Abstract
Purpose
In the C57BL/6J mouse retina, hyperoxia-induced degeneration of photoreceptors shows strong regional variation, beginning at a locus ~0.5 mm inferior to the optic disc. To identify gene expression differences that might underlie this variability in vulnerability, we have used microarray techniques to describe regional (superior-inferior) variations in gene expression in the retina.
Methods
Young adult C57BL/6J mice raised in dim cyclic illumination (12 h at 5 lx and 12 h in darkness) were exposed to hyperoxia (75% oxygen for two weeks). Retinas were collected from hyperoxia-exposed and control animals without fixation and divided into superior and inferior halves. RNA was extracted from each sample, purified, and hybridized to Mouse Gene 1.0 ST arrays (Affymetrix). The consistency of the microarray results was assessed using quantitative PCR for selected genes. Expression data were analyzed to identify genes and ncRNAs whose differential expression between the superior and inferior retina could be associated with relative vulnerability to hyperoxia.
Results
In control retinas, only two genes showed a fold difference in expression >2 between the superior and inferior retina; another 25 showed a fold difference of 1.5–2.0. Of these 27, the functions of six genes, including ventral anterior homeobox containing gene 2 (Vax2) and T-box 5 (Tbox5), are related to parameters of anatomic development and the functions of five are related to sensory perception. Among the latter, short-wave-sensitive cone opsin (Opn1sw) was more strongly expressed in the inferior retina and medium-wave-sensitive cone opsin (Opn1mw) in the superior retina. This is consistent with known differences in S- and M-cone distribution, confirming our separation of retinal regions. The highest fold difference was reported for membrane metalloendopeptidase (Mme), a member from the metallothionein group of cytoprotective proteins. To identify genes whose regulation by hyperoxia was significantly different between the inferior and superior retina, we calculated the “fold margin” (FM, the difference between hyperoxia-induced regulation in the inferior and superior retina) for each gene, and identified genes for which abs(FM) > 0.5. Genes thus identified numbered 112, and included many immune-, cell defense-, and inflammation- related genes.
Conclusions
Gene expression analysis revealed relatively subtle differences between inferior and superior regions of control C57BL/6J retinas, with only 27 genes showing an expression difference >1.5 fold. Among these, genes related to cytoprotection and apoptosis were included, along with genes related to central projections and cone-type differences. After hyperoxia-induced photoreceptor degeneration had begun, the number of genes that showed significant expression differences between the inferior and superior retina more than quadrupled, with genes related to immune processes, defense processes, and inflammation being numerically dominant.
Introduction
Tissue oxygen levels play an important role in the development of the retina and in the stability of photoreceptors. During development, the retina is initially avascular and is supplied with oxygen by diffusion from choroidal circulation. As photoreceptors develop and commence functioning in rodents during the second week of postnatal life [1], their consumption of oxygen increases steeply and their retinas become hypoxic. The onset of hypoxia is the stimulus for vasculogenesis [2], mediated by the potent hypoxia-induced angiogenic vascular endothelial growth factor (VEGF) [3]. The formation of retinal vessels relieves the hypoxia induced by photoreceptor activity. During this period, oxygen regulates a naturally occurring process of photoreceptor death in a linear way, hypoxia accelerating and hyperoxia decelerating the rate at which photoreceptors die [4]. As retinas mature, however, hyperoxia becomes toxic; both hypoxia and hyperoxia destabilize photoreceptors [5]. The toxic impact of hyperoxia on photoreceptors was first reported in adult rabbits [6] and subsequently in rodents [7,8]. Hyperoxia also induces the death of endothelial cells and the thinning of retinal vasculature and reduces VEGF expression (thus the opposite of the angiogenic effect of hypoxia) [9].
There is also evidence that hyperoxia can improve the stability of photoreceptors. In experimental retinal detachment, hypoxia caused by the separation of the outer retina from its normal source of nutrients is a factor in inducing the death and deconstruction of photoreceptors, as well as in reducing glial proliferation. Hyperoxia mitigates all these pathologies [10,11]. Hyperbaric oxygen therapy (HBO), which involves very high tissue oxygen levels for short periods, has been reported to have long-lasting benefits for patients with retinitis pigmentosa [12] and several other ophthalmic diseases, including glaucoma, macular detachment, and neovascular disease [13]. In recent work undertaken to resolve this apparent discrepancy [8,14], we have noted that the impact of hyperoxia on retinal gene expression is biphasic, initially regulating protective genes and later regulating genes related to photoreceptor death. This suggests that hyperbaric oxygen protects the retina because it is produced briefly, typically lasting only a few hours a day, and may regulate only the protective cohort of genes.
This study probes the mechanism of oxygen-induced destabilization of photoreceptors by exploiting known regional differences in photoreceptor vulnerability. A superior-inferior gradient of vulnerability is evident for hyperoxia, which in C57BL/6J mice affects photoreceptors in the inferior retina more severely [14,15]. Other works have described a periphero-central gradient in photoreceptor vulnerability to hyperoxia, both hyperbaric [16] and normobaric [7]. It is possible that these researchers noted the central location of vulnerable photoreceptors, without noting whether they were located in the superior or inferior retina.
Several gradients of vulnerability of the retina have been previously reported. For example, light-induced retinal damage can be more severe in the superior retina than the inferior retina of albino rats [17,18]. Degeneration induced by the photoreceptor toxin iodoacetic acid is more severe in the inferior retina and visual streak of rabbit retinas [19] and, in a genetically degenerative mouse model (the rd/rd mouse), significantly more cones survive in the inferior than superior hemisphere in most retinas [20]. Although various factors, such as light history, rhodopsin content, and fatty acid composition have been reported to influence the vulnerability of photoreceptors to damage, the mechanisms responsible for regional variations of photoreceptor vulnerability to hyperoxia remain unknown.
Methods
Animals and hyperoxic damage
C57BL/6J mice, a hyperoxia-vulnerable strain [21], were used for this study. Mice were raised in dim cyclic illumination (12 h at 5 lx and 12 h in dark) and maintained in this level of lighting for the duration of the experiment. At the postnatal age (P) of P83–P90 days, some mice were exposed to constant hyperoxia (75% O2) for 14 days. During exposure, the mice were given free access to food and water and were housed in plexiglass chambers, in which the oxygen level was controlled by a feedback device (OxyCycler; Reming Bioinstruments, Redfield, NY).
RNA isolation and purification
Oxygen-exposed animals were euthanized at the end of the period of hyperoxia; controls were euthanized at the same age. Retinas were removed and divided into superior and inferior halves. Retinal samples from two animals (one male and one female) were pooled for each condition (superior and inferior retina from the controls, superior and inferior retina exposed). RNA extraction was performed using TRIzol Reagent (Invitrogen™ Life Technologies, Carlsbad, CA) and an RNAqueous-Micro Kit (Ambion, Foster City, CA). TRIzol was used to isolate the RNA and the RNAqueous-Micro Kit was used to purify and DNase-treat the RNA. Retinas were quickly placed into a 1.5 ml tube containing 200 µl of TRIzol. Following the homogenization of retinas on ice, a further 660 µl of TRIzol and 160 µl of chloroform were added to the tube. The tube was vortexed for 20 s and allowed to stand for 7 min at room temperature. The tubes were centrifuged at 13,000× g for 10 min at 4 °C. The supernatant was then removed and placed into a clean 1.5 ml tube with half its volume of 100% ethanol. The tube was vortexed briefly before the contents underwent purification and DNase treatment, as detailed in the RNAqueous-Micro Kit manual. Purified DNAase-treated RNA was analyzed using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), to determine the quantity and quality of the sample. RNA samples were used only if the A260/A280 ratio was above 1.9 and the RIN (RNA integrity number) was greater than 8.5.
Affymetrix microarray analysis
The Affymetrix (Santa Clara, CA) Mouse Gene 1.0 ST Array was used; this is a whole transcript-based array that interrogates 28,853 well annotated genes. Staining, hybridization, washing, and scanning of the array were performed at the Biomolecular Resource Facility at the John Curtin School of Medical Research, Australian National University, following manufacturers’ protocols. Two experimental replicates were run independently, providing duplicates for each condition. Gene expression data were stored in .cel format files that could be interpreted by array analysis software.
Quality assessment and hierarchical clustering
Affymetrix® Expression Console™ software was used to supply probe set summarization and calculate quality assessment metrics. Gene expression data were uploaded into the Expression Console™ software to run a gene level core analysis. Once the analysis was complete, the numeric values of quality control metrics were summarized in a report and visualized in line graphs. Hierarchical clustering of data sets was performed using the heat map function in the Partek Genomics Suite (Partek Inc., St. Louis, MO). It uses log2 signal values. The following parameter settings were used: Method - average linkage; Dissimilarity - Euclidean; Clustering method-agglomerative.
Statistical analysis
Gene expression files in .cel format were imported into the Partek Genomics Suite, with the following normalization of their configuration: Probes to import - interrogating probes; Probe filtering - include core; Background correction - RMA background; Log base - 2; Probe set summarization - Median Polish. Statistical differences between experimental groups were examined using a t-test (post-hoc contrasts).
Gene expression in the inferior and superior regions of control retinas
Gene expression was compared between the superior and inferior regions of the control retinas using statistical and fold change analysis. In this comparison, there was no experimental procedure applied; therefore, the conventional term “fold change” seems inappropriate. Instead we use “fold difference.” Fold difference (FD) is then:
FD=E2/E1 when expression is greater in the second of two experimental (E) samples, FD=- E1/E2, when expression in the second sample is less.
In these equations, E1 is the level of expression in the first (usually the control) sample, and E2 is the level in the second (usually treated) sample. To apply this convention to the present samples, we chose the inferior retina as sample 2 (E2) and the superior retina as sample 1 (E1).
