Summary
Proteins delivered to the lysosome or the yeast vacuole via late endosomes are sorted by the ESCRT complexes and by associated proteins, including Alix and its yeast homolog Bro1. Alix, Bro1, and several other late endosomal proteins share a conserved 160 residue Bro1 domain whose boundaries, structure, and function have not been characterized. The crystal structure of the Bro1 domain of Bro1 reveals a folded core of 367 residues. The extended Bro1 domain is necessary and sufficient for binding to the ESCRT-III subunit Snf7 and for the recruitment of Bro1 to late endosomes. The structure resembles a boomerang with its concave face filled in and contains a triple tetratricopeptide repeat domain as a substructure. Snf7 binds to a conserved hydrophobic patch on Bro1 that is required for protein complex formation and for the protein-sorting function of Bro1. These results define a conserved mechanism whereby Bro1 domain-containing proteins are targeted to endosomes by Snf7 and its orthologs.
Introduction
Eukaryotic cells are defined by the presence of a complex set of organelles linked by vesicular trafficking pathways that sort transmembrane proteins and lipids. Multivesicular bodies (MVBs) are late endosomes that contain vesicles within their lumen that are formed by invagination of the limiting membrane (Gruenberg and Stenmark, 2004; Pelham, 2002; Piper and Luzio, 2001). The lumenal vesicles within MVBs are typically destined for delivery into the lysosome in metazoans and into the vacuole in yeast but may also be targeted for extra-cellular release. MVBs are critical loci for sorting in normal and pathogenic processes from yeast to man (Conibear and Stevens, 1995; Lemmon and Traub, 2000). These include the downregulation of cell-surface receptors such as growth factor receptors and yeast mating-pheromone receptors (Hicke, 2001; Katzmann et al., 2002), delivery of lysosomal and vacuolar hydrolases, antigen presentation, and budding of the enveloped viruses HIV and EIAV (Goila-Gaur et al., 2003; Martin-Serrano et al., 2003; Strack et al., 2003; von Schwedler et al., 2003; Goff et al., 2003). Monoubiquitination is a major signal for cargo entry into lumenal MVB vesicles, and many of the cargo proteins are so modified (Hicke, 2001; Katzmann et al., 2002).
The sorting of cargo and budding of vesicles into the lumen of the MVB depends on a network of approximately 18 proteins in yeast (Babst, 2005; Bowers et al., 2004) This network is expanded to roughly 26 proteins in humans (Strack et al., 2003; von Schwedler et al., 2003). In yeast, 12 of these proteins are organized into four large heterooligomeric complexes: the Vps27/Hse1 complex (Hrs/STAM in humans) (Bilodeau et al., 2002; Bilodeau et al., 2003; Raiborg et al., 2002; Katzmann et al., 2003) and the endosomal sorting complex required for transport complexes, ESCRT-I, II, and III (Babst et al., 2002a; Babst et al., 2002b; Katzmann et al., 2001). Several other monomeric or homooligomeric proteins are also required, including the AAA ATPase Vps4 (Babst et al., 1997; Babst et al., 1998), the deubiquitinating enzyme Doa4 (Amerik et al., 2000), and the protein known as Alix in human and Bro1 in yeast (Nikko et al., 2003; Odorizzi et al., 2003; Strack et al., 2003; von Schwedler et al., 2003). In a working model, the sorting of cargoes into MVB vesicles is initiated by Hrs, which is recruited to phosphatidylinositol 3-phosphate-bearing endosomes via its FYVE domain (Raiborg et al., 2001). The Hrs/STAM and ESCRT-I and II complexes are assembled in the cytosol and translocate to endosomes in the presence of appropriate recruitment signals (Babst et al., 2002b; Bache et al., 2003; Katzmann et al., 2001). Monoubiquitinated cargo is recruited by Hrs via its UIM motif (Shih et al., 2002; Swanson et al., 2003) and handed off to ESCRT-I, which binds monoubiquitin via the UEV domain of the Tsg101 (Vps23 in yeast) subunit (Katzmann et al., 2001; Sundquist et al., 2004; Teo et al., 2004). There is second hand-off to ESCRT-II. The yeast ESCRT-II Vps36 subunit contains a monoubiquitin binding NZF domain (Alam et al., 2004), while its mammalian counterpart contains a ubiquitin binding GLUE domain (Slagsvold et al., 2005). The ESCRT-III complex assembles on membranes and is intimately associated with the nascent invaginating vesicle (Babst et al., 2002a). ESCRT-III is involved in recruiting Bro1 (Bowers et al., 2004; Odorizzi et al., 2003; Peck et al., 2004; von Schwedler et al., 2003) as well as Vps4, which catalyzes the disassembly of ESCRT complexes from endosomal membranes (Babst et al., 1998).
The mammalian Bro1 ortholog, Alix, was first discovered as a Ca2+-dependent interactor of the apoptosis protein ALG-2 and was given the names ALG-2-interacting protein 1 (AIP1) (Vito et al., 1999) and Alix (Missotten et al., 1999). Here we use the term Alix because several unrelated proteins are also referred to as “AIP1.” Alix contains a C-terminal Pro-rich region that interacts with the endocytic proteins SETA (Chen et al., 2000; Schmidt et al., 2004), endophilins (Chatellard-Causse et al., 2002), and the ESCRT-I subunit Tsg101 (Strack et al., 2003; von Schwedler et al., 2003). Alix interacts with HIV-1 and other retroviral proteins containing the sequence motif YPXL (Goila-Gaur et al., 2003; Martin-Serrano et al., 2003; Strack et al., 2003; von Schwedler et al., 2003), and its Aspergillus homolog PalA interacts with a YPXL/I-containing host protein, PacC (Vincent et al., 2003). Finally, Alix interacts with the ESCRT-III subunit, CHMP4b (Katoh et al., 2003; Katoh et al., 2004; Strack et al., 2003; von Schwedler et al., 2003). MVBs have been reported to be extraordinarily rich in the unusual phospholipid 2,2′ lysobisphosphatidic acid (2,2′ LBPA), and Alix has attracted attention as a possible target for this lipid (Matsuo et al., 2004). The yeast homolog of Alix, Bro1, is involved in deubiquitination of the general amino acid permease Gap1 (Nikko et al., 2003) and is required for trafficking of carboxypeptidase S via the MVB pathway (Odorizzi et al., 2003). Bro1 recruits Doa4, which deubiquitinates MVB cargo proteins (Luhtala and Odorizzi, 2004). Yeast Bro1 lacks a PTAP sequence and does not interact strongly with Vps23 (Bowers et al., 2004), but Bro1 does bind to the yeast CHMP4 homolog Snf7 (Bowers et al., 2004; Odorizzi et al., 2003).
