Summary
Host- and pathogen-associated cytoplasmic double-stranded DNA triggers the activation of a NALP3 (also known as cryopyrin and NLRP3)-independent inflammasome 1, which activates caspase-1 leading to maturation of pro-interleukin-1β and inflammation. The nature of the cytoplasmic-DNA-sensing inflammasome is currently unknown. Here we show that AIM2 (absent in melanoma 2), an interferon-inducible HIN-200 family member that contains an amino-terminal pyrin domain and a carboxy-terminal oligonucleotide/oligosaccharide-binding domain 2, 3, senses cytoplasmic DNA by means of its oligonucleotide/oligosaccharide-binding domain and interacts with ASC (apoptosis-associated speck-like protein containing a CARD) through its pyrin domain to activate caspase-1. The interaction of AIM2 with ASC also leads to the formation of the ASC pyroptosome 4, which induces pyroptotic cell death in cells containing caspase-1. Knockdown of AIM2 by short interfering RNA reduced inflammasome/pyroptosome activation by cytoplasmic DNA in human and mouse macrophages, whereas stable expression of AIM2 in the non-responsive human embryonic kidney 293T cell line conferred responsiveness to cytoplasmic DNA. Our results show that cytoplasmic DNA triggers formation of the AIM2 inflammasome by inducing AIM2 oligomerization. This study identifies AIM2 as an important inflammasome component that senses potentially dangerous cytoplasmic DNA, leading to activation of the ASC pyroptosome and caspase-1.
A key innate immune response against infection with microbial or viral pathogens and tissue damage is the rapid activation of multiprotein complexes called inflammasomes 5. The inflammasomes activate caspase-1, a cysteine protease that processes the inactive pro-interleukin-1β (pro-IL1β) and pro-IL18 to their active pro-inflammatory cytokines IL1β and IL18, respectively. The most studied among these is the NALP3 inflammasome 5, which is activated by diverse stimuli perhaps by a lysosomal destablization mechanism 6, 7.
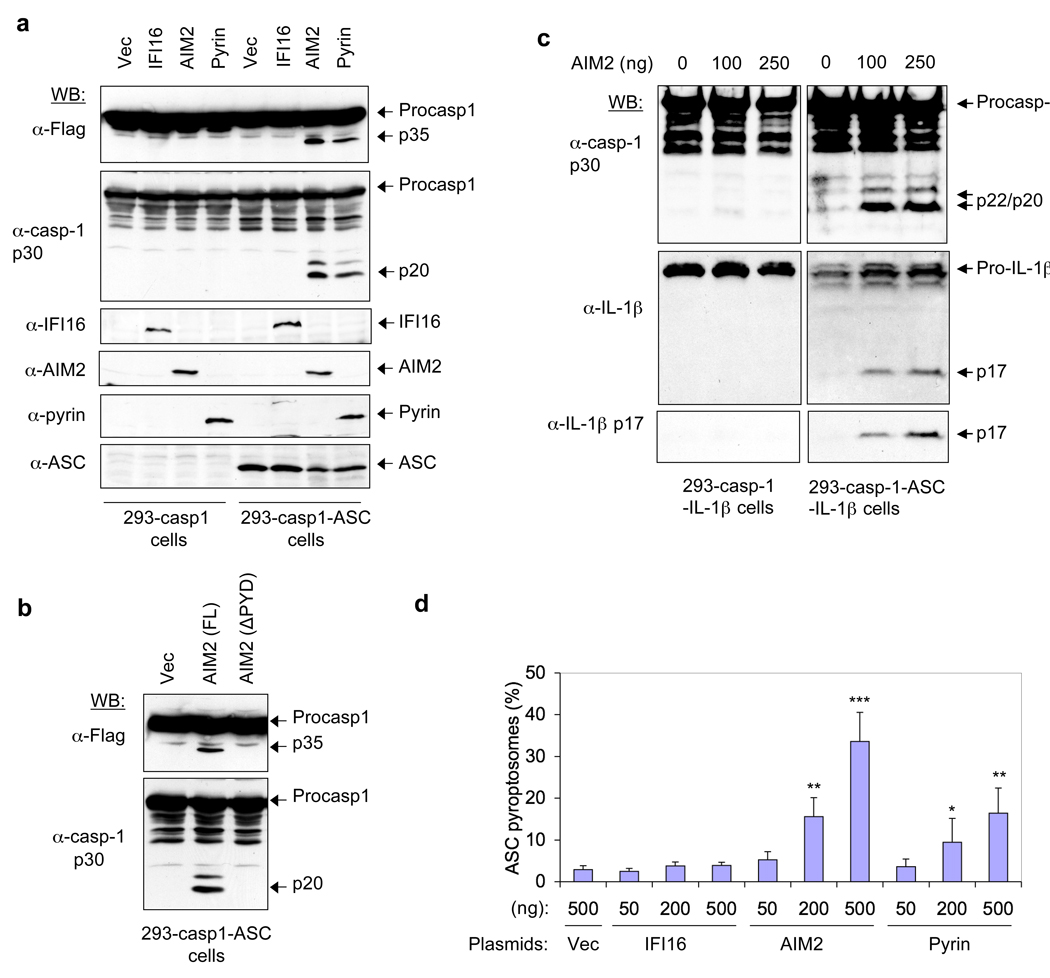
A recent study showed that DNA from different sources activates an ASC-dependent, but a NALP3-independent, inflammasome 1. To identify the DNA-sensing inflammasome, we searched the NCBI database for proteins with pyrin and oligonucleotide-binding domains. We identified four human proteins (IFI16, AIM2, IFIX and MNDA), which belong to the interferon-inducible HIN-200 family 2, 3, that meet these two criteria. Investigation of the ability of these proteins to activate caspase-1 when ectopically expressed in the stable 293T-caspase-1-ASC cell line8 (293T cell line containing caspase-1 and ASC) showed that AIM2 is the only member of the HIN-200 family capable of activating caspase-1 (Fig 1a and Supplementary Fig. 1). The activation of caspase-1 by AIM2 was dependent on an intact pyrin domain (PYD) because deletion of the PYD of AIM2 completely abrogated caspase-1 activation (Fig. 1b). This was also dependent on ASC, because no AIM2-induced caspase-1 activation was observed in the 293T-caspase-1 cells8, which lack ASC (Fig. 1a and Supplementary Fig. 1a). Additionally, expression of AIM2 induced secretion of activated caspase-1 and IL1β from stable 293T-caspase-1-ASC-pro-IL1β cells, which express ASC, but not from 293T-caspase-1-pro-IL1β cells, which lack ASC (Fig. 1c). Together, these results indicate that AIM2 can activate caspase-1 to produce the active IL1β cytokine in an ASC-dependent manner, perhaps by engaging ASC and inducing its oligomerization. Indeed, expression of AIM2 in 293T-ASC–EGFP-N1 cells 8 (293T cell line containing enhanced green fluorescent protein (EGFP)-tagged ASC) resulted in the formation of the oligomeric ASC pyroptosome that we showed recently to participate in caspase-1 activation during pyroptosis 4 (Fig. 1d and Supplementary Fig. 2).