Data were then analyzed using two-sample t-tests for significant differences (increases and decreases) between the inferior and superior retina in the expression of all probes.
Gene expression in the inferior and superior regions of hyperoxia-exposed retinas
To identify such genes, we analyzed the four samples arising from the two variables in this part of the study: treatment (hyperoxia versus control) and site (inferior versus superior). Each sample was available in duplicate. A 2-way ANOVA test applied to the eight data sets provided three p values for each gene, one giving the probability that the gene was not regulated by hyperoxia in the superior retina, the second giving the probability that the gene was not regulated in the inferior retina, and the third (the “combination” p value) giving the probability that the gene was not regulated in either region. For each gene, there were also available twofold ratio values, one that indicated the change in its expression induced by hyperoxia in the inferior retina, and a second that indicated the change in expression in the superior retina.
To identify genes regulated differently by hyperoxia in the two regions, we selected genes for which the 2-way ANOVA test gave a combination p value <0.05. Then we developed a specific measure of expression difference, called “fold margin” (FM).
We calculated a FM value for each gene, FM=FRi – FRs where FM is the fold margin, FRi is the fold ratio for that gene in the inferior retina, and FRs are the fold ratios for that gene in the superior retina.
Functional analysis
Analysis of gene ontology and molecular pathways was performed using the bioinformatics resource Database for Annotation, Visualization and Integrated Discovery (DAVID) and Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems Inc., Redwood City, CA). Using the Ingenuity Pathways Knowledge Base, IPA Core Analysis tools were used to assign genes to the most significant known molecular networks and biologic processes.
Quantitative polymerase chain reaction
Quantitative PCR (qPCR) was used to validate the expression changes of selected genes identified in the microarray experiment. Three biologic replicates were used for each condition; RNA was processed as for the microarray experiments. For each sample, 1 µg retinal RNA was reverse transcribed into cDNA using the Superscript III Reverse Transcriptase protocol. The resulting cDNA sample served as a template for real time qPCR using TaqMan® probes and accompanying Master Mix (Applied Biosystems, Foster City, CA). A typical 20 µl reaction is composed of 10 µl 2× TaqMan Gene Expression Master Mix, 1 µl TaqMan Gene Expression Assay, 0.4 µl cDNA templates and RNase-free water. All reactions were set up in duplicate and negative controls without any template or probe were included. The qPCR was performed on a Rotor-Gene 3000 and analyzed using the Rotor-Gene 6 software (Corbett Robotics, Mortlake, NSW, Australia). A typical qPCR program was used, including 2 min incubation at 50 °C and 10 min enzyme activation at 95 °C, followed by 40 cycles of 15 s denaturation at 95 °C plus 1 min probe annealing at 60 °C. The Ct (cycle threshold) means, representing the quantitation of the amount of cDNA in the original sample, were used to calculate fold change using the Pfaffl Equation [22]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as a reference gene against which relative expression values of other genes were calculated.
Results
Clustering analysis
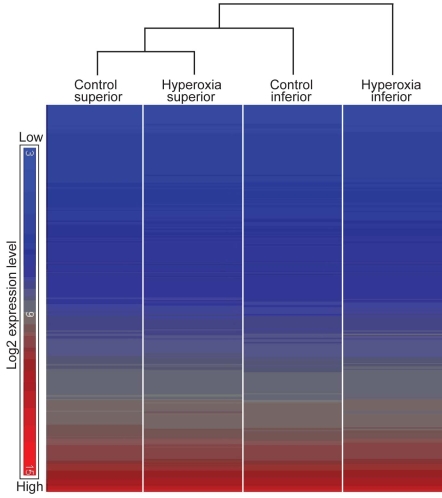
Hierarchical clustering analysis was performed using the heat map function in the Partek Genomics Suite (Figure 1). The replicates of each group clustered most closely to each other (GEO accession number GSE21246) and were therefore averaged; Figure 1 therefore shows the clustering of the averaged samples. Superior retina samples before and after hyperoxia clustered more closely to each other than to other groups, indicating that hyperoxia had little effect on the superior retina. Gene expression in the inferior retina diverged from the superior retina, with hyperoxia increasing the separation.
Figure 1.
Hierarchical clustering diagram. The study aims to identify the genes expressed differently in normoxic (control) and hyperoxic conditions in the superior and inferior halves of the retina because regional sensitivity to hyperoxia of the inferior was expected. Clustering analysis demonstrated similarity and correlation between replicates within an experimental group and between different groups. Each column comprises a set of horizontal lines with each line representing a single gene or noncoding RNA (ncRNA). The color given to the line depends on the level of expression of the gene, from blue (low expression) to red (high expression); the range of colors available is shown at left. In each column, the genes are arranged in low-to-high expression, going down the column. The program then compares the order of genes in the column and indicates, by the brackets at top, the similarities and differences between the four samples. The replicates of each sample clustered closely to each other (not shown) and were averaged to constitute the four samples shown. The samples from the superior retina before and after hyperoxia were most similar. The sample from the inferior retina after hyperoxia, in which photoreceptor degeneration was greatest, is most dissimilar from the other samples.
Control retina: genes differently expressed in inferior and superior regions
Comparison of gene expression levels in inferior and superior regions of control retinas identified expression changes in a limited cohort of sequences.
Identification
The FD analysis described in the Methods section identified only 31 sequences with an FD >1.5. Of these, 27 (listed in Table 1) are known genes. Approximately half of the genes were expressed more highly in the inferior retina, and the other half were expressed more highly in the superior retina. The FD was greater than two for only two of the 27 genes (T box 5 [Tbx5], membrane metalloendopeptidase [Mme]).
Table 1. Differentially expressed genes between inferior and superior retina of control (normoxic) C57BL/6J mice.
| Symbol | Description | RefSeq | p value | FD (I/S) |
|---|---|---|---|---|
|
Anatomic structure development (GO:0009653) | ||||
| Vax2 |
ventral anterior homeobox containing gene 2 |
NM_011912 |
0.021 |
1.840 |
| Sprr2b |
small proline-rich protein 2B |
NM_011469 |
0.012 |
1.630 |
| Vcan |
versican |
NM_001081249 |
0.001 |
1.598 |
| Slc26a4 |
solute carrier family 26, member 4 |
NM_011867 |
0.008 |
1.598 |
| Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
NM_009697 |
0.002 |
−1.564 |
| Efnb2 |
ephrin B2 |
NM_010111 |
0.015 |
−1.625 |
| Tbx5 |
T-box 5 |
NM_011537 |
0.0002 |
−2.087 |
|
Sensory perception (GO:0007600) | ||||
| Opn1sw |
opsin 1 (cone pigments), short-wave-sensitive |
NM_007538 |
0.038 |
1.762 |
| V1rb8 |
vomeronasal 1 receptor, B8 |
NM_053229 |
0.043 |
−1.508 |
| Olfr1416 |
olfactory receptor 1416 |
NM_147038 |
0.041 |
−1.577 |
| Opn1mw |
opsin 1 (cone pigments), medium-wave-sensitive |
NM_008106 |
0.010 |
−1.644 |
| Olfr652 |
olfactory receptor 652 |
NM_147048 |
0.017 |
−1.652 |
|
Catalytic activity | ||||
| Mme |
membrane metalloendopeptidase |
NM_008604 |
0.031 |
2.197 |
| Adh6b |
alcohol dehydrogenase 6B (class V) |
ENSMUST00000090166 |
0.017 |
1.516 |
| Rdh16 |
retinol dehydrogenase 16 |
NM_009040 |
0.004 |
−1.684 |
| Alpk2 |
alpha-kinase 2 |
ENSMUST00000035548 |
0.017 |
−1.979 |
|
G protein-coupled receptor | ||||
| Gpr137b |
G protein-coupled receptor 137B |
NM_031999 |
0.007 |
1.541 |
| Gprc5b |
G protein-coupled receptor C5B |
NM_022420 |
0.025 |
1.526 |
|
Amino acid transporter | ||||
| Slc7a11 |
solute carrier family 7 |
NM_011990 |
0.035 |
1.756 |
|
Cell migration | ||||
| Sst |
somatostatin |
NM_009215 |
0.016 |
1.666 |
|
Translation regulation | ||||
| Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
NM_183029 |
0.024 |
−1.544 |
|
Inductin of apoptosis | ||||
| Khdc1a |
KH domain containing 1A |
NM_183322 |
0.037 |
−1.575 |
|
Unknown function | ||||
| Snhg1 |
small nucleolar RNA host gene 1 |
AK051045 |
0.007 |
1.710 |
| predicted gene, EG624256 |
XR_034994 |
0.006 |
1.529 |
|
| predicted gene, EG668725 |
DQ386867 |
0.039 |
1.510 |
|
| Abi3bp |
ABI gene family, member 3 (NESH) binding protein |
NM_001014423 |
0.028 |
1.505 |
| RIKEN cDNA A530032D15Rik gene |
BC094285 |
0.007 |
−1.503 |
|
| Gm1381 |
gene model 1381, (NCBI) |
BC147167 |
0.005 |
−1.523 |
| Serpina1b |
serine preptidase inhibitor, clade A member 1B |
NM_009244 |
0.025 |
−1.557 |
| Gm1862 |
gene model 1862, (NCBI) |
XM_001478226 |
0.031 |
−1.576 |
| predicted gene, OTTMUSG00000001136 | XM_001005073 | 0.007 | −1.868 | |
Genes were grouped into several functional categories using gene ontology analysis. Categories with GO number are considered specific functional groups. Abbreviations: RefSeq represents NCBI reference sequence; FD represents fold difference; GO represents gene ontology.
