Abstract
Using REMD and united atom implicit solvent model we examine the role of backbone hydrogen bonds (HBs) in Aβ aggregation. The importance of HBs appears to depend on the aggregation stage. The backbone HBs have little effect on the stability of Aβ dimers or on their aggregation interface. The HBs also do not play a critical role in initial binding of Aβ peptides to the amyloid fibril. Their elimination does not change the continuous character of Aβ binding nor its temperature. However, cancellation of HBs forming between incoming Aβ peptides and the fibril disrupts the locked fibril-like states in the bound peptides. Without the support of HBs bound Aβ peptides form few long β-strands on the fibril edge. As a result the deletion of peptide-fibril HBs is expected to impede fibril growth. As for the peptides bound to Aβ fibril the deletion of interpeptide HBs reduces the β propensity in the dimers making them less competent for amyloid assembly. These simulation findings together with the backbone mutagenesis experiments suggest that a viable strategy for arresting fibril growth is the disruption of interpeptide HBs.
Introduction
Polypeptide chains show apparent generic propensity to assemble into supramolecular ordered structures called amyloid fibrils.1 Amyloid formation is a complex multistage conformational transition, which is initiated with the oligomerization of monomers and progresses with the development of protofibrils and mature amyloid fibrils as final products.1–3 Amyloid formation is linked to more than 20 various disorders, including Alzheimer’s, Parkinson’s, and Creutzfeldt-Jakob diseases.4 Biomedical studies have showed that oligomeric species5 and, to a lesser extent, fibrils6 have cytotoxic properties. Structural studies have uncovered a remarkable homogeneity of amyloid fibril cores based on β-sheet structure.7–12 Backbone hydrogen bonds (HBs) linking polypeptide chains into β-sheets and side chain interactions render amyloid fibrils remarkably stable against dissociation.13 In contrast to structurally ordered fibrils, oligomers sample multitude of conformations and exhibit a distribution of sizes starting with dimers.14–16
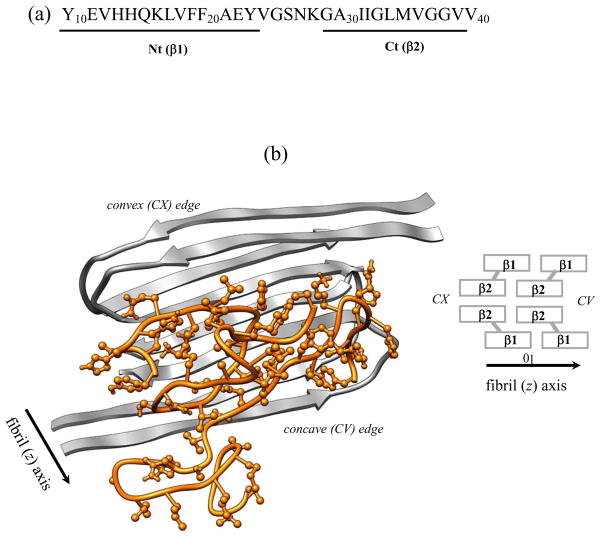
Aβ peptides, the fragments of amyloid precursor protein cleaved during cellular proteolysis, are the amyloidogenic species, which are linked to the onset of Alzheimer’s disease. The most common Aβ species are 40-mer Aβ1–40 fragments, which have been shown to form polymorphic amyloid fibrils.17 Among them is a two-fold symmetry structure of Aβ1–40 fibril, which is derived from the solid-state NMR experiments under agitated conditions9 (Fig. 1). This structure reveals that Aβ peptides are organized into parallel in-registry β-sheets laminated into four layers. 8,9 Elongation of Aβ fibrils was proposed to proceed via addition of monomers according to the two-stage “dock-lock” mechanism.18–21 During the first (docking) stage disordered incoming Aβ peptide binds to the fibril without integration into the fibril structure. During the second (locking) stage a bound Aβ monomer adopts an ordered fibril conformation through structural reorganization. Using computer simulations we have probed the thermodynamics of Aβ fibril growth by computing its free energy landscape.22 Our studies have suggested that the docking and locking stages are fundamentally different. The former occurs without detectable free energy barriers and resembles polymer adsorption on attractive walls. In contrast, locking is governed by rugged free energy landscape and is associated with the formation of β-sheets by Aβ peptides on the edges of amyloid fibrils.22
Fig. 1.
(a) The sequence of Aβ10–40 peptide and the allocation of the N-terminal Nt and C-terminal Ct regions. (b) Cartoon representation of the mutant Aβ10–40 hexamer. Fibril fragment includes four Aβ peptides in grey and consists of four parallel in-registry β-sheets formed by the β1 and β2 strands, which correspond to the Nt and Ct regions (panel (a)). Spacial allocation of β1 and β2 in the fibril gives rise to two distinct fibril edges - concave (CV) and convex (CX). Two incoming peptides in orange with side chains shown are bound to the CV fibril edge. The deletion of peptide-fibril backbone HBs in the mutant suppresses the formation of long β-strands in incoming peptides and disrupts their locking in fibril-like ordered conformations. The fibril structure is visualized using Chimera.58
It is natural to expect that backbone HBs provide an important contribution to fibril energetics. Indeed, hydrogen-deuterium exchange experiments have shown that roughly half of HBs in Aβ fibril remain protected even after 1000 hours of observation.23 These data imply that Aβ fibril has a rigid structural core shielded from water and stabilized by HBs. Direct experimental evaluation of the role of HBs can be accomplished by backbone mutagenesis, which replaces amide bonds with N-methyl, ester, or E-olefin peptide bonds.24–26 The experiments established that a deletion of a single HB may slow down the fibril assembly,26 whereas elimination of several HBs could prevent fibril formation all together.27 Although these experiments demonstrate the importance of HBs, it is not clear how their deletion affects the structure of Aβ peptides and fibrils and the mechanisms of their growth on a molecular level. In particular, (i) what are the roles played by the backbone HBs formed between incoming peptides and the fibril during docking and locking stages of fibril growth? (ii) What are the changes in the structure of Aβ peptides bound to the fibril or included in oligomer occurring in response to HB deletion? (iii) Are interpeptide HBs important for the stability of oligomers as they are for the fibrils?
Although it is challenging to address these questions experimentally, some insights can be obtained by means of all-atom computer simulations.28 Recently, all-atom molecular dynamics (MD) was used to explore the stability and energetics of fibril architectures29–32 and to study the fibril elongation.22,33–36 In this paper, we use exhaustive replica exchange MD (REMD) and implicit solvent model to study the effects of deletion of interpeptide HB on Aβ aggregation. Our overall goal is to probe the contribution of backbone HBs as a class of interactions to aggregation process. We show that the elimination of HBs forming between incoming peptides and the fibril has small destabilizing effect on Aβ binding to the fibril. By comparing the thermodynamics of Aβ docking in the systems with and without peptide-fibril HBs we concluded that their docking transitions are similar. In contrast, deletion of peptide-fibril HBs disrupts the locked state by drastically reducing the propensity of a bound Aβ peptide to form extended β-strands. As a result, the bound Aβ peptides experience a loss in β-structure. Finally, we report that the elimination of interpeptide HBs in Aβ dimers does not block their formation, but reduces the propensity for the β structure. We also discuss the comparison between our in silico data and experimental observations.
