Abstract
Cytoplasmic type I DnaJ/Hsp40 chaperones contain a Cys-rich domain consisting of four CXXCXG motifs that are in a reduced state and coordinate zinc, stabilizing the intervening sequence in a loop structure. However, the Cys-rich region of the endoplasmic reticulum localized HEDJ (ERdj3/ERj3p), is considerably different in sequence and arrangement. Unlike the typical type I molecule, the HEDJ CXC and CXXC motifs were demonstrated in this study to be predominantly oxidized in intramolecular disulfide bonds. In the native state, HEDJ bound to immobilized, denatured thyroglobulin. Unlike its binding partner GRP78, redox conditions affected the interaction of HEDJ with substrate. Substitution of the Cys-rich domain cysteine residues with serine diminished or abolished HEDJ binding in the in vitro assay. These findings suggest that the Cys-rich region of HEDJ and its oxidation state are important in maintaining the substrate interaction domain in a binding-competent conformation.
Keywords: chaperone, Hsp40, DnaJ, endoplasmic reticulum, GRP78, BiP, redox, HEDJ
INTRODUCTION
Within the ER1, proteins are folded, processed and sorted into pathways leading to expression at the cellular membrane, secretion or retrotranslocation and degradation. A number of resident ER chaperones contribute to the processing of protein folding intermediates. One chaperone of the DnaK/Hsp70 family, GRP78 has been shown to assist in the translocation of nascent protein chains into the ER, participate in protein folding and help retrotranslocate misfolded proteins to the proteasome for degradation [1–4]. GRP78 can be assisted by several proteins that have a J domain, a region identified by its homology to the J domain of DnaJ/Hsp40. Five such J domain-containing proteins have been identified in the mammalian ER, including MTJ1/HTJ1/ERdj1, hSec63/ERdj2, HEDJ/ERdj3/ERj3p, ERdj4 and ERdj5/JPDI [5–12]. The J domain from each of these proteins has been shown to bind to GRP78 and stimulate the ATPase activity of GRP78 [7–11, 13, 14]. The complete folding of client proteins interacting with GRP78 depends on cycles of ATP binding, hydrolysis and release [3].
DnaJ/Hsp40 proteins have been classified as type I, II or III. Type I proteins contain three regions in common: a J domain, a Gly/Phe-rich domain and a Cys-rich domain of four CXXCXG motifs [15]. Type I proteins bind substrates, prevent their aggregation and subsequently target them to the Hsp70 partner [16]. Type II proteins have the characteristic J and Gly/Phe domains and bind client proteins independently, but require an interaction with the Hsp70 partner in order to prevent aggregation of the client protein [16]. Type III proteins, with only the J domain in common, are most likely dedicated to specific client proteins [16]. Several ER, J-domain proteins contain a novel combination of motifs and are difficult to classify as strictly type I, II or III. For example, ERdj5/JPDI contains a J domain and Cys-rich domains that are PDI-like and thioredoxin-like [10, 11].
The Cys-rich domain of prototypic type I proteins such as the E. coli chaperone DnaJ, consist of four reduced, non-adjacent cysteine motifs that co-ordinate zinc and stabilize the intervening sequence in a loop structure [17]. In these chaperones, the Cys-rich domain can contribute to structural stability of the adjacent substrate-binding pocket and it is thought to be involved in the transfer of substrate to Hsp70 [18, 19]. Although HEDJ does contain both the typical J and Gly/Phe domains of the DnaJ/Hsp40 family, it has an atypical Cys-rich domain that contains cysteines arrayed as single CXC and CXXC motifs. Unlike DnaJ and other type I homologues, which are located in the relatively reducing cytoplasmic environment, HEDJ is located in the more oxidizing ER lumen. The redox state of the Cys residues in these motifs and their contribution to substrate interactions are not known.
Recent functional studies on HEDJ indicate that it is an important chaperone/co-chaperone for unfolded proteins. This ER stress protein is widely expressed, with higher levels in secretory tissues [8, 20]. It has been identified as part of a network of chaperones associated with the unassembled immunoglobulin heavy chain that is not completely folded and is one of the chaperones in a complex with the Shiga toxin subunit A [8, 21, 22]. It associates with an immunoglobulin κ light chain mutant and with the VSV-G ts045 mutant at the non-permissive temperature [20]. One recent study using a J domain mutant has suggested that HEDJ may bind to client proteins independently of GRP78 [20].