Functional analysis
The list of 27 genes was subjected to gene ontology analysis, using the bioinformatics resource DAVID, which organized the genes into several functional groups, including Anatomic structure development, Sensory perception, and Catalytic activity (Table 1). Genes related to retinal development formed the largest functional group; these relate to the organization of the retina’s projection to brain centers. A superior-inferior gradient is consistent with previous work on these genes [23-25]. Five of the 27 genes were in the Sensory perception group. Two genes are for short- and medium-wavelength cones; short-wave-sensitive cone opsin1 (Opsn1sw) is more strongly expressed in the inferior retina, and medium-wave-sensitive cone opsin1 (Opsn1mw) in the superior retina. These differences in expression match known regional differences in the retinal distributions of short- and medium-length cones [26-28], and confirm the present separation of inferior and superior retina samples.
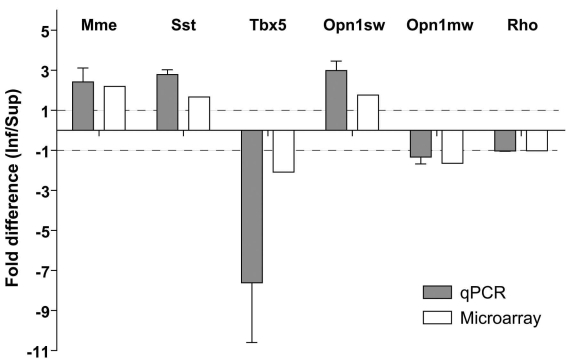
Quantitative PCR validation
Six genes were selected for confirmation of their differential expression in control retinas, using qPCR (Figure 2). The gene Tbx5 was chosen as an example of the Anatomic structure group, Opn1sw and Opn1mw were chosen as retina-relevant examples from the Sensory perception group, Mme and somatostatin (Sst) were chosen as a functionally inter-related pair, and rhodopsin (Rho) was chosen as a non-regulated, retina-relevant gene. Quantitative PCR confirmed the even expression of Rho and the differential regulation of Mme, Sst, Tbx5, and Opn1sw. For one gene, Opn1mw (encoding M-opsin), the FD assessed by qPCR was less than the threshold of 1.5 fold (−1.33 by PCR versus −1.64 by microarray). However, the difference was in the same direction in the qPCR and microarray assessments.
Figure 2.
Quantitative PCR for microarray validation. Array validation was performed by quantitative RT–PCR for 5 genes selected from the list in Table 1. Rhodopsin was used as a retina-relevant reference gene. qPCR is found in excellent agreement with the outcome of microarray analysis. The results of qPCR were averaged from three independent experiments, with the error bar indicating the standard error of the mean. Dashed lines at fold differences of 1 and −1 indicate no difference of gene expression between the inferior and superior regions.
Hyperoxia-exposed retina: genes differently regulated in inferior and superior regions
Genes whose regulation by hyperoxia differed significantly between the inferior and superior retina could include genes regulated in one region but not the other, genes upregulated in one region and downregulated in the other, or genes regulated in the same direction in the two regions, but much more strongly in one region.
Identification
Hyperoxia induced significant changes in gene expression in both the inferior (hyperoxia-vulnerable) and superior (hyperoxia-resistant) retina, but the number of hyperoxia-regulated genes was much higher in the inferior retina (Figure 3). For example, using the criterion p<0.01 on the 2-way ANOVA test, the expression of 281 genes and 110 ncRNAs changed significantly in the superior retina, while the expression of 618 genes and 231 ncRNAs changed in the inferior retina. Using p≤0.05 and requiring a fold ratio > 2 or < 0.5, the total number of genes differentially expressed was over six times greater in the inferior retina than in the superior retina (58 versus 9). Overall, more genes were upregulated than downregulated. This imbalance was particularly marked after applying both selection criteria; for example, 56 genes were upregulated in the inferior retina, compared to 2 that were downregulated.
Figure 3.
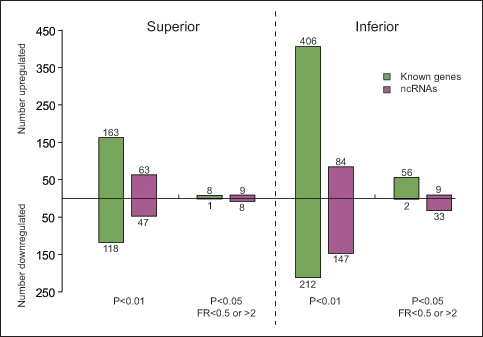
Significant more genes regulated in the inferior than the superior retina after hyperoxia. According to the different selection criteria, the number of genes and non-coding RNAs regulated by hyperoxia were assessed and separately elucidated for the superior and inferior regions. Fold ratio (FR) values < 0.5 indicate significant downregulation of expression (at least halved); FR values > 2 indicate significant upregulation of expression (more than doubled).
Combining the criteria of p<0.05 and abs (FM) >0.5 (Table 2), a total of 112 sequences with Affymetrix probe IDs were identified as differently regulated by hyperoxia between the inferior and superior regions. Of these 112, 97 matched known genes (Table 2). The gene with the most significant FM was Edn2 (endothelin 2), a possible stress response gene. The expression of gene Edn2 increased 7.02 fold in the inferior retina, but increased just 1.62 fold in the superior retina after being exposed to hyperoxia.
Table 2. Known genes and ncRNAs differentially regulated by hyperoxia between inferior and superior retina.
| Symbol | Description | RefSeq | P-value Treat*Site | FR Inf | FR Sup | FM |
|---|---|---|---|---|---|---|
| Edn2 |
endothelin 2 |
NM_007902 |
0.00001 |
7.02 |
1.62 |
5.40 |
| Gfap |
glial fibrillary acidic protein, mRNA |
NM_010277 |
0.00303 |
5.36 |
2.37 |
2.99 |
| C1qb |
complement component 1, beta polypeptide |
NM_009777 |
0.00085 |
4.20 |
1.45 |
2.75 |
| Bcl3 |
B-cell leukemia/lymphoma 3 |
NM_033601 |
0.00846 |
3.89 |
1.48 |
2.42 |
| C1qc |
complement component 1, subcomponent, C chain |
NM_007574 |
0.00468 |
4.21 |
1.93 |
2.28 |
| Mpeg1 |
macrophage expressed gene 1 |
NM_010821 |
0.00848 |
3.83 |
1.57 |
2.27 |
| C1qa |
complement component 1, subcomponent, alpha polypeptide |
NM_007572 |
0.01737 |
3.69 |
1.68 |
2.01 |
| Cd68 |
CD68 antigen |
NM_009853 |
0.01188 |
3.27 |
1.45 |
1.83 |
| Ly86 |
lymphocyte antigen 86 |
NM_010745 |
0.00329 |
3.95 |
2.16 |
1.78 |
| Fgf2 |
fibroblast growth factor 2 |
NM_008006 |
0.00038 |
3.10 |
1.45 |
1.64 |
| Timp1 |
tissue inhibitor of metalloproteinase 1 |
NM_001044384 |
0.00172 |
2.74 |
1.14 |
1.61 |
| Fcrls |
Fc receptor-like S, scavenger receptor |
NM_030707 |
0.00557 |
2.55 |
1.02 |
1.53 |
| Laptm5 |
lysosomal-associated protein transmembrane 5 |
NM_010686 |
0.00863 |
3.03 |
1.58 |
1.45 |
| Ccl3 |
chemokine (C-C motif) ligand 3 |
NM_011337 |
0.01929 |
2.46 |
1.06 |
1.40 |
| Ctss |
cathepsin S |
NM_021281 |
0.00330 |
3.03 |
1.68 |
1.35 |
| Lrrc2 |
leucine rich repeat containing 2 |
NM_028838 |
0.00324 |
2.71 |
1.48 |
1.23 |
| Crym |
crystallin, mu |
NM_016669 |
0.02834 |
2.74 |
1.52 |
1.22 |
| Slc25a37 |
solute carrier family 25, member 37 |
NM_026331 |
0.00087 |
2.43 |
1.21 |
1.22 |
| Prtg |
protogenin homolog (Gallus gallus) |
NM_175485 |
0.00036 |
2.35 |
1.19 |
1.16 |
| Lad1 |
ladinin |
NM_133664 |
0.00227 |
2.36 |
1.20 |
1.16 |
| Cst7 |
cystatin F (leukocystatin) |
NM_009977 |
0.03019 |
2.07 |
0.95 |
1.12 |
| Fcer1g |
Fc receptor, IgE, high affinity I, gamma polypeptide |
NM_010185 |
0.00232 |
2.44 |
1.33 |
1.11 |
| Gadd45b |
growth arrest and DNA-damage-inducible 45 beta |
NM_008655 |
0.01173 |
2.74 |
1.65 |
1.09 |
| Chi3l1 |
chitinase 3-like 1 |
NM_007695 |
0.02641 |
2.88 |
1.84 |
1.04 |
| Cd180 |
CD180 antigen |
NM_008533 |
0.02177 |
2.10 |
1.07 |
1.02 |
| Mt2 |
metallothionein 2 |
NM_008630 |
0.03497 |
2.41 |
1.39 |
1.02 |
| Klhl29 |
kelch-like 29 (Drosophila) |
BC145748 |
0.00162 |
2.20 |
1.18 |
1.02 |
| Socs3 |
suppressor of cytokine signaling 3 |
NM_007707 |
0.00892 |
2.12 |
1.13 |
1.00 |
| Cd53 |
