Abstract
Systems biology conceptualizes biological systems as dynamic networks of interacting elements, whereby functionally important properties are thought to emerge from the structure of such networks. Due to the ubiquitous role of complexes of interacting proteins in biological systems, their subunit composition and temporal and spatial arrangement within the cell are of particular interest. ‘Visual proteomics’ attempts to localize individual macromolecular complexes inside of intact cells by template matching reference structures into cryo electron tomograms. Here we have combined quantitative mass spectrometry and cryo electron tomography to detect, count and localize specific protein complexes within the cytoplasm of the human pathogen Leptospira interrogans. We describe a novel scoring function for visual proteomics and assess its performance and accuracy under realistic conditions. We discuss current and general limitations of the approach, as well as expected improvements in the future.
The biochemical processes of the living cell are catalyzed in large part by a multitude of functional modules, each of which is characterized by a specific cellular distribution in space and time. Frequently, such modules are complexes of interacting proteins. Quantitative mass spectrometry is commonly used to determine the composition of protein complexes and the proteome in general 1. However, since such mass spectrometric measurements are carried out on the combined lysates of multiple cells, spatial information is lost and properties unique to specific cells are averaged over the population of the lysed cells.
Cryo electron tomography (cryoET) is an imaging technique for the three-dimensional (3D) observation of cells in a close to life state 2. At the currently achievable resolution it should in principle be possible to identify and localize large protein complexes in frozen-hydrated specimens. This is accomplished by template matching 3 where the signals representing a specific protein complex are correlated with the signals acquired on the cell by cryoET. Thereby, an extensive search is performed that scans the entire tomogram for structural templates contained in a database. The combined structural signatures of multiple protein complexes, detected in the cell by cryoET, have the potential to describe the spatial proteome organization of a specific cell, a procedure referred to as ‘visual proteomics’ 4. The feasibility of such an approach has been discussed 3 and experimentally demonstrated for cell free systems 5; its application to intact cells has so far been limited to the unambiguous detection of ribosomes 6. The more general application of the technology has been hampered by the challenge to discriminate true from false positive template matches, a task that is complicated by the relatively low signal to noise ratio (SNR) of cryo electron tomograms. Structural diversity and the fact that different protein complexes may exhibit a similar shape and size but differ in abundance by over 4 orders of magnitude 7 further complicates detection.
To assess and alleviate the current limitations in template matching and to thus turn it into a generic method for suitable classes of templates, we realized, in this study, the following technical advances: i) we implemented in silico optimization of a novel scoring function under realistic conditions, namely with a large dynamic range of protein concentrations and various noise model scenarios, ii) we extracted a relevant number of cross correlation features from the tomograms and built reliable statistical models to distinguish true from false positive matches, and iii) we applied thorough statistical validation of template matching for different protein complexes localized in numerous tomograms of a large number of individual cells. Collectively, these steps allowed us to confidently detect and localize a range of complexes in single cells.
The human pathogen L. interrogans has a strongly elongated and helically coiled cell shape. The diameter of a cross section of a typical cell is no more than 100 to 180 nm while its length ranges from 6 to 20 μm. These properties make L. interrogans an ideal specimen for cryoET, as the cytoplasm of these bacteria can be observed with extra-ordinarily high contrast without sacrificing resolution. The narrow cross section allows excellent electron beam penetration and the elongated shape reduces the effects of molecular crowding 2. We therefore chose L. interrogans as a model system to apply the template matching method to detect, count and localize an array of different protein complexes in electron tomograms of frozen-hydrated, individual cells at different states. The robustness and the accuracy of our visual proteomics approach critically depends on prior knowledge of the absolute quantity of the targeted complexes in the cell, thus requiring the convergence of quantitative mass spectrometry and cryoET.
Results
Workflow and selection of target protein complexes
The general experimental workflow of this study consists of the synergistic use of quantitative mass spectrometry to select and quantify protein complexes suitable for visual proteomics and cryoET to detect and localize them in close-to-life, frozen-hydrated cells (Fig. 1). We used LC-MS/MS to generate an extensive proteome list for L. interrogans containing 2221 proteins, representing 61% of the proteome predicted from the 3658 open reading frames annotated in the L. interrogans genome (Supplementary Fig. 1). The data is available in PeptideAtlas. We performed a Psi-Blast analysis against protein sequences from all species and identified a set of 26 L. interrogans protein complexes that we initially considered suitable for template matching (Supplementary Table 1). The complexes in the set fulfilled the following criteria: i) the primary structures of the complex subunits are well conserved in bacterial species, ii) the 3D structure of bacterial homologues have been solved, and iii) the oligomeric assembly has the minimal mass and/or spatial elongation to make it detectable by cryoET.
Figure 1.
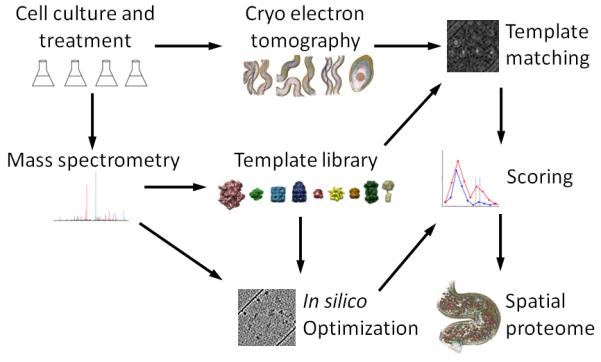
An integrated workflow for visual proteomics. Differently stimulated cells were subjected to shotgun MS and cryoET analysis. A template library was built that included the protein complexes identified in the proteome for which structures of satisfying homology were available. Targeted, quantitative mass spectrometry was employed to determine cellular concentration of the selected targets and to detect inducible changes in their abundance levels in different cellular states. Phantom cells were generated based on the quantitative Leptospira proteome in order to estimate the accuracy of template detection and to train a novel scoring function. The templates were cross correlated with the electron optical density in the tomograms by template matching as described earlier 4 and assigned into the spatial context of the cell using the statistically evaluated, optimized scoring function.
