likely that a difference in the mtDNA of these three inbred strains should account for this. Laboratory strains of mice have a high frequency of one type of Mus domesticus mtDNA, whose frequency in wild populations is only 0.04 (ref. 14). Nearly all of the ‘old’ inbred strains (established before 1922) were derived from females with this rare mtDNA type, which makes the number of expected sequence variations low (if any indeed occur).
In a search for putative differences between the mtDNA of the A/J mouse strain and the mtDNAs of the SKH2/J and NOD/LtJ mice, we sequenced the complete mitochondrial genome of all three strains and compared its sequence with the originally published mouse mtDNA sequence15. A single nucleotide change was found in the A/J strain but not in the NOD/LtJ and SKH2/J strains (Table 3). The change is an insertion of an adenine in a repeat of nine adenines in mt-Tr. The sequence change was confirmed by sequencing this region of the mtDNA from eight backcross mice with mitochondria inherited from the two A/J founder females, the four NOD/LtJ founder females and the two SKH2/J founder females. The length of this repeat appears to be polymorphic in mice, varying between 8 and 10 adenines. NOD/LtJ and SKH2/J mice repeats contain nine adenines. The mtDNA of the A/J, as well as of the MilP and NZB/B1NJ strains, contains 10 adenines. The published reference sequence contains eight adenines, and when we sequenced the tRNA-Arg gene from the CAST/Ei strain used in the backcross experiments described above, the length of the adenine repeat was also eight. The adenine repeat is located between nt 14 and 22 in the D stem of the 69-bp mouse mitochondrial tRNA-Arg and is not highly conserved through evolution (Fig. 2).
Table 3.
MtDNA sequence variation between the three inbred mouse strains and the published mouse sequence
| Position (bp) | Published sequence | A/J | NOD/LtJ | SKH2/J | Gene/RNA | Amino acid substitution |
|---|---|---|---|---|---|---|
| 1767 | A | G | G | G | mt-Rnr2 | |
| 3815 | T | del T | del T | del T | mt-Tg | |
| 3824 | ins T | ins T | ins T | mt-Tg | ||
| 4012 | A | G | G | G | mt-Nd2 | none |
| 4794 | T | C | C | C | mt-Nd2 | Ile→Thr |
| 5734 | C | T | T | T | mt-Co1 | Pro→Leu |
| 5737 | T | C | C | C | mt-Co1 | Val→Ala |
| 6238 | G | A | A | A | mt-Co1 | Cys→Tyr |
| 9348 | A | A | A | G | mt-Co2 | Ile→Val |
| 9553 | ins AAA | ins AAA | ins AAA | mt-Nd3 | ||
| 9826 | ins A | ins A | ins A | mt-Tr | ||
| 9827 | ins A | mt-Tr | ||||
| 9856 | C | A | A | A | mt-Tr | |
| 10059 | ins CAT | ins CAT | ins CAT | mt-Nd4 | ||
| 12042 | C | T | T | T | mt-Nd5 | Leu→Phe |
| 15823 | T | delT | delT | delT | D-loop | |
| 16119 | A | C | C | C | D-loop | |
| 16241 | A | delA | delA | delA | D-loop |
A total of 18 changes from the originally published mouse mtDNA sequence were identified15. All appeared homoplasmic within the level of detection of the sequencing gels. Sixteen of the changes were found in all three mouse strains. Fifteen of them, with the exception of the insertion of an adenine at position 9826 in mt-Tr, represent most likely errors in the published sequence. A BLAST search of the mouse EST database and non-redundant database revealed that all available ESTs, as well as two previously sequenced mouse strains, MilP and NZB/B1NJ (ref. 30), contained the same sequence changes. The A9348G sequence change was found only in the SKH2/J mouse. It appears to be a polymorphism, as the MilP and NZB/B1NJ strains, as well as 40 of the mouse ESTs, contained a guanine, whereas nine ESTs contained an adenine.
Fig. 2.
Evolutionary comparison and two-dimensional structure of mouse mt-Tr. a, Cloverleaf structure of mouse mt-Tr. The string of adenine residues in the D loop region is underlined. b, Sequence allignment of the tRNA-Arg from different species. Region corresponding to the adenines string in mouse is denoted by brackets.
