Abstract
BACKGROUND
We have developed a range of adenoviral (Ad) vectors based on human adenovirus serotype 5 (HAdV-5) displaying the fiber shaft and knob domains of species B viruses (HAdV-3, HAdV-11, or HAdV-35). These species B Ads utilize different cellular receptors than HAdV-5 for infection. We evaluated whether Ad vectors displaying species B fiber shaft and knob domains (Ad5F3Luc1, Ad5F11Luc1, and Ad5F35Luc1) would efficiently infect cancer cells of distinct origins, including prostate cancer.
METHODS
The fiber chimeric Ad vectors were genetically generated and compared with the original Ad vector (Ad5Luc1) for transductional efficiency in a variety of cancer cell lines, including prostate cancer cells and primary prostate epithelial cells (PrEC), using luciferase as a reporter gene.
RESULTS
Prostate cancer cell lines infected with Ad5F3Luc1 expressed higher levels of luciferase than Ad5Luc1, as well as the other chimeric Ad vectors. We also analyzed the transductional efficiency via monitoring of luciferase activity in prostate cancer cells when expressed as a fraction of the gene transfer in PrEC cells. In the PC-3 and DU145 cell lines, the gene transfer ratio of cancer cells versus PrEC was once again highest for Ad5F3Luc1.
CONCLUSION
Of the investigated chimeric HAdV-5/species B vectors, Ad5F3Luc1 was judged to be the most suitable for targeting prostate cancer cells as it showed the highest transductional efficiency in these cells. It is foreseeable that an Ad vector incorporating the HAdV-3 fiber could potentially be used for prostate cancer gene therapy.
Keywords: adenovirus, species B, fiber, prostate cancer, gene therapy
INTRODUCTION
Prostate cancer is one of the most common cancers in men worldwide. In the U.S, prostate cancer is not only the most frequently diagnosed cancer in men (approximately 25%), it is also one of the most common fatal cancers in men in recent years [1]. During the initial stages of this disease, tumor growth depends upon androgen; and thus androgen ablation or anti-androgen treatments can be therapeutically useful [2]. However, tumor growth eventually becomes androgen independent leading to decreased effectiveness of these treatments during this stage of locally advanced or metastatic disease [3]. These considerations rationalize the development of new treatment strategies for prostate cancer therapy.
One of the new treatment strategies is gene therapy. The U.S. National Institutes of Health has approved a number of gene therapy clinical trials for prostate cancer (www.clinicaltrials.gov). These approaches can be divided into at least five categories, i.e. tumor-suppressor gene therapy, suicide gene therapy, immunomodulatory gene therapy, anti-oncogene therapy, and oncolytic virus therapy. Vectors based on human adenovirus serotype 5 (HAdV-5), Ad5 vectors, have been widely employed as the vehicles for these therapies as they have several advantages over other gene delivery systems. They can deliver large therapeutic genes [4,5], up to approximately 37 kb [6]. Also, these vectors can be amplified to very high titers, which is critical for in vivo gene delivery and clinical applications [7]. Moreover, Ad5 vectors are relatively safe as the HAdV genome does not integrate into the host genome [8,9].
Currently, at least 51 serotypes of HAdVs have been identified, which have been grouped into 6 species, A through to F [10]. Of these, HAdV-5, a serotype from species C, is mainly used as a vector for in vitro and in vivo studies of cancer gene therapy. However, clinical trials utilizing Ad5 vectors have provided disappointing results [11–13], mainly due to inefficient gene delivery to human cancer cells [14–16]. One explanation for this observation is that the expression level of the coxsackie virus and adenovirus receptor (CAR), which mediates cell attachment via the fiber protein of the Ad5 vector [17], is low in many cancers [14,18]. For example, CAR expression is decreased in specimens of prostate tumors at varying degrees of progression when compared with normal prostate tissue [19]. Thus, new Ad vectors which can utilize receptors other than CAR should be used for prostate cancer gene therapy. Since the fiber protein mediates Ad binding to its receptor, the fiber protein of the Ad5 vector will require modification in order to achieve efficient gene delivery via a CAR-independent mechanism.
Previous studies have shown that the fibers of species A, C, D, E, and F bind to soluble recombinant CAR, but those of species B do not [20]. This result shows that HAdVs from species B achieve cell entry via a CAR-independent pathway. In addition, several recent studies have identified cellular receptors for species B HAdVs, including human adenovirus 3 (HAdV-3), 11 (HAdV-11), and 35 (HAdV-35). Our group has identified CD80 and CD86, which are co-stimulatory molecules providing T cell activation [21], as receptors for HAdV-3 [22]. The expression of CD80 and CD86 is, however, restricted to lymphoid cells [23,24]. The expression of CD80 and/or CD86 is rarely observed in cancer cells [25–27], making targeting via CD80 and CD86 a particularly unsuitable strategy for cancer therapy. Recent studies have shown that HAdV-3 also binds to CD46 as a cellular receptor for cell entry [28,29]. Further, it has been demonstrated that HAdV-11 and HAdV-35 utilize CD46 as a cellular receptor [30–32]. CD46 is ubiquitously expressed throughout the human body [33] but, more importantly, it is generally overexpressed on various human cancer cells when compared with their normal counterparts [34–38]. In addition to CD46, an as yet unidentified receptor X has been determined to act as an alternative receptor for some serotypes of species B, including HAdV-3 and HAdV-11 [39]. Of note is the discovery that receptor X is highly expressed on human cancer cell lines [39]. The binding to and subsequent infection in tumor cells can thus be facilitated by targeting CD46 or receptor X. During the initial cellular attachment phase of viral infection, the fiber protein plays a critical role. We therefore hypothesized that the incorporation of a species B fiber in a HAdV-5 based vector would confer CAR-independent tropism to this vector as well as increase its infectivity in CAR-deficient cancer cells.
In this study, the fiber shaft and knob domains of a HAdV-5 vector were genetically replaced with the corresponding domains from HAdV-3, HAdV-11, or HAdV-35, resulting in Ad5F3Luc1, Ad5F11Luc1, and Ad5F35Luc1 respectively. We examined the biological and physical properties of the fiber chimeric Ad vectors and evaluated the infectivity in cancer cell lines derived from different origins, with a focus on prostate cancer cell lines. The results highlight the potential utility of these chimeric Ad vectors in prostate cancer gene therapy approaches.
MATERIALS AND METHODS
Cell Lines
Chinese hamster ovary (CHO), human embryonic kidney (HEK) 293 [40], human lung epithelial A549, human breast epithelial MCF-7, human chronic myelogenous leukemia (CML) K562 cells and three human prostate epithelial cell lines, LNCaP, PC-3, and DU145 cells were all obtained from the American Type Culture Collection (ATCC; Manassas, VA) and HEK293A cells were purchased from Invitrogen (Carlsbad, CA). All cells except K562 cells were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham, (DMEM/F12; Sigma-Aldrich; St. Louis, MO) containing 10% fetal bovine serum, (FBS; Hyclone; Logan, UT), 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Mediatech, Inc; Herndon, VA). K562 cells were cultured in Iscove’s DMEM (Mediatech, Inc; Herndon, VA) containing 10% FBS, 4 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293F28 cells [41], which are derivatives of HEK293 cells that express the wild type HAdV-5 fiber, were used to upscale the fiber chimeric Ad vectors and were cultured in the same medium as HEK293 cells but supplemented with 100 μg/ml zeocin (Invitrogen; Carlsbad, CA). Human glioblastoma U118 and their derivatives U118-hCAR cells, which stably express human CAR were previously described [42]. CHO-CD80 and CHO-CD86 stably expressing human CD80 and human CD86, respectively, were derivatives of CHO [22,43]. These cell lines were cultured in DMEM/F12 as above. A CHO-derived cell line, CHO-C2, which stably expresses the C2 isoform of human CD46 was previously reported [44] and was cultured in Eagle’s minimum essential medium (MEM; Mediatech) containing 10% FBS, 2 mM L-glutamine, 10 mM MEM non-essential amino acids (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin. Primary prostate epithelial cells (PrEC; Lonza, Walkersville, MD), including the ones infected with Ad vectors, were cultured using the prostate epithelial cell medium bulletkit (Lonza, Walkersville, MD). All cells were propagated at 37°C in a 5% CO2 atmosphere. The cell lines infected with Ad vectors were maintained using the corresponding cell culture medium but containing 2% instead of 10% FBS.