Alix and Bro1 have in common a conserved 160 amino acid motif near their N terminus that is annotated as a “Bro1 domain” in the Pfam database (Bateman et al., 2002). Pfam lists 54 proteins containing this domain including Bro1, Alix, Rim20, Rhophilin, the protein tyrosine phosphatase HD-PTP, and Xp95, and their orthologs. Among these, Bro1, Alix, and Rim20 are known to associate with late endosomes and with ESCRT-III. The specific function of this highly conserved domain in Alix and Bro1 is not known. Mutation of residues within the Bro1 region of Rim20 interferes with its recruitment to endosomes and its interactions with Snf7 (Xu et al., 2004). Here we show that the folded core of the conserved Bro1 domain extends over nearly 370 residues and that a part of it has limited similarity to the tetratricopeptide repeat (TPR) domain. We find that the Bro1 domain is necessary and sufficient for the endosomal localization of Bro1 and is responsible for its interaction with Snf7. We go on to map the site of the Snf7 interaction on Bro1 to a hydrophobic surface region. This region is conserved in other Bro1 domain proteins, suggesting that this recruitment mechanism is a general property of Bro1 domains.
Results
Structure of the Bro1 Domain
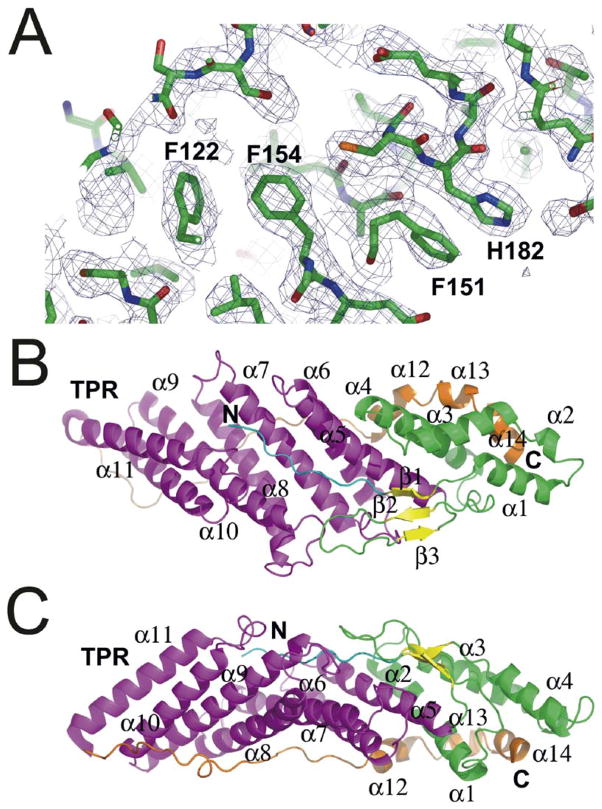
Seventeen constructs from Bro1 and its homologs Alix, Rim20, Rhophilin, and HD-PTP were made and tested for expression of soluble and folded proteins. A construct consisting of Bro1 residues 1–387 was soluble and yielded diffraction-quality crystals. The structure of Bro1 1–387 was determined by multiwavelength anomalous dispersion (MAD) from selenomethionyl protein at 2.3 Å (Figure 1A). The protein structure was refined against native data to a working R factor of 0.224 and a free R factor of 0.252 at 1.95 Å (Table 1). The entire structure could be visualized except for residues after 367, which are presumed disordered. There are two loosely associated protein monomers in the asymmetric unit. The limited interactions between them are consistent with a crystal contact and with the observation that this construct is a monomer in solution.
Figure 1. Overall Structure of Bro1.
(A) Electron density from MAD map contoured at 1.0 σ.
(B and C) Two views of the overall structure. The N-terminal region is colored cyan; the N-terminal (non-TPR) portion of the helical solenoid is colored green; the β sheet is colored yellow; the TPR domain is colored magenta; and the C-terminal region is colored orange.
Table 1.
Crystallographic Data, Phasing, and Refinement Statistics
| Native | Edge | Peak | Remote | ||
|---|---|---|---|---|---|
| Data Collection and Phasing | |||||
| Wavelength | 0.9999 | 0.9796 | 0.9790 | 0.9720 | |
| Space group | P21 | P21 | |||
| Unit cell | a | 76.4 | 75.4 | ||
| b | 58.3 | 57.7 | |||
| c | 120.8 | 115.4 | |||
| β | 90.79 | 91.176 | |||
| Resolution | 1.95 | 2.3 | 2.3 | 2.4 | |
| Rmerge | 0.065 | 0.064 | 0.066 | 0.056 | |
| Completeness | 93.3 | 94.7 | 95.0 | 95.2 | |
| Unique reflections | 73,172 | 44,455 | 44,530 | 39,403 | |
| Refinement | |||||
| Rwork | 0.224 | ||||
| Rfree | 0.252 | ||||
| Cross-validated Luzzati error | 0.30 | ||||
| Rmsd, bonds | 0.006 | ||||
| Rmsd, angles | 1.1 | ||||
| Mean B | 34.2 | ||||
| Wilson B | 18.6 | ||||
The overall structure of the Bro1 N-terminal half resembles a boomerang or banana shape with 30 × 38 × 100 Å dimensions (Figures 1B and 1C). The structure has a total of 14 α helices and three β sheets (Figure 2A). The great bulk of the structure, from residues 17 to 321, consists of a single integrated 11-helical solenoid, right-handed except for the first connection. The N-terminal eight residues fill in a pocket on the convex side of the banana. Residues 10–14, 90–96, and 106–110 form a 3-stranded antiparallel β sheet that packs against the N-terminal half of the helical solenoid. The concave face of the boomerang is filled in by the C-terminal residues 322–367. Residues 322–344 form an almost completely extended loop that spans ~50 Å. The last ordered residues, 345–367, form three short α helices, α12-α14. Pro-350 forms a kink between α12 and α13, which might otherwise form a single contiguous helix. Helix α14 packs roughly coaxially with the helices of the solenoid, while α12 and α13 are roughly orthogonal to the helices of the solenoid.