CD53 antigen |
NM_007651 |
0.01901 |
2.35 |
1.36 |
0.99 |
| Olfr43 |
olfactory receptor 43 |
NM_146711 |
0.03715 |
1.77 |
0.79 |
0.99 |
| S100a6 |
S100 calcium binding protein A6 (calcyclin) |
NM_011313 |
0.03199 |
2.06 |
1.08 |
0.98 |
| Tyrobp |
TYRO protein tyrosine kinase binding protein |
NM_011662 |
0.00115 |
2.25 |
1.27 |
0.98 |
| Ctla2a |
cytotoxic T lymphocyte-associated protein 2 alpha |
NM_007796 |
0.04101 |
1.88 |
0.91 |
0.97 |
| Csf1r |
colony stimulating factor 1 receptor |
NM_001037859 |
0.00009 |
2.37 |
1.40 |
0.97 |
| Stat3 |
signal transducer and activator of transcription 3 |
NM_213659 |
0.00102 |
2.40 |
1.44 |
0.96 |
| Antxr2 |
anthrax toxin receptor 2 |
NM_133738 |
0.00279 |
2.23 |
1.28 |
0.95 |
| C3ar1 |
complement component 3a receptor 1 |
NM_009779 |
0.01664 |
1.96 |
1.02 |
0.95 |
| Kremen1 |
kringle containing transmembrane protein 1 |
NM_032396 |
0.00444 |
2.02 |
1.07 |
0.95 |
| C3 |
complement component 3 |
NM_009778 |
0.03059 |
2.23 |
1.28 |
0.94 |
| B2m |
beta-2 microglobulin |
NM_009735 |
0.00093 |
2.38 |
1.44 |
0.94 |
| Lcn2 |
lipocalin 2 |
NM_008491 |
0.00238 |
2.13 |
1.19 |
0.94 |
| Irgm1 |
immunity-related GTPase family M member 1 |
NM_008326 |
0.02159 |
2.04 |
1.12 |
0.93 |
| Khdc1a |
KH domain containing 1A |
NM_183322 |
0.02946 |
1.73 |
0.87 |
0.86 |
| Chl1 |
cell adhesion molecule with homology to L1CAM |
NM_007697 |
0.01079 |
1.78 |
0.92 |
0.86 |
| H2-D1 (HLA-C) |
histocompatibility 2, D region locus 1 |
NM_010380 |
0.00124 |
2.00 |
1.16 |
0.84 |
| H2-K1 |
histocompatibility 2, K1, K region |
NM_001001892 |
0.00681 |
2.05 |
1.22 |
0.83 |
| Osmr |
oncostatin M receptor |
NM_011019 |
0.01265 |
2.70 |
1.91 |
0.79 |
| Fcgr3 (Fcrg2a) |
Fc receptor, IgG, low affinity III |
NM_010188 |
0.01391 |
1.79 |
1.01 |
0.78 |
| Rbm17 |
RNA binding motif protein 17 |
NM_152824 |
0.00686 |
1.88 |
1.10 |
0.78 |
| H2-Q7 |
histocompatibility 2, Q region locus 7 |
NM_010394 |
0.00093 |
1.89 |
1.14 |
0.75 |
| Lin7b |
lin-7 homolog B (C. elegans) |
NM_011698 |
0.00908 |
1.83 |
1.11 |
0.72 |
| Irf9 |
interferon regulatory factor 9 |
NM_008394 |
0.00479 |
2.21 |
1.50 |
0.70 |
| Gnb3 |
guanine nucleotide binding protein, beta 3 |
NM_013530 |
0.00035 |
1.83 |
1.14 |
0.69 |
| Spp1 |
secreted phosphoprotein 1 |
NM_009263 |
0.00216 |
1.77 |
1.10 |
0.68 |
| Moxd2 |
monooxygenase, DBH-like 2 |
NM_139296 |
0.01505 |
1.58 |
0.94 |
0.65 |
| Agtpbp1 |
ATP/GTP binding protein 1 |
NM_023328 |
0.00242 |
1.76 |
1.12 |
0.64 |
| Olfr652 |
olfactory receptor 652 |
NM_147048 |
0.02679 |
1.38 |
0.74 |
0.64 |
| Ly9 |
lymphocyte antigen 9 |
NM_008534 |
0.02946 |
1.59 |
0.95 |
0.64 |
| Myo10 |
myosin X |
NM_019472 |
0.00714 |
1.94 |
1.31 |
0.63 |
| V1rb8 |
vomeronasal 1 receptor, B8 |
NM_053229 |
0.04461 |
1.42 |
0.80 |
0.62 |
| Sec22c |
SEC22 vesicle trafficking protein-like C (S. cerevisiae) |
NM_178677 |
0.01512 |
1.65 |
1.04 |
0.61 |
| Sulf2 |
sulfatase 2 |
NM_028072 |
0.03293 |
1.58 |
0.97 |
0.61 |
| Cd33 |
CD33 antigen |
NM_001111058 |
0.00293 |
1.63 |
1.03 |
0.60 |
| Hexb |
hexosaminidase B |
NM_010422 |
0.00341 |
1.88 |
1.30 |
0.58 |
| Pld4 |
phospholipase D family, member 4 |
NM_178911 |
0.01563 |
1.84 |
1.27 |
0.57 |
| Skap2 |
src family associated phosphoprotein 2 |
NM_018773 |
0.00550 |
1.84 |
1.27 |
0.57 |
| Usp18 |
ubiquitin specific peptidase 18 |
NM_011909 |
0.02940 |
1.58 |
1.02 |
0.56 |
| Vmn2r68 |
vomeronasal 2, receptor 68 |
NM_001105181 |
0.00628 |
1.47 |
0.92 |
0.56 |
| Bst2 |
bone marrow stromal cell antigen 2 |
NM_198095 |
0.00535 |
1.60 |
1.05 |
0.55 |
| Wfs1 |
Wolfram syndrome 1 homolog (human) |
NM_011716 |
0.02286 |
1.54 |
0.99 |
0.54 |
| Rdh16 |
retinol dehydrogenase 16 |
NM_009040 |
0.00992 |
1.25 |
0.72 |
0.53 |
| V1rd6 |
vomeronasal 1 receptor, D6 |
NM_030738 |
0.04247 |
1.30 |
0.77 |
0.53 |
| Shbg |
sex hormone binding globulin |
NM_011367 |
0.01306 |
1.47 |
0.94 |
0.53 |
| H2-Q2 (HLA-F) |
histocompatibility 2, Q region locus 2 |
AB359227 |
0.00277 |
1.71 |
1.18 |
0.53 |
| Sulf1 |
sulfatase 1 |
NM_172294 |
0.00558 |
1.68 |
1.16 |
0.52 |
| Atf3 |
activating transcription factor 3 |
NM_007498 |
0.02131 |
1.63 |
1.11 |
0.52 |
| Mt1 |
metallothionein 1 |
NM_013602 |
0.00379 |
1.57 |
1.06 |
0.51 |
| V1re6 |
vomeronasal 1 receptor, E6 |
NM_134195 |
0.01502 |
1.37 |
0.86 |
0.51 |
| Dcps |
decapping enzyme, scavenger |
NM_027030 |
0.02157 |
1.56 |
1.06 |
0.50 |
| Tead3 |
TEA domain family member 3 |
NM_011566 |
0.01222 |
1.66 |
1.16 |
0.50 |
| Bmp2 |
bone morphogenetic protein 2 |
NM_007553 |
0.03978 |
0.71 |
1.21 |
−0.50 |
| Defb9 |
defensin beta 9 |
NM_139219 |
0.01100 |
0.81 |
1.32 |
−0.51 |
| Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, G protein), member a13 |
NM_144512 |
0.02354 |
0.92 |
1.43 |
−0.51 |
| Ankrd44 |
ankyrin repeat domain 44 |
NM_001081433 |
0.02845 |
0.91 |
1.42 |
−0.51 |
| Clec14a |
C-type lectin domain family 14, member a |
NM_025809 |
0.01571 |
0.88 |
1.40 |
−0.52 |
| Colec12 |
collectin sub-family member 12 |
NM_130449 |
0.00944 |
0.85 |
1.37 |
−0.52 |
| Wnt8a |
wingless-related MMTV integration site 8A |
NM_009290 |
0.00618 |
0.86 |
1.39 |
−0.53 |
| Kcnq5 |
potassium voltage-gated channel, subfamily Q, member 5 |
NM_023872 |
0.01549 |
0.82 |
1.36 |
−0.54 |
| Suclg2 |
succinate-CoA ligase, GDP-forming, beta subunit |
NM_011507 |
0.04602 |
0.86 |
1.42 |
−0.55 |
| Slc16a12 |
solute carrier family 16 member a12 |
NM_172838 |
0.01174 |
0.77 |
1.33 |
−0.56 |
| Snhg1 |
small nucleolar RNA host gene 1 |
AK051045 |
0.00764 |
0.52 |
1.08 |
−0.56 |
| Vcan |
versican |
NM_001081249 |
0.00164 |
0.91 |
1.48 |
−0.57 |
| Abpb |
androgen binding protein beta |
NM_001100464 |
0.02420 |
0.79 |
1.36 |
−0.57 |
| Mitf |
microphthalmia-associated transcription factor |
NM_001113198 |
0.00183 |
0.79 |
1.38 |
−0.58 |
| Nr2e3 |
nuclear receptor subfamily 2, group E, member 3 |
NM_013708 |
0.00803 |
0.84 |
1.45 |
−0.61 |
| Josd3 |
Josephin domain containing 3 |
BC056964 |
0.01926 |
0.64 |
1.28 |
−0.64 |
| Slc26a4 |
solute carrier family 26, member 4 |
NM_011867 |
0.00514 |
0.71 |
1.48 |
−0.77 |
| OTTMUSG00000007392 |
|
ENSMUST00000068009 |
0.02400 |
2.63 |
1.11 |
1.52 |
| ENSMUST00000105041 |
|
ENSMUST00000105041 |
0.00238 |
2.31 |
1.20 |
1.11 |
| LOC546230 |
|
ENSMUST00000103543 |
0.04064 |
2.04 |
1.06 |
0.99 |
| OTTMUSG00000001136 |
|
XM_001005073 |
0.01136 |
1.33 |
0.63 |
0.71 |
| A530032D15Rik |
|
BC094285 |
0.00291 |
1.32 |
0.64 |
0.69 |
|
BC094916 |
|
NM_001024721 |
0.04309 |
1.09 |
0.46 |
0.64 |
| A630098G03Rik |
|
EU099309 |
0.03236 |
1.34 |
0.74 |
0.60 |
| LOC544737 |
|
AK143218 |
0.02940 |
1.33 |
0.79 |
0.54 |
| 1110054P19Rik |
|
NM_026834 |
0.01399 |
1.36 |
0.85 |
0.51 |
|
EG668103 |
|
XM_994312 |
0.01637 |
1.37 |
0.87 |
0.50 |
| ENSMUSG00000071362 |
|
ENSMUST00000072014 |
0.01789 |
0.73 |
1.23 |
−0.50 |
| ENSMUST00000054825 |
|
ENSMUST00000054825 |
0.01130 |
0.66 |
1.23 |
−0.56 |
| ENSMUSG00000073593 |
|
ENSMUST00000097611 |
0.03139 |
0.73 |
1.30 |
−0.57 |
|
EG435337 |
|
NM_001013824 |
0.04833 |
0.66 |
1.26 |
−0.60 |
| E430024C06Rik | AK149411 | 0.04984 | 0.45 | 1.32 | −0.87 |
The selection of genes was made using the criteria p<0.05 and abs(FM) > 0.5. They are sorted by the value of fold margin. Unknown sequences were listed separately, following the known genes. Abbreviations: FR represents fold ratio; FM represents fold margin; inf represents inferior; sup represents superior; RefSeq represents NCBIreference sequence.