Figure 1. Activation of caspase-1 by AIM2.
a, b, The indicated cells in (1 × 106 cells/35 mm well) were transfected with 0.5 µg of an empty vector (vec) or constructs encoding human AIM2, IFI16 or pyrin (a), or vec, AIM2 or AIM2-ΔPYD plasmids (b) as indicated for 24 h. Lysates were analyzed by western blotting with anti-Flag antibody (1st panel from top), or anti-caspase-1 p30 polyclonal antibody (2nd panel from top). The blot in (a) was re-probed with IFI16, AIM2, pyrin or ASC antibodies (3rd-6th panel from top, respectively). c, Immunobloting for caspase-1 (upper panel), IL-1β (middle panel) and IL-1β p17 (lower panel) in culture supernatants of the indicated cell lines transfected with the indicated amounts of AIM2 expression plasmid. d, Percentages of ASC pyroptosomes in 293-ASC-EGFP-N1 cells following transfection with the indicated plasmids. Values represent mean ± S.D. (n = 4); *, P<0.05; **, P<0.01;***, P<0.005. casp1 or casp-1 refer to caspase-1; Procasp1 refers to Pro-caspase-1; 293 refers to 293T; α- refers to Anti-
To gain some insight into how AIM2 induces ASC-dependent caspase-1 activation, we tested whether AIM2 interacts directly with ASC to activate caspase-1. In vitro pull down experiments revealed that full-length AIM2, but not a truncated AIM2 lacking the PYD (AIM2-ΔPYD) can interact with ASC and its isolated PYD (Fig. 2a, b). Furthermore, immunoprecipitation of endogenous ASC from interferon γ-induced human THP-1 cell lysates with an ASC-specific antibody resulted in precipitation of endogenous AIM2 (Fig. 2c). No detectable interaction between ASC and endogenous IFI16, MNDA or IFIX was observed (Fig. 2c 4th to 6th panels from top), further supporting our earlier conclusion that only AIM2, among members of the HIN-200 family, can specifically interact with ASC to activate caspase-1. Together, these results indicate that AIM2 associates directly with ASC via PYD-PYD interactions. This interaction is reminiscent of the interaction of NALP3 or pyrin with the PYD of ASC, which induces ASC oligomerization leading to activation of caspase-1 in response to various stimuli 8, 9, 10.
Figure 2. AIM2 interacts with ASC to activate caspase-1.
a, b, Immunoblots showing in vitro association of ASC-GST and ASC-PYD-GST with full-length AIM2 (upper panel), but not with truncated AIM2-ΔPYD (middle panel) (a), and in vitro association of AIM2-GST with full-length ASC (b). The lower panels show Coomassie stained gel of the GST-tagged proteins used in the pull-down assay (see Supplementary Fig. 3). GST–ASC-CARD refers to the isolated CARD of ASC fused to GST. c, Association of endogenous ASC with endogenous AIM2 in THP-1 cell lysate detected by immunoprecipitation (IP, 1st lanes, left panels) with an ASC-specific monoclonal antibody as described in the “Methods”. IP with an Omi-specific monoclonal antibody was used as a negative control (2nd lanes, left panels). d, e, Immunoblots of lysates and culture supernatants of THP-1 cells (d) or mouse WT and NALP3−/− bone marrow macrophages (e) transfected with AIM2-targeting siRNA (siAIM2) or control non-specific siRNA (siCon) followed by transfection with poly (dA:dT) or treatment with MSU as indicated. The release of the active p20 subunit and some procaspase-1 in the culture supernatants is a result of cell death and lysis (see Fig. 6, and Supplementary movie 1).
Structural analysis of the C-terminal HIN-200 domain of AIM2 revealed that this domain contains two adjacent oligonucleotide/oligosaccharide-binding folds 11 that could potentially interact with DNA. Electrophoretic mobility shift assays with purified HIN-200 domain of AIM2 (AIM2-ΔPYD; Supplementary Fig. 4a, left panel) and the synthetic DNA poly(dA-dT)poly(dA-dT) (referred to throughout as poly(dA:dT)) showed formation of increasing amounts of slow-migrating AIM2–DNA complexes with increasing amounts of AIM2-ΔPYD (Supplementary Fig. 4a, right panel), indicating that this domain can indeed bind to DNA. As expected, a purified full-length AIM2, but not the isolated PYD, was also capable of binding to DNA and inducing a gel-shift of poly(dA:dT) (Supplementary Fig. 4b).
Given that AIM2 can interact with ASC and DNA through its PYD and HIN-200 domains, respectively, we next used AIM2-targeting short interfering RNA (siRNA) to demonstrate that AIM2 has an important role in caspase-1 activation by cytoplasmic DNA. Knocking down human AIM2 in THP-1 cells clearly reduced poly(dA:dT)-induced caspase-1 activation and IL1β processing (Fig. 2d, 4th lane, and Supplementary Fig. 5a, b). Knocking down AIM2 had little effect on caspase-1 and IL1β activation by monosodium urate (MSU), which activates caspase-1 through the NALP3 inflammasome (Fig. 2d, 5th and 6th lanes). Consistent with these results, knocking down mouse AIM2 in immortalized wild-type and NALP3−/− mouse bone marrow macrophages also significantly reduced poly(dA:dT)-induced caspase-1 activation (Fig. 2e and Supplementary Fig. 6a–c). The Aim2-targeting siRNAs had little effect on caspase-1 activation by lipopolysaccharide (LPS) plus ATP in mouse wild-type macrophages (Supplementary Fig. 6d and Supplementary Note 1). Together, these results indicate that AIM2 in human and mouse macrophages senses cytoplasmic DNA through its HIN-200 domain and is critical for caspase-1 activation by cytoplasmic DNA.