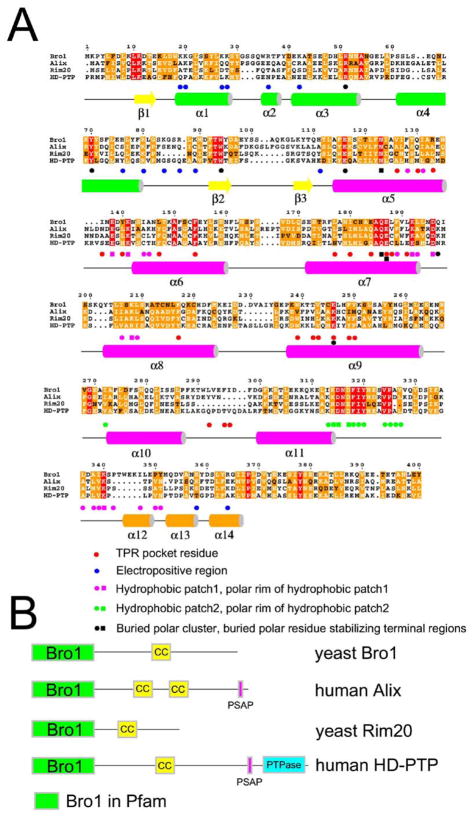
Figure 2. Homology of Bro1 Domains.
(A) Structural and sequence alignment of Bro1 domains. Key to symbols: red circle, TPR pocket residue; blue circle, electropositive region; magenta circles and squares, hydrophobic patch 1 and polar rim of hydrophobic patch 1, respectively; green circles and squares, hydrophobic patch 2 and polar rim of hydrophobic patch 2, respectively; black circle, buried polar cluster residues; black squares, buried polar residues stabilizing the N- or C-terminal regions, excluding TPR pocket residues.
(B) Domain structures of selected Bro1 domain proteins.
The entire assembly is stabilized by extensive hydrophobic interactions and by two buried charged and polar clusters. The first cluster centers on the conserved residues Arg-51, Tyr-70, Trp-94, and Glu-116, and the second on Glu-197 and Lys-246 (Figure 2A). The unpartnered main chain groups in the extended N and C termini are stabilized by interactions with some of the most conserved side chains in the Bro1 domain. The conserved residues Asn-123 and Glu-187 stabilize the extended N terminus, and Gln-186 stabilizes the extended C terminus. The critical roles of the N- and C-terminal extended regions explain why small deletions at either end of the domain lead to expression of insoluble proteins. The sequences of Bro1 homologs are well-conserved throughout the 367 ordered residues in the structure. In a structural and evolutionary sense, the Bro1 homology domain thus extends over 367 residues, rather than 160 as assigned previously (Bateman et al., 2002). Henceforth we will refer to this 367 residue unit, as opposed to the 160 residue unit, as the Bro1 domain (Figure 2B).
Bro1 Contains a TPR Substructure
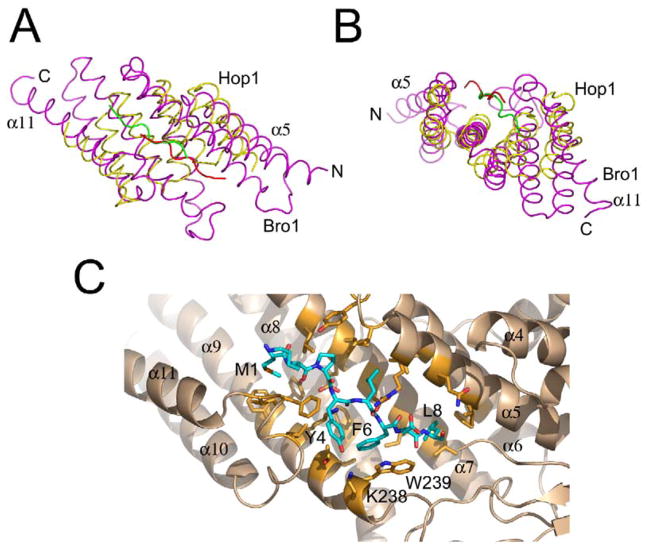
The top-scoring proteins identified by a search of the database of known structures with VAST (Gibrat et al., 1996) were all TPR domain proteins. TPR domains contain 2–16 repeats of 34 amino acids (Sikorski et al., 1990), each corresponding to two α helices (Das et al., 1998). The top hits, in order of Z score, were a synthetic designed TPR protein (Main et al., 2003), the TPR domain of O-linked GlnNac transferase (Jinek et al., 2004), the TPR1 domain of Hop1 (Scheufler et al., 2000), the peptide substrate binding domain of human type I collagen prolyl 4-hydroxylase (Pekkala et al., 2004), the third TPR domain of PEX5 (Gatto et al., 2000; Kumar et al., 2001), the TPR domain of the Ser/Thr protein phosphatase 5 (Das et al., 1998), and the Rac binding TPR domain of p67-Phox (Lapouge et al., 2000). A representative alignment was achieved with 68 Cα positions spanning Bro1 residues 123–259, which deviate by 1.9 Å rms from their counterparts in the designed TPR protein (Main et al., 2003). Taking a consensus of various alignments, the substructure of the Bro1 domain comprising residues 118–316 can be considered a TPR domain. The Bro1 TPR domain substructure contains three pairs of α helices (α5 and α6; α7 and α8; and α9 and α10) that correspond structurally to three TPR repeats. The Bro1 helices are 22–30 residues long, as compared to the 12–15 residues typical of TPR domains.
The TPR substructure of Bro1 contains a deep cleft that corresponds roughly to the Hsp90 peptide binding site in the TPR domain of Hop1 (Figures 3A and 3B; Scheufler et al., 2000) and the PTS1 binding site of PEX5 (Gatto et al., 2000). The cleft is at the center of the convex bend in the middle of the boomerang. The floor of the pocket is formed by α7 and the walls by α5, α9, and the α10-α11 linker. The pocket is filled in by the eight N-terminal residues (Figure 3C). Met-1 is half buried in a hydrophobic pocket made up by Leu-191, Phe-249, Tyr-250, and Ile-296. Pro-3 occupies a shallow pocket made of Ile-136, Tyr-140, Leu-188, and Leu-191. The Tyr-4 side chain hydroxyl makes a water-mediated hydrogen bond with Lys-238 and fills a hydrophobic pocket consisting of Thr-242, Leu-292, and Phe-295. Leu-5 fills a hydrophobic pocket consisting of the aliphatic part of Arg-133, Ile-136, Leu-147, and Leu-188. Phe-6 stacks with the conserved Trp-239 and also contacts Ile-180, Phe-217, and Val-243. Leu-8 fills a large hydrophobic pocket made of Val-126, Phe-154, Leu-177, and Trp-239. Main chain hydrogen bonds are formed between the backbone of the N terminus and the free carbonyl group of Phe-295 in the α10-α11 linker; waters linked to Glu-184 and Glu-187 on the pocket floor; and the side chains of Gln-130, Glu-184, and Lys-246. The details of binding in the Hop1 TPR domain/Hsp90 peptide complex (Scheufler et al., 2000) are quite different, apart from the gross similarity in the location of the sites. The Hsp90 peptide is bound in the opposite orientation and the main chain positions are about 5 Å apart.