Functional analysis
Ingenuity Pathway Analysis software was used to identify biologic functions, diseases, and well characterized pathways that are most relevant to the genes of interest. The program compared the genes identified as differentially regulated between the inferior and superior retina with a database of genes involved in particular functions, diseases, and pathways. It then calculated the probability that each function, disease, or pathway is not involved in the differential regulation. Using the criteria p<0.001 and a minimum involvement of five genes, the processes and diseases listed in Table 3 were identified. Some clusters of differentially regulated genes that possibly impact photoreceptor vulnerability and survival are listed in Table 4, arranged according to function, disease, or pathway.
Table 3. The “Biologic processes” and “Disease and disorder” groupings of genes identified as differentially expressed between superior and inferior retina after exposure to hyperoxia.
| Relevant Functions and Diseases | Number of molecules | p-value |
|---|---|---|
|
Biologic process | ||
| defense response |
22 |
5.9E-11 |
| immune response |
19 |
2.7E-10 |
| response to external stimulus |
13 |
7.9E-06 |
| immune effector process |
8 |
1.1E-05 |
| regulation of multicellular organismal process |
10 |
2.8E-05 |
| antigen processing and presentation |
7 |
4.0E-05 |
| regulation of immune system process |
7 |
4.6E-05 |
| activation of immune response |
6 |
7.1E-05 |
| response to stress |
14 |
3.2E-04 |
|
Disease and disorder | ||
| immunological disorder |
19 |
1.5E-08 |
| inflammatory disorder |
21 |
3.1E-08 |
| neurologic disorder |
26 |
2.6E-07 |
| genetic disorder |
38 |
1.1E-06 |
| infectious disorder |
10 |
5.7E-05 |
| respiratory disorder |
8 |
1.2E-04 |
| ophthalmic disorder | 7 | 5.1E-04 |
The two groupings shown met the criteria of significant relevance (p<0.001) and a minimum involvement of 5 genes from gene list of interest (Table 2).
Table 4. Clusters of genes differentially regulated between inferior and superior retina, with possible impact on photoreceptor vulnerability and survival.
| Symbol | Description | RefSeq | FR (inf) | FR (sup) | FM |
|---|---|---|---|---|---|
|
Complement system (p=1.66E-07) | |||||
| C1qb |
complement component 1, q subcomponent, beta polypeptide |
NM_009777 |
4.20 |
1.45 |
2.75 |
| C1qc |
complement component 1, q subcomponent, C chain |
NM_007574 |
4.21 |
1.93 |
2.28 |
| C1qa |
complement component 1, q subcomponent, alpha polypeptide |
NM_007572 |
3.69 |
1.68 |
2.01 |
| C3ar1 |
complement component 3a receptor 1 |
NM_009779 |
1.96 |
1.02 |
0.95 |
| C3 |
complement component 3 |
NM_009778 |
2.23 |
1.28 |
0.94 |
|
Acute phase response signaling, IL-10 signaling (p=1.8E-04–4.07E-03) | |||||
| Bcl3 |
B-cell leukemia/lymphoma 3 |
NM_033601 |
3.89 |
1.48 |
2.42 |
| Socs3 |
suppressor of cytokine signaling 3 |
NM_007707 |
2.12 |
1.13 |
1.00 |
| Stat3 |
signal transducer and activator of transcription 3 |
NM_213659 |
2.40 |
1.44 |
0.96 |
|
Ophthalmic disorders (p=5.10E-04) | |||||
| Timp1 |
tissue inhibitor of metalloproteinase 1 |
NM_001044384 |
2.74 |
1.14 |
1.61 |
| Ccl3 |
chemokine (C-C motif) ligand 3 |
NM_011337 |
2.46 |
1.06 |
1.40 |
| C3ar1 |
complement component 3a receptor 1 |
NM_009779 |
1.96 |
1.02 |
0.95 |
| C3 |
complement component 3 |
NM_009778 |
2.23 |
1.28 |
0.94 |
| Slc16a12 |
solute carrier family 16 (monocarboxylic acid transport) |
NM_172838 |
0.77 |
1.33 |
−0.56 |
| Vcan |
versican |
NM_001081249 |
0.91 |
1.48 |
−0.57 |
| Nr2e3 |
nuclear receptor subfamily 2, group E, member 3 |
NM_013708 |
0.84 |
1.45 |
−0.61 |
|
Oxidative stress response (p=1.66E-04) | |||||
| Mt2, Mt1e |
metallothionein 2 |
NM_008630 |
2.41 |
1.39 |
1.02 |
| Mt1, Mt1f | metallothionein 1 | NM_013602 | 1.57 | 1.06 | 0.51 |
P value accounts for the probability that each function, disease or pathway is not involved in the differential regulation. Abbreviations: FR represents fold ratio; FM represents fold margin; inf represents inferior; sup represents superior; RefSeq represents NCBI reference sequence.
Interestingly, four of the biologic processes identified in Table 3 are strongly related to the function of the immune system. Moreover, “immunological disorders” is the disease category most strongly related to the gene set identified as differentially regulated by hyperoxia between the inferior and superior retina.
Quantitative PCR validation
Expression changes of 11 genes were selected for confirmation by qPCR. From these, eight genes were chosen from Table 2: Edn2, glial fibrillary acidic protein (Gfap), B-cell leukemia/lymphoma 3 (Bcl-3), nuclear receptor subfamily 2, group E, member 3 (Nr2e3), wingless-related MMTV integration site 8A (Wnt8a), lin-7 homolog B (Lin7a), complement component 3 (C3), and growth arrest and DNA-damage-inducible 45 beta (Gadd45b). Three other genes were also chosen for validation: cyclic nucleotide gated channel alpha 2 (Cnga2), which has a p-value <0.05 but an FM <0.5, hypoxia inducible factor 1 alpha subunit (Hif1α), which has shown responses to hypoxia in previous studies [29,30], and medium-wave-sensitive cone opsin (Opn1mw), which is a gene known to be expressed by the retina and that is not hyperoxia-regulated [14].
The regulation changes of these genes observed by qPCR and in the microarray are shown in Figure 4. The direction of the change was confirmed in six of the eight genes that differentially regulate between the inferior and superior retina; the exceptions were Wnt8a and Nr2e3. The magnitude of the FM between the superior and inferior retina was more dramatic for genes Edn2, Gfap, and Bcl3. For Cnga2, the upregulation in the superior and downregulation in the inferior retina were more noticeable; however, the FM between them was < 0.5, which was consistent with the microarray data. For Hif1α and Opn1mw, which showed no significant change in the microarray, upregulation of approximately 30%–50% was observed by qPCR. However, such upregulation showed no regional differences. Upregulation of Hif1α by hyperoxia has been reported previously [14], but the upregulation was time-dependent, being prominent after three days of exposure, but not at the 14-day exposure used here.
Figure 4.
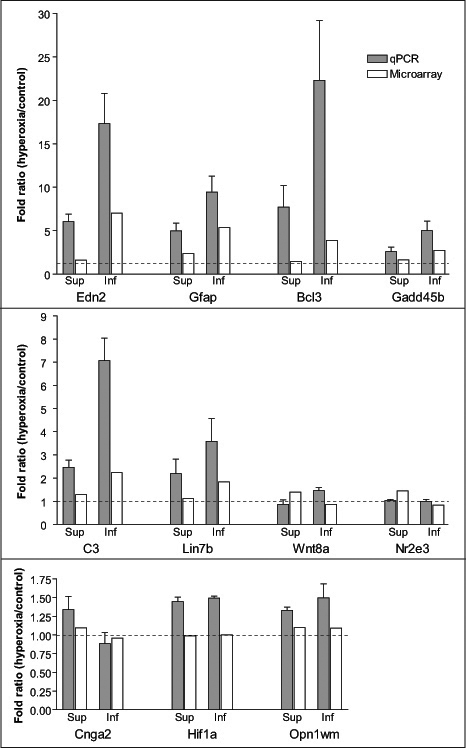
Quantitative PCR validation of expression changes induced by exposure to hyperoxia in a selected cohort of genes. For each gene, the change was estimated in both the superior and inferior retina by microarray analysis and qPCR. The results of qPCR were averaged from three independent experiments, with the error bar indicating the standard error of the mean. Fold ratio values >1 indicate an upregulation and values <1 indicate a downregulation. Where FR=1 (dotted line), expression was unchanged.