Construction of Recombinant Plasmids
All recombinant plasmids were constructed by “sticky-end” polymerase chain reactions (sePCR), which generate PCR products containing cohesive ends compatible with the intended restriction sites [45]. Table I summarizes the nucleotide sequences for the primers and the oligonucleotides used in this study. For the construction of the recombinant plasmid encoding the HAdV-3 fiber shaft and knob domains, the nucleotide sequence of these domains was amplified using primers SK3LF and 3KnR1 or primers SK3SF and 3KnR2 with pPIC9Ad3F (an unpublished construct) containing the Ad3 fiber gene as template. The resulting two PCR products were subsequently mixed, denatured, and annealed to generate a XmaI restriction enzyme overhang at the 5′ end of the HAdV-3 shaft domain (shown in bold for SK3LF and underlined for SK3LF and SK3SF in Table I) and four overhanging nucleotides at the 3′end of the HAdV-3 knob domain (shown in bold for 3KnR2 in Table I) corresponding to the overhang of BfuAI restriction enzyme site. The HAdV-5 fiber tail DNA sequence was amplified using plasmid pKAN3.1 [46] as template with primers SacSwa5T and Xma5T.R; SacI site for SacSwa5T and XmaI site for Xma5T.R are underlined in Table I. The PCR product for the HAdV-5 fiber tail domain was digested with SacI and XmaI. The two PCR products, i.e. the HAdV-3 fiber shaft and knob domains as well as the HAdV-5 fiber tail domain, were mixed and ligated simultaneously into the SacI and BfuA1 digested pNEB193/Swa5T/3ShaftBfu2x plasmid (an unpublished construct) containing only the nucleotide sequences for the HAdV-5 fiber tail and the HAdV-3 fiber shaft domains. This process resulted in plasmid pNEB193.5T3SK, which contained the nucleotide sequences for the HAdV-5 fiber tail, the HAdV-3 fiber shaft, as well as the HAdV-3 fiber knob domain.
TABLE I.
Primers used for cloning the recombinant plasmid DNA and PCR analysis
| Name of oligonucleotides | Direction | Sequence (5′ to 3′) | Position |
|---|---|---|---|
| SK3LF | Forward | 5′-ccGGGGTaTTAAGTCTTAAATGT-3′ | 31,497–31,517a,b |
| 3KnR1 | Reverse | 5′-ttcttcaGTCATCTTCTCTAATATAG-3′ | 32,324–32,306a |
| SK3SF | Forward | 5′-GGTaTTAAGTCTTAAATGTGTTAATCC-3′ | 31,499–31,525a,b |
| 3KnR2 | Reverse | 5′-acgattcttcaGTCATCTTCTCTAATATAG-3′ | 32,324–32,306a |
| SacSwa5T | Forward | 5′-taagagctcatttaAATGTCAGTTTCCTC-3′ | 30,948–30,998c |
| Xma5T.R | Reverse | 5′-gtaCCCCgGGtGGACTCTCTT-3′ | 31,177–31,160c, b |
| SK11/35LFf | Forward | 5′-ccggggtaCTTACTTTAAAATGT-3′ | 30,946–30,960d 30, 962–30,976e |
| 11KnR1 | Reverse | 5′-ttctTCAGTCGTCTTCTCTGATGTAG-3′ | 31,788–31,767d |
| SK11/35SFf | Forward | 5′-ggtaCTTACTTTAAAATGTTTAACCC-3′ | 30,946–30,967d 30, 962–30,983e |
| 11KnR2 | Reverse | 5′-acgattctTCAGTCGTCTTCTCTGATGTAG-3′ | 31,788–31,767d |
| 35KnR1 | Reverse | 5′-ttctTTAGTTGTCGTCTTCTGTAATG-3 | 31,798–31,777e |
| 35KnR2 | Reverse | 5′-acgattctTTAGTTGTCGTCTTCTGTAATG-3′ | 31,798–31,777e |
| MfeLF | Sense | 5′-TCGTTTGTGTTATGTTTCAACGTGTTTATTTTTCAATTGattt-3′ | 32, 792–32,830c |
| MfeSR | Anti-sense | 5′-aaatCAATTGAAAAATAAACACGTTGAAACATAACACAA-3′ | 32,830–32, 796c |
| SacCla5T | Forward | 5′-taagagctcatcgatTGTCAGTTTCCTC-3′ | 30,946–30,998c |
| Fiber −100 | Forward | 5′-CCTCCTGGCTGCAAACTTTCTC-3′ | 30,948–30,969c |
| Fiber +100 | Reverse | 5′-GAGGTGGCAGGTTGAATACTAG-3′ | 32,945–32,924c |
Position 1 refers to the beginning of the right arm of the HAdV-3 genome (accession No. DQ086466).
Point mutation(s) are in the nucleotide sequence corresponding to the HAdV genome
Position 1 refers to the beginning of the right arm of the HAdV-5 genome (accession No. AC_000008).
Position 1 refers to the beginning of the right arm of the HAdV-11 genome (accession No. AC_000015).
Position 1 refers to the beginning of the right arm of the HAdV-35 genome (accession No. AC_000019).
These primers anneal to the fiber genes for both HAdV-11 and HAdV-35.
Overhang is shown in bold.
Restriction enzyme sites that were used for cloning are underlined.
For the construction of the recombinant plasmids encoding the HAdV-11 or HAdV-35 fiber shaft and knob DNA sequences, a similar cloning strategy was utilized. HAdV-11 and HAdV-35 were purchased from the ATCC to amplify DNA sequences of their respective fiber shaft and knob domains utilizing two sets of primers: SK11/35LF and 11KnR1, or SK11/35SF and 11KnR2 for the HAdV-11 shaft and knob domains as well as SK11/35LF and 35KnR1 or SK11/35SF and 35KnR2 for the HAdV-35 shaft and knob domains. The resulting two PCR products for each Ad vector were subsequently mixed, denatured, and annealed to generate a XmaI restriction enzyme overhang at the 5′ end of the HAdV-11 or HAdV-35 shaft domain (shown in bold for SK11/35LF and underlined for SK11/35LF and SK11/35SF in Table I) and four overhanging nucleotides at the 3′ end of the HAdV-11 or HAdV-35 knob domain required for the ligation to another fragment containing a MfeI restriction enzyme site (shown for 11KnR2 or 35KnR2 in bold in Table I). Two kinds of oligonucleotides, MfeLF and MfeSR were synthesized and annealed to generate the double strand DNA fragment (overhanging four nucleotides at 5′ end to be used for the next step is shown in bold, a half site of nucleotides to be used for the next step recognized by SwaI is underlined, and the MfeI site to be used for cloning into the plasmid pKAN3.1, is also underlined in Table I). In addition, the HAdV-5 fiber tail DNA sequence was amplified using pKAN3.1 as template with primers SacCla5T and Xma5T.R, introducing a SacI site and a XmaI site (underlined for SacCla5T and Xma5T.R in Table I). The resulting PCR product for the HAdV-5 fiber tail domain was digested with the restriction enzymes SacI and XmaI. The PCR products for the shaft and knob domains, the DNA fragment containing the MfeI site, as well as the SacI and XmaI digested HAdV-5 fiber tail fragment were mixed and ligated simultaneously into the SacI and SwaI digested pNEB193/Cla5T/3ShaftBfu2x plasmid (an unpublished construct) containing the nucleotide sequences for the HAdV-5 tail and the HAdV-3 shaft. This process resulted in two plasmids, pNEB193.5T11SK and pNEB193.5T35SK. All three resulting plasmids (pNEB193.5T3SK, pNEB193.5T11SK, and pNEB193.5T35SK) were digested with AgeI and MfeI before ligation into the pKAN3.1 plasmid which was digested with the same enzymes. This additional cloning step was required for the subsequent homologous recombination step with a plasmid DNA carrying the HAdV-5 genome. The new plasmids were named pKAN5T3SK, pKAN5T11SK, and pKAN5T35SK. The nucleotide sequences of the DNA fragments cloned into all plasmids were confirmed by the UAB DNA sequencing core facility.
Construction of the Adenoviral Plasmids Encoding a Fiber Chimeric Gene
The constructed plasmids (pKAN5T3SK, pKAN5T11SK, and pKAN5T35SK) were used for homologous recombination with the SwaI-linearized pVK700 genomic plasmid. Plasmid pVK700 includes the HAdV-5 genome containing a firefly luciferase reporter gene controlled by the cytomegalovirus (CMV) promoter in the deleted early region 1 (E1) region [47]. The homologous recombination resulted in pMM703, pMM711, and pMM735 carrying chimeric fiber genes of HAdV-3, HAdV-11 and HAdV-35, respectively.
Adenovirus Generation, Propagation, Purification, and Titration
The recombinant plasmids pMM703, pMM711, and pMM735 were linearized by PacI and transfected into HEK293 cells using UniFECTOR transfection reagent (B-Bridge International Inc., Sunnyvale, CA) in order to generate Ad vectors, Ad5F3Luc1, Ad5F11Luc1, and Ad5F35Luc1. The cells were harvested in two to three weeks when cytopathic effect (CPE) was observed and disrupted by three freeze/thaw cycles. The cell lysates were centrifuged at 3,500 × g for 10 min at 4°C and used for reinfection of HEK293F28 cells. Subsequent to this, all Ad vectors were propagated by the two-step procedure developed by Von Seggern et al [48]. In brief, HEK293F28 cells were infected with the crude viral lysates to upscale mosaic Ad vectors incorporating wild type Ad5 fiber and fiber from species B HAdVs [41].