Figure 3. The TPR Domain Substructure.
(A and B) Structural overlay between Bro1 (magenta) and Hop1 (yellow), with the Hsp90 pentapeptide bound to Hop1 shown in green.
(C) Details of interactions between the N terminus of Bro1 and the TPR-like pocket.
In order to examine whether the association of the N terminus with the TPR-like pocket was a dynamic autoinhibitory mechanism or a “permanent” part of the folded Bro1 domain, we mutated the N terminus and the TPR-like pocket. A Bro1 construct in which the first 12 residues were deleted was insoluble when expressed, suggesting that this region is important for protein stability. The mutations K246A, W239A, and K246A/W239A targeted key interactions within the TPR-like pocket. These mutations had no effect on carboxypeptidase S sorting (data not shown). These data suggest that the TPR-like pocket is not involved in functionally essential interprotein interactions but is involved in stabilizing intraprotein interactions between different parts of the Bro1 domain.
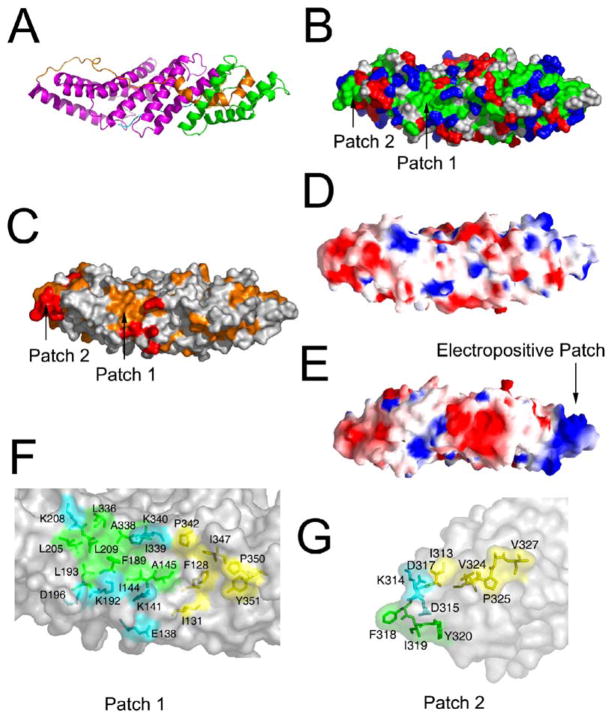
Functional Interaction Surfaces
In addition to the TPR-like pocket, the structure revealed two other prominent hydrophobic surfaces as potential interaction sites (Figures 4B and 4C). Surface 1 is in the middle of the filled-in concave side of the boomerang (Figures 4B and 4C). This patch includes parts of the back side of the TPR substructure and the C-terminal loop. The exposed side chains of Ile-144, Ala-145, Phe-189, Leu-193, Leu-206, Leu-209, Leu-336, Ala-338, and Ile-339 are contiguous with one another and make up a single unbroken hydrophobic surface (Figure 4F). The exposed side chains of Phe-128, Ile-131, Pro-342, Ile-347, Pro-350, and Tyr-351 are nearly contiguous with the above but are separated from the former by a single band of polar residues. Together, these residues form a gently concave groove some 30 Å across. Surface 1 is rimmed by several conserved and solvent-exposed polar residues: Glu-138, Lys-141, Lys-192, Asp-196, Lys-208, and Lys-340.
Figure 4. Functional Surfaces of the Bro1 Domain.
(A) Ribbon diagram of Bro1 domain shown in the same perspective as (B)–(D) for reference.
(B) Molecular surface of the Bro1 domain colored according to residue property: green, hydrophobic; blue, basic; red, acidic; white, uncharged polar.
(C) Bro1 domain surface colored according to residue conservation as in Figure 2A.
(D) Bro1 domain surface colored according to electrostatic potential, with blue electropositive and red electronegative.
(E) The same surface as in (D) is rotated by 180° about the x axis to show the opposite side of the boomerang.
(F) Close-up of hydrophobic patch 1.
(G) Close-up of hydrophobic patch 2.
Surface 2 is centered 50 Å away from surface 1. It is not as large as surface 1, but it is striking in that three hydrophobic side chains are hyperexposed at one tip of the boomerang, at the C-terminal end of the TPR substructure. The side chains of Phe-318, Ile-319, and Tyr-320 all project outward and are almost completely solvent exposed (Figure 4G). These residues adjoin several other exposed hydrophobic residues, including Ile-272, Ile-313, Val-324, Pro-325, Ala-326, and Val-327. Surface 2 has three conserved and exposed polar residues adjacent to it: Lys-314, Asp-315, and Asp-317. Surfaces 1 and 2 have the highest proportion of conserved residues of any region on the protein surface. In the crystal, surfaces 1 and 2 of two different Bro1 domains contact one another and thereby form the major contacts that hold the crystal lattice together. In solution, these surfaces would be exposed and available for protein:protein interactions.
Most of the protein surface is weakly electronegative or electroneutral. However, the N terminus of the helical solenoid is highly electropositive (Figures 4D and 4E). Lys-19, −20, −27, −28, −42, −84, −89, and −358, Arg-36, −80, −87, and −364, and His-76 and −111 are all in this region, with few acidic residues to neutralize them. This feature is at the opposite tip and on the opposite side of the boomerang relative to hydrophobic surfaces 1 and 2.
A Bro1-Snf7 Complex
Hexahistidine-tagged Bro1 1–387 and full-length untagged Snf7 were coexpressed in E. coli in order to determine whether they can form binary complex in vivo in a heterologous expression system. When coexpressed, both proteins are highly soluble, although more Bro1 1–387 than Snf7 is produced. Both proteins copurify on Talon-Co2+ affinity resin (Figure 5A). Bro1 1–387 and Snf7 therefore interact directly and form a complex together. The complex migrates on an analytical gel filtration column with an apparent native molecular weight of 104 kDa. This is larger than the calculated molecular weight of a 1:1 complex of 73.7 kDa. The Bro1 domain has an apparent native molecular weight of 45 kDa by gel filtration, consistent with the monomer observed in the crystal. Snf7 is predicted to consist largely of coiled coils and is therefore likely to be elongated. Due to aggregation of isolated Snf7, we were unable to determine the native MW of Snf7 alone. The elevated apparent MW could be consistent with a 1:1 complex of the two components, given a highly elongated Snf7, although it could also be consistent with a 1 Bro1:2 Snf7 complex.
Figure 5. The Bro1 Domain and Snf7 Form a Complex through Surface Patch 1.