Discussion
The microarray analysis presented provides an overview of regional variations in gene expression in the normal retina before and after hyperoxic stress. Further, immunohistochemical studies will be needed to identify the sites at which gene products are expressed. Although hyperoxia is specifically toxic to photoreceptors, hyperoxia may regulate genes in any of the several classes of retinal neurons and neuroglia, and in the endothelium of blood vessels.
Gene expression before hyperoxia
Comparing gene expression between the superior and inferior retina under normoxic (control) conditions, two genes showed an FD in expression of greater than two, and 27 showed an FD of greater than 1.5. These differences in gene expression are relatively limited, but may be significant in determining the relative invulnerability of the superior retina to hyperoxic damage and, conversely, the vulnerability of the inferior retina.
Seven out of the 27 genes (listed in Table 1) are involved in anatomic structure development (gene ontology GO-term 0009653). Vax2, Tbx5, and ephrin B2 (Efnb2) are important in eye morphogenesis and are topographic determinants of the visual projections between the retina and tectum. In vertebrate development, the polarity of the dorsal-ventral (dorsal-superior, ventral-inferior) eye axis is determined by spatially restricted expression of transcription factors Vax2 in the inferior retina and Tbx5 in the superior retina [24,25]. Efnb2 is an axon guidance molecule expressed in a low-to-high superior-inferior gradient in the ganglion cell layer [31]. The expression of Efnb2 and its receptor is regulated by Vax2 and Tbx5, which in turn control the dorsal-ventral polarity of retinotectal projection [23]. These developmental patterns were detected in the adult retina in the present experiments, providing evidence that our separation of inferior-superior retina was accurate.
Five genes are associated with sensory perception (GO-term 0007600). They belong to the rhodopsin-like superfamily of G-protein-coupled receptors, although they are involved in distinct sensory systems. Opn1mw encodes the medium-wave-sensitive cone opsins (expressed in M-cones) and Opn1sw encodes the short-wave-sensitive cone opsins (S-cones or “blue” cones). The uneven distribution of cone receptors, with S-cones more numerous in the inferior retina and M-cones in superior retina, has been described in mice [27,28] and cats [26]. The expression differences observed (1.76 for Opn1sw in an inferior-superior comparison and −1.64 for Opn1mw) confirm the differences in cone topography detected by immunohistochemistry.
The gene showing the largest FD between the superior and inferior regions of the normoxic retina is Mme, which is expressed more than twofold in the inferior retina. The product of this gene is a membrane-bound metalloendopeptidase, commonly known as neutral endopeptidase or neprilysin. It has been found in various cell lines, including endothelial cells and vascular smooth muscle cells. Mme belongs to a family of zinc metalloproteases [32].
Another member of this family is endothelin converting enzyme-1 (ECE-1), which shares 58% amino acid homology with Mme [33]. ECE-1 catalyzes big endothelin to peptide endothelin and plays a key role in proteolytic endothelin activation [34]. However, the conversion of big endothelins to their respective endothelin peptides may not solely depend on ECE-1. In guinea pig lung parenchyma, Mme was suggested to play a role in the conversion to endothelin-1 and endothelin-2 [35]. Given their structural homology, it is not surprising that they share functions. As reported previously [14], endothelin-2 is the most upregulated gene in the whole retina after two weeks of hyperoxia. In the present study, endothelin-2 is the most upregulated gene in the vulnerable inferior retina after hyperoxic exposure. This correlates with the overexpression of Mme in the inferior retina. Perhaps Mme is responsible for or at least represents one pathway of the biosynthesis of endothelin-2 in the inferior retina after hyperoxic exposure.
Somatostatin is uniquely capable of stimulating Mme activity in primary cortical neurons [36]. Most interestingly, the gene encoding somatostatin (Sst) is also expressed more in the inferior than the superior retina (Table 1). The relatively high expression of both Mme and Sst may provide some insight into the vulnerability of the inferior mouse retina. One possibility is that there is an unknown regulatory peptide that protects photoreceptors against oxidative damage and is also, like amyloid beta (Aβ), a substrate of Mme. Upregulation of Mme might degrade this peptide, reduce its protective action, and cause the vulnerability of the inferior retina to hyperoxic stress. Other possibilities include regional variations in retinal metabolism, which in turn may be observed in regional variations in retinal blood supply. In the rabbit and monkey, for example, regional variations in the choroidal circulation (the major source of oxygen supply to photoreceptors) have been described, matching retinal specializations [37]. An inferior-superior difference in the retinal circulation has also been observed in humans [38].
Another established difference between the superior and inferior retina is environmentally determined. When animals are raised (as in the present experiments) in light from sources in the ceiling of a room, the inferior retina is more exposed to ambient light and is more resistant to subsequent light stress [17]. Exposure to modest ambient light can “pre-condition” the retina, making it resistant to subsequent hyperoxic stress [39]. However, the relative vulnerability of the inferior retina clearly cannot be explained by such a mechanism. The current results provide a starting point for further detailed study and are a first step in describing the mechanisms involved.
Gene expression after hyperoxia
Comparison with previous microarray studies
The present data show that hyperoxia-induced regulation of genes occurred significantly more in the inferior retina in the C57BL/6J mouse retina (Figure 3); the number of significantly regulated known genes identified was approximately sevenfold greater in the inferior retina (58 versus nine). The present data can be compared to the all-retina study of the Natoli group [14], who reported that following hyperoxic exposure for 14 days, 77 known genes were significantly regulated, using the same fold change criterion. Of the 9 genes significantly regulated by hyperoxia in the superior retina in the present study, five (55%) are also on Natoli et al. and coworkers list of all-retina regulated genes. Of the 58 genes significantly regulated by hyperoxia in the inferior retina, 26 (45%) are also on the list of all-retina regulated genes. It was also of interest to compare the inferior-specific regulated genes (Table 2, 97 known genes) with those identified by Natoli group as hyperoxia-regulated in their all-retina analysis. Of the 77 genes identified by Natoli group, 21 (27%) were also in the inferior-specific list, including Bcl3, C3, Edn2, Gfap, and Gadd45b.
One previous microarray study has described the genes regulated by light damage in mice using the light-vulnerable BALB/cJ mouse strain [40]. The authors of that study reported that 70 genes were upregulated at least twofold. Fourteen of these 70 genes (20%) also displayed differential expression change in this study. It is possible that the light vulnerability of retina in BALB/cJ mice and the hyperoxia vulnerability of the inferior retina of C57BL/6J mice share some common features and mechanisms.
Endothelin-2
The gene Edn2 that encodes endothelin-2 is the most strongly regulated gene in the inferior retina after hyperoxic exposure and is also the most differentially regulated gene between the inferior and superior retina after hyperoxic exposure (Table 2). Its expression increased sevenfold in the inferior retina compared to 1.6 fold in the superior retina after exposure to hyperoxia.
Endothelin-2 belongs to a family of peptides comprising endothelin-1, endothelin-2, and endothelin-3 [41]. The endothelins are stress-responsive regulators that work in paracrine and autocrine fashion in a variety of organs, with either beneficial or detrimental results. For example, endothelin-1 is a potent vasoconstrictor in blood vessels. It also acts as a mitogen on the vascular smooth muscle and has been implicated in several vascular diseases, such as hypertension and atherosclerosis [42].
Endothelin-2 has been found to be produced in endothelial cells, the heart, and the kidneys [43,44]. In the retina, the expression of Edn2 is highly induced in models of retinitis pigmentosa, retinal detachment, and light damage and the increased production of Edn2 is localized to photoreceptors [45]. Photoreceptor-derived Edn2 is suggested to serve as a stress signal in the signaling cascade between the photoreceptor and Müller glia cells. In the presence of damaging stress such as excessive light exposure and gene mutation, Müller glia cells sense the photoreceptor damage and react by upregulating leukemia inhibitory factor (LIF) [46]. An increase in leukemia inhibitory factor in turn induces the expression of Edn2 in photoreceptors. The signal is then passed back to Müller glia cells by binding Edn2 to endothelin receptor B (Ednrb), which is expressed on Müller cells. This positive feedback loop then stimulates the production of survival factors, such as fibroblast growth factor 2 (FGF2), in Müller cells and/or photoreceptors to protect visual cells from further damage [45,46].
Involvement of the immune system
Misregulation of the innate immune response and other immune-mediated processes has been suggested to play a role in the development and progression of retinal degenerative diseases, such as age-related macular degeneration (AMD), in human patients [47,48]. The involvement of inflammation and immune-mediated processes has been suggested, for example, in the formation of drusen and inflammation triggered by the debris generated by dying retinal pigment epithelial (RPE) cells, as a factor in photoreceptor cell death [48].