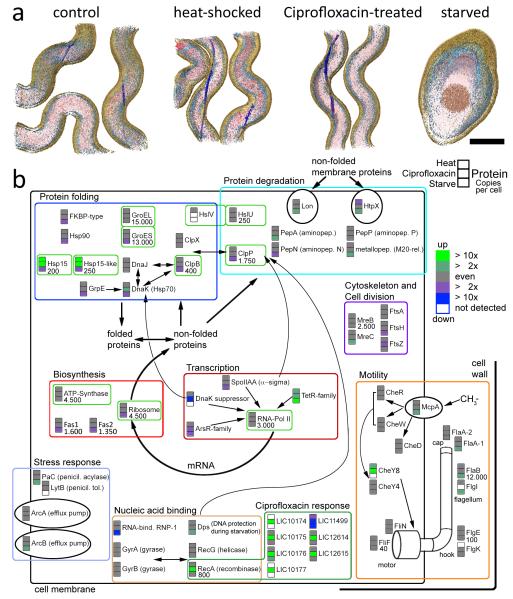
We then used label free quantitative proteomics based on inclusion list guided LC-MS/MS 8 to identify components of the protein complexes on the target list. We analyzed extracts from L. interrogans cells in four states, (i) exponentially growing, unperturbed cells, (ii) cells subjected to heat shock, simulating fever, (iii) cells treated with the antibiotic Ciprofloxacin, and (iv) starved cells. The data indicated the quantitative behavior of the L. interrogans proteome in general, and specifically of proteins associated with the relevant pathways and protein complexes (Fig. 2, Supplementary Results). Apart from the starvation state which was associated with significant morphological changes of L. interrogans cells (Fig. 2a), the antibiotics treatment caused the most significant change in abundance of the protein complexes targeted for template matching. After 24h of antibiotic treatment with 5 μg ml−1 Ciprofloxacin we observed an increase of ATP-Synthase by 35% and ClpB by 50%, whereas the concentration of the ribosome decreased by 25% and Hsp15 proteins by thirty- to forty-fold (Fig. 2b). Therefore we chose to investigate cells primarily in the unperturbed and antibiotics treated condition.
Figure 2.
Stress response of L. interrogans cells in the context of the protein complexes selected as templates for template matching (bright green boxes). Cells of exponentially growing cultures were incubated for 1 h at 42 °C, for 24 h in the presence of 5 μg μl−1 Ciprofloxacin or for 7 d in the absence of nutrients, respectively, and compared against non-treated control cultures. 3D surface rendered volumes are shown in (a) with the cytoplasm colored in red, membranes in bright blue, periplasmic flagella in dark blue and the cell wall in brown (scale bar: 200 nm) (b) Up and down regulation for each protein is shown in green and blue color, respectively, for the heat shock (fever), antibiotics (Ciprofloxacin treatment) and starvation condition versus the control condition. Proteins are grouped based on their function. The relative abundance of many proteins is reduced under starvation and heat shock proteins are strongly up-regulated under both stress conditions (heat shock and antibiotic treatment). While the abundance of the Ciprofloxacin target DNA-gyrase stayed level upon treatment, recombinase A and a cluster of (so far) hypothetical proteins showed a very strong response (dark green box). Copy numbers per cell, as determined by SRM, are given for the control condition (if applicable).
Out of the initial 26 protein complexes, five are abundant in the cytoplasm and occur at more than 1000 copies per cell, another seven are of medium abundance and occur at least at 100 copies per cell, while the remaining complexes are present at less than 100 copies per cell (Supplementary Table 1). We used the quantitative proteomic data to select nine protein complexes from the initial set that cover a matrix of different molecular weights and cellular abundances (Table 1, Supplementary Figs. 1 and 2). Three complexes, ATP-Synthase, RNA-Polymerase II and the ribosome are part of central cellular pathways and their abundance was modulated under different conditions (Supplementary Results). The remaining six selected complexes are involved in protein folding and degradation. Specifically, the GroEL and GroEL-ES complex are chaperones that assist protein folding; ClpB, ClpP and HslU/HslV are unfoldases and proteases; and Hsp15 is a heat shock protein. In contrast to other organisms where Hsp15 is involved in the recycling of 50S ribosomal units, Hsp15 and Hsp15-like in L. interrogans are homologous to Hsp20 9, a family of small stress induced proteins that form large, oligomeric assemblies 10,11.
Table 1. Cellular abundance of template protein complexes.