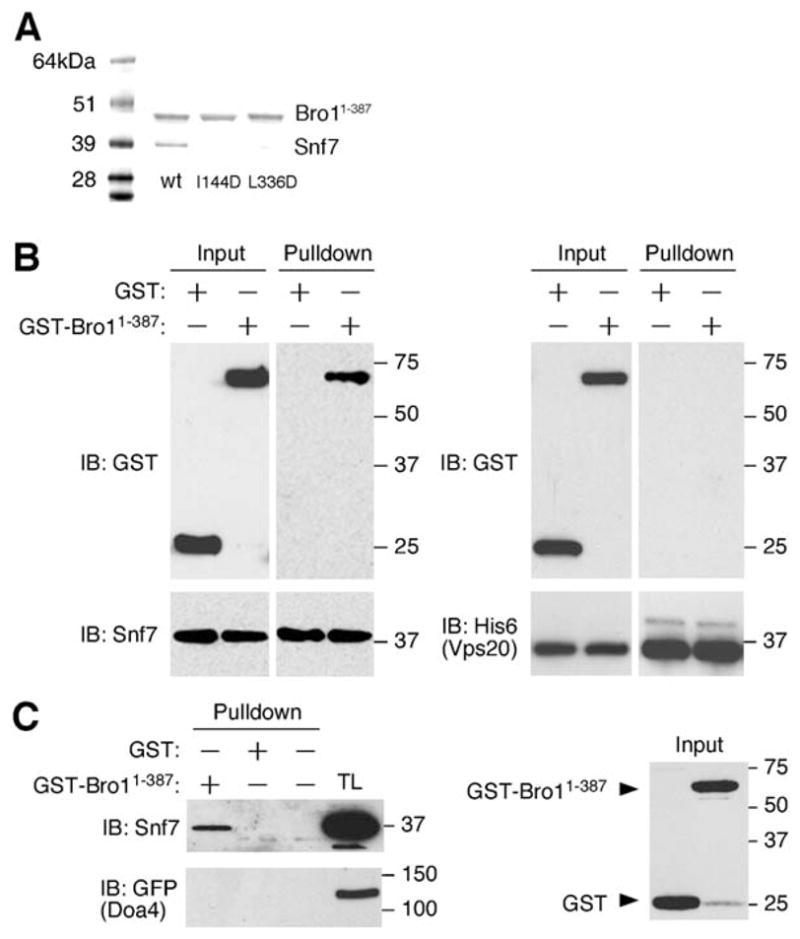
(A) His6-tagged Bro1 1–387 and untagged Snf7 were coexpressed in E. coli and copurify on Ni-NTA and gel filtration chromatography and visualized by SDS-PAGE and Coomassie staining. Bro1 1–387 mutants I144D and L336D in surface patch 1 express at normal levels, but coexpressed Snf7 does not copurify with them.
(B) Ni-NTA agarose beads were used to isolate purified His6-tagged Snf7 or His6-tagged Vps20 that was mixed either with GST-tagged Bro1 1–387 or GST alone.
(C) Glutathione-sepharose beads were used to isolate either GST-tagged Bro1 1–387 or GST alone that had been mixed with a total yeast extract.
The interaction between Bro1 1–387 and Snf7 was also observed when recombinant forms of each protein were individually expressed in E. coli and purified from bacterial lysates. After being mixed with hexahistidine-tagged Snf7, GST-tagged Bro1 1–387, but not GST alone, was isolated using Ni2+-NTA affinity resin (Figure 5B). In contrast, purified GST-Bro1 1–387 was not pulled down in similar experiments using His-tagged Vps20 (Figure 5B), which is a related ESCRT-III subunit that binds Snf7 (Babst et al., 2002a). Similarly, purified recombinant Snf7 (but not Vps20) was isolated using glutathione-sepharose when combined with GST-Bro1 1–387, but not with GST alone (data not shown). Furthermore, the addition of purified GST-Bro1 1–387 to yeast cell extracts resulted in the isolation of native Snf7 (Figure 5C). However, GST-Bro1 1–387 was unable to pull out a Doa4-GFP fusion protein from yeast cell extracts (Figure 5C), indicating that this region of Bro1 is not responsible for interacting with Doa4, a deubiquitinating enzyme that was recently found to be recruited to endosomes by binding Bro1 (Luhtala and Odorizzi, 2004).
In order to locate the site of interaction on the surface of Bro1, residues in the two hydrophobic surface patches were mutated and the effects on complex formation with Snf7 were tested when the mutant Bro1 constructs were coexpressed with Snf7 in E. coli. Ile-144, Phe-189, Leu-209, and Leu-336 of patch 1 and Asp-315, Asn-316, Phe-318, and Ile-319 of patch 2 were tested alone or in combination. I144D and L336D blocked the formation of the complex with Snf7 (Figure 5A). These mutants had no effect on the overall levels of soluble Bro1 1–387 produced, so we conclude that the mutations act by disrupting the interaction with Snf7 rather than by destabilizing the fold of Bro1. Other mutations in patch 1 and all mutations tested in patch 2 did not block complex formation (data not shown). This demonstrates that hydrophobic patch 1 comprises at least part of the Snf7 binding site on Bro1.