Complement cascade proteins have been shown to be upregulated in the AMD retina, showing a strong presence along the RPE-choroid interface and in drusen, the hallmark of AMD, suggesting that the activation of the complement pathway has a potential role in drusen biogenesis and the etiology of AMD [47]. A murine model of laser-induced choroidal neovascularization (CNV) in C57BL/6J mice revealed the deposition of C3 and membrane attack complex in the neovascularizing tissue. Complement depletion in C3−/− mice resulted in the inhibition of CNV and a marked reduction of angiogenic factors [49]. C3a is the bioactive fragment of complement component C3; a blockade of the C3a receptors (C3aR) also reduces CNV [50]. In addition, in the degenerative mouse model rd1, the gene ontology groups, “defense response,” “complement activity,” and “stress response,” were significantly over-represented in the differentially expressed genes in the degenerative retina. The classical complement pathway was found to be upregulated in the rd1 mouse [51]. The complement pathway has also been implicated in the development of glaucoma and in a model of ocular hypertension [52,53].
Immune-related factors and pathways may thus be important in the process of photoreceptor cell death induced by hyperoxic stress. The present data increase previous understanding of this involvement, but are arguably still inadequate to determine whether immune misregulation causes or results from cell death induced by hyperoxia.
Retinal disease relevant genes
Seven genes listed in Table 2 were identified as associated with ophthalmic diseases and three of them correspond to known retinal disease loci listed on the RetNet website (C3, versican [Vcan], and Nr2e3). Previous studies reported a significant association between a polymorphism in the C3 gene and susceptibility to AMD [54,55]. Wagner syndrome can be caused by a mutation in the human VCAN gene, which encodes versican, a proteoglycan present in the vitreous body [56]. The syndrome features peculiar lesions of the vitreous and retina, retinal detachment, and progressive chorioretinal dystrophy [56,57]. Versican is widely distributed in the extracellular matrix of a variety of tissues and plays an important role in reducing oxidant injury by enhancing cell-matrix interaction [58]. Mutations in the human NR2E3 gene, which encodes a retina-specific nuclear receptor, lead to enhanced S-cone syndrome and retinitis pigmentosa [59].
Antioxidant genes
Metallothioneins (MTs) are a class of low molecular weight, cysteine-rich proteins that bind with copper and zinc. The metallothionein family is composed of four separately encoded isoforms, known as MT-I, -II, -III, and -IV [60]. They serve as important regulators of metal homeostasis and provide cytoprotection in tissues subjected to oxidative stress as a result of their free radical scavenging ability [61]. Increased expression of MT has been reported in various induced models of retinal degeneration, including light [40], hyperoxia [14], and hypoxia [62]. Metallothioneins have been shown to protect RPE cells against toxic levels of cadmium, heme- and iron-induced oxidation, and UV light-induced apoptosis [63]. As endogenous antioxidants, they also protect ganglion cells from oxidative stress caused by the glutamate analog N-methyl-D-aspartic acid (NMDA) [64]. In this study, expression of Mt1 and Mt2 is more elevated in the inferior retina that has more tissue damage. The prominence of antioxidants in the retina’s response to hyperoxia indicates the importance of free radicals in the damage caused [14]. Conversely, the vulnerability of the inferior retina does not seem to be due to lack of antioxidants in that region. However, the upregulation of Mt1 and Mt2 are not sufficient for protecting the inferior retina against hyperoxia-induced cell death. This is consistent with previous findings that Mt1 and Mt2 alone are insufficient for protecting photoreceptors against hyperbaric oxygen-induced cell death [16].
Acknowledgments
This research was supported by the Australian Research Council through the ARC Centre of Excellence in Vision Science (CE0561903) and by a grant-in-aid from the Sir Zelman Cowen Universities Fund. We thank Mr. Stephen Ohms for statistical advice and Dr. Kaiman Peng from the ACRF Biomolecular Resource Facility in John Curtin School of Medical Research at The Australian National University.
References
- 1.Graymore C. Metabolism of the developing retina. Br J Ophthalmol. 1959;43:34–9. doi: 10.1136/bjo.43.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan-Ling T, Gock B, Stone J. The effect of oxygen on vasoformative cell division. Evidence that “physiological hypoxia” is the stimulus for normal retinal vasculogenesis. Invest Ophthalmol Vis Sci. 1995;36:1201–14. [PubMed] [Google Scholar]
- 3.Stone J, Itin A, Alon T, Pe'er J, Gnessin H, Chan-Ling T, Keshet E. Development of retinal vasculature is mediated by hypoxia-induced vascular endothelial growth factor (VEGF) expression by neuroglia. J Neurosci. 1995;15:4738–47. doi: 10.1523/JNEUROSCI.15-07-04738.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maslim J, Valter K, Egensperger R, Hollander H, Stone J. Tissue oxygen during a critical developmental period controls the death and survival of photoreceptors. Invest Ophthalmol Vis Sci. 1997;38:1667–77. [PubMed] [Google Scholar]
- 5.Wellard J, Lee D, Valter K, Stone J. Photoreceptors in the rat retina are specifically vulnerable to both hypoxia and hyperoxia. Vis Neurosci. 2005;22:501–7. doi: 10.1017/S0952523805224112. [DOI] [PubMed] [Google Scholar]
- 6.Noell WK. Visual cell effects of high oxygen pressures. American Physiology Society: Federation Proceedings. 1955;14:107–8. [Google Scholar]
- 7.Yamada H, Yamada E, Ando A, Esumi N, Bora N, Saikia J, Sung CH, Zack DJ, Campochiaro PA. Fibroblast growth factor-2 decreases hyperoxia-induced photoreceptor cell death in mice. Am J Pathol. 2001;159:1113–20. doi: 10.1016/S0002-9440(10)61787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natoli R, Provis J, Valter K, Stone J. Expression and role of the early-response gene Oxr1 in the hyperoxia-challenged mouse retina. Invest Ophthalmol Vis Sci. 2008;49:4561–7. doi: 10.1167/iovs.08-1722. [DOI] [PubMed] [Google Scholar]
- 9.Yamada H, Yamada E, Hackett SF, Ozaki H, Okamoto N, Campochiaro PA. Hyperoxia causes decreased expression of vascular endothelial growth factor and endothelial cell apoptosis in adult retina. J Cell Physiol. 1999;179:149–56. doi: 10.1002/(SICI)1097-4652(199905)179:2<149::AID-JCP5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Mervin K, Valter K, Maslim J, Lewis G, Fisher S, Stone J. Limiting photoreceptor death and deconstruction during experimental retinal detachment: the value of oxygen supplementation. Am J Ophthalmol. 1999;128:155–64. doi: 10.1016/s0002-9394(99)00104-x. see comments. [DOI] [PubMed] [Google Scholar]
- 11.Lewis G, Mervin K, Valter K, Maslim J, Kappel P, Stone J, Fisher S. Limiting the proliferation and reactivity of retinal müller cells during detachment: the value of oxygen supplementation. Am J Ophthalmol. 1999;128:165–72. doi: 10.1016/s0002-9394(99)00103-8. [DOI] [PubMed] [Google Scholar]
- 12.Vingolo EM, Rocco M, Grenga P, Salvatore S, Pelaia P. Slowing the degenerative process, long lasting effect of hyperbaric oxygen therapy in retinitis pigmentosa. Graefes Arch Clin Exp Ophthalmol. 2008;246:93–8. doi: 10.1007/s00417-007-0652-z. [DOI] [PubMed] [Google Scholar]
- 13.Oguz H, Sobaci G. The use of hyperbaric oxygen therapy in ophthalmology. Surv Ophthalmol. 2008;53:112–20. doi: 10.1016/j.survophthal.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Natoli R, Provis J, Valter K, Stone J. Gene regulation induced in the C57BL/6J mouse retina by hyperoxia: a temporal microarray study. Mol Vis. 2008;14:1983–94. [PMC free article] [PubMed] [Google Scholar]
- 15.Smit-McBride Z, Oltjen SL, LaVail MM, Hjelmeland LM. A strong genetic determinant of hyperoxia-related retinal degeneration on mouse chromosome 6. Invest Ophthalmol Vis Sci. 2007;48:405–11. doi: 10.1167/iovs.06-0854. [DOI] [PubMed] [Google Scholar]
- 16.Nachman-Clewner M, Giblin FJ, Dorey CK, Blanks RH, Dang L, Dougherty CJ, Blanks JC. Selective degeneration of central photoreceptors after hyperbaric oxygen in normal and metallothionein-knockout mice. Invest Ophthalmol Vis Sci. 2008;49:3207–15. doi: 10.1167/iovs.07-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers F, Valter K, Chan S, Walsh N, Maslim J, Stone J. Effects of Oxygen and bFGF on the Vulnerability of Photoreceptors to Light Damage. Invest Ophthalmol Vis Sci. 2001;42:804–15. [PubMed] [Google Scholar]
- 18.Tanito M, Kaidzu S, Ohira A, Anderson RE. Topography of retinal damage in light-exposed albino rats. Exp Eye Res. 2008;87:292–5. doi: 10.1016/j.exer.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Liang L, Katagiri Y, Franco LM, Yamauchi Y, Enzmann V, Kaplan HJ, Sandell JH. Long-term cellular and regional specificity of the photoreceptor toxin, iodoacetic acid (IAA), in the rabbit retina. Vis Neurosci. 2008;25:167–77. doi: 10.1017/S0952523808080401. [DOI] [PubMed] [Google Scholar]
- 20.LaVail MM, Matthes M, Yasumura D, Steinberg R. Variability in rate of cone degeneration in the retinal degeneration (rd/rd) Mouse. Exp Eye Res. 1997;65:45–50. doi: 10.1006/exer.1997.0308. [DOI] [PubMed] [Google Scholar]
