Abstract
Purpose
The role of SRY-Box 2 (SOX2) in anophthalmia/microphthalmia (A/M) is well known, with 10%–20% of A/M explained by mutations in SOX2. SOX2 plays roles in the development of both the posterior and anterior segment structures of the eye and relies on interactions with tissue-specific partner proteins to execute its function, raising the possibility that SOX2 mutations may result in varying ocular phenotypes. Recent data has identified a missense mutation in SOX2 in an extended pedigree with phenotypes as varied as A/M, isolated iris hypoplasia, iris and chorioretinal coloboma, pupil defects, and hypermetropia, suggesting a broader phenotypic spectrum associated with SOX2 mutations.
Methods
Screening of SOX2 was completed in 89 patients with a variety of ocular anomalies, including 28 with A/M and 61 with normal eye size and anterior segment dysgenesis (28), cataract (14), isolated coloboma (5), or other eye disorders (14).
Results
The recurrent de novo frameshift mutation c.70del20 was identified in one patient with microphthalmia and syndromic anomalies consistent with SOX2 anophthalmia syndrome; the mutation frequency in our A/M population (4%) was lower than previously reported; it is likely that extensive utilization of clinical SOX2 testing has led to a bias toward SOX2-negative A/M cases in our research cohort. No disease-causing mutations were identified in patients with non-microphthalmia phenotypes.
Conclusions
The recurrent c.70del20 mutation accounts for 21% of all independent SOX2 mutations reported to date. Due to the increased use of clinical SOX2 testing, the frequency of SOX2 mutations identified in research A/M populations will likely continue to decrease. Mutations in SOX2 do not appear to be a common cause of ocular defects other than anophthalmia/microphthalmia.
Introduction
SOX2 is a High Mobility Group (HMG) DNA binding domain-containing transcription factor with a well established role as a common cause of dominant anophthalmia/microphthalmia (A/M). SOX2/Sox2 expression begins early in eye development and is critical for the formation of different ocular tissues [1-4]. In mice, Sox2 is expressed within the optic vesicle and stalk, the developing neural retina, the placodal area of the surface ectoderm, and throughout lens development starting with the lens placode and continuing through differentiating lens fibers [1,2,4]. Studies of human fetal tissues are consistent with the mouse data; SOX2 expression is seen in the developing neural retina and optic stalk as well as during all stages of lens development [3].
SOX proteins typically contain three functional domains: the High Mobility Group (HMG) domain that is responsible for DNA binding, an activation or repression domain, and a partner-factor interaction domain. SOX2, like other SOX proteins, requires interaction with partner factors to successfully regulate target genes, likely due to low-affinity DNA-binding by SOX proteins alone [5]. Partner factors bind to the regulatory sequence near the site of SOX binding and also interact with the SOX protein, thus forming a ternary complex. This complex stabilizes the interaction between the SOX protein and DNA and allows specific regulation of diverse target genes [5]. For example, SOX2 has been shown to partner with δ-crystallin enhancer factor 3 (δEF3) as well as paired box gene 6 (PAX6) to activate δ-crystallin expression during lens formation [6,7] and with octomer-binding transcription factor 3/4 (Oct3/4) to regulate fibroblast growth factor 4 (FGF4) or undifferentiated embryonic cell transcription factor 1 (UTF1) target genes in embryonic stem cells [5]. Thus, depending on the affected site/domain, it is possible that some SOX2 mutations may affect certain protein interactions more than others and therefore result in variable and tissue-specific phenotypes.
Mutations in SOX2 account for 10%–20% of A/M [8-11]. Individuals with SOX2 mutations often have associated systemic anomalies, termed SOX2 anophthalmia syndrome, consisting of ocular, brain, pituitary, genitourinary, and gastresophageal anomalies, although eye defects can be isolated as well [8-11]. The anophthalmia/microphthalmia phenotype in patients with SOX2 mutations can be bilateral, unilateral, or, occasionally, absent; 14 of the 71 individuals reviewed previously [11] or subsequently reported [12] do not have anophthalmia/microphthalmia of either eye. Severe SOX2 mutations (whole gene deletion/nonsense) which most likely produce complete loss-of-function alleles almost uniformly result in anophthalmia/microphthalmia (47 out of 50 cases [11]), suggesting that these mutations result in a major disruption of eye development, while missense mutations are more likely to be associated with non-microphthalmia phenotypes (11 of 21 cases [11,12]).