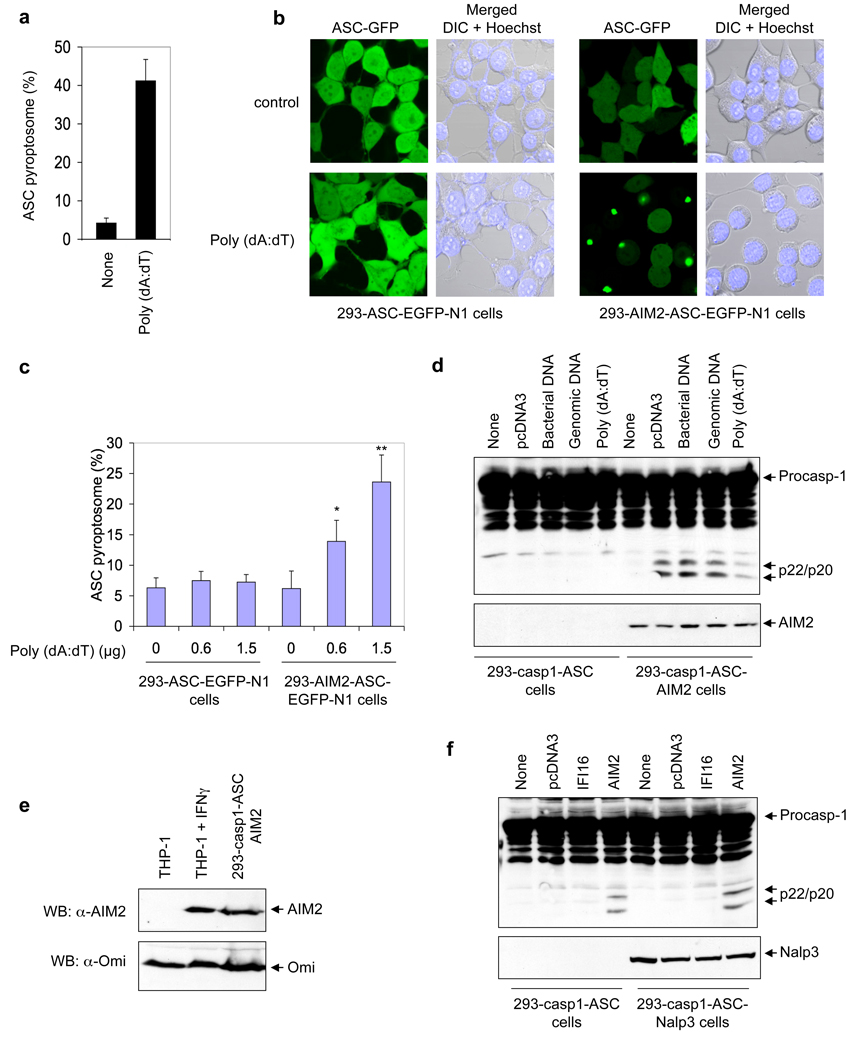
A pyroptosome formation assay in THP-1-ASC–EGFP-N1 cells 4 revealed that cytoplasmic DNA induces robust ASC oligomerization (Fig. 3a). To provide direct evidence that AIM2 is required for DNA-induced ASC oligomerization, we used 293T cells, which do not express endogenous AIM2 (Supplementary Fig. 7a), to generate a stable 293T cell line that co-expresses AIM2 and ASC–EGFP proteins (designated 293T-AIM2-ASC–EGFP-N1). Transfection with poly(dA:dT) induced ASC pyroptosome formation in 293T-AIM2-ASC–EGFP-N1 cells within 2 h after transfection (Fig. 3b, lower right panel), but not in control 293T-ASC–EGFP-N1 cells, which do not express AIM2 (Fig. 3b, lower left panel). Quantification of the ASC pyroptosomes revealed a dose-dependent effect of poly(dA:dT) in these cells (Fig. 3c). Consistent with these results, DNA from different sources including poly(dA:dT) (Supplementary Note 2) was able to induce caspase-1 activation in a stable 293T-caspase-1-ASC-AIM2 cell line that co-expresses pro-caspase-1, ASC and AIM2, but not in the parental 293T-caspase-1-ASC cell line, which lacks AIM2 (Fig. 3d). The expression level of AIM2 in the 293T-caspase-1-ASC-AIM2 cells is comparable to the expression of endogenous AIM2 in interferon-γ-induced THP-1 cells (Fig. 3e). Transfection of plasmid DNA into a stable 293T-caspase-1-ASC-NALP3 cell line that co-expresses pro-caspase-1, ASC and NALP3 did not induce caspase-1 activation, indicating that, unlike AIM2, NALP3 does not participate directly in sensing cytoplasmic DNA (Fig. 3f). These results underscore the essential role of AIM2 in cytoplasmic DNA sensing and DNA-induced ASC oligomerization and caspase-1 activation, and indicate that binding of cytoplasmic DNA to AIM2 generates the molecular signal necessary for this process to occur.
Figure 3. Cell-based reconstitution of the AIM2 inflammasome.
a, Percentages of ASC pyroptosomes in untransfected (none) or poly (dA:dT)-transfected THP-1 cells. Values represent mean and s.d. (n = 3). b, Fluorescence confocal images showing formation of the speck-like ASC pyroptosomes only in 293-AIM2-ASC-EGFP-N1 cells (right panels), but not in 293-ASC-EGFP-N1 cells (left panels), 2 h after transfection with vehicle (control, upper panels) or poly (dA:dT) (lower panels). DIC, Differential Interference Contrast. c, Percentages of ASC pyroptosomes in the indicated cell lines following transfection with poly (dA:dT) for 2h. Values represent mean ± S.D. (n = 7); *, P<0.005; **, P<0.001. d, Immunoblot for caspase-1 in lysates from 293-caspase-1-ASC (1st to 5th lanes) or 293-caspase-1-ASC-AIM2 (6th to 10th lanes) cells following transfection with the indicated types of DNA (0.5 µg/1 × 106 cells) for 24 h. e, Immunoblot for AIM2 (upper panel) or Omi (lower panel) in lysates from uninduced (1st lane) or interferon γ-induced (2nd lane) THP-1 cells, or 293-caspase-1-ASC-AIM2 cells (3rd lane). f, Immunoblot for caspase-1 in lysates from 293-caspase-1-ASC (1st to 4th lanes) or 293-caspase-1-ASC-NALP3 (5th to 8th lanes) cells following transfection with the indicated plasmid DNA (0.5 µg /1 × 106 cells) for 24 h.