- 21.Walsh N, Bravo-Nuevo A, Geller S, Stone J. Resistance of photoreceptors in the C57BL/6-c2J, C57BL/6J, and BALB/cJ mouse strains to oxygen stress: evidence of an oxygen phenotype. Curr Eye Res. 2004;29:441–7. doi: 10.1080/02713680490522416. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin T, O'Leary DD. Molecular gradients and development of retinotopic maps. Annu Rev Neurosci. 2005;28:327–55. doi: 10.1146/annurev.neuro.28.061604.135714. [DOI] [PubMed] [Google Scholar]
- 24.Barbieri AM, Lupo G, Bulfone A, Andreazzoli M, Mariani M, Fougerousse F, Consalez GG, Borsani G, Beckmann JS, Barsacchi G, Ballabio A, Banfi S. A homeobox gene, vax2, controls the patterning of the eye dorsoventral axis. Proc Natl Acad Sci USA. 1999;96:10729–34. doi: 10.1073/pnas.96.19.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koshiba-Takeuchi K, Takeuchi JK, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, Ogura T. Tbx5 and the retinotectum projection. Science. 2000;287:134–7. doi: 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- 26.Linberg KA, Lewis GP, Shaaw C, Rex TS, Fisher SK. Distribution of S- and M-cones in normal and experimentally detached cat retina. J Comp Neurol. 2001;430:343–56. doi: 10.1002/1096-9861(20010212)430:3<343::aid-cne1035>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 27.Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–23. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 28.Szel A, Rohlich A, Caffe B, Aguirre J, Van Veen T. Unique topographic separation of two spectral classes of cones in the mouse retina. J Comp Neurol. 1992;325:327–42. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- 29.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshet E. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 30.Grimm C, Wenzel A, Groszer M, Mayser H, Seeliger M, Samardzija M, Bauer C, Gassmann M, Reme CE. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat Med. 2002;8:718–24. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 31.Hindges R, McLaughlin T, Genoud N, Henkemeyer M, O'Leary DD. EphB forward signaling controls directional branch extension and arborization required for dorsal-ventral retinotopic mapping. Neuron. 2002;35:475–87. doi: 10.1016/s0896-6273(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 32.Turner AJ, Isaac RE, Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays. 2001;23:261–9. doi: 10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Xu D, Emoto N, Giaid A, Slaughter C, Kaw S, deWit D, Yanagisawa M. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994;78:473–85. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi M, Matsushita Y, Iijima Y, Tanzawa K. Purification and characterization of endothelin-converting enzyme from rat lung. J Biol Chem. 1993;268:21394–8. [PubMed] [Google Scholar]
- 35.Lebel N, D'Orleans-Juste P, Fournier A, Sirois P. Role of the neutral endopeptidase 24.11 in the conversion of big endothelins in guinea-pig lung parenchyma. Br J Pharmacol. 1996;117:184–8. doi: 10.1111/j.1476-5381.1996.tb15172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Iwata N, Tsubuki S, Takaki Y, Takano J, Huang SM, Suemoto T, Higuchi M, Saido TC. Somatostatin regulates brain amyloid beta peptide Abeta42 through modulation of proteolytic degradation. Nat Med. 2005;11:434–9. doi: 10.1038/nm1206. [DOI] [PubMed] [Google Scholar]
- 37.Nork TM, Kim CB, Shanmuganayagam D, Van Lysel MS, Ver Hoeve JN, Folts JD. Measurement of regional choroidal blood flow in rabbits and monkeys using fluorescent microspheres. Arch Ophthalmol. 2006;124:860–8. doi: 10.1001/archopht.124.6.860. [DOI] [PubMed] [Google Scholar]
- 38.Chung HS, Harris A, Halter PJ, Kagemann L, Roff EJ, Garzozi HJ, Hosking SL, Martin BJ. Regional differences in retinal vascular reactivity. Invest Ophthalmol Vis Sci. 1999;40:2448–53. [PubMed] [Google Scholar]
- 39.Zhu Y, Valter K, Stone J. Environmental Damage to the Retina and Preconditioning: Contrasting Effects of Light and Hyperoxic Stress. Invest Ophthalmol Vis Sci. 2010 doi: 10.1167/iovs.09-5050. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Wu W, Dentchev T, Zeng Y, Wang J, Tsui I, Tobias JW, Bennett J, Baldwin D, Dunaief JL. Light damage induced changes in mouse retinal gene expression. Exp Eye Res. 2004;79:239–47. doi: 10.1016/j.exer.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Inoue A, Yanagisawa M, Kimura S, Kasuya Y, Miyauchi T, Goto K, Masaki T. The human endothelin family: three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc Natl Acad Sci USA. 1989;86:2863–7. doi: 10.1073/pnas.86.8.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agapitov AV, Haynes WG. Role of endothelin in cardiovascular disease. J Renin Angiotensin Aldosterone Syst. 2002;3:1–15. doi: 10.3317/jraas.2002.001. [DOI] [PubMed] [Google Scholar]
- 43.Howard PG, Plumpton C, Davenport AP. Anatomical localization and pharmacological activity of mature endothelins and their precursors in human vascular tissue. J Hypertens. 1992;10:1379–86. doi: 10.1097/00004872-199211000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Plumpton C, Champeney R, Ashby MJ, Kuc RE, Davenport AP. Characterization of endothelin isoforms in human heart: endothelin-2 demonstrated. J Cardiovasc Pharmacol. 1993;22(Suppl 8):S26–8. doi: 10.1097/00005344-199322008-00009. [DOI] [PubMed] [Google Scholar]
- 45.Rattner A, Nathans J. The genomic response to retinal disease and injury: Evidence for endothelin signaling from photoreceptors to glia. J Neurosci. 2005;25:4540–9. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joly S, Lange C, Thiersch M, Samardzija M, Grimm C. Leukemia inhibitory factor extends the lifespan of injured photoreceptors in vivo. J Neurosci. 2008;28:13765–74. doi: 10.1523/JNEUROSCI.5114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–32. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 48.Johnson LV, Leitner WP, Rivest AJ, Staples MK, Radeke MJ, Anderson DH. The Alzheimer's A beta -peptide is deposited at sites of complement activation in pathologic deposits associated with aging and age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:11830–5. doi: 10.1073/pnas.192203399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bora PS, Sohn JH, Cruz JM, Jha P, Nishihori H, Wang Y, Kaliappan S, Kaplan HJ, Bora NS. Role of complement and complement membrane attack complex in laser-induced choroidal neovascularization. J Immunol. 2005;174:491–7. doi: 10.4049/jimmunol.174.1.491. [DOI] [PubMed] [Google Scholar]
- 50.Terai K, Walker D, McGeer E, McGeer P. Neurons express proteins of the classical complement pathway in Alzheimer disease. Brain Res. 1997;769:385–90. doi: 10.1016/s0006-8993(97)00849-4. [DOI] [PubMed] [Google Scholar]
- 51.Rohrer B, Demos C, Frigg R, Grimm C. Classical complement activation and acquired immune response pathways are not essential for retinal degeneration in the rd1 mouse. Exp Eye Res. 2007;84:82–91. doi: 10.1016/j.exer.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stasi K, Nagel D, Yang X, Wang RF, Ren L, Podos SM, Mittag T, Danias J. Complement component 1Q (C1Q) upregulation in retina of murine, primate, and human glaucomatous eyes. Invest Ophthalmol Vis Sci. 2006;47:1024–9. doi: 10.1167/iovs.05-0830. [DOI] [PubMed] [Google Scholar]
- 53.Kuehn MH, Kim CY, Ostojic J, Bellin M, Alward WL, Stone EM, Sakaguchi DS, Grozdanic SD, Kwon YH. Retinal synthesis and deposition of complement components induced by ocular hypertension. Exp Eye Res. 2006;83:620–8. doi: 10.1016/j.exer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 55.Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–61. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 56.Miyamoto T, Inoue H, Sakamoto Y, Kudo E, Naito T, Mikawa T, Mikawa Y, Isashiki Y, Osabe D, Shinohara S, Shiota H, Itakura M. Identification of a novel splice site mutation of the CSPG2 gene in a Japanese family with Wagner syndrome. Invest Ophthalmol Vis Sci. 2005;46:2726–35. doi: 10.1167/iovs.05-0057. [DOI] [PubMed] [Google Scholar]
- 57.Wagner DH. Spivack's Gastrostomy: An Analysis of Thirteen Cases with a Review of the Technic. Ann Surg. 1938;107:1005–21. doi: 10.1097/00000658-193806000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y, Wu J, Lee DY, Yee A, Cao L, Zhang Y, Kiani C, Yang BB. Versican protects cells from oxidative stress-induced apoptosis. Matrix Biol. 2005;24:3–13. doi: 10.1016/j.matbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol. 2003;121:1316–23. doi: 10.1001/archopht.121.9.1316. [DOI] [PubMed] [Google Scholar]
- 60.Palmiter RD. The elusive function of metallothioneins. Proc Natl Acad Sci USA. 1998;95:8428–30. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–94. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- 62.Thiersch M, Raffelsberger W, Frigg R, Samardzija M, Wenzel A, Poch O, Grimm C. Analysis of the retinal gene expression profile after hypoxic preconditioning identifies candidate genes for neuroprotection. BMC Genomics. 2008;9:73. doi: 10.1186/1471-2164-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu H, Hunt DM, Ganti R, Davis A, Dutt K, Alam J, Hunt RC. Metallothionein protects retinal pigment epithelial cells against apoptosis and oxidative stress. Exp Eye Res. 2002;74:83–92. doi: 10.1006/exer.2001.1101. [DOI] [PubMed] [Google Scholar]
- 64.Suemori S, Shimazawa M, Kawase K, Satoh M, Nagase H, Yamamoto T, Hara H. Metallothionein, an endogenous antioxidant, protects against retinal neuron damage in mice. Invest Ophthalmol Vis Sci. 2006;47:3975–82. doi: 10.1167/iovs.06-0275. [DOI] [PubMed] [Google Scholar]