Several families with missense mutations and non-microphthalmia phenotypes have been previously reported including two probands with optic nerve hypoplasia and their unaffected fathers [13], a father and son with iris and chorioretinal uveal colobomas [14], and, more recently, a large four-generation pedigree with diverse phenotypes, including microphthalmia in some individuals [12]. The latest report describes a mutation in a four-generation pedigree that results in the substitution of a highly conserved amino acid within the partner-factor interaction region of the SOX2 protein and is the first mutation reported in this domain. While the proband was affected with bilateral anophthalmia, the other eight family members carrying the mutation were affected with milder ocular phenotypes. Five of the affected individuals did not have anophthalmia or microphthalmia. The proband’s mother had bilateral iris and chorioretinal colobomata, the maternal aunt was found to have a small inferior retinal tuft in one eye along with hypermetropia and astigmatism, and the proband’s brother was affected with hypermetropia only. Finally, the maternal grandmother was affected with iris hypoplasia and hypermetropia while a maternal great-aunt had microcornea only. In addition to the two cases above with isolated anterior segment defects (iris hypoplasia and microcornea), two individuals demonstrated anterior segment defects (posterior embryotoxon and an iris pupillary defect) in the presence of microphthalmia, further suggesting that SOX2 plays a role in anterior segment development. Several family members who did not carry the mutation had normal eye examinations [12].
As discussed above, the variability of phenotypes associated with, in particular, missense changes in SOX2 may be explained by the fact that these mutations result in alteration of its interactions with tissue-specific protein partners rather than a complete loss-of-function [5,12-14]. In addition to this, the genetic makeup of these partner factors, other ocular development genes, or yet unknown factors are likely to play role in the phenotypic expression of SOX2 mutations. Since expression during lens development represents an important site of SOX2 activity, it is possible that some mutations may lead to phenotypes associated with specific loss of SOX2-related activity in the lens, such as cataracts and anterior segment defects.
Mutations in SOX2 may result in a broader phenotypic spectrum consistent with its role in both posterior and anterior segment development and dependent on the affected partner interaction domain. The role of SOX2 in A/M has been well characterized while its potential involvement in other eye conditions has not yet been investigated. To further explore the role of SOX2 in variable ocular diseases, we undertook screening of this gene in our population of patients with a variety of ocular defects.
Methods
This human study was approved by the Institutional Review Boards of Children's Hospital of Wisconsin and Albert Einstein Healthcare Network. Cases were identified and clinical data collected through the Genetic Studies of Human Developmental Disorders at the Medical College of Wisconsin, Milwaukee, WI or the A/M Clinical Registry at the Albert Einstein Medical Center, Philadelphia, PA. Genomic DNA was extracted using the Autopure LS system (Qiagen, Valencia, CA) from blood or buccal samples and the SOX2 coding region was amplified using primers: set 1 forward, 5'-AGT CCC GGC CGG GCC GAG-3', and set 1 reverse, 5'-GGT AGC CCA GCT GGT CCT G-3', and set 2 forward, 5'-CAA GAC GCT CAT GAA GAA GG-3', and set 2 reverse, 5'-TAC TCT CCT CTT TTG CAC CC-3'. PCR products were sequenced in both directions using Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA) with 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). Chromatograms were examined manually and using Mutation Surveyor software (SoftGenetics, State Collge, PA). All initially identified changes were confirmed by additional independent PCR and sequencing experiments.
The screening included 89 patients with a variety of ocular disorders (Table 1). Twenty-eight patients were affected with anophthalmia/microphthalmia (17 bilateral, 17 syndromic). An additional 28 patients had anterior segment dysgenesis (22 bilateral, 20 syndromic); eight of these patients were specifically noted to have iris hypoplasia (two with full Axenfeld-Rieger syndrome), four were noted to have pupil anomalies, 11 had Peters’ anomaly (one with iris coloboma in addition), and the remaining five had other anterior segment defects. There were five patients with isolated coloboma (three were iris and chorioretinal). Finally, six patients had isolated high myopia, four patients had glaucoma with no other ocular anomalies (three congenital, one adult-onset), 14 had cataract with no other ocular anomalies (11 congenital, two juvenile, and one adult-onset), and the remaining four had other eye defects (including microspherophakia, Septo-optic dysplasia, and Morning Glory disc anomaly).