Cellular abundance of the template protein complexes as determined by selected reaction monitoring (SRM). Copy numbers per cell are given for all relevant conditions, the values in brackets are standard deviations. In contrast to numbers given in Fig. 2, stochiometric relations have been taken into account. For Hsp15 the sum of both closely related gene products is given.
| Template | Non-stimulated | Heat-shocked | Antibiotics-stimulated |
|---|---|---|---|
| Ribosome | 4500 (500) | 3500 (700) | 3400 (300) |
| RNA Polymerase II | 3000 (200) | 3000 (200) | 3000 (400) |
| ATP Synthase | 1500 (500) | 1500 (400) | 2300 (600) |
| GroEL | 1100 (100) | 1300 (150) | 1300 (150) |
| GroES | 1900 (200) | 1900 (300) | 1700 (350) |
| ClpB | 70 (10) | 70 (10) | 100 (40) |
| ClpP | 140 (30) | 110 (60) | 140 (70) |
| Hsp15 | 40 (5) | 310 (60) | 1300 (150) |
| HslUV | 20 (5) | 15 (5) | 15 (10) |
Generation of a scoring function and in silico test data
The accurate detection, quantification and localization of the selected templates in the L. interrogans cells by cryoET depends on a statistically reliable determination of true positive discovery rates. The ensuing problem of correctly discriminating true from false cross correlations is similar to the problem of discriminating true from false assignments of fragment ion spectra to peptide sequences in mass spectrometry based proteomics. We therefore adapted the statistical concepts established in proteomics and implemented in the PeptideProphet 12 software tool to generate a discriminate scoring function for visual proteomics. It consists of a linear combination of the following three knowledge-based, empirical sub-scores: (i) distribution of cross-correlation scores from matching templates to background; (ii) distribution of cross-correlation scores from matching templates to competing templates (local score compared to the score of all other templates in the database); and (iii) distribution of cross-correlation scores from matching templates to three different decoy templates (arbitrarily chosen geometrical shapes: cube, ellipsoid, cylinder). Reliable detection by cryoET is further complicated by the fact that the features to which the templates are assigned are of vastly different abundance. Consequently, false matches of a low abundance complex to a highly abundant competing structure might dramatically inflate the false positive error rate for the low abundance complex. We therefore measured the absolute average cellular concentration of the targeted protein complexes under all the investigated conditions by selected reaction monitoring (SRM)-mass spectrometry 13 in order to generate test data sets containing the target protein complexes at their actual cytoplasmic concentration and to validate the signals derived from real data with an independent method (Supplementary Table 2). The number of copies per cell of the targeted complexes ranged from 4500 copies per cell for the ribosome in non-stimulated cells to 15 copies per cell HslU/V complex in antibiotic treated cells, respectively (Table 1).
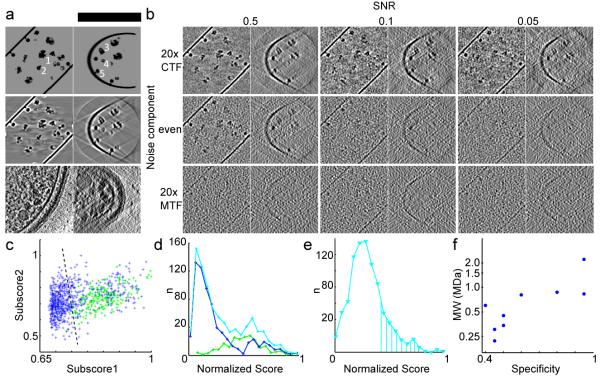
To optimize the relative weight of each subscore in the scoring function and to test whether the selected templates can, in principle, be detected at the electron optical properties of the acquired tomograms, we generated test data sets (phantom cells) in silico that contained two independent noise components 14 at various levels: contrast transfer function (CTF) convoluted (quantum) noise to simulate the interaction of the electron beam with the specimen and modulation transfer function (MTF) convoluted (detector) noise to simulate the imperfection of the charge-coupled device (CCD) camera (Fig. 3, Supplementary Results).
Figure 3.
Generation of in silico test data and development of a scoring function for template matching in cryo electron tomograms. (a) An unprocessed phantom cell (top, scale bar: 150 nm) and tomographic reconstruction without noise (middle) are shown in comparison to a real data set (bottom). The positions of a Ribosome (1), RNA-Polymerase (2), GroEL (3), Hsp (4) and ATP-Synthase are indicated. (b) Reconstructions with an SNR of 0.5, 0.1 and 0.05 are shown with predominant quantum (top), even (middle) and predominant detector noise component (bottom). The noise models in the middle left (conservative) and top right (optimistic) are discussed in text. All panels in (a) and (b) are slices through reconstructions of 5 nm in thickness displayed along the Z-axis (left) and X-axis (right). (c-e) Performance of RNA-Polymerase II detection. (c) Linear discrimination of the subscores 1 and 2 in the in silico test data sets; true and false positives in green and blue, optimal discrimination threshold as dashed line. (d) Score distribution in the in silico test data sets; true and false positives in green and blue, cumulative curve in cyan. (e) Score distribution from real data. The marked area under the curve corresponds to the absolute abundance expectation value for the given cellular volume as determined by SRM. The curve shape is very similar to the theoretical distribution shown in (d). (f) Logarithm of the molecular weight plotted against the specificity achieved in silico at 50% sensitivity (conservative noise model).
We then adjusted the total SNR by comparing the cross-correlation intensity obtained by template matching ribosomes in real tomograms to phantom data sets. To account for the structural noise component during this process 15, we used two different conformations of the ribosomal ratchet-like motion 16 (PDBs 1P85, 1P86, 1P87, 1P6G) for the generation of phantom cells and used a third structure as template (PDBs 2AW7, 2AWB). Two scenarios developed above showed cross-correlation intensities (0.3-0.45 for top ribosome hits) that roughly matched the real data (0.30-0.35): (i) a total SNR of 0.5 with even quantum and detector noise contribution and (ii) a total SNR of 0.05 with a predominant quantum noise contribution. In both cases the phantom data look similar to real tomograms (Fig. 3). We therefore concluded that these two noise scenarios match real data the best and investigated the performance of template matching further in both cases.