Model and Simulation Methods
Molecular dynamics simulations
Simulations of Aβ peptides were performed using CHARMM MD program37 and united atom force field CHARMM19 coupled with the SASA implicit solvent model.38 Full description, applicability, and testing of the CHARMM19+SASA model can be found in our previous studies.32,39,40 In particular, we have shown that CHARMM19+SASA force field accurately reproduces the experimental distribution of chemical shifts for Cα and Cβ atoms in Aβ monomers.39,41 Combination of CHARMM19 force field and SASA model has been used to fold α-helical and β-sheet polypeptides42,43 and to study aggregation of amyloidogenic peptides.22,44
Simulation system
We used Aβ10–40 peptides, which are the N-terminal truncated fragments of the full-length Aβ1–40 (Fig. 1). Solid-state NMR studies have shown that the two-fold symmetry fibril structures of Aβ1–40 and Aβ10–40 peptides are similar.9,45 Similarities in oligomerization pathways of Aβ1–40 and Aβ10–40 were reported experimentally 14 and computationally. 46 It is also known that the first nine N-terminal residues in the Aβ1–40 fibril are disordered.9 Consequently, we use Aβ10–40 as a model of the full-length Aβ1–40 peptide.
Two Aβ10–40 systems, hexamer and dimer, were considered. Because these systems were introduced in our previous studies,22,39 we provide only their brief description. The hexamer includes four peptides forming a fibril fragment and two incoming peptides interacting with the fibril (Fig. 1). The structure of the Aβ10–40 fibril fragment is modeled using the coordinates of backbone atoms determined from the solid-state NMR measurements.9 To emulate the stability of large fibril sample, the backbones of fibril peptides were constrained to their experimental positions using soft harmonic potentials with the constant kc = 0.6kcal/(molÅ2). The harmonic constraints permit backbone fluctuations with the amplitude of about 0.6Å at 360K, which are comparable with the fluctuations of atoms on the surface of folded proteins.47 Constraints were not applied to the side chains of fibril peptides or to the atoms in incoming peptides. The latter were free to dissociate and reassociate with the fibril. The constraints capture the rigidity of amyloid fibril and eliminate the necessity to simulate large fibril systems to achieve their stability. Throughout the paper the peptides in grey in Fig. 1b are referred to as fibril and the colored peptides are termed incoming.
The second system includes two Aβ10–40 peptides and is designed to investigate the formation of the simplest oligomer, dimer.39 No constraints were applied to this system. Hexameric and dimeric systems were subject to a spherical boundary condition with the radius Rs = 90Å and the force constant ks = 10kcal/(molÅ2). The concentration of Aβ peptides is ~ 1mM.
To study the contribution of hydrogen bonds (HB) to aggregation we deleted interpeptide electrostatic interactions between the NH and CO backbone groups in the incoming peptides and the fibril. This modification of the energy function affects only electrostatic interactions between the incoming peptides and the fibril and do not perturb any other electrostatic interactions. In the dimer, all interpeptide electrostatic interactions between the NH and CO backbone groups were canceled. The deletion of these interactions eliminates interpeptide HBs and is similar in spirit to N-methyl, ester, or E-olefin backbone mutagenesis.24–26 However, it is important to keep in mind that experimental backbone mutagenesis is site specific, whereas in our study the backbone mutation spans the entire chain. Technically, the selected electrostatic interactions were canceled using the BLOCK functionality in CHARMM. The hexamer and dimer systems with intact or canceled HBs are referred to as wild-type (WT) and mutant (MT), respectively.
Replica exchange simulations
Conformational sampling was performed using replica exchange molecular dynamics (REMD).48 The REMD implementation is described in our previous studies.22,39 Briefly, 24 replicas were distributed linearly in the temperature range from 330 to 560K (hexamer) or from 300 to 530K (dimer) with the increment of 10K. The temperature ranges span the spectrum of conformational states of Aβ peptides from aggregated to completely dissociated. Small temperature increment ensures significant overlap of energy distributions from neighboring temperatures, a prerequisite for efficient conformational sampling. The exchanges were attempted every 20 (hexamer) or 80ps (dimer) between all neighboring replicas with the average acceptance rate of 37% (hexamer) and 53% (dimer). Five REMD trajectories were produced for the mutant hexamer resulting in a cumulative simulation time of 24μs. We also obtained five mutant dimer trajectories with the cumulative simulation time of 81 μs. Between replica exchanges the system evolved using NVT underdamped Langevin dynamics with the damping coefficient γ= 0.15ps−1 and the integration step of 2 fs. Because the initial parts of REMD trajectories are not equilibrated and must be excluded from thermodynamic analysis, the cumulative equilibrium simulation time is reduced to τsim ≈ 16μs (hexamer) and ≈ 66μs (dimer). The REMD trajectories were started with random distributions of (incoming) peptides in the sphere.
Computation of structural probes
To probe the interactions between Aβ peptides, we computed the number of side chain contacts. A side chain contact is formed, if the distance between the centers of mass of side chains is less than 6.5Å. If both amino acids in contact are apolar, the contact is considered hydrophobic. Backbone HBs between NH and CO groups were assigned according to Kabsch and Sander.49 In all, we defined two classes of interpeptide backbone HBs. The first includes any HB between incoming peptide and the fibril (hexamer) or between the peptides in the dimer. The second class corresponds to parallel β-sheet HBs. A parallel HB (pHB) is formed between the residues i and j, if at least one other HB is also present between i+2 and j or j +2 (or between i − 2 and j or j − 2). This definition of pHB follows from the structural analysis of parallel β-sheets. A peptide is considered bound, if it forms at least one hydrophobic side chain contact with the fibril.
In our previous studies bound states of incoming peptides with large number of pHBs were termed “locked”. Because in this study interpeptide HBs are canceled, we describe the locked state using parallel side chain contacts. Interpeptide parallel contact between the residues i and j occurs, if at least one other side chain contact is formed between adjacent residues i+k and j +k (k = ±2). Parallel contacts take place, if the incoming peptide binds to the fibril in parallel registry similar to that occurring in parallel β-sheets.
The secondary structure content was computed using the distribution of (φ,ψ) dihedral angles as described in our previous study.39 The thickness D of the layer formed by incoming peptides bound to the fibril edges is computed using the approach introduced earlier.22 Throughout the paper angular brackets <..> imply thermodynamic averages. Because hexamer system includes two indistinguishable incoming peptides, we report averages over two peptides. The same applies to the dimer system. The distributions of states produced by REMD were analyzed using multiple histogram method.50 The convergence of REMD simulations and error analysis are presented in Supporting Information.