Table 1. Summary of ocular conditions and SOX2 mutations reported in this study.
| Ocular condition | Number of cases | Bilateral | Syndromic | Number of mutations |
|---|---|---|---|---|
| Anophthalmia |
8 |
6/8 |
6/8 |
0 |
| Microphthalmia |
20 |
11/20 |
11/20 |
1 |
|
A/M total |
28 |
17/28 |
17/28 |
1/28 |
| Anterior segment dysgenesis |
28 |
22/28 |
20/28 |
0 |
| Cataract |
14 |
14/14 |
1/14 |
0 |
| High myopia |
6 |
6/6 |
0/6 |
0 |
| Coloboma (isolated) |
5 |
3/5 |
0/5 |
0 |
| Glaucoma |
4 |
4/4 |
2/4 |
0 |
| Other eye defect |
4 |
4/4 |
4/4 |
0 |
|
Non-A/M total |
61 |
53/61 |
27/61 |
0/61 |
| Total number | 89 | 70/89 | 44/89 | 1/89 |
Paired homeodomain transcription factor 2 (PITX2) was previously screened in 26 of the 28 patients with anterior segment dysgenesis with no mutations identified. Forkhead box E3 (FOXE3) was previously screened in 20 of the 28 patients with anterior segment dysgenesis (including 9 of 11 with Peters’ anomaly) with no mutations identified. Two patients with mutations in SOX2 identified via clinical testing were known to decline enrollment in this research study during this period of patient recruitment.
Results and Discussion
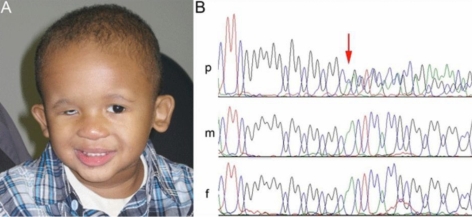
Screening of these 89 patients identified a disease-causing mutation in one patient. The recurrent frameshift mutation in SOX2, c.70del20, was identified in Patient 1 of African American decent who was affected with microphthalmia and optic nerve hypoplasia in the right eye but had a normal left eye; in addition to ocular defects, the patient displayed micropenis, cryptorchidism, prostatic utricle, pancreatic deficiency, low-set prominent ears, umbilical hernia, feeding disorder, and global developmental delay, consistent with SOX2 anophthalmia syndrome (Figure 1A,B). This mutation is predicted to result in a frameshift and truncation of normal SOX2 protein at amino acid 23 (7% of its total length), before the HMG DNA-binding domain. The parents of Patient 1 are unaffected and were found to carry normal SOX2 alleles consistent with the de novo phenotype observed in their child (Figure 1B). The recurrent c.70del20 mutation has now been reported in eleven patients from ten unrelated pedigrees, typically resulting in a severe phenotype characterized by bilateral anophthalmia/microphthalmia and systemic anomalies (Table 2). The family reported in this study represents only the second case of unilateral eye defects associated with the c.70del20 mutation; the previously described pedigree includes a patient with unilateral ocular phenotype and a sibling with normal eyes bilaterally [15]. Also, to the best of our knowledge, this is the first reported occurrence of the c.70del20 mutation in an African American patient, but several previous reports did not include race/ethnicity data. The c.70del20 mutation appears to be the major recurrent mutation in SOX2; one other mutation, c.529C>T, has been noted in three unrelated families [8,13] while the remaining 34 reported mutations are unique [11]. Two similar deletions, a 17-base pair deletion at the same nucleotide position, and a 23-base pair deletion in the codon before, have each been reported in one family [9,16]. Slipped-strand mispairing has been implicated as the likely mechanism of this recurrent mutation due to the presence of a GGCGGC repeat sequence flanking the region [17]. These 17 to 23-base pair deletions account for 25% of all SOX2 mutations in probands (12/48) with the c.70del20 mutation contributing 21% (10/48). This warrants special attention to this region in A/M patients being evaluated for SOX2 mutations.