Assessment of the performance of visual proteomics in silico
Next, we used the in silico test data sets to optimize the subscores by a linear discriminate analysis and to validate the true- from false positive discrimination (Fig. 3c-e). The two simulation conditions that best matched the real data comprise a conservative and a more optimistic scenario, since the detector noise component substantially limits the attainable resolution by damping higher frequencies. In case of the conservative scenario (even contribution of quantum and detector noise at a total SNR of 0.5), at a sensitivity of 75%, the following specificities were obtained for the targeted complexes: Ribosome >90%, RNAPolymerase II ~50%, GroEL ~60%, GroELS ~70%, undistinguished GroEL or GroELS >90%, HslUV >90% and ATP-Synthase ~50% (Supplementary Fig. 3). The smaller templates ClpB, ClpP and Hsp barley or never reached this sensitivity level, but when the sensitivity was set to 50%, they were discovered with a specificity of: ClpB ~40%, ClpP ~45% and Hsp ~45%. In the case of the optimistic scenario with predominant quantum noise, the specificity for most of the templates was at least equal to the more conservative scenario at the 75% sensitivity but at a ten-fold lower total SNR. Exceptions were RNA-Polymerase II and Hsp which reached ~40% and ~25% at 50% sensitivity, respectively. This is quite remarkable given the lower SNR of 0.05 but can be explained with the low-pass filter applied during reconstruction that can reduce quantum noise more effectively than detector noise. We also found that the performance of visual proteomics, particularly in case of the smaller templates is strongly dependent on template abundance. For example, when phantom cells were investigated that mimic the untreated cell state in which Hsp is present at very low abundance, the specificity dropped to less than 10%. Generally, we can conclude from the analysis of the in silico test data that template matching can theoretically detect protein complexes of distinct shape and sufficient size (Fig. 3f) over a dynamic range of maximal two orders of magnitude in cellular abundance.
Assessment of the performance of visual proteomics in cells
The quantitative mass spectrometry data reflect the protein abundance as an average over a large number of cells. To detect cell to cell variations and variations in the local concentration of the targeted complexes, we acquired six tomograms using an identical experimental setup for each, the non-stimulated, heat-shocked and antibiotics-treated condition that collectively contained subvolumes of 37 individual L. interrogans cells, each covering ~10% of the average cell volume. To detect, localize and quantify the targeted protein complexes (Table 1) we applied the optimized template matching method. In order to compensate for potential variations in the local protein concentration, we selected tomograms with similar SNR from the larger data pool and applied the optimized scoring function to the selected volumes of each cellular state as a whole. We set the anticipated discovery rate to 80% of the cytoplasmic concentration of the template protein complexes. When the conservative noise model was used as a base for estimating the confidence for visual proteomics in real data sets, the following specificities are achieved in average: Ribosome >90%, RNA-Polymerase II ~40%, GroEL ~80%, GroELS ~70%, undistinguished GroEL or GroELS >90%, ATP-Synthase ~50% (including angular bias correction, see Supplementary Results) and Hsp 45% (in the Ciprofloxacin treated state). The detection of the low abundant target protein complexes turned out to be very challenging in real data sets: When the number of single particles that can be expected per tomogram ranged from 0 to 5, the resulting statistical models were noisy and not straight-forward to interpret. We therefore omitted HslU/V, clpB and clpP from our analysis.
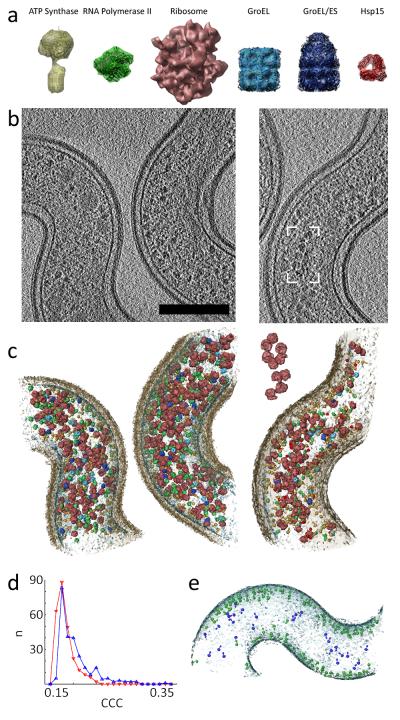
The resulting score distributions of several real datasets were in good agreement with the abundance expectation value for the given template in the particular fraction of the cytoplasmic volume (as determined by SRM), demonstrating the power of SRM to function as an independent method for validating template matching (Fig. 3d-e, Supplementary Fig. 4). In addition, some templates provided auxiliary information for the orthogonal validation of the data: (i) Ribosome handedness: when ribosomes with inverted handedness were used as decoy templates, they could be clearly discriminated from their native counterparts (Fig. 4d). (ii) Ribosome spatial arrangement: A group of ribosomes that resembles the pseudo-planar relative orientation of poly-ribosomes reported recently for bacterial lysates 17 could be found (Fig. 4 b-c). (iii) ATP-Synthase cellular localization: ATP-Synthase is membrane-embedded and points inwards from the cytoplasmic membrane. We therefore asked what fraction of the high quality template matches conformed to these highly restrictive features. The ATPSynthase complexes matched in a tomogram of a L. interrogans cell using the optimized scoring function are shown in Fig. 4e. Plausible positioning and orientation was discriminated manually from non-plausible matches. These data indicate a false positive discovery rate in real tomograms (~10%) that is more optimistic than in test data sets (50%), demonstrating that topological accuracy is not necessarily in agreement with positional correctness (Supplementary Results).