Results
Using REMD we probed the interactions of the mutant (MT) Aβ10–40 peptides with the amyloid fibril and in the dimers (Fig. 1). The Aβ10–40 mutation is accomplished by switching off inter-peptide hydrogen bonds (HBs). Because we have previously investigated the fibril growth and dimer formation for the wild-type (WT) Aβ10–40 peptide,22,32,39 the mutation effect is considered by comparing the MT and WT. Following experimental Aβ fibril structure,9 we distinguish two sequence regions in Aβ10–40, the N-terminal (Nt, residues 10 to 23) corresponding to the first fibril β-strand β1 and the C-terminal (Ct, residues 29 to 39) corresponding to the second fibril β-strand β2 (Fig. 1a). The MT thermodynamic quantities are obtained at the temperature 360K, at which the WT Aβ peptide locks into fibril-like state as reported earlier.22,32
Binding of Aβ peptides to amyloid fibril
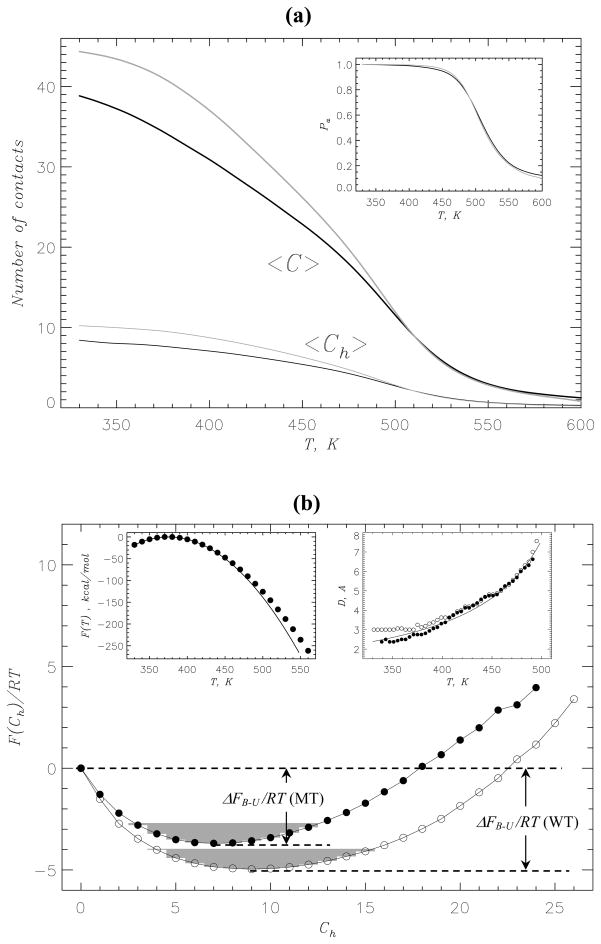
To investigate binding of the MT Aβ10–40 peptides to amyloid fibril, we computed the thermal averages of the number of peptide-fibril side chain contacts <C(T)> as a function of temperature T. Fig. 2a shows that at all temperatures <C(T)> (and the number of hydrophobic peptide-fibril contacts <Ch(T)>) are smaller than those computed for the WT. For example, at 360K the MT peptide forms <C>≈ 36.2 side chain contacts with the fibril that is about 15% smaller than for the WT (≈ 42.7). The average number of hydrophobic contacts <Ch> at 360K is decreased by 20% (from ≈ 9.8 to 7.9). Although the peptide-fibril HBs are energetically canceled, we can formally consider their “formation”. The HB definition demonstrates that the number of peptide-fibril HBs <Nhb> present in the MT system is ≈ 1.5. For comparison, for the WT <Nhb>≈ 10.5. It is also instructive to consider the probability of binding of MT Aβ peptide to the fibril, Pa. (Operationally, Pa is defined as a probability of forming any hydrophobic peptide-fibril side chain contact.) It follows from the inset to Fig. 2a that for the MT Pa ≈ 1.0 at T ≲ 450K and the temperature dependencies of Pa for the WT and MT are almost indistinguishable.
Fig. 2.
(a) Binding of Aβ10–40 peptides to amyloid fibril is probed by the thermal averages of the numbers of peptide-fibril side chain contacts <C(T)> and hydrophobic contacts <Ch(T)> as a function of temperature. The inset displays the probability of peptide binding to the fibril, Pa. The data in black and grey correspond to the MT and WT, respectively. (b) Free energy of incoming Aβ10–40 peptide F(Ch) as a function of the number of peptide-fibril hydrophobic side chain contacts Ch: the MT (filled circles), the WT (open circles). The free energy of Aβ binding to the fibril is ΔFB–U = FB − FU, where FB and FU = 0 are the free energies of the bound (B) and unbound (U, Ch = 0) states. FB is obtained by integrating over the B states (shaded in grey), for which F(Ch) ≤ Fmin + 1.0RT, where Fmin is the minimum in F(Ch). The free energies F(Ch) are computed at 360K. Left inset: Temperature dependence of the MT hexamer free energy F(T). Quadratic fitting function F(T) ≃ −α(T − Td)2 (α = −0.009kcal/(molK2) and Td = 375K) is shown by black continuous curve. Maximum value of F(T) is set to zero. Right inset: The thickness D(T) of the layer formed by Aβ peptides bound to the fibril edge as a function of temperature (filled and open circles represent the MT and WT, respectively). Continuous line marks the fit to the MT data with the inverse temperature function D0/(Tu − T), where D0 =592KÅ and Tu = 578K. The panels (a) and (b) suggest that the deletion of peptide-fibril backbone HBs has a minor impact on Aβ binding to the fibril.
Further insight into MT binding energetics is obtained by computing the free energy of Aβ incoming peptide as a function of the number of peptide-fibril hydrophobic contacts, F(Ch) (Fig. 2b). Compared to the WT, the MT free energy profile F(Ch) is more shallow and the stability of the MT bound state is reduced by ΔΔFB–U = ΔFB–U (MT) − ΔFB–U (WT) ≈ 1.4RT, where ΔFB–U (MT) and ΔFB–U (WT) are the binding free energies of the MT and WT. More importantly, the MT free energy profile F(Ch) does not reveal binding barriers and is qualitatively similar to that of the WT. For the latter we have shown that the docking (binding) to amyloid fibril is consistent with the continuous transition.22 Additional information about the MT binding can be extracted from the temperature dependence of the hexamer free energy F(T) (inset to Fig. 2b). If the underlying structural transition is continuous, F(T) is expected to follow a quadratic dependence on temperature F(T) ~ −(T − Td)2, where Td is the docking temperature. 51 The inset to Fig. 2b demonstrates that F(T) can be fit with the quadratic function with Td = 375K. Interestingly, for the WT Td is about the same (380K).22
In our previous studies of Aβ fibril elongation we considered the thickness D of the layer formed by the incoming peptides bound to the fibril edge.22 The temperature dependence D(T) is indicative of the nature of Aβ binding transition. According to the theory of adsorption of polymers on attractive walls,52 if the binding transition is continuous, the thickness of adsorbed layer D(T) follows the inverse temperature dependence (Tu − T)−1, where Tu is dissociation temperature. The inset to Fig. 2b shows that in the temperature range of binding (340 ≲ T ≲ 490K) D(T) indeed scales as (Tu − T)−1. Furthermore, the temperature dependencies D(T) for the WT and MT are almost identical except for the low temperatures T ≲ 370K ≈ Td (see Discussion).