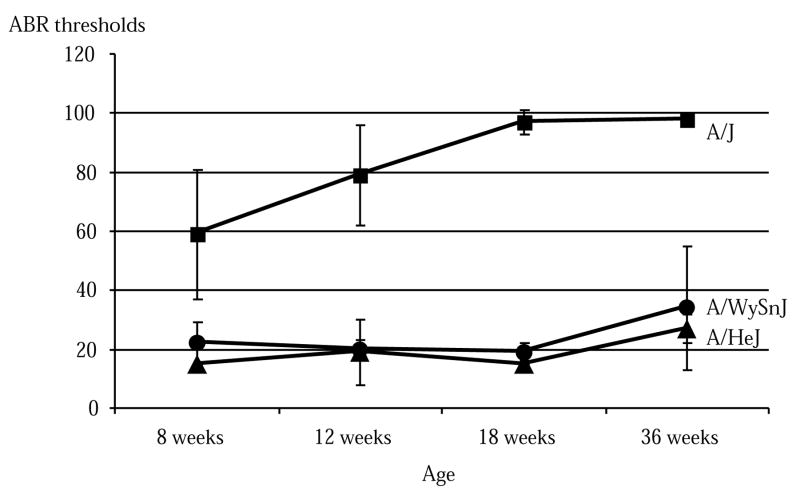
Several lines of evidence support the hypothesis that this insertion of an adenine in mt-Tr of the A/J strain is responsible for the worsening of the hearing deficit in mice homozygous for a susceptibility allele at the Ahl locus (Tables 1 and 2). First, this is the only difference in the sequence of the mtDNA of the A/J strain that is not present in the NOD/LtJ and SKH2/J strains. Second, we sequenced the mitochondrial genome of the A/J strain and the two similar substrains A/HeJ and A/WySnJ, which have significantly milder and later-onset hearing impairment (Fig. 3). The only difference in sequence was the lack of the adenine insertion in the two substrains (nine adenines). Third, when sequencing mt-Tr in the diabetic NOD/LtJ progenitor to the diabetes-resistant NOD.NON-H2nb1 congenic strain used in our experiments, NOD/LtJ had heteroplasmy for the 9-bp and 10-bp adenine repeat (10 being preponderant). Diabetes in humans is associated with several mtDNA mutations, and heteroplasmy is generally associated with pathogenic mutations (http://www.gen.emory.edu/mito-map.html). Fourth, in similarity to these mouse results, all the mtDNA mutations predisposing to hearing deficits in humans involve ribosomal or tRNA genes, require additional environmental or genetic factors for phenotypic expression and have relatively subtle biochemical effects2,4,16,17. Fifth, the fact that this insertion has also been observed in other mouse strains without apparent clinical impact is not surprising, given that the clinical phenotype is subtle, dependent on the genotype of the Ahl locus and limited to an additional hearing loss of less than 20 db. Finally, a single base pair change in the D loop of tRNA-Arg has been shown to be functionally important. A base change from cytosine to adenine at position 20 of the wild-type yeast tRNA-Arg transcript increased sixfold the aminoacylation by Escherichia coli arginyl-tRNA synthetase18.
Fig. 3.
Comparison of age-related hearing loss among A/J, A/HeJ and A/WySnJ inbred strains of mice. Averages and standard errors of ABR threshold measurements (16 kHz stimulus) are shown for each strain and time point. The number of mice tested per strain varied from 4 to 11 at 8 weeks, 8 to 18 at 12 weeks, 3 to 8 at 18 weeks and 8 to 14 at 36 weeks.
The interaction between the mitochondrial and nuclear genome described here mimicks in many ways the maternally inherited hearing deficits in humans2,5,16,17. It is, however, necessary to verify the role of the mitochondrial genome by generating and characterizing additional crosses of inbred mouse strains. Once the interaction has been verified, several experimental approaches will provide opportunities to study the pathophysiological basis of the hearing deficit and the role of the mtDNA mutation in it. The identification of Ahl and of other nuclear modifier genes, as well as the biochemical analysis of the effects of the mtDNA change in different mouse tissues, may be an example of such approaches. In the interim, it is possible to speculate about the possible functional interactions, based on the known role of Ahl and its chromosomal location.
First, it has been shown that Ahl is the major gene causing non-syndromic AHL in at least 10 inbred mice strains19. Independently, it has also been shown that, compared with controls, at least a proportion of people with hearing loss associated with aging, that is, presbycusis, have a significant load of acquired mitochondrial DNA mutations in their auditory tissue20,21. Our data may link these two findings, allowing speculation that a common pathway of age-related hearing loss is oxidative phosphorylation dysfunction.