Figure 4.
Template matching in subvolumes of L. interrogans cells. (a) Template library of protein complexes which are shown scaled to each other and superimposed with amino acid chain traces (in black, if applicable). The surface rendering has been done at the relevant resolution of the references applied to template matching. (b-c) The localization of the targeted protein complexes by template matching is shown for representative subvolume of the non-stimulated (left) and antibiotics-treated condition (right). Scale bar: 200 nm. (b) Slices through the reconstructed volumes of 7 nm in thickness without post-processing, (c) surface rendered model of the assigned templates in context with the cell wall (transparent brown) and membrane (transparent blue). The Box and inset show a group of ribosomes resembling the pseudo-planar relative orientation of poly-ribosomes reported recently for bacterial lysates 17. (d) Distribution of the top 250 cross correlation coefficients (CCCs) extracted a tomograms with the ribosome (blue) as template and mirrored ribosome as decoy template (red). The cross-correlation intensity is lower in case of the decoy template and the curve shape changes. (e) Matches of ATP Synthase in a L. interrogans cell in context with the cell membrane. Singles particles with a plausible positioning and orientation (membrane embedded and pointing into the cytoplasm) are colored in green, non-plausible false positives in blue.
The combined quantitative proteomics-cryoET method described here allowed us to link abundance measurements obtained from the combined cell lysate of many cells with the sub-cellular distribution of selected protein complexes in single cells and to detect variations from cell to cell and even sub-cellular volumes. Cells in the non-stimulated state displayed an average ribosome concentration of ~20 μM (~40 mg/ml) in the cytoplasm, but the local concentration ranged from 5-30 μM (~10-65 mg/ml) independent of the different conditions investigated. The local fluctuations in case of total GroEL together with GroEL-ES were larger and ranged from ~8-100 μM (~0.5-6.5 mg/ml). In some cases these local fluctuations can be explained by the following phenomena: Some regions of L. interrogans cells are occupied by large spherical structures (~50-100 nm) of relatively homogenous electron optical density (Supplementary Fig. 5). The nature of these structures is unknown, but if present, they cause a local decrease of protein complexes by displacement. The most obvious response to stress in the proteome was the up-regulation of Hsp: the cytoplasmic concentration of Hsp increased from 0.06 μM in non-stimulated cells to 30 μM (~45 μg/ml) during stress. However, even slight variations in the total SNR between different tomograms as well as in the local SNR can severely hamper the template detection, particularly in case of the ‘small’ protein complexes.
Discussion
The visual proteomics concept holds great promise for the description of the spatial proteome and the observation of proteins at work under close to live conditions. In particular, because labeling strategies for electron microscopy inside intact cells are not applicable in a straightforward manner, the interpretation of cryo electron tomograms at the single molecule level may ultimately lead to pseudo-atomic maps of cells 2. The synergistic method described here tests the accuracy of the visual proteomics concept and allows us to discuss its potential, as well as current and general limitations.
We showed that the noise generated by the detectors widely-used for cryoET (CCD cameras), but not by the interaction of the electron beam with the specimen itself, is currently a critical technical limitation. It reduces the attainable resolution to a level at which only ‘very large’ protein complexes such as the ribosome or GroEL can be discovered with high confidence. The development of alternative detection concepts is an active field of research and might enable a superior image digitization for cryoET in the near future, thereby pushing the size threshold of detectable complexes towards smaller molecular weights (Supplementary Table 1, Fig. 3f).
Our study shows that the very large molecular machines are, in most cases, of low abundance in the cytoplasm and therefore will be difficult to detect in general. Notable exceptions are protein complexes involved in transcription, translation and protein folding that comprise the most abundant cytoplasmic proteins 7. This problem will be even more severe for higher organisms, which have a larger dynamic range of protein concentrations than bacteria 7,18. Much higher throughput in cryoET data acquisition, but also in the computational postprocessing, will be essential to detect low abundant protein complexes with higher confidence.
We demonstrate that molecular crowding can hamper the detection of protein complexes by template matching (Supplementary Fig. 2). This problem might be addressed by more sophisticated image classification techniques in the future. In addition, further development of the preparation techniques used for cryoET, particularly specimen thinning, might enable the investigation of less crowded biological systems.
Another important limitation is structural diversity and protein complex oligomeric states: Most templates used in this study form very stable assemblies. For example, in the case of the ribosome it is reasonable to assume that the predominant species in exponentially growing L. interrogans cells is engaged in translation and therefore fully assembled, as has been shown in vivo for yeast 19. The expansion of template libraries in order to cover as many structural species as possible is, however, an important focus for further development.
The biological system investigated here is certainly exceptional in terms of specimen thickness as well as the targeted protein complexes, and therefore represents the currently feasible state of our visual proteomics approach that combines quantitative proteomics and cryoET. Nevertheless, a quantitative assessment of different structural species of short-lived protein complexes could enable the integration of other targets showing dynamic structural changes, while further technical improvements increasing the resolution and signal to noise of cryoET might enable the application of the technology to a wide range of biological systems and facilitate structure based modeling in systems biology. The method described here can be applied generally to estimate confidence values of cross-correlation based feature extraction in cryoET and therefore will also be useful for structure determination by subtomogram averaging 20. In silico optimization of the data acquisition parameters up-front can be envisioned as well as in silico testing of novel classifiers.