Taken together, our results suggest that the docking (binding) of the MT and WT are similar and the peptide-fibril backbone HBs make minor contribution to binding.
Peptide-fibril hydrogen bonds and locked state
We now examine the impact of canceling peptide-fibril HBs on the locked state formed by the Aβ peptides. In our previous study we have showed that the locked state is associated with the formation of parallel β-sheets by the WT peptides bound to the fibril edge.22,32 For the WT the occupancy of the locked state exceeds 0.5 at the locking temperature Tl ≈ 360K. Appropriate progress variable mapping locking transition is the number of peptide-fibril parallel HBs, Nphb (see Methods). For the WT, <Nphb>≈ 6.0 at 360K.22 Even though the peptide-fibril HBs in the MT, including the parallel ones, are switched off, it is still instructive to analyze them. We found that at 360K the MT “forms” virtually no parallel HBs (<Nphb>≈ 0.2).
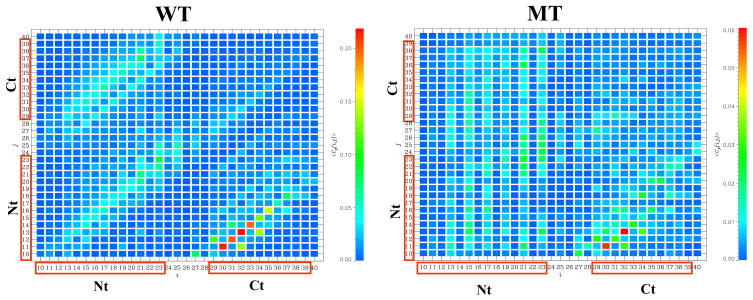
To provide further evidence that the MT locked state is disrupted, we compute the number of peptide-fibril parallel side chain contacts <Cp> (see Methods). At 360K the WT peptide forms <Cp>≈ 12.0 contacts with the fibril. When HBs are switched off, <Cp> decreases almost three-fold to 4.4. Additional observations concerning the changes in the locked state follow from the parallel contact maps <Cp(i, j)> (Fig. 3). The WT map <Cp(i, j)> indicates that parallel contacts tend to occur along the diagonals (including the main one) implying that the incoming Aβ peptide forms parallel in- or off-registry conformations with the fibril. The WT contact map <Cp(i, j)> is also consistent with the formation of long β -strands. In contrast, the MT map <Cp(i, j)> shows a disordered pattern, which seems to disfavor extended parallel arrangements between incoming peptides and the fibril.
Fig. 3.
The contact maps <Cp(i, j)> display the probabilities of forming side chain parallel contacts between the fibril residue i and the residue j from incoming peptide: the WT (left panel), the MT (right panel). The maps <Cp(i, j)> are computed at 360K and color coded according to the scales. The residues from the Nt and Ct sequence regions are boxed (Fig. 1a). The figure suggests that the deletion of peptide-fibril backbone HBs in the MT disfavors the formation of β-strand structure in the bound Aβ peptides.
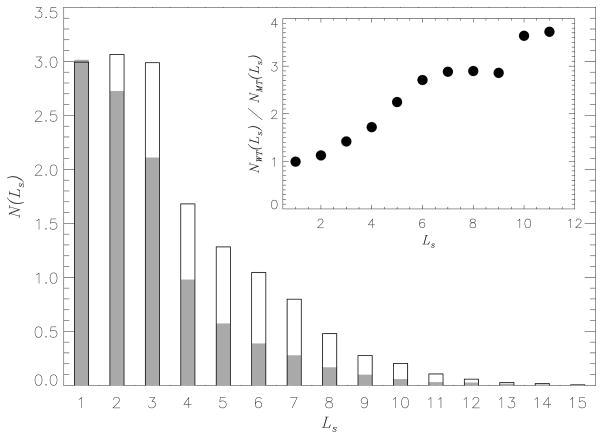
The lack of cooperativity in the formation of MT parallel contacts is supported by the distribution of β-strand lengths. Fig. 4 displays the thermal distribution of the number of residues N(Ls) belonging to the β -strands of the length Ls. It is seen that long β -strands are formed less frequently in the MT than in the WT. For example, the number of residues participating in short β-strands (Ls < 3) is similar for the MT and the WT (5.7 and 6.1, respectively). However, the number of residues in long β-strands (Ls ≥ 3) is reduced in half in the MT compared to the WT (4.7 vs 9.0, respectively). This observation is confirmed by the inset to Fig. 4, which shows the ratio NWT (Ls)/NMT (Ls) as a function of Ls. The number of residues occurring in the short β -strands is about the same (NWT (Ls)/NMT (Ls) ~ 1). In contrast, the probability of occurrence of long β-strands in the MT is suppressed. For instance, there is an almost four-fold decrease in the number of residues involved in the β-strands of the length Ls = 10.
Fig. 4.
Thermal distribution of the number of residues N(Ls) involved in β -strands of the length Ls in bound Aβ peptides. Filled and empty bars correspond to the MT and the WT, respectively. The inset shows the ratio NW T (Ls)/NMT (Ls) as a function of Ls, where NW T (Ls) and NMT (Ls) are computed for the WT and MT. The figure indicates that long β-strands occur less frequently in the MT Aβ peptides bound to the fibril than in the WT. The distributions N(Ls) are computed at 360K.