Figure 1.
Identification of a c.70del20 mutation in a patient with unilateral microphthalmia. A: Photograph of Patient 1 with SOX2 anophthalmia syndrome. Note right microphthalmia (prosthesis in place) and prominent ears. B: Sequence fragments showing the c.70del20 region in the patient (p), his mother (m) and his father (f). The position of the deletion is indicated with a red arrow. Note normal SOX2 sequence in the patient’s parents consistent with their unaffected status.
Table 2. Clinical findings in patients with the recurrent c.70del20 mutation in SOX2.
| Reported case | 1 | 2 | 3 (sibling) | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Right Eye |
AN |
NOR |
NOR |
MI |
AN |
AN |
AN |
MI |
AN |
AN |
MI, ONH |
| Left Eye |
AN |
AN |
NOR |
AN |
ASD, CA, COL, GL |
AN |
AN |
MI |
AN |
AN |
NOR |
| Other Anomalies |
Yes |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
Yes |
| Reference | [17] | [15] | [15] | [12] | [10] | [10] | [3] | [11] | [11] | [11] | This study |
The reported cases are numbered based upon date of publication. In the table AN indicates Anophthalmia; MI indicates Microphthalmia; ASD indicates Anterior segment dysgenesis; CA indicates Cataract; COL indicates Coloboma; GL indicates Glaucoma; NOR indicates Normal; and ONH indicates Optic nerve hypoplasia.
The frequency of SOX2 mutations within our population of patients with anophthalmia/ microphthalmia (1/28=4%) was much lower than has been previously reported [8-11], likely due to increased clinical testing of SOX2 in affected patients before enrollment in a research study. If the two patients known to have a clinically identified mutation in SOX2 are included, the frequency of SOX2 mutations increases to 10% (3/30), which is consistent with previous reports. Given the well known role of SOX2 in anophthalmia/microphthalmia, the easily recognizable associated features, and the availability of clinical sequencing, it seems likely that many physicians may order clinical testing before referring a family to a research study. Thus, the frequency of SOX2 mutations observed in current and future research populations will likely be significantly lower than the actual frequency of mutations in A/M.
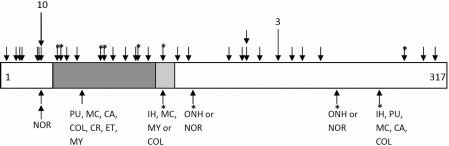
Previously reported mutations in SOX2 associated with A/M or non-A/M phenotypes are scattered throughout the gene with no particular pattern identified (Figure 2). No disease-causing mutations were identified in 61 patients with ocular anomalies other than anophthalmia/microphthalmia, including 20 with phenotypes which specifically match those seen in the family reported by Mihelec et al. [12]: iris hypoplasia, pupil anomalies, coloboma, and optic nerve hypoplasia. In all, 28 patients with anterior segment anomalies and an additional five patients with coloboma were screened with no disease-causing mutations identified. The SOX2 sequence appears to be extremely conserved with only one synonymous polymorphism (c.453G>A) observed in our patient group (also seen in controls).
Figure 2.
Schematic drawing of the SOX2 protein with mutations indicated. Mutations resulting in A/M phenotype (unilateral or bilateral) are shown on the top while mutations resulting in non-microphthalmia phenotypes (bilateral) are indicated below; missense mutations are indicated with asterisks. Each specific mutation is marked with a single arrow, unless the mutation resulted in both A/M and non-A/M ocular phenotypes, in which case the mutation is marked with an arrow above and below. Recurrent mutations are indicated with longer arrows with the number of families listed above. The dark gray box represents the homeodomain and the light gray box a partner-factor interaction domain. In the image, NOR indicates Normal; PU indicates Pupillary abnormality; MC indicates Microcornea; CA indicates Cataract; COL indicates Coloboma; CR indicates Chorioretinal dystrophy; ET indicates Esotropia; MY indicates High myopia; IH indicates Iris hypoplasia; ONH indicates Optic nerve hypoplasia.