Second, the chromosomal location of Ahl is very close to the autosomal recessively transmitted waltzer mutation, which causes circling behavior and deafness22,23. This has suggested possible allelism, especially because the cochlear pathology of hair cell loss and spiral ganglion cell degeneration observed in old mice from the inbred strains with AHL is similar but less severe than that seen in mice homozygous for waltzer22,24–26. It should also be noted that the modifier of the deaf waddler gene in mice maps to the same region, and the recessive non-syndromic deafness gene DFNB12 and the Usher syndrome type ID gene map to the syntenic region on human chromosome 10q21–q22 (refs. 27–29). This suggests that Ahl may be involved in several forms of deafness and that mitochondrial mutations may be modifiers of phenotypic expression in several forms of hearing loss. It is hoped that additional mouse models for mtDNA disorders can be identified or experimentally induced, and that this will provide a basis for understanding and eventually treating these diseases.
Methods
Mice
All the mice used in these studies were reared and tested under modified barrier conditions in the Mouse Mutant Resource at the Jackson Laboratory. To alleviate husbandry difficulties encountered with diabetic mice, we used the resistant NOD.NON-H2nb1 congenic strain rather than its diabetic NOD/LtJ progenitor, but for brevity we have designated these mice as NOD/LtJ. The care and use of the animals reported on in this study were approved by the Animal Care and Use Committee of The Jackson Laboratory. The Jackson Laboratory is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Assessment of hearing in mice
Hearing in mice was assessed by ABR thresholds with equipment from Intelligent Hearing Systems using described methods and equipment13. The ABR thresholds of all the mice were measured for broadband clicks and pure-tone frequencies of 8 kHz, 16 kHz and 32 kHz. Evoked responses to the 16 kHz stimulus were used to assess hearing because the mouse ear is most sensitive to sounds at that frequency and because these responses showed the greatest elevation above normal values.
Genotyping the nuclear Ahl gene
DNA was extracted from spleen tissue of backcross mice. The closely linked microsatellite marker D10Mit138 was used to deduce genotypes at the Ahl locus on chromosome 10. All backcross mice were typed for D10Mit138 by standard PCR methods19. PCR studies were carried out for 30 cycles, the products being separated on 3% agarose gels (Metaphor; FMC BioProducts) and visualized by ethidium bromide staining. Primer pairs for D10Mit138 were purchased from Research Genetics.
Statistical analyses
Probability values were calculated by non-parametric methods using the Mann–Whitney test for unpaired means, performed with the StatView computer program. Non-parametric statistics were used because of the non-normal distribution of ABR thresholds in these mice.
mtDNA sequencing
Synthetic oligonucleotides were designed using Oligo 6.3 program (Molecular Biology Insights) to amplify the whole mtDNA in 21 overlapping PCR products with an average size of 914 bp. PCR reactions contained 400 ng DNA, 50 pmol of each primer, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs and 1 U Taq DNA polymerase (PE Applied Biosystems) in a total volume of 50 μl. The DNA was initially denatured at 94 °C for 5 min, followed by 35-step cycles of denaturing at 94 °C for 30 s, annealing at 49–53 °C (according to the conditions for each primer pair recommended by the Oligo program) for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 10 min in the GeneAmp PCR System 9700 (PE Applied Biosystems).
PCR products were extracted from 1% agarose gel using Concert Rapid Gel Extraction System (Life Technologies) or directly purified using Concert Rapid PCR Purification System (Life Technologies).
The complete sequence was obtained by sequencing each of the PCR products in one direction with the primers previously used for the PCR amplifications and specifically designed internal sequencing primers. The dideoxynucleotide chain termination method was employed by using the dsDNA Cycle Sequencing System (Life Technologies) with γ-[33P]-dATP (NEN). The mtDNA region around mt-Tr was sequenced in both orientations in all the subjects.
Acknowledgments
This work was supported by grants and a contract from the National Institutes of Health/National Institute of Deafness and Other Communication Disorders. Jackson Laboratory institutional shared services are supported by a National Cancer Institute support grant.