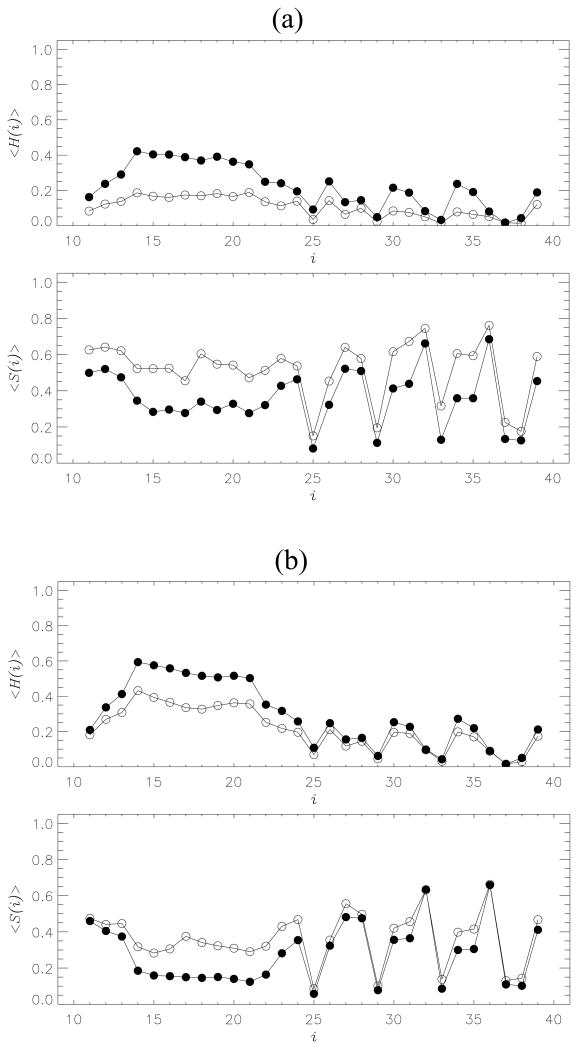
If there are few long β-strands in the MT, then one would expect that the deletion of interpeptide HBs changes the secondary structure in the bound Aβ peptides (Fig. 5a). Indeed, the average fraction of β-strand structure <S> is decreased one-third, from ≈ 0.52 (the WT) to ≈ 0.36 (the MT). Simultaneously, the helix fraction <H> increases two-fold, from 0.11 (the WT) to 0.22 (the MT). It follows from Fig. 5a that the most significant changes in the MT secondary structure are observed in the N-terminal Nt, in which the β-strand and helix contents, <S(Nt)> and <H(Nt)>, become almost equal (0.36 and 0.33, respectively). For comparison, for the WT <S(Nt)> and <H(Nt)> are 0.55 and 0.15.22
Fig. 5.
(a) Thermal distributions of the fractions of helix <H(i)> and β-strand <S(i)> structure formed by the residues i in Aβ10–40 peptides bound to the fibril: the MT (filled circles), the WT (open circles). (b) Distributions <H(i)> and <S(i)> computed for Aβ10–40 dimer: the MT (filled circles), the WT (open circles). The deletion of backbone HBs in the MT induces strand-to-helix structural conversion in the dimers and incoming peptides. The distributions <H(i)> and <S(i)> are obtained at 360K.
In the previous studies we have reported that the Aβ10–40 N-terminal represents the primary aggregation interface engaged in the fibril growth.32 To probe the MT aggregation interface, we analyze the distribution of peptide-fibril side chain contacts <C(s1,s2)> formed between sequence regions in the fibril s1 and incoming peptides s2. It follows from Table 1 that the Nt and Ct regions in the WT fibril form 18.3 and 14.7 contacts with incoming peptides, whereas the Nt and Ct regions in the WT incoming peptide form 20.8 and 12.1 contacts with the fibril.32 In the MT fibril, the Nt and Ct regions form 13.7 and 14.2 contacts with incoming peptides. There are 17.2 and 10.7 contacts formed by the Nt and Ct regions of the MT incoming peptide. For both systems, the Ct-Ct interactions are the least frequent. Overall, for the MT N-terminal the preference to be involved in the aggregation interface appears to be less pronounced compared to the WT. In fact, the standard deviation in the distribution of <C(s1,s2)> is 3.0 for the WT and 1.8 for the MT.
Table 1.
Distribution of side chain contacts <C(s1,s2)>‡ between incoming peptide and the fibril in the MT system
| fibril Nt | fibril Ct | |
|---|---|---|
| peptide Nt | 7.8(9.5)† | 9.4(11.3) |
| peptide Ct | 5.9(8.8) | 4.8(3.3) |
s1 and s2 denote sequence regions Nt and Ct in the fibril and incoming peptide.
Numbers in parenthesis refer to <C(s1,s2)> computed for the WT. Both distributions <C(s1,s2)> are obtained at 360K.
The results presented above argue that the deletion of peptide-fibril HBs destabilizes the locked state by disrupting the formation of long β-strands in Aβ peptides bound to the fibril. As a consequence an increase in the helix propensity and concurrent decrease in the β-strand content are observed in the bound Aβ peptides.
Interpeptide hydrogen bonds in Aβ dimers
To study the importance of backbone HBs for Aβ10–40 dimer formation we computed the thermal average of the number of interpeptide side chain contacts <Cd> in the MT dimer. At 360K <Cd> is ≈ 25.4 that is about 15% smaller than <Cd> in the WT dimer (≈ 30.0). The deletion of HBs has no impact on the probability of dimer formation Pd (i.e., the probability of forming any hydrophobic interpeptide side chain contact), which remains ≈ 1.0. However, the MT dimer has a smaller fraction of residues <S> sampling β-structure and an elevated helix content <H> (Fig. 5b). At 360K <S> is decreased from 0.37 (the WT) to 0.28 (the MT), whereas <H> reaches 0.29 in the MT compared to 0.21 in the WT. Therefore, upon deletion of HBs the helix and β-strand propensities become similar. Interestingly, the change in β-structure is mainly observed in the Nt region (Fig. 5b), where <S(Nt)> decreases from 0.36 in the WT to 0.22 in the MT, but <H(Nt)> raises from 0.32 to 0.46, respectively. As a result, in the MT <H(Nt)> exceeds <S(Nt)> more than two-fold. For comparison, the Ct region mostly retains its secondary structure (<S(Ct)> and <H(Ct)> change from 0.36 and 0.11 in the WT to 0.31 and 0.14 in the MT). The secondary structure changes are consistent with the analysis of the dimer free energy landscape (Fig. S2 in Supporting information). The free energy of the β rich state increases by ΔΔFS = 1.7RT, from ΔFS = −7.8RT in the WT to −6.1RT in the MT.
To check if canceling interpeptide HBs alters the dimer aggregation interface, we computed the distribution of interpeptide side chain contacts <Cd(s1, s2)> formed between Aβ10–40 sequence regions (Table 2). It is seen that the numbers of Nt-Nt contacts <Cd(Nt, Nt)> in the MT and WT are considerably larger than <Cd(s1, s2)> formed between other regions. For both systems <Cd(Nt, Nt)> constitutes about 41% of all <Cd(s1, s2)> contacts. Furthermore, it follows from Table 2 and Fig. S3 (Supporting Information) that for the MT the number of interpeptide contacts formed by the Nt <Cd(Nt)> (≈ 13.6) is almost twice as large as <Cd(Ct)> (≈ 7.2), the number of contacts formed by the Ct. For comparison, for the WT <Cd(Nt)>≈ 2 <Cd(Ct)> also. In addition, Table 2 shows that the values of <Cd(s1, s2)> for the MT and WT differ by no more than 20%. These findings suggest that the deletion of HBs mostly preserves the distribution of interpeptide interactions in the dimer and except for the changes in secondary structure has minor impact on its formation.
Table 2.
Distribution of interpeptide side chain contacts <Cd(s1, s2)>‡ in the MT dimer
| Nt | Ct | |
|---|---|---|
| Nt | 7.0(8.3)† | 4.1(4.8) |
| Ct | 1.9(2.3) |
s1 and s2 denote sequence regions Nt and Ct.