Our data suggests that SOX2 mutations are not a common cause of ocular defects other than anophthalmia/microphthalmia. Further studies with larger numbers of different phenotypes may reveal additional mutations, but there is no evidence at this time to suggest that routine screening of SOX2 in patients with other ocular phenotypes is warranted.
Acknowledgments
We are grateful to the families for their participation in this study and to Gabriella Nassif for assistance in screening SOX2 during her training in the laboratory. This project was supported by awards EY015518 and EY015518–04S1 from the National Eye Institute (E.V.S.) and Children’s Research Institute Foundation at Children’s Hospital of Wisconsin grant, GCRC Grant M01-RR00058 from NIH, as well as two grants from Mellon Mid-Atlantic Charitable Trusts, the Albert B. Millett Memorial Fund and the Rae S. Uber Trust, and a grant from the Gustavus and Louis Pfeiffer Research Foundation.
References
- 1.Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–32. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 2.Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–75. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, Gregory JW, Taylor D, Cavallo L, Faienza MF, Fischetto R, Achermann JC, Martinez-Barbera JP, Rizzoti K, Lovell-Badge R, Robinson IC, Gerrelli D, Dattani MT. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93:1865–73. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–32. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–7. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 6.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–86. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–9. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–3. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 9.Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, de Sanctis U, Salt A, Collin JR, Vivian AJ, Free SL, Thompson P, Williamson KA, Sisodiya SM, van Heyningen V, Fitzpatrick DR. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. [DOI] [PubMed] [Google Scholar]
- 10.Bakrania P, Robinson DO, Bunyan DJ, Salt A, Martin A, Crolla JA, Wyatt A, Fielder A, Ainsworth J, Moore A, Read S, Uddin J, Laws D, Pascuel-Salcedo D, Ayuso C, Allen L, Collin JR, Ragge NK. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br J Ophthalmol. 2007;91:1471–6. doi: 10.1136/bjo.2007.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider A, Bardakjian T, Reis LM, Tyler RC, Semina EV. Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am J Med Genet A. 2009;149A:2706–15. doi: 10.1002/ajmg.a.33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mihelec M, Abraham P, Gibson K, Krowka R, Susman R, Storen R, Chen Y, Donald J, Tam PP, Grigg JR, Flaherty M, Gole GA, Jamieson RV. Novel SOX2 partner-factor domain mutation in a four-generation family. Eur J Hum Genet. 2009;17:1417–22. doi: 10.1038/ejhg.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelberman D, Rizzoti K, Avilion A, Bitner-Glindzicz M, Cianfarani S, Collins J, Chong WK, Kirk JM, Achermann JC, Ross R, Carmignac D, Lovell-Badge R, Robinson IC, Dattani MT. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116:2442–55. doi: 10.1172/JCI28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Liang X, Yi J, Zhang Q. Novel SOX2 mutation associated with ocular coloboma in a Chinese family. Arch Ophthalmol. 2008;126:709–13. doi: 10.1001/archopht.126.5.709. [DOI] [PubMed] [Google Scholar]
- 15.Zenteno JC, Perez-Cano HJ, Aguinaga M. Anophthalmia-esophageal atresia syndrome caused by an SOX2 gene deletion in monozygotic twin brothers with markedly discordant phenotypes. Am J Med Genet A. 2006;140:1899–903. doi: 10.1002/ajmg.a.31384. [DOI] [PubMed] [Google Scholar]
- 16.Chassaing N, Gilbert-Dussardier B, Nicot F, Fermeaux V, Encha-Razavi F, Fiorenza M, Toutain A, Calvas P. Germinal mosaicism and familial recurrence of a SOX2 mutation with highly variable phenotypic expression extending from AEG syndrome to absence of ocular involvement. Am J Med Genet A. 2007;143:289–91. doi: 10.1002/ajmg.a.31524. [DOI] [PubMed] [Google Scholar]
- 17.Zenteno JC, Gascon-Guzman G, Tovilla-Canales JL. Bilateral anophthalmia and brain malformations caused by a 20-bp deletion in the SOX2 gene. Clin Genet. 2005;68:564–6. doi: 10.1111/j.1399-0004.2005.00518.x. [DOI] [PubMed] [Google Scholar]