To understand how cytoplasmic DNA induces AIM2 activation, we tested the ability of purified recombinant human AIM2 to induce activation of a chimaeric PYD–caspase-1 protein, which contains an N-terminal PYD derived from ASC instead of the CARD, in vitro (see Supplementary Note 3). Incubation of purified AIM2 or ASC pyroptosome with PYD-caspase-1 resulted in its activation (Fig. 4a, 2nd and 4th lanes, respectively). No activation of PYD-caspase-1 was seen when it was incubated with AIM2-ΔPYD which lacks the PYD (3rd lane), indicating that the interactions of the PYD of AIM2 with the PYD of PYD-caspase-1 is required for activation. AIM2 was also able to activate a chimeric WT PYD-caspase-9, but not an active site mutant PYD-caspase-9 C287A (Fig. 4b). Since initiator procaspases are activated by dimerization, these results indicate that AIM2 activates the procaspase 1 and 9 chimeras by inducing the dimerization of their inactive monomeric forms. Unexpectedly, incubation of AIM2 with poly (dA:dT) did not increase the ability of AIM2 to activate PYD-caspase-9 or PYD-caspase-1 (Fig. 4c, and data not shown). This suggests that the purified AIM2 might have already been activated by bacterial DNA during expression in bacteria, or self-oligomerized during purification. Indeed, chemical cross-linking experiments in the absence of DNA showed that the purified AIM2 preparation contains dimeric and oligomeric forms of AIM2 (Fig. 4d, second lane). Interestingly, cross-linking in the presence of DNA increased the oligomeric forms of AIM2 (Fig. 4d, 3rd and 4th lanes), in a dose dependent manner, as evidenced by the formation of very large AIM2 oligomers that failed to enter the gel. This result indicates that binding of dsDNA to AIM2 induces its oligomerization, which explains the ability of cytoplasmic DNA to activate the AIM2 inflammasome in macrophages and in AIM2-reconstituted 293T cells.
Figure 4. In vitro reconstitution of the AIM2 inflammasome.
a, Immunoblot for caspase-1 (upper panel) or IL-1β (lower panel) following in vitro incubation of PYD-caspase-1 and pro-IL-1β with purified GST-AIM2 (2nd lane), GST-AIM2-ΔPYD (3rd lane) or ASC pyroptosome (4th lane). b, c, Autoradiograms of 35S-labeled WT or C287A chimeric PYD-caspase-9 following in vitro incubation with purified GST-AIM2 (2nd and 6th lanes), GST-AIM2-ΔPYD (3rd and 7th lanes) or ASC pyroptosome (4th and 8th lanes) (b) or with the indicated amounts of purified GST-AIM2 in the presence or absence of the indicated amounts of poly (dA:dT) (c). d, Immunoblot for AIM2 after cross-linking of purified AIM2 (2 µg) with glutaraldehyde in the presence of the indicated amounts of 64-mer dsDNA. A longer exposure of this gel is shown in Supplementary Fig. 8.
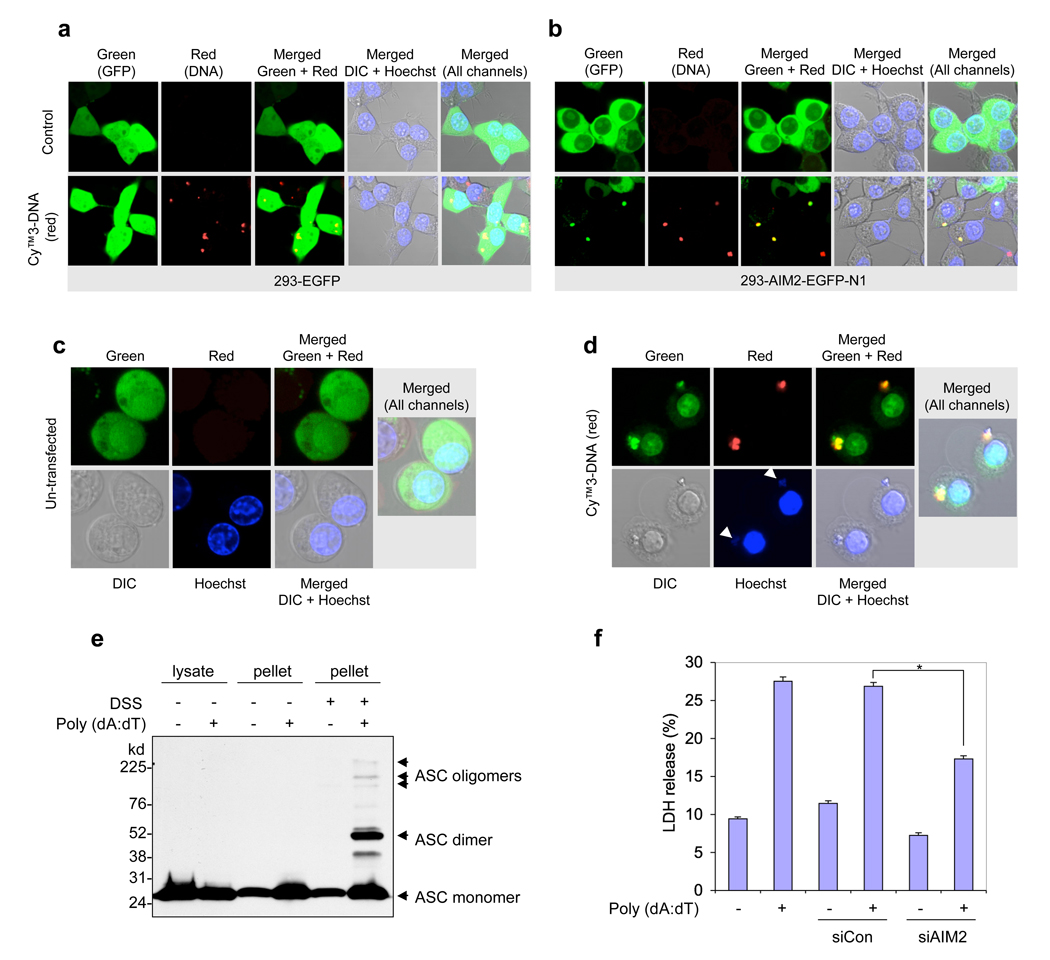
To provide direct evidence that cytoplasmic DNA can indeed induce AIM2 oligomerization, we transfected 293T-AIM2–EGFP-N1 cells or NALP3−/−-AIM2–EGFP-N1 mouse macrophages cells that stably express human or mouse EGFP-tagged AIM2, respectively, with a Cy3-labelled plasmid DNA. Confocal live cell imaging revealed that DNA induced notable clustering of the cytoplasmic AIM2–EGFP around the transfected DNA in both 293T cells and NALP3−/− macrophages (Fig. 5b, d and Supplementary Fig. 9). DNA did not induce clustering of free EGFP in the control 293T-EGFP cells (Fig. 5a), indicating that AIM2 is responsible for the clustering of AIM2–EGFP in the 293T and NALP3−/− macrophages. Interestingly, DNA-induced clustering of AIM2–EGFP caused pyroptotic cell death with characteristic plasma membrane swelling in the NALP3−/−-AIM2–EGFP-N1 cells (Fig. 5d, Supplementary Fig. 10 and Supplementary Movie 1), but not in the 293T-AIM2–EGFP-N1 cells (Fig. 5b), because these cells lack ASC and caspase-1. Pre-treatment of the NALP3−/−-AIM2–EGFP-N1 cells with the caspase inhibitor zVAD-FMK (Z-Val-Ala-Asp-Fluoromethyl ketone) did not affect DNA-induced clustering of AIM2–EGFP in these cells, but prevented cell death (Supplementary Fig. 9b). Taken together, these results provide direct evidence that cytoplasmic DNA binds to AIM2 and induces its oligomerization in live cells. This is the first demonstration of an inflammasome bound to its ligand in live cells.