HEK293 cells were then infected with cell lysates containing mosaic Ad vectors producing the vectors incorporating only one type of fiber. The resulting Ad vectors were Ad5F3Luc1 incorporating the HAdV-5 fiber tail domain and the HAdV-3 fiber shaft and knob domains, Ad5F11Luc1 incorporating the HAdV-5 fiber tail domain and the HAdV-11 fiber shaft and knob domains, and Ad5F35Luc1 incorporating the HAdV-5 fiber tail domain and the HAdV-35 fiber shaft and knob domains (Table II). A control Ad vector, Ad5Luc1 is a replication-defective E1-deleted HAdV-5 based vector containing a firefly luciferase reporter gene driven by a CMV promoter [49]. All vectors produced in HEK293 cells were purified by two rounds of CsCl gradient ultracentrifugation [50]. CsCl was removed by dialysis using 10 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl containing 10% glycerol (dialysis buffer). The Ad vectors were stored at −80°C prior to the next experiments. The infectious titer (plaque forming units [PFU]/ml) of the purified Ads were determined by triplicate 50% Tissue Culture Infective Dose (TCID50) assays using HEK293A cells, as previously described [50]. The physical titer (viral particles [VP]/ml) were determined by Maizel’s method with a conversion factor of 1.1 × 1012 VP/ml per absorbance unit at 260 nm [51].
TABLE II.
Composition of the fiber domain of chimeric Ad vectors
| Name of an Ad vector | Fiber |
||
|---|---|---|---|
| Tail | Shaft | Knob | |
| Ad5Luc1 | 5 | 5 | 5 |
| Ad5F3Luc1 | 5 | 3 | 3 |
| Ad5F11Luc1 | 5 | 11 | 11 |
| Ad5F35Luc1 | 5 | 35 | 35 |
All numbers refer to serotype of human adenovirus
Polymerase Chain Reaction (PCR) Analysis of the Fiber Region
To confirm the presence of chimeric fiber genes in the Ad genomes, 107 VP of purified Ads were boiled at 95°C for 5 min and analyzed by PCR reactions using the TaqPCR Master Kit (Qiagen Inc., Valencia, CA). The sequences for primers and oligonucleotides used in this study are summarized in Table I. Forward (Fiber −100) and reverse (Fiber +100) primers were used to amplify the region containing the full length of the nucleotide sequences for all four types of fibers. Forward (SK3LF) and reverse (3KnR1) primers were used to amplify the region containing the HAdV-3 fiber shaft and knob nucleotide sequence. Forward (SK11/35LF) and reverse (11KnR1) primers were used to amplify the region containing the HAdV-11 fiber shaft and knob nucleotide sequence. Forward (SK11/35LF) and reverse (35KnR1) primers were used to amplify the region containing the HAdV-35 fiber shaft and knob nucleotide sequence (Table I). The following PCR conditions were applied: 1 min denaturation at 96°C, 1 min annealing at 60°C, and 3 min extension at 72°C to amplify the region of the whole fiber gene, or 1 min extension at 72°C, to amplify the nucleotide sequence of the shaft and knob domain.
Analysis of Adenoviral Proteins by GelCode Blue Staining and Western Blotting
Aliquots of purified Ad vectors equal to 1010 VP were denatured by boiling in Laemmli sample buffer (Bio-Rad laboratories Inc., Hercules, CA) at 95°C for 10 min. The proteins were separated by electrophoresis in sodium dodecyl sulfate 12% polyacrylamide gel (SDS-PAGE) and stained with GelCode Blue Stain Reagent according to the manufacturer’s instructions (Pierce, Rockford, IL).
For western blot analysis of the fiber proteins, aliquots of purified Ad vectors equal to 5 × 109 VP were denatured, and the viral proteins were separated by the same method described above. The separated viral proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and detected with a mouse monoclonal anti-HAdV-5 fiber tail domain antibody (4D2) (Lab Vision Corp., Fremont, CA), followed by a horseradish peroxidase-conjugated secondary anti-mouse antibody (Dako North America, Inc. Carpinteria, CA). The blot was developed using an ECL Plus Western Blotting Detection Kit (Amersham Biosciences, Piscataway, NJ). Pre-stained protein ladder of Kaleidoscope Standards (Bio-Rad Laboratories, Inc.) was used in the stained gel and the western blot.
Virus Thermostability Assay
Thermostability of Ad vectors was examined by the method described by Ugai et al. [52,53]. For each purified Ad, 1010 VP were incubated for three and seven days in the dialysis buffer at −80°C, 4°C, and 37°C. The resulting infectious titers of Ad vectors were determined by TCID50 using HEK293A cells.
One-step Growth Curve Analysis of Adenoviral Vectors
HEK293 cells, grown to 80% confluence in 6-cm dishes, were infected with Ad vectors at a multiplicity of infection (MOI) of 10 PFU/cell. The infected cells were maintained in 3 ml of medium containing 2% FBS. The culture medium and infected cells were scraped and collected in a 15-ml tube at various times (1, 12, 24, and 48 hours) post-infection. The cell suspension was centrifuged at 1,000 × g for 5 min at 4°C, and the cell pellet was resuspended in 2 ml of medium. The cells in the suspension were disrupted by three freeze/thaw cycles. The lysates were centrifuged at 3,500 × g for 10 min at 4°C and the supernatants were used for the subsequent infection.
Gene Transfer Assay
U118, U118-hCAR, HEK293, A549, MCF-7, DU145, LNCaP, PC-3, and PrEC cells grown in 24-well plates were infected with Ad vectors at a MOI of 10 PFU/cell. For blocking experiments, 0.2 ml of the recombinant HAdV-5 knob protein [54] diluted to the final concentration 0.5 and 5 μg/ml in FBS-free medium was added to the wells. No blocking agent was added to the control wells. Cells were incubated with the recombinant HAdV-5 knob protein at 4°C for one hour, subsequently Ad vectors at an MOI of 10 PFU/cell were added to the cells in 0.2 ml of medium containing 4% FBS. After incubation at 37°C for 24 hours, infected cells were washed with phosphate buffered saline (PBS; pH 7.4) and lysed in passive lysis buffer (Promega, Madison, WI). The cell lysates’ luciferase activities were measured according to the manufacturer’s protocol. Each data point was measured in triplicate and calculated as the mean of these three determinations.
To evaluate gene transfer efficiency of the Ad vectors at various incubation times (0, 12, 24, and 48 hours) at 37°C, the cells were infected with Ad vectors at an MOI of 10 PFU/cell in 0.4 ml of medium containing 2% FBS. After various incubation times the cell lysates were prepared, and the luciferase activities were measured as described above. Each data point was measured in triplicate and calculated as the mean of these three determinations.
The protein concentration was determined using the DC protein assay kit (Bio-Rad laboratories Inc., Hercules, CA) according to the manufacturer’s instructions, and the luciferase activity was normalized for protein concentration.
Flow Cytometry
Adherent cells (CHO, CHO-C2, CHO-CD80, CHO-CD86, U118, U118-hCAR, HEK293, A549, MCF-7, LNCaP, PC-3, and DU145,) were detached from a 75-cm2 tissue culture flask by treatment with Versene solution (0.53 mM EDTA in PBS [pH 7.4], UAB Media Preparation Shared Facility, Birmingham, AL) and washed with SM buffer (PBS [pH 7.4] containing 0.1% bovine serum albumin [BSA] and 0.01% sodium azide, stored at 4°C). K562 cells grown in suspension were harvested by centrifugation and washed with SM buffer. Subsequent to washing, aliquots of 5 × 105 cells were incubated for 1 hour at 4°C with either the anti-CD46 MAb J4-48 (Beckman Coulter, Fullerton, CA), the anti-CD80 MAb L307.4 (BD Pharmingen, San Jose, CA), the anti-CD86 MAb 2331 (BD Pharmingen), the anti-CAR MAb RmcB (a kind gift from Dr. Douglas [42]), or the anti-αv integrin antibody L230 (Axxora, San Diego, CA). Following this incubation, the cells were washed three times with SM buffer and incubated with Alexa fluor 488-conjugated goat anti-mouse immunoglobulin G (Invitrogen, Carlsbad, CA). After two additional washing steps, the cells were fixed with 0.5% paraformaldehyde in PBS (pH 7.4) and analyzed with a FACScan (Becton Dickinson, Mountain Vies, CA) at the UAB Cell Sorting Facility.
Determination of the Gene Transfer Ratio in Cancer versus PrEC Cells
As a means of showing transductional selectivity of the vectors for prostate tumor cells versus normal prostate, we divided the luciferase activity values in prostate cancer cells (LNCaP, PC-3, and DU145) with each vector’s luciferase activity in PrEC cells at 12 hours post-infection. Each vector’s gene expression ratios were that normalized to the control vector by dividing by the ratio of Ad5Luc1 in cancer cell lines compared with PrEC cells. This results in a value of 1 for the control Ad5Luc1 vector. Values greater than 1 indicate transductional selectivity for prostate cancer cells compared to Ad5Luc1.