Online Methods
Cell culture and treatment
The Leptospira interrogans serovar Copenhageni of the strain Fiocruz L1-130 were obtained from the American Type Culture Collection (ATCC No. BAA-1198) und cultivated at room temperature in EMJH medium. Cultures of 30 ml were grown at 30 °C to a density of 2*10^7 ml−1 and then stimulated (or left untreated as a control). The antibiotics treatment was done for 24 h with 5 μg ml−1 Ciprofloxacin and the heat shock treatment for 1 h at 42 °C. For starving, cells were pelleted by centrifugation at 3000xg, resuspended in phosphate buffered saline (PBS) and cultivated for further 7 d. Afterwards, the cells were harvested by centrifugation at 3000xg, washed once in PBS, counted, pelleted again, resuspended in 2 ml denaturation buffer (100 mM HEPES pH 7.6, 6 M urea), sonicated for 5 min and stored at −80 °C.
Shotgun mass spectrometry
Proteins obtained from cultures of the non-stimulated, heat-shocked, antibiotics-treated and starved condition were methanol/chloroform precipitated. The samples were resolubilized in 6 M urea, 100 mM HEPES at pH 8.5. The proteins were reduced with 5 mM DTT for 45 min at 37°C and alkylated with 25 mM iodoacetamide for 45 min in the dark before diluting the sample with 100 mM HEPES at pH 8.5 to a final urea concentration below 1.5 M urea. Proteins were digested by incubation with trypsin (1/100, w/w) for at least 6 hours at 37 °C. The peptides were cleaned up by C18 reversed-phase spin columns according to the manufacturer’s instructions (Harvard Apparatus). The dried down peptides were resolubilized to a final concentration of 1 mg ml−1 in off-gel electrophoresis buffer containing 6.25% glycerol and 1.25% IPG buffer (GE Healthcare). The peptides were separated on both pH 3-10 IPG strips and pH 3-7 IPG strips (GE Healthcare) with a 3100 OFFGEL fractionator (Agilent) using a protocol of 1 hour rehydration at maximum 500 V, 50 μA and 200 mW followed by the separation at maximum 8000V, 100 μA and 300 mW until 50 kVh were reached. After iso-electric focusing the fractions were concentrated and cleaned up by C18 reversed-phase spin columns according to the manufacture’s instructions (Harvard Apparatus).
ESI-based LC-MS/MS (LTQ Thermo Finnigan) analyses were carried out using an Agilent 1100 series (Agilent Technologies) on a 75 μM x 10.5 cm fused silica microcapillary reversed phase column. After sample loading, the sample were separated by a 65 minute linear gradient of 5 to 35 % acetonitrile in water, containing 0.1% formic acid, with a flow rate of 200 nl/min. Peptides eluting from the capillary column were selected for CID by the mass spectrometer using a protocol that alternated between one MS scan and three MS/MS scans. The specific m/z value of the peptide fragmented by CID was excluded from reanalysis for 2 min using the dynamic exclusion option.
For quantitative MS analysis a hybrid LTQ-FT-ICR mass spectrometer was interfaced to a nanoelectrospray ion source (both Thermo Fisher Sientific). Chromatographic separation of peptides was achieved on an Agilent Series 1100 LC system (Agilent Technologies) equipped with a 11 cm fused silica emitter, 100 μm inner diameter (BGB Analytik), packed in-house with a Magic C18 AQ 5 μm resin (Michrom BioResources). Peptides were separated by a 120 minute linear gradient of 5 to 40 % acetonitrile in water, containing 0.1 % formic acid, with a flow rate of 0.95 μl/min. Three MS/MS spectra were acquired in the linear ion trap per each FT-MS scan which was acquired at 100,000 FWHM nominal resolution settings with an overall cycle time of approximately 1 second. Charge state screening was employed to select for ions with at least two charges and rejecting ions with undetermined charge state.
Data processing and compilation of PeptideAtlas
MS/MS spectra were searched using the SEQUEST search tool against the predicted proteome from Leptospira interrogans serovar Copenhageni str, complete genome NCBI genome number NC_005823 and NC_005824 (http://www.ncbi.nlm.nih.gov/entrez), consisting of 3658 proteins as well as known contaminants such as porcine trypsin and human keratins (Non-Redundant Protein Database, National Cancer Institute Advanced Biomedical Computing Center, 2004, ftp://ftp.ncifcrf.gov/pub/nonredundant). The search was performed with semi-tryptic cleavage specificity, mass tolerance of 3 Da, methionine oxidation as variable modification and cysteine carbamidomethylation as fixed modification. The database search results were further processed using the PeptideProphet program modified to include the pI information.
Directed mass spectrometry
For relative quantification based on MS1 intensities, samples were prepared and analyzed as described above with a few modifications: Tryptic digests of cell extracts from the respective cultures were analyzed by LCMS/MS, whereby the mass spectrometer was instructed to select for collision activated dissociation (CAD) precursors ions present on a rolling inclusion mass list. This mass list consisted of precursor ion masses of proteotypic peptides (PTPs) selected from ~500 proteins linked to stress response. The PTPs and their respective m/z, RT and fragment ion coordinates were extracted from the L. interrogans instance of PeptideAtlas. For quantification the SuperHirn peak extraction and alignment algorithm was used and peptide quantities were determined from the ion current of the specific signal. The post processing, particularly ratio calculation and statistical analysis was done in Matlab: All ratios were at first calculated on peptide level. After integration onto protein level outliers were removed that deviated more than 2 standard deviations from the mean value (Supplementary Table 3). The described label-free relative quantification has been verified using isobaric tagged reagents (ITRAQ) for ~400 high abundant proteins on an Applied Bio Systems 4800 MALDI TOFTOF mass spectrometer (not shown) and the resulting data were integrated into the L. interrogans instance of PeptideAtlas.