Figure 5. Cytoplasmic DNA-induced AIM2 oligomerization and pyroptosis.
a, b, Confocal live cell images of 293-EGFP cells (a) or 293-AIM2-EGFP-N1 (b) following transfection with vehicle (control, upper panels) or Cy™3-labeled DNA (red, lower panels). c, d, Confocal live cell images of NALP3−/− -AIM2-EGFP-N1 bone marrow macrophages following transfection with vehicle (c) or Cy™3-labeled DNA (d). Notice the oligomerization of AIM2-GFP by the red DNA in the 293-AIM2-EGFP-N1 cells (b, bottom panels) and NALP3−/− -AIM2-EGFP-N1 macrophages (d). The green (AIM2-EGFP), red (Cy™3-labeled DNA), gray (DIC), and blue (Hoechst stain, nucleus) channels, and the merged channels are indicated. The two white arrows (lower-second panel from left in (d)) indicate staining of the cytoplasmic DNA with the blue Hoechst stain, which specifically stains DNA. Notice the pyroptotic cell death features induced by the Cy™3-labeled DNA in NALP3−/− -AIM2-EGFP-N1 macrophages (d), but not in the 293-AIM2-EGFP-N1 (b). e, Immunoblot for ASC showing the presence of the oligomeric ASC pyroptosomes only in the pellets of poly (dA:dT)-transfected (6th lane), but not in the pellets of untransfected (5th lane), NALP3−/− macrophages. A shorter exposure and a more detailed legend of this gel is shown in Supplementary Fig. 12. f, Percentages of LDH release into the culture medium of NALP3−/− macrophages transfected with vehicle (1st and 2nd columns), non-specific control siRNA (siCon, 3rd and 4th columns) or mouse AIM2-specific siRNA (siAIM2, 5th and 6th columns) followed by transfection with or without poly (dA:dT) (1 µg/1 × 106 cells) as indicated. Values represent mean ± S.D. (n = 3); *, P < 0.001.
The putative tumour-suppressive activity of AIM2 (ref. 12) might be attributed to its ability to induce pyroptosis in cells that co-express ASC and pro-caspase-1. Indeed, transient expression of AIM2 in 293T-ASC-caspase-1 cells resulted in pyroptotic cell death as measured by the release of lactate dehydrogenase (LDH) in the culture medium (Supplementary Fig. 11a). No AIM2-induced cell death was observed in cells that lack caspase-1 or ASC (such as 293T-caspase-1, 293T-ASC–EGFP-N1 or 293T-AIM2–EGFP-N1 cells; Fig. 5b, Supplementary Fig. 11a and data not shown). Consistent with this, transfection of poly(dA:dT) or plasmid DNA into the NALP3−/− bone marrow macrophages induced ASC pyroptosome formation (Fig. 5e), cell death with characteristic features of pyroptosis (Supplementary Fig. 11b, lower panels, and Supplementary Movie 1) and release of LDH into the culture medium (Fig. 5f). Knocking down AIM2 by Aim2-specific siRNA significantly reduced cell death and LDH release in these cells (Fig. 5f). These data indicate that AIM2 is a bona fide activator of inflammation and caspase-1-dependent cell death (pyroptosis) in cells that express both ASC and caspase-1.
Our work clearly identifies AIM2 as a major activator of caspase-1 in response to cytoplasmic DNA. As demonstrated in vitro and in live cells, DNA binds directly to AIM2 and induces its oligomerization. We suggest that the oligomeric AIM2–DNA complex serves as a molecular platform, much like the oligomeric NALP3 platform, to recruit ASC and facilitate the self-association of its PYD to form ASC dimers, which subsequently oligomerize with other ASC dimers to form a large structure we called recently the “ASC pyroptosome” 4. This is supported by our observation that engagement of AIM2 by cytoplasmic DNA leads to the formation of ASC pyroptosomes in THP-1 and NALP3−/− macrophages and in 293T-AIM2-ASC–EGFP-N1 cells (Fig. 3 and Fig 5). Cytoplasmic DNA, whether derived from infection with viral or microbial pathogens, tissue damage or delivered by cationic liposomes, is a strong activator of type I interferon response. AIM2 is an interferon-inducible protein and we have observed upregulation of AIM2 in THP-1 macrophages after treatment with interferon-, LPS or transfection with the synthetic DNA poly(dA:dT). Our data indicate that the induction of AIM2 serves as a backup innate immune response to the NALP3 inflammasome, capable of sensing cytoplasmic DNA and of triggering a strong pro-inflammatory response. The perceived redundancy in the response to DNA by the NALP3 and AIM2 inflammasomes could be explained by differences in the mechanisms by which these two inflammasomes are activated. Recent studies indicate that the NALP3 inflammasome is activated by many stimuli that cause phagosomal/lysosomal destabilization 6, 7, 13, 14, 15. When DNA viruses (for example, adenovirus) infect macrophages, the DNA-containing viral particles accumulate in the phagosomes 16, which could lead to destablization of the phagosome thereby resulting in activation of NALP3 as has recently been shown 1. In contrast, when DNA escapes the phagosomes it could be sensed directly by AIM2, which is induced by the initial type I interferon response to viral entry, by means of direct interaction with its oligonucleotide/oligosaccharide-binding-fold-containing domain, thereby leading to AIM2 oligomerization and activation. It is therefore probable that the NALP3 and AIM2 inflammasomes represent two important lines of defence to ensure a full pro-inflammatory response to potentially dangerous cytoplasmic DNA. Additional discussion is provided in the Supplementary Discussion.