Numbers in parenthesis refer to <Cd(s1, s2)> computed for the WT. Due to exhaustive REMD sampling <Cd(Nt,Ct)>≈<Cd(Ct, Nt)>. Both distributions <Cd(s1, s2)> are obtained at 360K.
Discussion
Deletion of hydrogen bonds has different effect on docked and locked states
In this paper we examined the effect of deletion of HBs forming between Aβ incoming peptides and the fibril. By comparing the thermodynamics of fibril growth for the WT and MT fibrils we arrived at the following conclusions.
Docked state
Switching off peptide-fibril HBs causes minor destabilization of the bound state of Aβ peptides. In particular, the binding free energy for the MT increases ΔΔFB–U ≈ 1.4kcal/mol relative to the WT and the number of side chain contacts linking bound peptide to the fibril decreases 15 to 20% (Fig. 2). The deletion of HBs does not change the continuous character of Aβ binding to the fibril. The MT free energy profile F(Ch) in Fig. 2b remain barrierless and the temperature dependencies of the hexamer free energy F(T) and the thickness of the layer formed by bound peptides D(T) are similar for the WT and MT. From the fit to F(T) we inferred that the docking temperature Td is not affected by the backbone mutation.
Following our previous studies we have computed the probabilities of binding of the MT peptides to the concave (CV) and convex (CX) fibril edges, PCV and PCX (Fig. 1b and Fig. S4 in Supporting Information). To this end, we assumed that the peptide is bound to the CV (CX) edge, if the z-coordinate of its center of mass, zcm, is positive (negative). (Because peptide binding to the fibril sides is negligible, these definitions capture the binding to the edges.22) The MT incoming peptides demonstrate a strong preference to bind to the CV edge (PCV ≈ 0.92), whereas the CX edge has low affinity (PCX = 1 − PCV ≈ 0.08). Strikingly, the binding of the WT Aβ10–40 peptide is characterized by identical PCV and PCX values. To further substantiate this observation we have analyzed the free energy of the MT incoming peptide F(z) along the fibril axis z. The free energy F(z) has two well-defined minima associated with the CV and CX binding (data not shown). Importantly, the free energy gap between the two minima ΔFCV–CX = FCV − FCX ≈ −2.6RT is almost the same as that obtained earlier for the WT (≈ −2.5RT).22 Therefore, the deletion of HBs does not alter the preference of incoming Aβ peptides to bind to the CV edge.
Taken together, the results of our study suggest that the peptide-fibril HBs do not play a critical role in binding (docking) of Aβ peptides to the amyloid fibril.
Locked state
In contrast to the docked state, the locked state is disrupted by the cancellation of HBs. Due to the backbone mutation parallel HBs are eliminated and the number of parallel peptide-fibril side chain contacts <Cp> decreases three-fold implicating that parallel alignment of incoming peptides with the fibril is compromised. More importantly, Fig. 4 demonstrates that without HBs few long β-strands can be formed by the MT incoming peptides upon binding to the fibril edge. As a result compared to the WT the MT parallel contact map <Cp(i, j)> is poorly organized (Fig. 3). In line with these findings the deletion of HBs leads to partial conversion of the β-strand structure into helix in the bound Aβ peptides. These observations demonstrate that the extension and the formation of β-strands in the bound Aβ peptides are primarily induced by the peptide-fibril backbone HBs. Similar conclusion has been reached by us in the previous study, in which we compared the secondary structure in Aβ monomers, dimers, and the incoming peptides bound to the fibril.39
The temperature dependence of the bound layer thickness D(T) also supports the disruption of the MT locked state. The plots of D(T) obtained for the WT and MT diverge at low temperatures T ≲ 370K. In this temperature interval the WT D(T) levels off due to the formation of the locked state at Tl = 360K. In contrast, for the MT D(T) continues its monotonic decrease suggesting the lack of the rigid structure in the bound peptide.
To complement structural analysis we scrutinize the energetics of the MT binding to the fibril at 360K. Elimination of HBs increases the average effective energy <Eint> of interactions between incoming peptide and the fibril by <ΔEint>=<Eint(MT)> − <Eint(WT)>= 30.5kcal/mol (from −89.5 to −59.0kcal/mol). In particular, due to the backbone mutation peptide-fibril electrostatic and van-der-Waals interactions, <Eel> and <Evdw>, increase by 14.8 and 21.7kcal/mol, but these are partially offset by the decrease in the solvation energy <Esolv> by ≈ 5.9kcal/mol. Importantly, the cancellation of HBs directly contributes only to the change in electrostatic energy <Eel>. Therefore, changes in other energy terms implicate a structural reorganization in the bound Aβ peptides. This conclusion is consistent with the disruption of the β-rich locked state.
Finally, it is worth noting that the deletion of HBs reduces the polarization of the aggregation interface involved in the fibril growth. Most of peptide-fibril interactions for WT Aβ10–40 peptides involve the N-terminal Nt. However, this preference becomes muted for the MT. One may conjecture that the lack of ordered β-structure in the MT peptide reduces the differences in the binding affinities of the Nt and Ct sequence regions.
Thus, our results suggest that backbone HB are critically important for the locking stage in fibril growth. Elimination of HBs disrupts the locked state, but does not prevent the binding of incoming Aβ peptides to the amyloid fibril. Therefore, the deletion of HBs is expected to impede fibril elongation.
Deletion of hydrogen bonds has minor impact on Aβ10–40 dimer
We have explored the consequences of canceling backbone HBs formed between Aβ peptides in the dimer. Our data suggest that the deletion of HBs has a minor destabilizing effect on the dimer. The number of interpeptide side chain contacts in the MT dimer decreases about 15% compared to the WT. Furthermore, the distribution of interpeptide side chain contacts remains virtually intact in the MT, i.e., the respective changes do not exceed 20% (Fig. S3 and Table 2). As in the WT the N-terminal of the MT forms most of interpeptide contacts and constitutes the primary aggregation interface.39 Therefore, the interpeptide backbone HBs contribute relatively little to the dimer aggregation.
This conclusion is supported by the analysis of dimer energetics. At 360K the average effective energy <Eint> of interpeptide interactions is −61.8kcal/mol, which represents 69% of <Eint> associated with the peptide-fibril interactions. The average energies of van-der-Waals and electro-static interpeptide interactions <Evdw> and <Eel> in the dimer constitute approximately 70% of the respective peptide-fibril energies (−17.8 and −57.0kcal/mol for the dimer and −27.5 and −81.5kcal/mol for the peptide-fibril interactions). The same relation applies when the changes in solvation energy <Esolv> upon dimer formation or binding of incoming peptide are considered. However, the energy of interpeptide HBs <Ehb> in the dimer (−4.5kcal/mol) represents only 36% of the energy of peptide-fibril HBs (−12.6kcal/mol). These calculations illustrate small contribution of HBs to the aggregation interface in the dimer compared to that involved in the fibril growth. As a result, the deletion of HBs makes no significant change in the Aβ dimer stability.