Statistical Analysis
Statistical analysis was performed with two-tailed unpaired t-tests among groups. P values <0.05 were considered statistically significant.
RESULTS
Generation of Fiber Chimeric Adenoviral Vectors
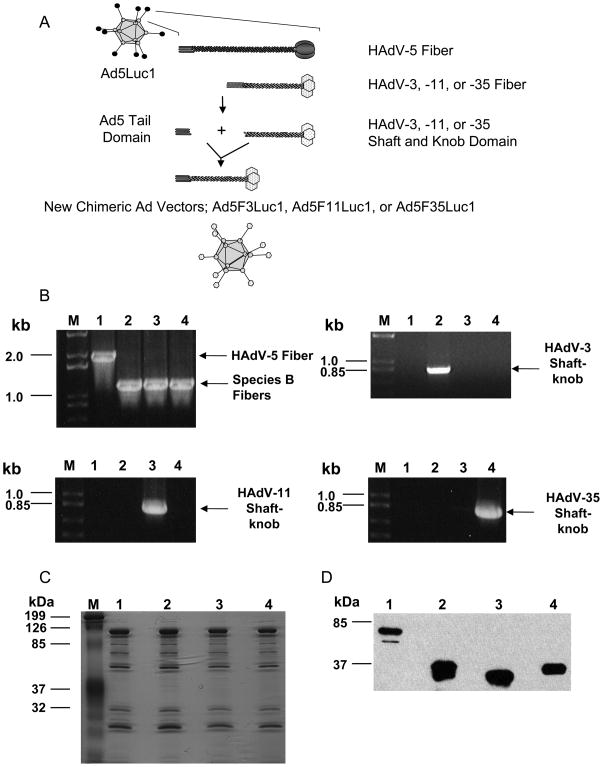
Plasmids pKAN5T3SK, pKAN5T11SK, and pKAN5T35SK encoding chimeric fibers were constructed by replacing the nucleotide sequence of the HAdV-5 fiber shaft and knob domains with the corresponding sequences from HAdV-3, HAdV-11, or HAdV-35 as described in the Materials and Methods section. Plasmids containing the recombinant Ad genomes with the chimeric fiber genes were subsequently derived by homologous recombination between the above plasmids and pVK700 [47]. The Ad vectors were generated in HEK293 cells, upscaled in HEK293F28 cells [41], and finally amplified in HEK293 cells to yield Ad vectors displaying only one type of fiber. Figure 1A shows a schematic representation of the fiber chimeric Ad vectors generated in this study. The composition of the fiber domain of the fiber chimeric Ad vectors is summarized in Table II. To confirm the replacement of the chimeric fiber gene in each vector, PCR analyses were done with the purified Ad vectors. The PCR analyses were performed with HAdV-5-specific primer sets, which anneal at 100 base pairs (bp) upstream and downstream of the HAdV-5 fiber gene, as well as serotype-specific primers for the fiber shaft and knob domains. Using the HAdV-5 specific primers, the PCR amplification produced a 1990 bp long HAdV-5 fiber product and approximately 1200 bp short fiber products for species B fiber (Fig. 1B, Upper left panel). The PCR analyses using serotype-specific primers demonstrated the amplification of the corresponding fiber region in each of the fiber chimeric Ad vector (Fig. 1B, upper right panel for Ad5F3Luc1, Fig. 1B, lower left panel for Ad5F11Luc1, and Fig. 1B, lower right panel for Ad5F35Luc1).
Fig. 1.
Characterization of chimeric Ad vectors. A: Schematic representation of a chimeric fiber of HAdV-3, HAdV-11, and HAdV-35 with HAdV-5 tail domain. B: Validation of the replacement of an HAdV-5 fiber gene with that of either HAdV-3, HAdV-11, or HAdV-35 in the HAdV-5 genome by PCR; all panels: M: DNA size markers; Lane 1: PCR using DNA from purified Ad5Luc1 as a template; Lane 2: PCR using DNA from purified Ad5F3Luc1 as a template; Lane 3: PCR using DNA from purified Ad5F11Luc1 as a template; Lane 4: PCR using DNA from purified Ad5F35Luc1 as a template. Upper left panel: PCR with pan fiber-specific primers; upper right panel: PCR with HAdV-3 fiber shaft knob-specific primers; lower left panel: PCR with HAdV-11 fiber shaft knob specific-primers; and lower right panel: PCR with HAdV-35 fiber shaft knob-specific primers. C: Analysis of protein composition of viral particles by GelCode blue staining of a 12% SDS-PAGE gel. A total of 1010 VP of purified Ad5Luc1 and chimeric Ad vectors was loaded per lane. M: protein molecular mass markers in kilodalton (kDa); Lane1: Ad5Luc1; Lane2: Ad5F3Luc1; Lane3: Ad5F11Luc1; Lane 4: Ad5F35Luc1. D: Detection of fiber proteins incorporated into purified viral particles by Western blot with an antibody against HAdV-5 fiber tail. A total of 5 × 109 VP of the purified Ad vectors were run on a 12% SDS-PAGE; separated viral proteins were transferred to a PVDF membrane and probed by an antibody against the HAdV-5 fiber tail (4D2). Lane1: Ad5Luc1; Lane2: Ad5F3Luc1; Lane3: Ad5F11Luc1; Lane 4: Ad5F35Luc1; protein molecular mass markers (in kDa) are indicated on the left.
To confirm that the viral structural proteins were not affected by the exchange of the fiber shaft-knob domains, the protein composition of the purified Ad5Luc1 and the chimeric Ad vectors was analyzed by protein staining analysis. The capsid proteins were separated by SDS-PAGE and stained with GELCODE blue. The fiber chimeric Ad vectors contained the Ad structural proteins in proportions similar to Ad5Luc1 (Fig. 1C).
It was also confirmed by western blot analysis with anti-HAdV-5 fiber tail domain 4D2 MAb that the viral particles incorporated the fiber shaft and knob domains from HAdV-3, HAdV-11, or HAdV-35 (Fig. 1D). The western blot analysis indicated that the molecular mass of the fiber monomer of Ad5F3Luc1, Ad5F11Luc1, and Ad5F35Luc1 is approximately 35–37 kDa, corresponding to the mass predicted by the amino acid sequences. These results clearly demonstrated the incorporation of the species B fiber proteins into the Ad5Luc1 viral particles. In addition, wild type HAdV-5 fiber, expressed by HEK293F28 cells which were used for first step of virus propagation, was not detected by western blot analysis of the purified viral particles (Fig. 1D).
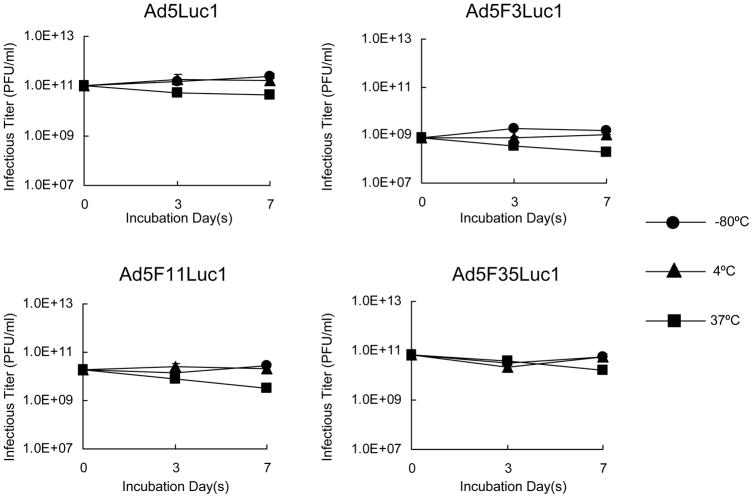
Thermostability of Fiber Chimeric Adenoviral Vectors
To determine if the thermostability of the resulting fiber chimeric Ad vectors was affected, a thermostability assay was performed [52,53]. Equal numbers of Ad vectors (1010 VP) containing either HAdV-5 or chimeric fibers were incubated at various temperatures (−80°C, 4°C, and 37°C) for 3 or 7 days followed by a TCID50 assay to analyze the remaining infectivity. All the Ad vectors demonstrated similar infectivity profiles subsequent to various time and temperature incubations, with all the Ad vectors displaying a decrease in infectivity after 7 days incubation at 37°C (Fig. 2). This data indicated that replacement of the fiber shaft-knob domain from species B HAdVs into the HAdV-5 viral particle does not influence the stability of the viral particles.
Fig. 2.
Thermostability of Ad5Luc1 and fiber chimeric Ad vectors. Purified viral particles (1010 VP) were incubated at indicated temperatures for 3 and 7 days, and the resulting infectivity was examined by titration in a triplicate TCID50 on HEK293 cells. −80°C (●), 4°C (▲), and 37°C (■). Data points represent mean ± standard deviation (n = 3).