Recruitment of Bro1 to Endosomes
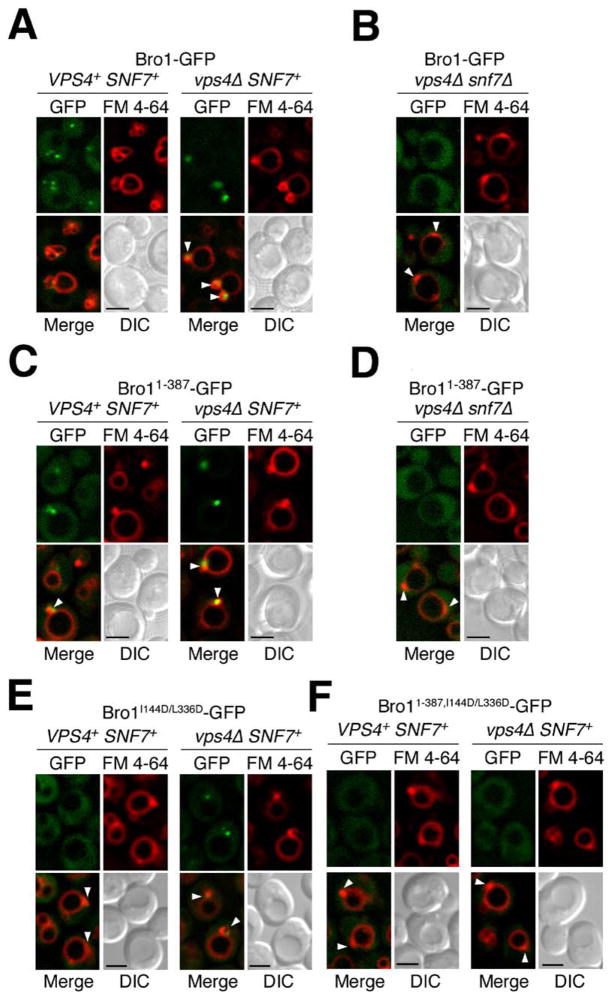
The association of Bro1 with endosomes is dependent upon Snf7 (Odorizzi et al., 2003), whereas endosomal dissociation of Bro1 (along with other components of the MVB machinery) requires the Vps4 ATPase (Babst et al., 1998; Odorizzi et al., 2003). As shown previously (Luhtala and Odorizzi, 2004), wild-type cells expressing full-length Bro1-GFP exhibited diffuse cytoplasmic fluorescence in addition to multiple fluorescent punctate structures that were also labeled by FM 4–64 (Figure 6A), a fluorescent lipophilic dye that stains endosomal and vacuolar membranes (Vida and Emr, 1995). In the absence of Vps4 (vps4Δ), however, both Bro1-GFP and FM 4–64 were concentrated at class E compartments (Figure 6A, arrowheads), which are abnormal late endosomal structures that occur upon mutation of Vps4 or other components of the class E Vps machinery (Katzmann et al., 2002). Like full-length Bro1-GFP, a truncated Bro1 1–387-GFP fusion protein was colocalized with FM 4–64 at punctate structures both in cells containing wild-type Vps4 and in vps4Δ cells (Figure 6C), indicating that amino acids 1–387 of Bro1 are sufficient for endosomal association. Upon deletion of both SNF7 and VPS4, Bro1-GFP and Bro1 1–387-GFP were entirely cytoplasmic (Figures 6B and 6D, respectively). Similarly, both the full-length and the truncated Bro1-GFP fusion proteins were completely cytoplasmic in snf7Δ cells expressing wild-type Vps4 (data not shown), consistent with a role for Snf7 in the recruitment of Bro1 to endosomes (Odorizzi et al., 2003).
Figure 6. The Bro1 Domain Mediates Bro1 Localization.
The indicated strains were stained with FM 4–64 and examined by fluorescence and DIC microscopy. Arrowheads indicate class E compartments. Scale bars equal 2 μm.
To determine whether the endosomal association of Bro1 is dependent upon its interaction with Snf7, we tested whether Bro1 localization was affected upon mutation of hydrophobic patch 1. As shown in Figure 6E, full-length Bro1-GFP containing the I144D/L336D substitution was entirely cytoplasmic in VPS4+ cells but was partially associated with the class E compartment in vps4Δ cells. The weak endosomal localization of the full-length Bro1 I144D/L336D mutant in the absence of Vps4 function may be attributed to interactions that could occur between the C-terminal portion of Bro1 and endosome-associated proteins other than Snf7 that are concentrated at the class E compartment in vps4Δ cells. Indeed, the truncated Bro1 1–387-GFP fusion protein containing the I144D/L336D substitution was completely cytoplasmic both in VPS4+ and vps4Δ cells (Figure 6F). Thus, we conclude that the association of Bro1 with Snf7, which is mediated by hydrophobic patch 1 in the Bro1 domain, is important for the localization of Bro1 to endosomal membranes.
Cargo Sorting in the MVB Pathway
To determine whether the MVB pathway is dependent upon the interaction between Bro1 and Snf7, we examined the sorting of carboxypeptidase S (CPS) in yeast cells expressing wild-type BRO1 versus mutant bro1 alleles. CPS is a vacuolar hydrolase that is transported from the Golgi to endosomes, where CPS is sorted into lumenal MVB vesicles that are subsequently delivered into the vacuole lumen (Odorizzi et al., 1998). Thus, in wild-type cells examined by fluorescence microscopy, a GFP-CPS fusion protein was observed entirely within the lumen of the vacuole (Figure 7A). In contrast, deletion of the BRO1 gene (bro1Δ), resulted in the mislocalization of GFP-CPS to the vacuole membrane (Figure 7B), which is indicative of a block in the MVB pathway (Odorizzi et al., 2003). GFP-CPS was also mis-localized to the vacuole membrane in cells expressing bro1I144D/L336D (Figure 7C), indicating that hydrophobic patch 1 of Bro1, which is involved in binding Snf7, is critical for protein sorting via the MVB pathway. Hydrophobic patch 2, however, which is not required for the Snf7 interaction, was not required for the function of Bro1 in MVB sorting, as GFP-CPS was localized correctly within the vacuole lumen in cells expressing bro1Y320D (Figure 7D). In addition, simultaneous substitution of alanines in place of both Lys-246 and Trp-239 did not block the sorting of GFP-CPS (data not shown), suggesting that the TPR-like domain does not function in MVB sorting.
Figure 7. The Bro1 Domain and Surface Patch 1 Are Required for Cargo Sorting.
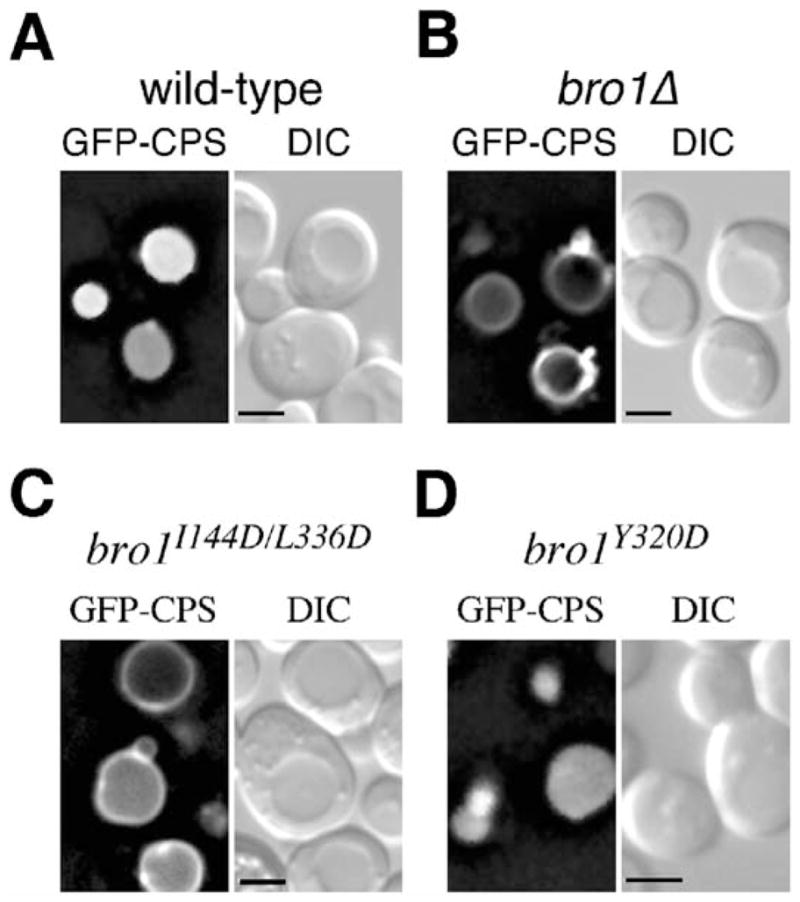
The indicated strains expressing GFP-CPS were examined by fluorescence and DIC microscopy. Scale bar equals 2 μm.