Deletion of hydrogen bonds changes secondary structure propensities
It is important to note that switching off backbone HBs does change the secondary structure propensities in Aβ peptides. The average fraction of β-strand structure <S> is decreased 30% and 24% in the incoming peptides and dimer, respectively. Simultaneously, there is an increase in the helix fraction <H> in both Aβ systems. Interestingly, the most pronounced changes <ΔS>=<SMT> − <SWT> and <ΔH>=<HMT> − <HWT> occur in the Nt sequence region (Fig. 5, the subscripts refer to the MT and WT). For example, <ΔS(Nt)> and <ΔS(Ct)> are −0.14 and −0.05 for the dimer or −0.19 and −0.15 for the hexamer. Similarly, the changes in helix structure <ΔH(Nt)> and <ΔH(Ct)> are 0.14 and 0.03 for the dimer or 0.18 and 0.07 for the hexamer. For the dimer the ratio of β-strand to helix fractions, <S(Nt)> / <H(Nt)>, decreases from 1.1 in the WT to 0.5 in the MT. For the hexamer, <S(Nt)> / <H(Nt)> changes from 3.6 to 1.1, respectively. As a result in the Nt the helix propensity exceeds the β one (dimer) or becomes comparable with it (hexamer).
Because the formation of β-structure is a hallmark of locking Aβ peptides into the fibril, it is not unexpected that the deletion of HBs has a major impact on the fibril growth, but only minor impact on the dimer formation. Broadly speaking, the interpeptide HBs appear to drive the acquisition of β-strand content in aggregated Aβ species. Although their elimination does not block the dimer assembly or binding of incoming peptides to the amyloid fibril, without HBs the Aβ aggregated species become less competent to form amyloids.
Comparison with the experiment and other simulation studies
The contribution of HBs to aggregation has been studied experimentally by backbone mutagenesis.25,26,53 In these experiments, the amide backbone groups are replaced with N-methyl or ester groups, effectively eliminating one HB. It is also possible to introduce E-olefin backbone modifications, in which both, donor (NH) and acceptor (CO), are deleted that cancels up to three HBs. It has been shown that short amyloidogenic Aβ fragments, Aβ16–20, in which two ester bonds are introduced, do not assemble into amyloid fibrils and remain monomeric even at elevated concentrations (~ 1mM).24 Ester mutations have similar albeit not so dramatic effect on the full-length Aβ1–40 peptides. Single site amide-ester substitution in the Aβ central hydrophobic cluster (residues 17 to 20) may slow down aggregation.26 Interestingly, the WT amyloid fibril can still seed the Aβ1–40 mutant with ester backbone modification. These observations show that elimination of a single HB is not sufficient to prevent Aβ1–40 fibril formation. However, the E-olefin substitution at Phe19 and Phe20 in Aβ1–40, which disrupts several HBs per peptide in the fibril structure (Fig. 1), blocks fibril formation.27 Importantly, the WT Aβ1–40 fibril can no longer act as a deposition template for the E-olefin mutant peptides. Therefore, if sufficient number of backbone HBs is deleted, the fibril formation and growth can be inhibited.
Similar observations follow from the experiments on N-methylated Aβ mutants.25 Double backbone modifications at the positions Leu17 and Phe19 or at Gly37 and Val39 do not prevent fibril formation, but slows down aggregation by a factor of two. However, simultaneous insertion of N-methyl groups at these four sequence positions blocks aggregation and renders Aβ monomeric. Although N-methyl backbone mutations are not as conservative as ester or E-olefin substitutions, they are still indicative of the critical role played by backbone HBs in fibrillogenesis. Hence, the experiments on backbone mutagenesis are qualitatively consistent with our in silico findings.
The importance of backbone peptide-fibril HBs for fibril elongation also follows from the recent MD studies,54,55 which investigated the binding of short N-methyl peptides, Aβm, to Aβ fibril. According to these studies there are several modes of inhibiting fibril elongation. One includes Aβm binding to the fibril edges by forming backbone HBs. Because N-methyl mutations in Aβm backbone are introduced at alternating sequence positions, bound Aβm creates a “cap” with the outward “face” incapable of forming HBs with incoming peptides. Other modes occur when Aβm chains bind to the fibril in disordered orientation on its edges. Although those MD studies 55 were too short to observe fibril dissociation, they nevertheless revealed the destabilization of Aβ fibril fragments when coincubated with Aβm. These findings support our conclusion that the deletion of peptide-fibril HBs primarily destabilizes the locked state.
Finally, it is important to discuss the accuracy of implicit solvent model. We have previously showed that this model reproduces a number of experimental observations, 40 including the distribution of chemical shifts, dissociation temperature, and fibril growth mechanism. To provide additional test of the model accuracy we compare the energy of a backbone HB determined from the experiments and our simulations. The backbone ester and E-olefin substitutions afford an accurate estimate of the strength of backbone HB.56 Kelly and coworkers have found that a HB formed between the β-strands in the Pin WW domain has the energy of −1.3kcal/mol. Because this estimate is obtained in the apolar environment, it is likely to be in the upper end of HB stability.57 Our computational analysis of the WT binding energetics reveals that the electrostatic energy associated with the peptide-fibril HBs <Ehb> is −12.6kcal/mol. Because, on an average, the WT Aβ peptide forms <Nhb>≈ 10.5 HBs with the fibril, the average energy of a single peptide-fibril HB is −1.2kcal/mol. This estimate is very close to the experimental one suggesting that the implicit solvent model fairly well captures the energetics of fibril growth.
Conclusions
Using REMD and united atom implicit solvent model we examined the role of interpeptide backbone HBs in Aβ fibril growth and dimer formation. The importance of HBs appears to depend on the aggregation stage. Backbone HBs have little effect on the stability of Aβ dimers or on the distribution of dimeric interpeptide interactions. The HBs also do not play a critical role in binding (docking) of Aβ peptides to the amyloid fibril. Elimination of peptide-fibril HBs does not change the continuous character of Aβ docking transition or the temperature, at which it occurs. In contrast, cancellation of peptide-fibril HBs disrupts the locked states in the bound Aβ peptides. Without HBs incoming Aβ peptides form few long β-strands upon binding to the fibril edge. Because the bound Aβ peptides fail to form the fibril-like locked states, the deletion of peptide-fibril HBs is expected to impede fibril growth. As for the peptides bound to the fibril the deletion of interpeptide HBs reduces the β propensity in the dimers making them less competent for amyloid assembly. Taken together these findings suggest that interpeptide HBs play increasingly important role along the amyloidogenic assembly pathway becoming essential for the formation of fibril-like state. Our findings are consistent with the backbone mutagenesis experiments suggesting that a viable strategy for arresting fibril growth is the disruption of HBs between incoming peptides and the fibril. The energies of a typical backbone HB obtained from our simulations and estimated experimentally are in good agreement.