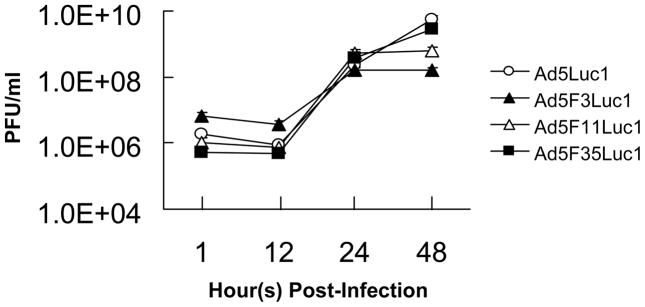
Analysis of Viral Replication
Since the fiber of Ad5Luc1 was replaced with species B fibers, we sought to determine whether this change affected the replication of the chimeric Ad vectors. A one-step growth curve analysis was performed in HEK293 cells using the lysate harvested and prepared from the infection of each Ad vector at an MOI of 10 PFU/cell in HEK293 cells. The kinetics of viral replication of each chimeric Ad vector were similar to that of Ad5Luc1 during 12 to 24 hours (Fig. 3). Therefore, replacement of the fiber shaft and knob domains did not affect viral replication.
Fig. 3.
Comparison of a one step growth curve obtained by infection of HEK293 cells with Ad5Luc1 (○), Ad5F3Luc1 (▲), Ad5F11Luc1 (△), and Ad5F3Luc1 (■). HEK293 cells were infected with the vectors above at an MOI of 10 PFU/cell. Data points represent mean ± standard deviation (n = 3).
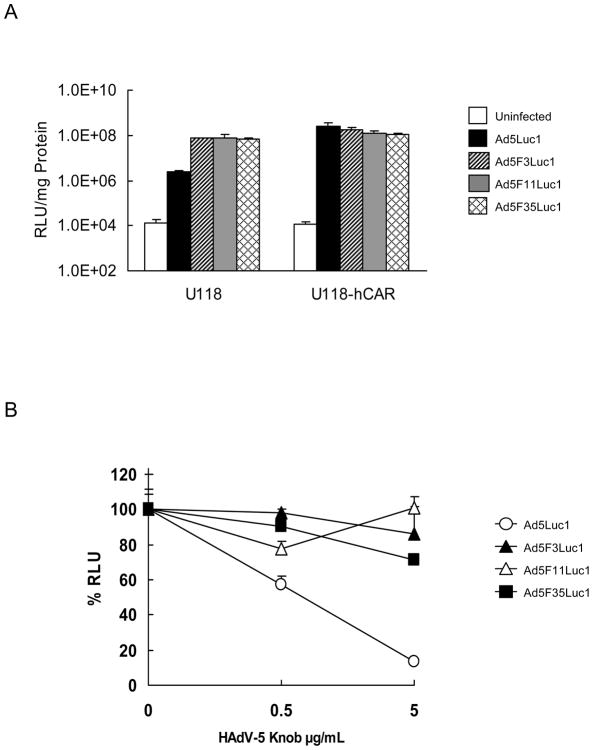
The Fiber of the Chimeric Adenoviral Vectors Dictates CAR-independent Tropism
As shown previously, the luciferase activity of Ad5Luc1-infected U118-hCAR cells, a cell line that stably overexpresses CAR [42], was 100-fold higher than that of CAR-negative U118 cells (Fig. 4A). These data indicate that gene transfer of Ad5Luc1 is dependent on the expression level of CAR. To verify whether the fiber chimeric Ad vectors achieve CAR-independent gene transfer, a well-established gene transfer experiment with HAdV-5 knob-blocking was performed [14,54,55]. Figure 4B demonstrates that the recombinant HAdV-5 knob protein inhibits transduction of Ad5Luc1, but not that of the chimeric Ad vectors. When blocking with the recombinant HAdV-5 knob protein at a final concentration of 0.5 μg/ml, the luciferase activity of Ad5Luc1-infected cells, but not that of cells infected by fiber chimeric Ad vectors, was lowered by more than 50% when compared to the activity in the absence of the blocking protein (Fig. 4B). Likewise, a final concentration of 5 μg/ml achieved an approximate 86% reduction in the luciferase activity of the cells infected with Ad5Luc1 when compared to unblocked controls. Some reduction in luciferase activity was observed for the fiber chimeric Ad vectors during the blocking experiment (Fig. 4B), but it was much lower compare to Ad5Luc1. These findings confirm that the fiber chimeric Ad vectors achieve cell entry via a CAR-independent route.
Fig. 4.
Gene transfer of Ad5Luc1 and chimeric Ad vectors in CAR-negative U118 and CAR expressing U118-hCAR cells, and gene transfer in A549 cells with HAdV-5 knob blocking. A: Luciferase activities in the lysates of CAR-negative U118 and CAR expressing U118-hCAR cells infected with no virus (white bar), Ad5Luc1 (black bar), Ad5F3Luc1 (diagonal bar), Ad5F11Luc1 (gray bar), and Ad5F35Luc1 (cross-hatch bar) at an MOI of 10 PFU/cell. The luciferase activity was measured 24 hours post-transduction. Data points represent mean ± standard deviation (n = 3). B: The percentage of the luciferase activities in the lysates of A549 cells infected with Ad5Luc1 (○), Ad5F3Luc1 (▲), Ad5F11Luc1 (△), and Ad5F35Luc1 (■) at an MOI of 10 PFU/cell after being blocked by the recombinant HAdV-5 knob protein at various concentrations. Final concentration of the recombinant HAdV-5 knob protein used to block transduction is indicated in μg/ml. The luciferase activity was measured at 24 hours post-transduction. Bars represent the mean ± S.E.M. (n = 3).
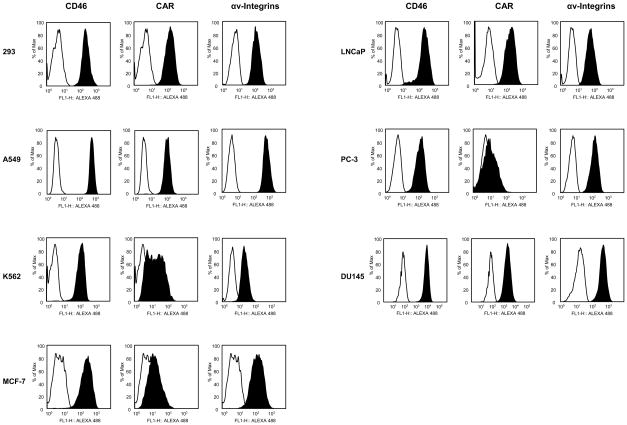
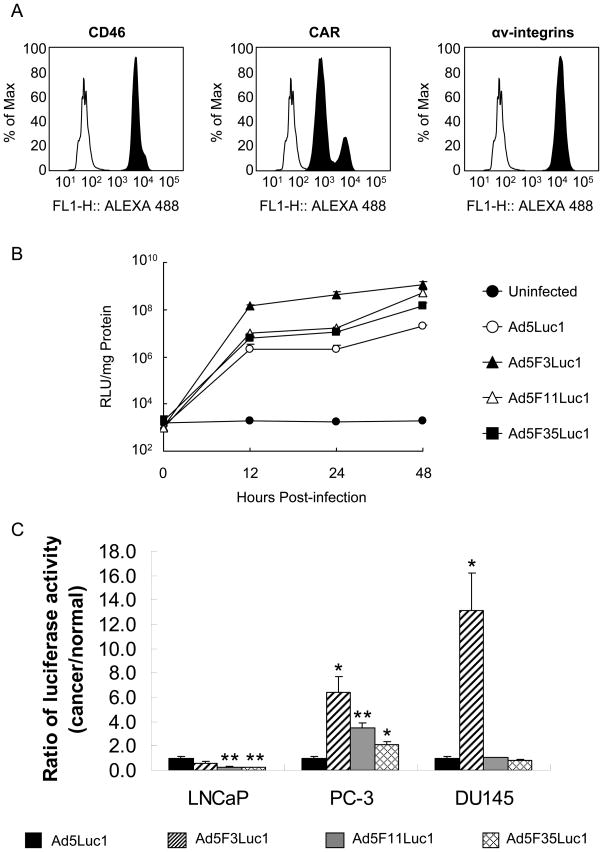
Expression Analysis of Adenoviral Receptors in Cancer Cell lines
Previous studies have identified CD46 as a primary receptor for human adenovirus species B [28–32]. Flow cytometry was performed to determine the expression levels of CD46 on the following cell lines: HEK293, A549, K562, MCF-7, DU145, LNCaP, and PC-3; with CHO-C2 cells utilized as a positive control (data not shown) for CD46 expression [44]. All cancer cell lines highly expressed CD46 (Fig. 5). In addition to the CD46 expression levels, the expression levels of CAR and αv-integrins, which are required for the HAdV-5 infection were analyzed [56]. For this analysis, U118-hCAR cells [42] were used as a positive control for CAR expression and HEK293 cells were used as a positive control for the expression of αv-integrins [57]. The cell lines A549, HEK293, DU145, and LNCaP expressed very high levels of CAR and the cell line K562 expressed medium levels of CAR (Fig. 5). MCF-7 and PC-3 cells, however, expressed very low levels of CAR (Fig. 5). Expression of αv-integrins was high in all cancer cell lines except K562, which expressed only moderate levels (Fig. 5). The expression levels of CD80 and CD86 were also determined, as it has been shown that CD80 and CD80 are the receptors for HAdV-3 [22]. The flow cytometry data showed that K562 cells expressed medium levels of CD80 (41% of cells were positive), and A549 cells expressed very little CD80 (3 % of cells were positive). All other cell lines were CD80 negative (less than 1 % of cells were positive). All cell lines were CD86 negative (less than 1 %of cells were positive) except for K562, which expressed very low levels (1.7% of cells were positive) (data not shown).