Discussion
The ESCRT complexes, Bro1, and other class E Vps proteins form a network that sorts cargo and directs the invagination of endosomal membranes into the lumen of the MVB. Systematic interaction studies have painted a broad-brush outline of the network (Bowers et al., 2004; von Schwedler et al., 2003), but the precise mechanisms of sorting and membrane remodeling remain largely unknown. Bro1 and Alix are a critical nexus for such interactions. We have found that the Bro1 domain of Bro1 comprises a single folded domain that mediates at least one of its functions, its binding to Snf7 and targeting to endosomes. This finding is consistent with results reported for Alix and Rim20, in which N-terminal deletions have been shown to abrogate Snf7-dependent targeting (Peck et al., 2004; Xu et al., 2004). The Bro1 interaction with Snf7 is conserved in humans, where the corresponding interaction is between Alix and CHMP4b. We have found that Snf7 interacts with hydrophobic patch 1 on the middle of the filled-in concave side of the Bro1 domain.
The topology of MVB budding presents a negatively curved concave pit to the cytosol. The curvature is the opposite of that presented to the cytosol during endocytosis. In endocytosis, preferential interactions between protein domains and positively curved membranes are important for the timed recruitment of key factors and even for the induction of curvature (Ford et al., 2002; Hurley and Wendland, 2002). The banana-shaped BAR domain binds to positively curved membranes with its concave face (Peter et al., 2004). It is tempting to speculate that the MVB pathway should contain similar factors that bind negatively curved membranes by using a convex face of the protein surface. Alix has been found to be enriched in the lumenal vesicles of MVBs and has been postulated to be such a factor (Gruenberg and Stenmark, 2004). The Bro1 domain is an attractive candidate for a component of a negative curvature sensing system. However, the isolated Bro1 domain is cytosolic in the absence of Snf7. To the extent that it interacts with curved membranes, it is likely to do so in a Snf7-dependent manner. The convex face of the Bro1 boomerang contains a highly electropositive region at one end, which would favor binding to acidic phospholipids. The site does not overlap with the Snf7 binding site, consistent with simultaneous binding to ESCRT-III and the anionic membrane surface.
In summary, our results define the Bro1 domain as a single conserved 370-amino acid residue structure. Given the endosomal localization of Bro1 domain proteins Alix and Rim20, inferences from previous deletion analyses of Alix and Rim20, and the conservation of most residues in the surface identified as the Bro1 binding site for Snf7, it is reasonable to infer that the Bro1 domain is a conserved ESCRT-III targeting domain. This information elucidates one more cog in the complex cellular machinery of cargo sorting into multivesicular bodies. It is a significant step toward understanding the overall 3-dimensional organization of the protein network of MVB sorting and HIV budding.
Experimental Procedures
Bro1 Domain Expression and Purification
DNA coding for Bro1 residues 1–387 was subcloned into the pGST-parallel2 vector (Sheffield et al., 1999). Overexpression of the GST-tagged protein in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Stratagene, La Jolla, CA) was induced with 0.5 mM isopropyl-β-D-thiogalactoside at OD600 = 1.0 at 20°C for 24 hr. The protein was purified by using glutathione sepharose (Amersham-Pharmacia), the GST tag was removed with His6-TEV protease, and the protein was subjected to a second purification with Talon-Co2+ resin (Clontech, Mountain View, CA) to remove TEV protease. The protein was further purified on a Superdex 200 preparative grade gel filtration column (Amersham Biosciences, Piscataway, NJ). The cleaved protein contains the vector-derived sequence GAMGS followed by Bro1 residues 1–387. The protein was stored at −80°C in elution buffer.
Bro1 Domain:Snf7 Complex Coexpression and Purification
The DNA sequences encoding Bro1 residues 1–387 and the complete gene for Snf7 were amplified by PCR to generate cassettes containing the Shine-Delgarno translational start signal and cloned directly into the polycistronic pST39 expression vector (Tan, 2001; Hierro et al., 2005). The Bro1 domain was tagged with a C-terminal hexahistidine-tag and cloned into the vector using Xbal/BamHI cloning site. Snf7 was untagged and cloned into the vector using BspEI/MIul cloning site. The plasmid was transformed into Escherichia coli strain BL21(DE3) Codon Plus RIL and expressed by inducing with 0.5 mM IPTG at OD600 = 0.8 and grown at 20°C for 30 hr. Cells were lysed by high-pressure homogenization in 1× PBS (pH 7.5) buffer and 100 (μl of protease inhibitor cocktail (Sigma) per liter of culture medium. The Bro1 1-387-Snf7 complex was isolated using Talon Co2+ affinity chromatography (Clontech) and was further purified with a Superdex S 200 gel filtration column.
Crystallization and Data Collection
Crystals of Bro1 residues 1–387 were obtained using hanging drop vapor diffusion by mixing equal amounts of 4–6 mg/ml protein solution and a reservoir solution of 20% polyethylene glycol 3350/0.2 M tri-potassium citrate monohydrate (pH 8.3) and incubating for 1 week. Crystals were cryo-protected in mother liquor supplemented with 25% glycerol and flash-frozen in liquid nitrogen. Selenomethionyl protein crystals were obtained under similar conditions except that potassium citrate was replaced by 0.2 M ammonium iodide. Native data were collected at beamline ID-22 at the Advanced Photon Source, using 0.5° oscillations, 1 s exposures, and a Mar225 CCD detector. The Se-MAD data were collected at beamline X-9B at the NSLS (Brookhaven, NY). Data were indexed and reduced using HKL2000 (HKL Research).
Structure Determination
Ten selenium sites were located in an automated Patterson search using SOLVE (Terwilliger and Berendzen, 1999). The resulting density synthesis was improved by density modification in RESOLVE (Terwilliger, 2000). The map was interpreted manually using the interactive graphics program O (Jones et al., 1991) and refined against the native data by using CNS (Brunger et al., 1998). The final model has no residues in disallowed regions of the Ramachandran plot.