Supplementary Material
Acknowledgments
This work was supported by the grant R01 AG028191 from the National Institute on Aging (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or NIH. Fig. 1 was produced using the UCSF Chimera package.58 Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Kirkitadze MD, Bitan G, Teplow DB. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 3.Murthy RM. Ann Rev Biomed Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe DJ. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 5.Haass C, Selkoe DJ. Nature Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 6.Yoshiike Y, Akagi T, Takashima A. Biochemistry. 2007;46:9805–9812. doi: 10.1021/bi700455c. [DOI] [PubMed] [Google Scholar]
- 7.Serpell LC. Biochim Biophys Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 8.Burkoth TS, Benzinger T, Urban V, Morgan DM, Gregory DM, Thiyagarajan P, Botto RE, Meredith SC, Lynn DG. J Am Chem Soc. 2000;122:7883–7889. [Google Scholar]
- 9.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luhrs T, Ritter C, Adrian M, Loher B, Bohrmann DR, Dobeli H, Schubert D, Riek R. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paravastua AK, Leapman RD, Yaua WM, Tycko R. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meersman F, Dobson CM. Biochim Biophys Acta. 2006;1764:452–460. doi: 10.1016/j.bbapap.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Bitan G, Vollers SS, Teplow DB. J Biol Chem. 2003;278:34882–34889. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- 15.Jablonowska A, Bakun M, Kupniewska-Kozak A, Dadlez M. J Mol Biol. 2004;344:1037–1049. doi: 10.1016/j.jmb.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 16.Narayanan S, Reif B. Biochemistry. 2005;44:1444–1452. doi: 10.1021/bi048264b. [DOI] [PubMed] [Google Scholar]
- 17.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 18.Tseng BP, Esler WP, Clish CB, Stimson ER, Ghilardi JR, Vinters HV, Mantyh PW, Lee JP, Maggio JE. Biochemistry. 1999;38:10424–10431. doi: 10.1021/bi990718v. [DOI] [PubMed] [Google Scholar]
- 19.Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 20.Cannon MJ, Williams AD, Wetzel R, Myszka DG. Anal Biochem. 2004;328:67–75. doi: 10.1016/j.ab.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 21.O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 22.Takeda T, Klimov DK. Biophys J. 2009;96:442–452. doi: 10.1016/j.bpj.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kheterpal I, Zhou S, Cook KD, Wetzel R. Proc Natl Acad Sci USA. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon DJ, Meredith SC. Biochemistry. 2003;42:475–485. doi: 10.1021/bi0259857. [DOI] [PubMed] [Google Scholar]
- 25.Sciarretta KL, Boire A, Gordon DJ, Meredith SC. Biochemistry. 2006;45:9485–9495. doi: 10.1021/bi0605585. [DOI] [PubMed] [Google Scholar]
- 26.Bieschke J, Siegel SJ, Fu Y, Kelly JW. Biochemistry. 2008;47:50–59. doi: 10.1021/bi701757v. [DOI] [PubMed] [Google Scholar]
- 27.Fu Y, Bieschke J, Kelly JW. J Amer Chem Soc. 2005;127:15366–15367. doi: 10.1021/ja0551382. [DOI] [PubMed] [Google Scholar]
- 28.Ma B, Nussinov R. Curr Opin Struct Biol. 2006;10:445–452. doi: 10.1016/j.cbpa.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Buchete NV, Tycko R, Hummer G. J Mol Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 30.Zheng J, Jang H, Ma B, Tsai CJ, Nussinov R. Biophys J. 2008;93:3046–3057. doi: 10.1529/biophysj.107.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchete NV, Hummer G. Biophys J. 2007;92:3032–3039. doi: 10.1529/biophysj.106.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda T, Klimov DK. Biophys J. 2009;96:4428–4437. doi: 10.1016/j.bpj.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, Lei H, Duan Y. J Amer Chem Soc. 2005;127:13530–13537. doi: 10.1021/ja050767x. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen PH, Li MS, Stock G, Straub JE, Thirumalai D. Proc Natl Acad Sci USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krone MG, Hua L, Soto P, Zhou R, Berne BJ, Shea J-E. J Amer Chem Soc. 2008 doi: 10.1021/ja8017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda T, Klimov DK. Biophys J. 2008;95:1758–1772. doi: 10.1529/biophysj.108.131698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoler RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comp Chem. 1982;4:187–217. [Google Scholar]
- 38.Ferrara P, Apostolakis J, Caflisch A. Proteins Struct Funct Bioinform. 2002;46:24–33. doi: 10.1002/prot.10001. [DOI] [PubMed] [Google Scholar]
- 39.Takeda T, Klimov DK. Proteins Struct Funct Bioinf. 2009;77:1–13. doi: 10.1002/prot.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda T, Klimov DK. J Phys Chem B. 2009;113:11848–11857. doi: 10.1021/jp904070w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou L, Shao H, Zhang Y, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJL, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. J Amer Chem Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 42.Ferrara P, Caflisch A. Proc Natl Acad Sci USA. 2000;97:10780–10785. doi: 10.1073/pnas.190324897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiltpold A, Ferrara P, Gsponer J, Caflisch A. J Phys Chem B. 2000;104:10080–10086. [Google Scholar]
- 44.Cecchini M, Rao F, Seeber M, Caflisch A. J Chem Phys. 2004;121:10748–10756. doi: 10.1063/1.1809588. [DOI] [PubMed] [Google Scholar]
- 45.Paravastu AK, Petkova AT, Tycko R. Biophys J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeda T, Klimov DK. J Phys Chem B. 2009;113:6692–6702. doi: 10.1021/jp9016773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Vitkup D, Karplus M. J Mol Biol. 1999;285:1371–1375. doi: 10.1006/jmbi.1998.2374. [DOI] [PubMed] [Google Scholar]
- 48.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;114:141–151. [Google Scholar]
- 49.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 50.Ferrenberg AM, Swendsen RH. Phys Rev Lett. 1989;63:1195–1198. doi: 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- 51.Landau LD, Lifshitz EM. Course of Theoretical Physics. Vol. 5. Butterworth-Heinemann; Oxford: 1984. Statistical Physics. [Google Scholar]
- 52.Grosberg AY, Khokhlov AR. Statistical Physics of macromolecules. AIP Press; Woodbury: 1994. [Google Scholar]
- 53.Yang X, Wang M, Fitzgerald MC. Bioorg Chem. 2004;32:438–449. doi: 10.1016/j.bioorg.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Soto P, Griffin MA, Shea JE. Biophys J. 2007;93:3015–3025. doi: 10.1529/biophysj.107.112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chebaro Y, Derreumaux P. Proteins Struct Funct Bioinform. 2009;75:442–452. doi: 10.1002/prot.22254. [DOI] [PubMed] [Google Scholar]
- 56.Fu Y, Gao J, Bieschke J, Dendle MA, Kelly JW. J Amer Chem Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J, Bosco DA, Powers ET, Kelly JW. Nature Struct Mol Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.