METHODS SUMMARY
Assay of caspase-1 activation
All cells were seeded in 35 mm 6-well plates. Stimulation with DNA [(poly (dA:dT), plasmid, genomic or bacterial] were performed in serum-free (SF) OPTI-MEM® I by cationic liposome transfection using Lipofectamine 2000 (3.5 µl.ml−1), as per the manufacturer's protocol (Invitrogen). After stimulation, cell culture supernatants and cell pellets were analyzed by immunobloting with anti-caspase-1 and anti-IL-1β specific antibodies.
siRNA knockdown
THP-1, or immortalized WT or NALP3−/− macrophages were attached to 35 mm 6-well plates. Cells were then transfected with siRNA oligonucleotides (50 nM) for 24 h using Lipofectamine 2000 in 2 ml of SF OPTI-MEM® I. The following day, cells were washed with SF OPTI-MEM® I and then transfected with poly (dA:dT) (1 µg/ml) for 6 hours with Lipofectamine 2000 in 2 ml of SF OPTI-MEM® I. The culture supernatants and cells were separated and then processed for immunoblot analysis with anti-caspase-1 and anti-IL-1β specific antibodies.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Cell culture
Human 293T and THP1 cell lines were purchased from ATCC. 293T cells were maintained in DMEM/F12 (GIBCO) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. THP1 cells were grown in RPMI 1640 (GIBCO) supplemented with 10% FBS, 2 mM L-glutamine, 10 mM Hepes, 1 mM Sodium pyruvate, 1.5 gm/L Sodium bicarbonate, 0.05 mM 2-Mercaptoehanol and 100 U/ml penicillin and streptomycin. The v-myc and v-raf immortalized mouse WT, ASC−/− and NALP3−/− macrophages (kind gift from Dr. Eicke Latz, University of Massachusetts Medical School) were grown in DMEM (GIBCO) supplemented with 10% FBS, 100 U/ml penicillin and streptomycin. All cells were grown at 37° C with 5% CO2. Before stimulation, THP-1 cells were differentiated for 3 h with 0.5–1 µM PMA, washed and allowed to attach to culture dishes for 24 h. All experiments for immunoblot analysis of cell culture supernatants used SF OPTI-MEM® I medium (GIBCO). Transfection experiments with the synthetic poly (dA:dT) DNA or other types of DNA (plasmid, genomic or bacterial) were performed using Lipofectamine 2000 (7 µl/ml−1) as per the manufacturer's protocol (Invitrogen).
Antibodies and reagents
Mouse polyclonal anti-human AIM2 antibody was purchased from Abnova. This antibody can detect overexpressed AIM2 but not endogenous AIM2. The mouse monoclonal anti-human AIM2 antibody (3B10) used to detect endogenous AIM2 in THP-1 cell lysates was obtained from Dr. Ricky Johnstone (The Peter MacCallum Cancer Institute, Victoria, Australia). The anti-human IFI16 monoclonal (sc-8023) and anti-human MNDA polyclonal (sc-6051) antibodies were obtained from Santa Cruz Biotechnology. The anti-human IFIX polyclonal antibody was obtained from Dr. Mien-Chie Hung (The University of Texas M. D. Anderson Cancer Center). The anti-IL-1β monoclonal antibody (32D) was obtained from the NCI preclinical repository, Biological resource branch. Rabbit polyclonal anti-human mature (17 kDa) IL-1β (Asp116) was obtained from Cell Signaling. Other antibodies used against human ASC (anti-human ASC monoclonal; from Dr. Junji Sagara), human caspase-1 p30 (anti-human caspase-1, residues 100–404; our lab) and mouse caspase-1 p20 (anti-mouse caspase-1 p20; from Dr. Junying Yuan) have been described before 4, 8. Poly (dA:dT) sodium salt was from Sigma-Aldrich. CytoTox96 LDH-release kit was from Promega. The Cy™3-labeled plasmid DNA (MIR 7904) was obtained from Mirus Bio, LLC (Madison, WI).
Full-length cDNAs and expression constructs
The pCMV-sport6 mammalian expression plasmids for human full-length AIM2 (IMAGE:30367168), IFIX (IMAGE:5212268) and MNDA (IMAGE:5223430) were obtained from Open Biosystems and their sequences were verified by automated DNA sequencing. pcDNA3-IFI16 plasmid was a kind gift from Toru Ouchi. pcDNA3-AIM2-T7 and pcDNA3-AIM2-ΔPYD-T7 were generated by cloning PCR-generated full-length AIM2 or AIM2-ΔPYD cDNAs, respectively, in a modified pcDNA3-T7 plasmid in frame with a C-terminal T7 tag. For stable expression of a C-terminal EGFP-tagged human or mouse AIM2 in 293T cells or in mouse NALP3−/− immortalized macrophages, the AIM2 cDNAs were fused in frame with EGFP cDNA in the pEGFP-N1 plasmid. The AIM2-EGFP fusion cDNAs were then excised from the pEGFP-N1 plasmids and subcloned into the pMSCVpuro retroviral transfer vector to generate the pMSCVpuro-hAIM2-EGFP-N1 and pMSCVpuro-mAIM2-EGFP-N1 vectors. For expression of AIM2 or AIM2-ΔPYD with a His6 or GST tag in bacteria, PCR-generated full-length AIM2 or AIM2-ΔPYD cDNAs were inserted into the bacterial expression vector pET-21a (+) in-frame with a C-terminal His6 or GST tag. Bacterial expression constructs for GST-tagged ASC, ASC-PYD or ASC-CARD were described previously 17. The mammalian expression plasmids for full-length human pyrin was described before (pcDNA-pyrin-myc-HisB) 9.