Footnotes
References
- 1.Prezant TR, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nature Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 2.Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mutat. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Jaber L, et al. Sensorineural deafness inherited as a tissue specific mitochondrial disorder. J Med Genet. 1992;29:86–90. doi: 10.1136/jmg.29.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan M, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–972. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 5.Bykhovskaya Y, et al. Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am J Hum Genet. 2000;66:1905–1910. doi: 10.1086/302914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham BH, et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nature Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 7.Melov S, et al. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nature Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 8.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 10.Levy SE, Waymire KG, Kim YL, MacGregor GR, Wallace DC. Transfer of chloramphenicol-resistant mitochondrial DNA into the chimeric mouse. Transgenic Res. 1999;8:137–145. doi: 10.1023/a:1008967412955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sligh JE, et al. Mouse models of mitochondrial disease. Am J Hum Genet. 2000;67 (suppl 2):A53. [Google Scholar]
- 12.Inoue K, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nature Genet. 2000;26:176–181. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- 13.Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hearing Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris SD, Sage RD, Wilson AC. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature. 1982;295:163–165. doi: 10.1038/295163a0. [DOI] [PubMed] [Google Scholar]
- 15.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 16.Fischel-Ghodsian N. Mitochondrial RNA processing and translation—link between mitochondrial mutations and hearing loss? Mol Genet Metab. 1998;65:97–104. doi: 10.1006/mgme.1998.2751. [DOI] [PubMed] [Google Scholar]
- 17.Guan M, et al. Deafness-associated mtDNA 7445 mutation has pleiotropic effects, affecting tRNASer(UCN) precursor processing and expression of NADH dehydrogenase ND6 subunit gene. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, et al. A single base substitution in the variable pocket of yeast tRNAArg eliminates species-specific aminoacylation. Biochim Biophys Acta. 1999;1473:356–362. doi: 10.1016/s0304-4165(99)00143-9. [DOI] [PubMed] [Google Scholar]
- 19.Johnson KR, Zheng QY, Erway LC. A major gene on chromosome 10 affecting age-related hearing loss is common to at least ten inbred strains of mice. Genomics. 2000;70:171–180. doi: 10.1006/geno.2000.6377. [DOI] [PubMed] [Google Scholar]
- 20.Fischel-Ghodsian N, et al. Temporal bone analysis of patients with presbycusis reveals high frequency of mitochondrial mutations. Hearing Res. 1997;110:147–154. doi: 10.1016/s0378-5955(97)00077-4. [DOI] [PubMed] [Google Scholar]
- 21.Bai U, Seidman MD, Hinojosa R, Quirk WS. Mitochondrial DNA deletions associated with aging and possibly presbycusis: a human archival temporal bone study. Am J Otol. 1997;18:449–453. [PubMed] [Google Scholar]
- 22.Deol MS. The anatomy and development of the mutants pirouette, shaker-1 and waltzer in the mouse. Proc R Soc Lond B Biol Sci. 1956;145:206–213. doi: 10.1098/rspb.1956.0028. [DOI] [PubMed] [Google Scholar]
- 23.Bryda EC, Ling H, Flaherty L. A high-resolution genetic map around waltzer on mouse chromosome 10 and identification of a new allele of waltzer. Mamm Genome. 1997;8:1–4. doi: 10.1007/s003359900336. [DOI] [PubMed] [Google Scholar]
- 24.Li HS, Hultcrantz M. Age-related degeneration of the organ of Corti in two genotypes of mice. ORL J Otorhinolaryngol Relat Spec. 1994;56:61–67. doi: 10.1159/000276611. [DOI] [PubMed] [Google Scholar]
- 25.Spongr VP, Flood DG, Frisina RD, Salvi RJ. Quantitative measures of hair cell loss in CBA and C57BL/6 mice throughout their lifespans. J Acoust Soc Am. 1997;101:3546–3553. doi: 10.1121/1.418315. [DOI] [PubMed] [Google Scholar]
- 26.Willott JF, Erway LC. Genetics of age-related hearing loss in mice. IV. Cochlear pathology and hearing loss in 25 BXD recombinant inbred mouse strains. Hearing Res. 1998;119:27–36. doi: 10.1016/s0378-5955(98)00029-x. [DOI] [PubMed] [Google Scholar]
- 27.Chaib H, et al. Mapping of DFNB12, a gene for non-syndromal autosomal recessive deafness, to chromosome 10q21-22. Hum Mol Genet. 1996;5:1061–1064. doi: 10.1093/hmg/5.7.1061. [DOI] [PubMed] [Google Scholar]
- 28.Noben-Trauth K, Zheng QY, Johnson KR, Nishina PM. mdfw: a deafness susceptibility locus that interacts with deaf waddler (dfw) Genomics. 1997;44:266–272. doi: 10.1006/geno.1997.4869. [DOI] [PubMed] [Google Scholar]
- 29.Wayne S, et al. Localization of the Usher syndrome type ID gene (Ush1D) to chromosome 10. Hum Mol Genet. 1996;5:1689–1692. doi: 10.1093/hmg/5.10.1689. [DOI] [PubMed] [Google Scholar]
- 30.Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen in mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]