Mutagenesis
Bro1 mutations were generated using QuikChange II or II XL site-directed mutagenesis kits (Stratagene). All mutations were confirmed by DNA sequencing.
In Vitro Binding of Snf7 and Bro1 1–387
To examine the interaction between purified Bro1 1–387 and Snf7, the SNF7 gene was amplified by PCR using an oligonucleotide that eliminated the endogenous stop codon and, instead, placed hexahistidine (6xHis) in frame with the 3′ end of the Snf7 coding sequence. The resulting SNF7-6xHis gene fusion was subcloned into pGEX-4T1, transformed into E. coli, overexpressed, and purified using glutathione sepharose as described above. The GST tag was removed using thrombin protease (Novagen), and the Snf7-6xHis protein was subjected to a second purification with glutathione sepharose to remove GST. Purified Snf7-6xHis was incubated by rotation at 4°C for 1 hr with purified GST-Bro1 1–387 or with GST (5 μg of each protein) in binding buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, 0.05% Triton X-100). Afterward, 25 (μl of Ni2+-NTA agarose beads equilibrated in binding buffer was added, and the mixture was incubated similarly for 1 hr at 4°C. The beads were harvested by centrifugation for 1 min at 10,000 × g at 4°C, then subjected to 3 washes with ice-cold binding buffer followed by 2 washes with binding buffer in the absence of Triton X-100. The beads were dried by rotary evaporation, resuspended in SDS-PAGE sample buffer, then analyzed by SDS-PAGE and Western blotting using anti-GST antibodies and anti-Snf7 antibodies (Babst et al., 1998).
To examine the interaction between Bro1 1–387 and native Snf7 expressed in yeast, cell extracts were prepared from the strain GOY23 as described previously (Luhtala and Odorizzi, 2004) and incubated by rotation at 4°C for 1 hr with 5 μg purified GST-Bro1 1–387 or purified GST alone together with 25 μl of glutathione sepharose beads equilibrated in lysis buffer. The beads were subsequently harvested by centrifugation, washed, and examined by SDS-PAGE followed by Western blotting as described above.
Fluorescence Microscopy
GFP and FM 4–64 fluorescence and differential interference contrast (DIC) microscopy was performed using a Leica DMRXA microscope equipped with a Cooke Sensicam digital camera (Applied Scientific Instruments). Images were deconvolved using Slidebook software (Intelligent Imaging Innovations) and processed using Adobe Photoshop 7.0 software (Adobe Systems Incorporated). Cells were stained with FM 4–64 using a pulse-chase procedure at 30°C as previously described (Odorizzi et al., 2003). To observe the localization of GFP-CPS, cells were transformed with pGO47, which contains the GFP-CPS fusion (Odorizzi et al., 1998).
Acknowledgments
We thank Peter Zwart, Zbigniew Dauter, and Greg Miller for assistance with data collection at beamline X9B, National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL); the SER-CAT staff for assistance at beamline ID-22, Advanced Photon Source (APS), Argonne National Laboratory (ANL); Rodolfo Ghirlando for analytical ultracentrifugation analysis; and Scott Emr and Jenny Hinshaw for discussions. This research was supported by IATAP and NIDDK intramural support to J.H.H. and by extramural NIH support (GM065505) to G.O. Research carried out at the NSLS, BNL was supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886, and at the APS, ANL by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38.
Footnotes
Accession Numbers
Structural coordinates of the Bro1 domain of Bro1 have been deposited in the Protein Data Bank with accession number 1ZB1.
References
- Alam SL, Sun J, Payne M, Welch BD, Black BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- Chen B, Borinstein SC, Gillis J, Sykes VW, Bogler O. The glioma-associated protein SETA interacts with AIP1/Alix and AZIG-2 and modulates apoptosis in astrocytes. J Biol Chem. 2000;275:19275–19281. doi: 10.1074/jbc.M908994199. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast–sorting out the sorting proteins. Cell. 1995;83:513–516. doi: 10.1016/0092-8674(95)90088-8. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Goff A, Ehrlich LS, Cohen SN, Carter CA. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J Virol. 2003;77:9173–9182. doi: 10.1128/JVI.77.17.9173-9182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J Virol. 2003;77:6507–6519. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles–ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Hierro A, Kim J, Hurley JH. Polycistronic expression and purification of the ESCRT-II endosomal trafficking complex. Methods Enzymol. 2005 doi: 10.1016/S0076-6879(05)03027-2. in press. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Wendland B. Endocytosis: driving membranes around the bend. Cell. 2002;111:143–146. doi: 10.1016/s0092-8674(02)01044-9. [DOI] [PubMed] [Google Scholar]
- Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Hatta K, Maki M. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch Biochem Biophys. 2004;421:159–165. doi: 10.1016/j.abb.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor down-regulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Roach C, Hirsh IS, Turley S, deWalque S, Michels PAM, Hol WGJ. An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J Mol Biol. 2001;307:271–282. doi: 10.1006/jmbi.2000.4465. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Smith SJM, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K. Structure of the TPR domain of p67(phox) in complex with Rac • GTP. Mol Cell. 2000;6:899–907. doi: 10.1016/s1097-2765(05)00091-2. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166:717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main ERG, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul M, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999;6:124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- Nikko E, Marini AM, Andre B. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J Biol Chem. 2003;278:50732–50743. doi: 10.1074/jbc.M306953200. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Peck JW, Bowden ET, Burbelo PD. Structure and function of human Vps20 and Snf7 proteins. Biochem J. 2004;377:693–700. doi: 10.1042/BJ20031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkala M, Hieta R, Bergmann U, Kivirikko KI, Wierenga RK, Myllyharju J. The peptide-substrate-binding domain of collagen prolyl 4-hydroxylases is a tetratricopeptide repeat domain with functional aromatic residues. J Biol Chem. 2004;279:52255–52261. doi: 10.1074/jbc.M410007200. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Insights from yeast endosomes. Curr Opin Cell Biol. 2002;14:454–462. doi: 10.1016/s0955-0674(02)00352-6. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Piper RC, Luzio JP. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic. 2001;2:612–621. doi: 10.1034/j.1600-0854.2001.20904.x. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D’Arrigo A, Stang E, Stenmark H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshush IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schmidt MHH, Hoeller D, Yu JH, Furnari FB, Cavenee WK, Dikic I, Bogler O. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol Cell Biol. 2004;24:8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Prot Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiano D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-inter-acting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Prot Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J Biol Chem. 2004;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Rainbow L, Tilburn J, Arst HN, Penalva MA. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Xu WJ, Smith FJ, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]