Fig. 5.
Analysis of expression of CD46, CAR, and αv-integrins on various cancer cell lines by flow cytometry. Filled, black histograms indicate stained cells; open, white histograms indicate unstained control cells.
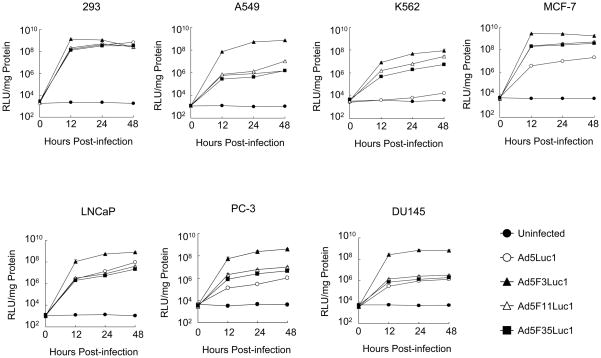
Gene Delivery by the Fiber Chimeric Adenoviral Vectors in Cancer Cell Lines
The level of luciferase gene expression in the chimeric Ad vectors-infected cells was compared with that of Ad5Luc1-infected cells in the same cell lines used for receptor analysis by flow cytometry (Fig. 6). In HEK293 cells, which are positive for both CAR and CD46, all Ad vectors, including Ad5Luc1, provided the same expression pattern of luciferase. The levels of luciferase in all CD46-positive cells that had low to medium levels of CAR (K562, MCF-7 and PC-3), were higher for cells infected with the fiber chimeric Ad vectors compared to Ad5Luc1 (Fig. 6). Except for HEK293 cells, the expression levels obtained with Ad5F3Luc1 were higher than those obtained with the other vectors in CAR- and CD46-positive cells (A549, LNCaP, and DU145). The expression levels obtained with Ad5F11Luc1, AdF35Luc1, and Ad5Luc1, however, were equally low in these same cells as compared to Ad5F3Luc1. Of note, the Ad5F3Luc1 vector provided higher gene delivery in prostate cancer cell lines than did Ad5Luc1 or the other chimeric Ad vectors. Further, the luciferase activities in all cells reached a plateau by 12 hours post-infection.
Fig. 6.
Comparison of transductional efficiency in various human cancer cells (HEK293, A549, K562, and MCF-7) and prostate cancer cells (LNCaP, DU145, and PC-3) with Ad5Luc1 and chimeric Ad vectors at various times. The luciferase activities in the lysates of cells infected with Ad5Luc1 (○) and chimeric Ad vectors Ad5F3Luc1 (▲), Ad5F11Luc1 (△), and Ad5F35Luc1 (■) at an MOI of 10 PFU/cell were measured at various time points (0, 12, 24, and 48 hours post-transduction) and normalized for protein concentration. Data points represent mean ± standard deviation (n = 3). Closed circles (●) indicate background luciferase activity.
Transduction Efficiency of the Fiber Chimeric Adenoviral Vectors in Normal Human Prostate Cells (PrEC)
Ad5F3Luc1 demonstrated a higher transduction efficiency in prostate cancer cell lines when compared with the other Ad vectors. We then sought to determine the transduction efficiency of these Ad vectors in primary prostate epithelial cells (PrEC). To this end, we initially determined the expression of CD46, CAR, and αv-integrins on these cells. As shown in Fig. 7A, PrEC cells express high levels of CD46, CAR and αv-integrins. At 12 hours post-infection, Ad5F3Luc1 achieved 14 to 70-fold higher transduction than the other fiber chimeric Ad vectors or Ad5Luc1 in PrEC cells. Luciferase activity reached a plateau at 12 hours post-infection, similar to what was observed in the cancer cell lines (Figs. 6 and 7B).
Fig. 7.
Analysis of gene transfer in PrEC cells. A: Analysis of CD46, CAR, and αv-integrins expression in prostate epithelial cells (PrEC) by flow cytometry. Filled, black histograms indicate stained cells; open, white histograms indicate unstained control cells. B: Comparison of transduction efficiency in primary prostate epithelial cells with Ad5Luc1 and chimeric Ad vectors. The luciferase activities in the lysates of cells infected with Ad5Luc1 (○) and the chimeric Ad vectors Ad5F3Luc1 (▲), Ad5F11Luc1 (△), and Ad5F35Luc1 (■) at an MOI of 10 PFU/cell were measured at various time points (0, 12, 24, and 48 hours post-transduction) and normalized for protein concentration. Data points represent mean ± standard deviation (n = 3). Closed circles (●) indicate background luciferase activity. C: Gene transfer in prostate cancer cells (LNCaP, PC-3, and DU145) expressed as a ratio of the gene transfer in primary prostate epithelial cells at 12 hours post transduction, normalized by that of Ad5Luc1; Ad5Luc1 (black bar), Ad5F3Luc1 (diagonal bar) Ad5F11Luc1 (gray bar), and Ad5F35Luc1 (cross-hatch bar). Bars represent the mean ± S.E.M. A t-test statistical analysis was performed with respect to Ad5Luc1 and significance is indicated by * P < 0.05; ** P < 0.01 (n = 3).
The viral infectivity was further evaluated by comparing the ratio of the luciferase activity in prostate cancer cells to that in PrEC cells. Figure 7C shows the ratios of luciferase activity in prostate cancer cell lines versus PrEC cells at 12 hours post-infection relative to the luciferase activity of Ad5Luc1, with increased values being indicative of greater cancer cell gene transductional specificity. The chimeric vectors other than Ad5F3Luc1 had significantly lower specificity for LNCaP cells (CD46+/CAR+) compared to Ad5Luc1 (p = 0.0045 and p = 0.0042 respectively), and the ratios of Ad5F3Luc1 and Ad5Luc1 were not significantly different (p = 0.1172) for LNCaP cells. Ad5F3Luc1 demonstrated the highest specificity in PC-3 (CD46+/CAR−) and DU145 (CD46+/CAR+) cell lines, with respective luciferase activity ratios of approximately 6 and 13 fold higher than that of Ad5Luc1 (p = 0.0142 and p = 0.0168 respectively). Ad5F11Luc1 and Ad5F35Luc1 also showed significantly higher specificity in CD46+/CAR− PC-3 cell lines with respective luciferase activity ratios of approximately 3.5 and 2, (p = 0.0057 and p = 0.0274 respectively) but no specificity in DU145 cells (CD46+/CAR+) compared to Ad5Luc1. These results clearly indicate that Ad5F3Luc1 has the most favorable prostate tumor-selective tropism.
DISCUSSION
This study clearly demonstrates that chimeric Ad vectors incorporating a fiber from species B (HAdV-3, -11, or -35) have distinct infectivity in different types of cancer cells. In particular, Ad5F3Luc1, which incorporates the HAdV-3 fiber shaft and knob domains, seems to be a very suitable vector for prostate cancer. These conclusions are based on the observed biological and physical properties of the fiber chimeric Ad vectors, as well as their infectivity in cancer cell lines derived from different tumors. In fact, many groups have replaced the fiber shaft and knob domains or only the fiber knob domain of HAdV-5 with that of other human Ads, including those of species B [54,58–60] and non-human Ads [14,55], in order to develop HAdV-5 based vectors that achieve CAR-independent infectivity. However, the effect on viral particle structure by the replacement of the fiber shaft and knob domains of HAdV-5 with the species B fiber shaft and knob domains has not been thoroughly evaluated. We first investigated whether the replacement of the HAdV-5 fiber would negatively impact the Ad biological and physical properties. It was found that all the fiber chimeric Ad vectors had the same protein composition as Ad5Luc1 (Fig. 1C), and appeared to be equally thermostable (Fig. 2). The fiber chimeric Ad vectors had kinetics of viral replication similar to that of Ad5Luc1 from 12 to 24 hours, although the replacement of the HAdV-5 fiber with the chimeric fiber of HAdV-3 or HAdV-11 appeared to reduce the total production of adenovirus at 48 hours post-infection in 293 cells (Fig. 3). Altogether, our results indicate that the incorporation of species B fiber shaft and knob domains into the Ad5 vector has no significant effects on the viral particles, their thermostability or infectious dynamics.