Generation of stable cell lines
293T-caspase-1, 293T-caspase-1-ASC and 293T-ASC–EGFP-N1 cell lines have been described previously 4, 8, 9. Stable 293T-caspase-1-pro-IL1β or 293T-caspase-1-ASC-pro-IL1β were generated by transfecting 293T-caspase-1 or 293T-caspase-1-ASC, respectively, with dual expression plasmid pMSCVgfp-pro-IL1β. Stable cell lines were obtained after multiple cell sorting over a period of one month by flow cytometry as described before 8. Stable 293T-caspase-1-ASC-AIM2 cells were generated by transfecting 293T-caspase-1-ASC with dual expression plasmid pRSC-GFP-AIM2 followed by sorting as above. Stable 293T-AIM2-ASC–EGFP-N1 cell line was generated by co-transfecting 293T cells with pMSCVpuro-ASC–EGFP-N1 8 and pcDNA3-AIM2-T7 plasmids at a 1:4 ratio, followed by cell sorting as above and selection in Geneticin (G418, GIBCO)- and puromycin-containing medium. Stable 293T-AIM2–EGFP-N1 cells, expressing the human AIM2–EGFP fusion protein, were generated by transfecting 293T cells with pMSCVpuro-hAIM2–EGFP-N1 expression plasmid followed by cell sorting as described previously and selection with puromycin-containing medium. Stable NALP3−/−-AIM2–EGFP-N1, which express the mouse AIM2–EGFP fusion protein, were generated by infecting the NALP3−/− immortalized macrophages with pMSCVpuro-mAIM2–EGFP-N1 retroviral vector followed by cell sorting as above and selection with puromycin containing medium.
siRNA knockdown
Human AIM2-targeting siRNA oligonucleotides (AIM2 Stealth™ Select RNAi, Catalog # 1299003) were obtained from Invitrogen. The sequences of each siRNA oligonucleotide in this pool are as follows: hAIM2-HSS114049 , 5 ’-UAUGGUGCUAUGAACUCCAGAUGUC-3 ’ ; hAIM2-HSS114050,5 ’-UUUCAGCUUGACUUAGUGGCUUUGG-3 ’ , hAIM2-HSS114051 , 5 ’-UUCUCUGAUAGAUUCCUGCUGGGCC-3 ’ . Mouse AIM2-targeting siRNA oligonuleotides (ON-TARGETplus SMARTpool) were obtained from Dharmacon. The sequences of each siRNA oligonucleotide in this pool are as follows: mAIM2 J044968-09 , 5 ’-ACAUAGACACUGAGGGUAU-3 ’ ; mAIM2 J044968-1 0 , 5 ’-UGUCUAAGGCUUGGGAUAU-3 ’ ; mAIM2 J044968-1 1 , 5 ’-CUACCUGAGGAUAGCAUUU-3 ’ ; mAIM2 J044968-1 2 , 5 ’-AGUACUAAGAAAUCAGUGA-3 ’ . N on-targeting control siRNA oligonucleotides (AllStars Neg. Control siRNA, catalog # 1027281) were obtained from QIAGEN. For AIM2 knockdown experiments in the human THP-1 cell line, cells were seeded in 35 mm 6-well plates at a density of 2 × 106 cells/well in the presence of 1 µM PMA and allowed to attach to culture dishes for 3 h. Cells were then washed and treated with LPS (2 µg/ml) for 16 h to induce differentiation. The differentiated cells were then transfected with siRNA oligonucleotides (50 nm) for 24 h using Lipofectamine 2000 (3.5 µl/ml) in 2 ml of SF OPTI-MEM® I medium. The following day, cells were washed with SF OPTI-MEM ® I medium and then transfected with poly (dA:dT) for 6 hours with Lipofectamine 2000 in 2 ml of SF OPTI-MEM® I. The culture supernatants and cells were separated and then processed for immunoblot analysis. For AIM2 knockdown experiments in the immortalized mouse macrophages, cells were seeded in 6-well plates at a density of 2 × 106 cells/well in SF OPTI-MEM® I medium and allowed to attach for 2 hours. The cells were then transfected with siRNA oligonucleotides (50 nM) for 24 h using Lipofectamine 2000 in 2 ml of SF OPTI-MEM® I medium. The following day, cells were washed with SF OPTI-MEM® I medium and then transfected with poly (dA:dT) (1 µg/ml) for 6 hours with Lipofectamine 2000 in 2 ml of SF OPTI-MEM® I. The culture supernatants and cells were separated and then processed for immunoblot analysis.
Immunoblot analysis
Cell culture supernatants were precipitated by the addition of an equal volume of methanol and 0.25 volumes of chloroform as described before 7. The supernatant/methanol/chloroform mixtures were vortexed and then centrifuged for 10 min at 20,000g. The upper phase was discarded and 500 µl methanol was added to the interphase. This mixture was centrifuged for 10 min at 20,000g and the protein pellet was dried at 55 °C, resuspended in Laemmli buffer and boiled for 5 min at 99 °C. Samples were separated by 12.5 % SDS-PAGE and were transferred onto nitrocellulose membranes. Blots were probed with rabbit polyclonal antibody to the human caspase-1 p30, rabbit polyclonal antibody to human cleaved IL-1β (Asp116; Cell Signaling) or rat monoclonal-anti-mouse caspase-1 p20 antibody (a kind gift fro Dr. Junying Yuan, Harvard university). Total cell lysates were mixed with SDS sample buffer, fractionated on 12.5 % SDS-PAGE and then immunoblotted as described above.
Immunoprecipitation and In vitro GST pull-down assays
THP1 cells (107 cells) were treated with IFN-γ (75 ng/ml) for 24 h and then lysed in 250 µl buffer A (20 mM Hepes, pH 7.5, 50 mM KCl, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 0.5% NP-40, 1 mM DTT, 0.2 mM PMSF, 2 µg ml−1 aprotinin, 5 mM NaF, 1 mM Na3VO4). The cell lysate was centrifuged at 20,000g to remove cell debris. The supernatant was diluted 2X with a hypotonic lysis buffer (10 mM Hepes, pH 7.8, 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and protease inhibitor) and then immunoprecipitaed with anti-human ASC or anti-human Omi mouse monoclonal antibodies at 4°C. The protein-antibody complexes were then precipitated with protein G-sepharose bead, and the bead-bound complexes and lysates were then fractionated by SDS-PAGE followed by immunobloting with anti-AIM2 (3B10) or anti-IFI16 mouse monoclonal antibodies, or anti-ASC, anti-Omi, anti-MNDA, or anti-IFIX rabbit polyclonal antibodies. In vitro pull down assays were performed with recombinant bacterially expressed GST-tagged full length human ASC, ASC-PYD or ASC-CARD and C-terminal His6-tagged human AIM2 or AIM2-ΔPYD. The GST-tagged proteins were isolated from bacterial lysates by glutathione-affinity purification on glutathione-agarose beads. The bead-bound proteins were then incubated for 2 h at 4°C with bacterially produced His6-tagged AIM2 or AIM2-ΔPYD proteins. After incubation, the complexes were fractionated by SDS-PAGE and detected by immunobloting with anti-His6-HRP antibody. Similarly GST-tagged AIM2 or AIM2-ΔPYD was allowed to interact with bacterially produced ASC and the bound ASC was assayed by immunobloting with anti-ASC monoclonal antibody.