Targeted mass spectrometry
For 20 proteins covering the entire abundance range, absolute quantification was done using selected reaction monitoring (SRM), based on heavy labeled reference peptides that served as an internal standard and carried out for 5 biological replicates (Supplementary Table 2). The A hybrid quadrupole-linear ion trap mass spectrometer (4000 QTRAP) was operated with a beta release of Analyst 1.4.2 supporting scheduled experiments (Applied Biosystems/MDS Sciex). The instrument was coupled to a Tempo nano-LC system (Applied Biosystems/MDS Sciex) for peptide separation using a 30-min gradient from 5 to 30% acetonitrile (0.1% formic acid) at 300 nl/min flow rate. A fused silica emitter of 75-μm inner diameter was packed in house with 13 cm of Reprosil-Pur 120 ODS-3.3 μm (Dr. Maisch GmbH).
Based on the PeptideAtlas data we generated MRM transitions specific for the proteotypic peptides using TIQAM. We restricted the peptides to those in the mass range of 800–2400 Da and not containing methionine. For each precursor we calculated the transitions with precursor charges 2+ and 3+ and the four smallest y-ions with m/z > precursor m/z + 30. Collision energies (CE) were calculated according to the following formulas: CE = 0.044 × m/z + 5.5 (2+) and CE = 0.051 × m/z + 0.5 (3+). Then MRM-triggered MS/MS experiments were performed with 100 transitions per run (dwell time: 20 ms/transition). Results were imported into TIQAM, and the three transitions with the best signal-to-noise ratio were selected for quantitative analysis if the corresponding MS/MS spectrum was in accordance with the targeted peptide. To calculate the copies per cell of the targeted protein complexes, the cell number in each sample and the protein complex stochiometry was taken into account.
Cryo electron tomography and image processing
Cells from stimulated or untreated cultures, respectively, were pipetted onto R2/1, 200 mesh copper grids (Quantifoil), rapidly plunge frozen into liquid Ethan and then introduced into a Technai F20 cryo electron microscope (FEI) equipped with image filter (Gatan). Tomograms were acquired with an underfocus of 6.5μm, typically covering a tilt range from −63° to 63° with tilt increments of 1.5° for the high tilts and 2° between −40° and 40° at nominal magnification was 34.000x, corresponding to an object pixel size of 0.63 nm at the specimen level. Six Tomograms of at least 10 cells were acquired for each of the conditions: the untreated, heat shock and antibiotics treatment; Four Tomograms of cells originating from the starvation condition that have not been used for template matching (see below) were acquired at a nominal magnification of 27.500x corresponding to an object pixel size of 0.75 nm. All image processing operations were performed with either the EM-package or TOM-package for Matlab (The MathWorks). The 3D surface rendered representations were calculated with either Chimera (UCSF) or Amira (TGS). Tomograms were initially reconstructed with a binning factor of 2 by weighted back projection.
Template matching and scoring
For template matching, manually pre-selected subvolumes were reconstructed with a binning factor of 1 (corresponding to an object pixel size of 1.26 nm). In order to determine cellular protein concentrations, the cell volume must be determined: To calculate the average cell volume, the subvolume of a cell covered in a tomogram was extrapolated to the entire cell length (Supplementary Fig. 6). The structural templates were downloaded from the RCSB and EMD structural databases (see Supplementary Table 1 for accession numbers), sampled to the relevant voxel size and convoluted with the given contrast transfer function using the TOM package for Matlab (Supplementary Fig. 2a). In case of ATP-Synthase a disc shaped density (5 nm radius) was added around the transmembrane domain. Three different geometrical shapes (flat cylinder, cube and ellipsoid) served as decoy templates and were convoluted with the same envelope functions. Template matching was carried out by parallel computing, as described in earlier by calculating the local cross correlation function as implemented in Molmatch. Three Phantom cells (in silico test data sets, Fig. 3a) for both, the untreated and ciprofloxacin-treated condition, were created by pasting each template at a number corresponding to the cytoplasmic concentration into a volume of 148×148×148 voxels containing an artificial membrane environment. Each cell was rotated by 45° and 90° around the Z-axis to account for three different missing wedge conditions. To test the influence of molecular crowing spherical densities of random radius were pasted to the template periphery simulating 2 conditions: (i) a moderate crowding and (ii) intensive crowding with partially higher intensity than the templates (Supplementary Fig. 2b), making the total number of test volumes 54. The test data sets were generated using the following sequential steps: (i) The template structures were sampled to the relevant pixel size and pasted into a phantom cell at the cytoplasmic concentration determined in the control or antibiotics-treated state, respectively (Fig. 3a). ATP-Synthase complexes were added to the artificial membrane environment taking into account the correct topology. (ii) The thus generated phantom cells were rotated around the z-axis to simulate different missing wedge directions (not shown) and afterwards projected due to the angular increment scheme of real data sets. (iii) Two independent noise components were sequentially added to the resulting two-dimensional projection images: CTF-convoluted (quantum) noise to simulate the interaction of the electron beam with the specimen and MTF-convoluted (detector) noise to simulate the imperfection of the CCD camera. Thereby, the relative contribution of both noise components was varied accounting for three different scenarios (Fig. 3b): a predominant particle (20x excess of CTF-convoluted noise), a predominant detector (20x excess of MTF-convoluted noise) and an even noise contribution of both components. (vi) In the final step projections were low-pass filtered to 4 nm resolution (1st zero of CTF) and 3D reconstructions were calculated by weighted back-projection. The thus generated test data sets were subjected to template matching. From the resulting cross correlation volumes (in silico test data sets and measured data), the top CCCs were extracted in four-fold excess (based on the absolute abundance measurements by SRM) in order to build appropriate statistical models (Supplementary Fig. 7). Cell areas been masked before peak extraction by means of semi-automatic segmentation as implemented in Amira to remove background. The CCCs extracted from all tomograms arising of a certain stimulation condition (either untreated, heat shocked or antibiotics treated) were subjected to the scoring function (1) as a whole, that consist of a linear combination of three empirical, knowledge-based subscores:
| (1) |
whereby CCCPar is the local cross correlation coefficient at a given orientation; CCCTopComp and CCCTopDecoy are the highest cross correlation coefficients of any of the other template or decoy templates within the very same position, respectively. These three values have been normalized by dividing through the standard deviation in order to rely on curve shapes rather than absolute values. A, B, C are weighting factors for the subscores. The optimal weighting of the subscores was determined for each template separately by linear discrimination analysis of the in silico test data sets through maximization of the following function (2)
| (2) |
whereby Disc is the discrimination; nTrueIdentified the number of identified true positives; nTruePositive the theoretically achievable maximum number of true positive identifications and nidentified the number of identifications. The relative weights of subscores were very similar for the different noise scenarios.
This evaluation yielded an average double hit rate of up to 10% in the in silico test data sets and real data sets (positions in the tomograms that have been assigned to more than 1 template). Double hits were reassigned to the most likely template due to their relative score within the total score distribution of all templates. This procedure resulted in a false positive discovery rate of maximal 5% in the in silico test data sets.
EdSumm AOP
Protein complexes can be detected, counted and localized within the bacterium Leptospira interrogans by combining quantitative mass spectrometry-based proteomics analysis with cryo electron tomography, with the aid of an improved template matching method.
EdSumm issue
Protein complexes can be detected, counted and localized within the bacterium Leptospira interrogans by combining quantitative mass spectrometry-based proteomics analysis with cryo electron tomography, with the aid of an improved template matching method.
Supplementary Material
Acknowledgements
This project has been funded in part by ETH Zurich, the Swiss National Science Foundation (grant 31000-10767), Federal funds from the National Heart, Lung, and Blood Institute, the National Institutes of Health (contract no. N01-HV-28179), by SystemsX.ch the Swiss initiative for systems biology, in part by the PROSPECTS (proteomics in time and space) European network of excellence, and with funds from the ERC project ‘Proteomics V3.0’. M.B. was supported by a long-term fellowship of the European Molecular Biology Organization and a Marie Curie fellowship of the European Commission, J.A.M. was supported by a fellowship from the Swedish society for medical research (SSMF), A.S. and V.L. were supported by the Competence Center for Systems Physiology and Metabolic Diseases. We thank O. Medalia, W. Baumeister, the electron microscopy facility of ETH Zurich (EMEZ) for continued support and F. Förster for critical reading of the manuscript.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annual review of biochemistry. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 3.Best C, Nickell S, Baumeister W. Localization of protein complexes by pattern recognition. Methods in cell biology. 2007;79:615–638. doi: 10.1016/S0091-679X(06)79025-2. [DOI] [PubMed] [Google Scholar]
- 4.Nickell S, Kofler C, Leis AP, Baumeister W. A visual approach to proteomics. Nature Reviews Molecular Cell Biology. 2006;7:225–230. doi: 10.1038/nrm1861. [DOI] [PubMed] [Google Scholar]
- 5.Frangakis AS, et al. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc. Natl. Acad. Sci.USA. 2002;99:14153–14158. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortiz JO, Forster F, Kurner J, Linaroudis AA, Baumeister W. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol. 2006;156:334–341. doi: 10.1016/j.jsb.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Malmstrom J, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt A, et al. An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics. 2008;7:2138–2150. doi: 10.1074/mcp.M700498-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nally JE, Artiushin S, Timoney JF. Molecular characterization of thermoinduced immunogenic proteins Q1p42 and Hsp15 of Leptospira interrogans. Infect Immun. 2001;69:7616–7624. doi: 10.1128/IAI.69.12.7616-7624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennaway CK, et al. Dodecameric structure of the small heat shock protein Acr1 from Mycobacterium tuberculosis. J. Biol. Chem. 2005;280:33419–33425. doi: 10.1074/jbc.M504263200. [DOI] [PubMed] [Google Scholar]
- 11.Kim R, Kim KK, Yokota H, Kim SH. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc. Natl. Acad. Sci. USA. 1998;95:9129–9133. doi: 10.1073/pnas.95.16.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 13.Stahl-Zeng J, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Forster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol. 2008;161:276–286. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Baxter WT, Grassucci RA, Gao H, Frank J. Determination of signal-to-noise ratios and spectral SNRs in cryo-EM low-dose imaging of molecules. J Struct Biol. 2009;166:126–132. doi: 10.1016/j.jsb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H, et al. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 17.Brandt F, et al. The Native 3D Organization of Bacterial Polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 19.Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- 20.Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.