We have demonstrated that the chimeric Ad vectors incorporating species B fiber shaft and knob domains achieve cell entry without utilizing CAR (Fig. 4B). Moreover, we analyzed the transductional efficiency of the fiber chimeric Ad vectors in various prostate cancer cell lines with known CAR, CD46 and integrin expression levels (Fig. 5). In this regard, the results of our receptor expression analyses were similar to the findings previously reported by Sandberg et al. [27]. Our group previously identified CD80 and CD86 as receptors for HAdV-3 attachment; however, neither of these two receptor types were expressed on the prostate cancer cell lines we examined (data not shown). Despite these findings, we found that Ad5F3Luc1 had significantly increased transductional efficiency in prostate cancer cell lines compared to the other Ad vectors (Fig. 6). Therefore, the transduction of Ad5F3Luc1 into the prostate cancer cells was mediated via receptors other than CD80 and CD86.
It has also been shown that HAdV-3 can utilize CD46 as a cellular attachment receptor [28,29], which is overexpressed in cancer cells [34–38]. However, the binding affinity of HAdV-3 for CD46 is lower than that of HAdV-11 and HAdV-35 [28,31,39]. In addition to the findings regarding HAdV-3 receptor binding, Tuve et al. concluded that HAdV-3 utilized an as yet unknown receptor called receptor X [39]. These results indicate that HAdV-3 may utilize a broad range of receptors to achieve the high transduction observed.
It would be advantageous to develop a vector with a high infectivity of prostate cancer cells, but a low infectivity of normal cells. We therefore examined the transductional efficiency of species B fiber chimeric Ad vectors in primary normal prostate epithelial cells (PrEC). PrEC cells express CD46, CAR, and αv-integrins, as determined by flow cytometry (Fig. 7A). Once again, Ad5F3Luc1 had the highest transductional efficiency in PrEC when compared to the other vectors (Fig. 7B). However, as stated before, the transductional efficiency of Ad5F3Luc1 was also the highest in all prostate cancer cell lines (Fig. 6). To gauge the relative transductional efficiency in prostate cancer versus normal prostate cells, we calculated the ratio of the gene transfer obtained in prostate cancer cells to the gene transfer obtained in PrEC cells. In the PC-3 and DU145 cell lines, the gene transfer ratio of prostate cancer versus normal prostate cells for Ad5F3Luc1 was the greatest among the tested vectors (Fig. 7C). The result indicates that, of the investigated subgroup B chimeric Ad vectors, Ad5F3Luc1 is the best vector to target prostate cancer cells. Of note, both PC-3 and DU145 cell lines are androgen-insensitive and can be considered as examples of more progressed cancer than LNCaP, which is androgen-sensitive [61]. Since androgen-insensitive prostate cancer is especially in need of alternative treatments than the currently available methods, the development of Ad5F3Luc1-based vectors is more clinically relevant.
Using prostate samples from patients, Loberg et al. demonstrated that the expression level of CD46 was not significantly different among normal prostate tissue, prostatic intraepithelial neoplasia, localized prostate cancer or metastatic prostate cancer specimens [62]. The replacement of the HAdV-5 fiber with that of HAdV-11 and HAdV-35 may not be a good strategy to target only prostate tumor for clinical application as CD46 is a high affinity cellular receptor for HAdV-11 and HAdV-35.
In summary, replacement of the fiber shaft and knob domains of HAdV-5 did not affect viral growth properties, and the transduction efficiency of these fiber chimeric Ad vectors dramatically increased in CAR-negative cancer cell lines. Furthermore, Ad5F3Luc1 achieved the highest level of gene transfer in prostate cancer cells. Of note, Ad5F3Luc1 achieved the greatest ratio of gene transfer in progressed prostate cancer to normal prostate cells. Also, a future extension of Ad5F3Luc1 vector usage for clinical application could be to generate a conditionally replicative adenoviral vector (CRAd) containing the HAdV-3 fiber shaft-knob. Such a CRAd would more efficiently target and kill prostate cancer cells compared to normal prostate cells, thus contributing to the development of new treatments for prostate cancer.
Acknowledgments
We thank Enid F. Keyser and Marion L. Spell for FACS analysis. We also thank Drs. Masato Yamamoto, George C. Dobbins, Ruan van Rensburg and Joel N. Glasgow for useful advice and careful review of manuscript. This work was supported by a grant from the National Institute of Health: 7PO1CA104177.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury S, Burbridge S, Harper PG. Chemotherapy for the treatment of hormone-refractory prostate cancer. Int J Clin Pract. 2007;61(12):2064–2070. doi: 10.1111/j.1742-1241.2007.01551.x. [DOI] [PubMed] [Google Scholar]
- 4.Fisher KJ, Choi H, Burda J, Chen SJ, Wilson JM. Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217(1):11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 5.Kochanek S, Clemens PR, Mitani K, Chen HH, Chan S, Caskey CT. A new adenoviral vector: Replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and beta-galactosidase. Proc Natl Acad Sci U S A. 1996;93(12):5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer DJ, Ng P. Helper-dependent adenoviral vectors for gene therapy. Hum Gene Ther. 2005;16(1):1–16. doi: 10.1089/hum.2005.16.1. [DOI] [PubMed] [Google Scholar]
- 7.Wivel NA, Gao G, Wilson JM. Adenovirus vectors. In: Friedmann T, editor. The Development of Human Gene Therapy, Cold Spring Harbor Monograph Series. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 87–110. [Google Scholar]
- 8.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 9.Verma IM, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 10.Kojaoghlanian T, Flomenberg P, Horwitz MS. The impact of adenovirus infection on the immunocompromised host. Rev Med Virol. 2003;13(3):155–171. doi: 10.1002/rmv.386. [DOI] [PubMed] [Google Scholar]
- 11.Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Ther. 2001;8(2):89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- 12.Khuri FR, Nemunaitis J, Ganly I, Arseneau J, Tannock IF, Romel L, Gore M, Ironside J, MacDougall RH, Heise C, Randlev B, Gillenwater AM, Bruso P, Kaye SB, Hong WK, Kirn DH. a controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6(8):879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds PN, Curiel DT. New generation adenoviral vectors improve gene transfer by coxsackie and adenoviral receptor-independent cell entry. Kidney Int. 2002;61(1 Suppl):S24–31. doi: 10.1046/j.1523-1755.2002.0610s1024.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama M, Both GW, Banizs B, Tsuruta Y, Yamamoto S, Kawakami Y, Douglas JT, Tani K, Curiel DT, Glasgow JN. An adenovirus serotype 5 vector with fibers derived from ovine atadenovirus demonstrates CAR-independent tropism and unique biodistribution in mice. Virology. 2006;350(1):103–115. doi: 10.1016/j.virol.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 15.Leon RP, Hedlund T, Meech SJ, Li S, Schaack J, Hunger SP, Duke RC, DeGregori J. Adenoviral-mediated gene transfer in lymphocytes. Proc Natl Acad Sci U S A. 1998;95(22):13159–13164. doi: 10.1073/pnas.95.22.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickham TJ, Roelvink PW, Brough DE, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14(11):1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 17.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 18.Tsuruta Y, Pereboeva L, Glasgow JN, Luongo CL, Komarova S, Kawakami Y, Curiel DT. Reovirus sigma1 fiber incorporated into adenovirus serotype 5 enhances infectivity via a CAR-independent pathway. Biochem Biophys Res Commun. 2005;335(1):205–214. doi: 10.1016/j.bbrc.2005.07.054. [DOI] [PubMed] [Google Scholar]
- 19.Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, Schmitt LD, McCormick F. Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res. 2002;62(13):3812–3818. [PubMed] [Google Scholar]
- 20.Roelvink PW, Lizonova A, Lee JG, Li Y, Bergelson JM, Finberg RW, Brough DE, Kovesdi I, Wickham TJ. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72(10):7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultze J, Nadler LM, Gribben JG. B7-mediated costimulation and the immune response. Blood Rev. 1996;10(2):111–127. doi: 10.1016/s0268-960x(96)90040-5. [DOI] [PubMed] [Google Scholar]