Confocal microscopy
The 293-ASC-EGFP-N1 or 293-AIM2-ASC-EGFP-N1 cells, or NALP3−/− macrophages were seeded on 35 mm cover glass bottom culture dishes and allowed to attach for 24 h. Next day cells were transfected with poly (dA:dT) (1 µg/dish) for 2–3 hours using Lipofectamine 2000 and then stained with Hoechst 33342 for 30 min. Cells were then observed using a Zeiss LSM 510 Meta confocal microscope. In the experiments using the Cy™3-labeled plasmid DNA, 293-hAIM2-EGFP-N1 cells or NALP3−/− -mAIM2-EGFP-N1 macrophages were seeded on 35 mm cover glass bottom culture dishes and allowed to attach for 24h. Next day cells were transfected with Cy™3-labeled plasmid DNA (0.5 µg/dish) for 2–3 hours using Lipofectamine 2000 and then stained with Hoechst 33342 for 30 min. Cells were then observed using a Zeiss LSM 510 Meta confocal microscope. The EGFP (green) was excited with the 488 nm argon laser. The nuclear Hoechst 33342 stain (blue) was excited with the 405 nm diode laser. The Cy™3 (red) was excited with the 543 nm He/Ne Laser.
In vitro caspase-1 activation and IL-1β cleavage Assays
Lysates containing PYD-caspase-1 and pro-IL-1β were prepared from 293-PYD-caspase- 1 or 293-pro-IL-1β stable cell lines in CHAPS buffer (20 mM Hepes-KOH, pH 7.5, 5 mM MgCl2, 0.5 mM EGTA, 0.1 mM PMSF, 0.1 % CHAPS). GST-AIM2 or ASC pyroptosomes were produced in bacteria and purified by standard procedures 18. Purified GST-AIM2 or ASC pyroptosomes were incubated with PYD-caspase-1-containing lysates together with pro-IL-1β in CHAPS buffer for 60 min at 37° C. The reaction mixtures were then fractionated by SDS-PAGE and analyzed by western blotting with anti-caspase-1 p30 and anti-IL-1β antibodies. In some experiments, in vitro translated 35S-labeled and nuclease treated PYD-caspase-9 (WT or C287A) was incubated with purified GST-AIM2 or ASC pyroptosomes in the presence or absence of poly (dA:dT) for 60 min at 37°C and the reaction products then analyzed by SDS-PAGE followed by autoradiography.
Pyroptosome isolation from poly (dA:dT) transfected macrophages
NALP3−/− macrophages in 10 cm culture dishes were transfected with 6 µg of poly (dA:dT) or vehicle (H2O) using Lipofectamine 2000 in SF OPTI-MEM®I for 4 hours. The cells were then pelleted by centrifugation and then lysed in buffer A (20 mM Hepes-KOH, pH7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 320 mM sucrose, 0.1 mM PMSF). ASC pyroptosomes were then isolated from the lysates and cross-linked with DSS as described in detail recently 18.
Purification and cross-linking of AIM2
Full-length AIM2 with a C-terminal His6 tag was expressed in bacteria and then purified on Talon® beads (Clontech) according to standard procedures. Purified AIM2 (2 µg) was incubated with or without a 64-mer synthetic dsDNA (0.5 and 1 µg) for 15 min at room temperature in 50 µl PBS. The reaction mixtures were then cross-linked with 1 µl of 1% glutaraldehyde which covalently cross-links lysine residues in close proximity to each other in oligomeric proteins. Glutaraldehyde does not cross-link proteins to DNA. The reactions were terminated by addition of 2 µl of 2 M Tris-HCl, pH 8.0. Cross-linked AIM2 was solubilized by addition of an equal volume of 2X Laemmli SDS sample buffer and fractionated on a 12.5% SDS-polyacrylamide gel.
Statistics
All values are expressed as the mean s.d. of individual samples. Samples were analysed using the student's t-test.
Supplementary Material
Supplementary Information is linked to the on line version of the paper at www.nature.com/nature
Acknowledgments
This work was supported by NIH grants AG14357 and AR055398 to ESA. We thank M. McCormick and D. Wang for technical assistance, X. Jiao for help with the confocal microscopy, J. Sagara for the anti-human ASC antibody, J. Yuan for the anti-mouse caspase-1 antibody, R. Johnstone for the anti-human AIM2 antibody, M-C Hung for the anti-human IFIX antibody, T. Ouchi for the IFI16 cDNA and E. Latz for the immortalized mouse bone marrow macrophages.
REFERENCES
- 1.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 2.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu JW, et al. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol. Cell. 2007;28:214–227. doi: 10.1016/j.molcel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 10.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 11.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 12.DeYoung KL, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 13.Dostert C, et al. Innate immune activation through NALP3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the NALP3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franchi L, Nunez G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur J Immunol. 2008;38:2085–2089. doi: 10.1002/eji.200838549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier O, Gastaldelli M, Boucke K, Hemmi S, Greber UF. Early steps of clathrin-mediated endocytosis involved in phagosomal escape of Fcgamma receptor-targeted adenovirus. J Virol. 2005;79:2604–2613. doi: 10.1128/JVI.79.4.2604-2613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasula SM, et al. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. Epub 2002 Apr 19. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes-Alnemri T, Alnemri ES. Chapter Thirteen Assembly, Purification, and Assay of the Activity of the ASC Pyroptosome. Methods Enzymol. 2008;442:251–270. doi: 10.1016/S0076-6879(08)01413-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the on line version of the paper at www.nature.com/nature