- 22.Short JJ, Pereboev AV, Kawakami Y, Vasu C, Holterman MJ, Curiel DT. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology. 2004;322(2):349–359. doi: 10.1016/j.virol.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Collins M, Ling V, Carreno BM. The B7 family of immune-regulatory ligands. Genome Biol. 2005;6(6):223. doi: 10.1186/gb-2005-6-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 25.Mutti L, Valle MT, Balbi B, Orengo AM, Lazzaro A, Alciato P, Gatti E, Betta PG, Pozzi E. Primary human mesothelioma cells express class II MHC, ICAM-1 and B7-2 and can present recall antigens to autologous blood lymphocytes. Int J Cancer. 1998;78(6):740–749. doi: 10.1002/(sici)1097-0215(19981209)78:6<740::aid-ijc12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Yu L, Takenobu H, Shimozato O, Kawamura K, Nimura Y, Seki N, Uzawa K, Tanzawa H, Shimada H, Ochiai T, Tagawa M. Increased infectivity of adenovirus type 5 bearing type 11 or type 35 fibers to human esophageal and oral carcinoma cells. Oncol Rep. 2005;14(4):831–835. [PubMed] [Google Scholar]
- 27.Sandberg L, Papareddy P, Silver J, Bergh A, Mei YF. Replication-competent Ad11p vector (RCAd11p) efficiently transduces and replicates in hormone-refractory metastatic prostate cancer cells. Hum Gene Ther. 2009;20(4):361–373. doi: 10.1089/hum.2007.124. [DOI] [PubMed] [Google Scholar]
- 28.Fleischli C, Sirena D, Lesage G, Havenga MJ, Cattaneo R, Greber UF, Hemmi S. Species B adenovirus serotypes 3, 7, 11 and 35 share similar binding sites on the membrane cofactor protein CD46 receptor. J Gen Virol. 2007;88(Pt 11):2925–2934. doi: 10.1099/vir.0.83142-0. [DOI] [PubMed] [Google Scholar]
- 29.Sirena D, Lilienfeld B, Eisenhut M, Kalin S, Boucke K, Beerli RR, Vogt L, Ruedl C, Bachmann MF, Greber UF, Hemmi S. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol. 2004;78(9):4454–4462. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segerman A, Atkinson JP, Marttila M, Dennerquist V, Wadell G, Arnberg N. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol. 2003;77(17):9183–9191. doi: 10.1128/JVI.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9(11):1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 32.Marttila M, Persson D, Gustafsson D, Liszewski MK, Atkinson JP, Wadell G, Arnberg N. CD46 Is a Cellular Receptor for All Species B Adenoviruses except Types 3 and 7. J Virol. 2005;79(22):14429–14436. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnstone RW, Loveland BE, McKenzie IF. Identification and quantification of complement regulator CD46 on normal human tissues. Immunology. 1993;79(3):341–347. [PMC free article] [PubMed] [Google Scholar]
- 34.Hara T, Kojima A, Fukuda H, Masaoka T, Fukumori Y, Matsumoto M, Seya T. Levels of complement regulatory proteins, CD35 (CR1), CD46 (MCP) and CD55 (DAF) in human haematological malignancies. Br J Haematol. 1992;82(2):368–373. doi: 10.1111/j.1365-2141.1992.tb06431.x. [DOI] [PubMed] [Google Scholar]
- 35.Thorsteinsson L, O’Dowd GM, Harrington PM, Johnson PM. The complement regulatory proteins CD46 and CD59, but not CD55, are highly expressed by glandular epithelium of human breast and colorectal tumour tissues. Apmis. 1998;106(9):869–878. doi: 10.1111/j.1699-0463.1998.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 36.Kinugasa N, Higashi T, Nouso K, Nakatsukasa H, Kobayashi Y, Ishizaki M, Toshikuni N, Yoshida K, Uematsu S, Tsuji T. Expression of membrane cofactor protein (MCP, CD46) in human liver diseases. Br J Cancer. 1999;80(11):1820–1825. doi: 10.1038/sj.bjc.6690604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray KP, Mathure S, Kaul R, Khan S, Carson LF, Twiggs LB, Martens MG, Kaul A. Expression of complement regulatory proteins-CD 35, CD 46, CD 55, and CD 59-in benign and malignant endometrial tissue. Gynecol Oncol. 2000;76(2):176–182. doi: 10.1006/gyno.1999.5614. [DOI] [PubMed] [Google Scholar]
- 38.Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol Immunol. 2003;40(2–4):109–123. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 39.Tuve S, Wang H, Ware C, Liu Y, Gaggar A, Bernt K, Shayakhmetov D, Li Z, Strauss R, Stone D, Lieber A. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J Virol. 2006;80(24):12109–12120. doi: 10.1128/JVI.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 41.Belousova N, Korokhov N, Krendelshchikova V, Simonenko V, Mikheeva G, Triozzi PL, Aldrich WA, Banerjee PT, Gillies SD, Curiel DT, Krasnykh V. Genetically targeted adenovirus vector directed to CD40-expressing cells. J Virol. 2003;77(21):11367–11377. doi: 10.1128/JVI.77.21.11367-11377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M, Sumerel LA, Belousova N, Lyons GR, Carey DE, Krasnykh V, Douglas JT. The coxsackievirus and adenovirus receptor acts as a tumour suppressor in malignant glioma cells. Br J Cancer. 2003;88(9):1411–1416. doi: 10.1038/sj.bjc.6600932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasu C, Wang A, Gorla SR, Kaithamana S, Prabhakar BS, Holterman MJ. CD80 and CD86 C domains play an important role in receptor binding and co-stimulatory properties. Int Immunol. 2003;15(2):167–175. doi: 10.1093/intimm/dxg017. [DOI] [PubMed] [Google Scholar]
- 44.Oglesby TJ, White D, Tedja I, Liszewski K, Wright L, Van den Bogarde J, Atkinson JP. Protection of mammalian cells from complement-mediated lysis by transfection of human membrane cofactor protein and decay-accelerating factor. Trans Assoc Am Physicians. 1991;104:164–172. [PubMed] [Google Scholar]
- 45.Zeng G. Sticky-end PCR: new method for subcloning. Biotechniques. 1998;25(2):206–208. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]
- 46.Borovjagin AV, Krendelchtchikov A, Ramesh N, Yu DC, Douglas JT, Curiel DT. Complex mosaicism is a novel approach to infectivity enhancement of adenovirus type 5-based vectors. Cancer Gene Ther. 2005;12(5):475–486. doi: 10.1038/sj.cgt.7700806. [DOI] [PubMed] [Google Scholar]
- 47.Belousova N, Krendelchtchikova V, Curiel DT, Krasnykh V. Modulation of adenovirus vector tropism via incorporation of polypeptide ligands into the fiber protein. J Virol. 2002;76(17):8621–8631. doi: 10.1128/JVI.76.17.8621-8631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Von Seggern DJ, Kehler J, Endo RI, Nemerow GR. Complementation of a fibre mutant adenovirus by packaging cell lines stably expressing the adenovirus type 5 fibre protein. J Gen Virol. 1998;79 (Pt 6):1461–1468. doi: 10.1099/0022-1317-79-6-1461. [DOI] [PubMed] [Google Scholar]
- 49.Krasnykh V, Belousova N, Korokhov N, Mikheeva G, Curiel DT. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J Virol. 2001;75(9):4176–4183. doi: 10.1128/JVI.75.9.4176-4183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanegae Y, Makimura M, Saito I. A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J Med Sci Biol. 1994;47(3):157–166. doi: 10.7883/yoken1952.47.157. [DOI] [PubMed] [Google Scholar]
- 51.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36(1):115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 52.Ugai H, Watanabe S, Suzuki E, Tsutsui-Nakata H, Yokoyama KK, Murata T. Stability of a recombinant adenoviral vector: optimization of conditions for storage, transport and delivery. Jpn J Cancer Res. 2002;93(5):598–603. doi: 10.1111/j.1349-7006.2002.tb01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ugai H, Borovjagin AV, Le LP, Wang M, Curiel DT. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J Mol Biol. 2007;366(4):1142–1160. doi: 10.1016/j.jmb.2006.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krasnykh VN, Mikheeva GV, Douglas JT, Curiel DT. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70(10):6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glasgow JN, Kremer EJ, Hemminki A, Siegal GP, Douglas JT, Curiel DT. An adenovirus vector with a chimeric fiber derived from canine adenovirus type 2 displays novel tropism. Virology. 2004;324(1):103–116. doi: 10.1016/j.virol.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 56.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 57.Shayakhmetov DM, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol. 2000;74(22):10274–10286. doi: 10.1128/jvi.74.22.10274-10286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stecher H, Shayakhmetov DM, Stamatoyannopoulos G, Lieber A. A capsid-modified adenovirus vector devoid of all viral genes: assessment of transduction and toxicity in human hematopoietic cells. Mol Ther. 2001;4(1):36–44. doi: 10.1006/mthe.2000.0410. [DOI] [PubMed] [Google Scholar]
- 59.Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J Virol. 2000;74(6):2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizuguchi H, Hayakawa T. Adenovirus vectors containing chimeric type 5 and type 35 fiber proteins exhibit altered and expanded tropism and increase the size limit of foreign genes. Gene. 2002;285(1–2):69–77. doi: 10.1016/s0378-1119(02)00410-9. [DOI] [PubMed] [Google Scholar]
- 61.Liu AY. Differential expression of cell surface molecules in prostate cancer cells. Cancer Res. 2000;60(13):3429–3434. [PubMed] [Google Scholar]
- 62.Loberg RD, Wojno KJ, Day LL, Pienta KJ. Analysis of membrane-bound complement regulatory proteins in prostate cancer. Urology. 2005;66(6):1321–1326. doi: 10.1016/j.urology.2005.06.094. [DOI] [PubMed] [Google Scholar]