In the present study, the Cys-rich domain redox state and its contribution to substrate interactions were examined. In native HEDJ, the Cys residues were found to be predominantly oxidized, forming intramolecular disulfide-bridges. The reduction of Cys residues altered the conformation of HEDJ as judged by migration on native gels. HEDJ was capable of binding to denatured substrate molecules independently of GRP78. Disruption of its disulfide bonds, either by the addition of reducing agents or by site directed mutagenesis of the Cys-rich domains, significantly altered the conformation of HEDJ and diminished its ability to bind to client proteins. This study highlights the importance of the novel combination of domains in the ER localized, DnaJ/Hsp40 homologue, HEDJ and suggests possible functions for its Cys-rich motifs within the ER compartment.
MATERIALS AND METHODS
Materials, Antibodies
Immobilized thyroglobulin conjugated to 4% crosslinked-beaded agarose, goat anti-mouse IgG1-agarose, equine myoglobulin, dithiothreitol (DTT), and diamide were purchased from Sigma (St. Louis, MO). Antibodies to GRP78, GRP94 and HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibody to HA was purchased from Covance Research Products (Denver, PA), and rabbit anti-HA antibody was from Sigma. Antibody to Sec61β was custom made (Cocalico Biologicals, Reamstown, PA) to the N terminal sequence: PGPTPSGTNC [14]. Antibody to HEDJ was prepared in rabbits (Cocalico Biologicals) to a peptide representing the C-terminal 19 residues and purified by affinity chromatography on immobilized peptide. Methoxy poly(ethylene glycol) maleimide (PEG-Mal, mw 5000) was from Nektar Therapeutics-Shearwater (Huntsville, AL). Other reagents were from Sigma or Fisher Scientific unless noted.
COS-1, HepG2 and HeLa cells were cultured in DMEM with 4.5g/L glucose and L-glutamine (Cambrex BioScience, Walkersville, MD), 10% fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin and 100 μg/ml streptomycin and incubated at 5% CO2.
Modified microsomal preparation and binding assay
A post-nuclear microsomal fraction was freshly prepared for each binding assay. Cell cultures (100 mm plates) were washed 2 times with PBS and scraped into 0.9 ml aliquots of cold hypotonic lysis buffer (25 mM HEPES, pH 8, 11 mM KCl) and homogenized in a Dounce homogenizer with 50 strokes. The ionic composition of the lysates was adjusted by adding 0.1 ml of cold 1 M KCl. To remove nuclear and cellular debris, two successive centrifugations at 4°C were done: a 10 min centrifugation was done at 1200 × g and the tubes were re-centrifuged for 15 min at approximately 5000 × g. Supernatants were centrifuged at 140,000 × g in a Beckman Type 70.1 Ti rotor for 30 min at 4°C. The pellets were suspended in HK buffer (HEPES, 25 mM, pH 8, 125 mM KCl) with 2 mM MgCl2, 0.5 mM CaCl2, 0.5% Triton X-100 and protease inhibitors (Complete mini, EDTA-free, Roche Diagnostics). Lysed pellets were clarified by centrifugation at 12,000 × g for 10 min at 4 °C. The total protein concentration of the preparation was determined by the BCA protein assay (Pierce, Rockford, IL). Some microsomal lysates were treated with 50 U/ml apyrase for 30 min at RT to lower ATP levels. Others received 10 mM glucose and 5 U/ml hexokinase for 1 h [23].
The binding assay was a modification of an earlier method [24]. Immobilized thyroglobulin or immunoglobulin was denatured (dTg and dIg) in 6 M urea, 1 M β mercaptoethanol (urea denaturation) for 40 min at RT with rocking. Alternatively, immobilized Tg or Ig was denatured in 4 M GdHCl, 100 mM Tris HCl, pH 8, 20 mM DTT. After 10 min incubation at RT with rocking, iodoacetamide (IA) was added (100 mM final concentration) and the solution incubated for an additional 30 min (GdHCl denaturation). dTg or dIg beads were washed 4 times in HK buffer with 2.5 mM EDTA. Beads were washed and equilibrated in HK buffer containing 2 mM MgCl2 and 0.5 mM CaCl2 (binding buffer). Aliquots of dTg or dIg were incubated with microsomal lysate (50 μl packed beads to 50 μg protein in 50 μl binding buffer adjusted to a final concentration of 0.25% Triton X-100) for 2–3 h at 4°C with rocking. The slurry was transferred to a Handee Mini-spin Column (Pierce) and the flow-through collected and combined with a subsequent wash of 2.5 ml of binding buffer. This fraction is labeled F. dTg or dIg beads were washed with an additional 0.5 ml of binding buffer and this fraction (W) was collected and analyzed separately to demonstrate that the initial wash had been sufficient to remove unbound protein. After 10 min of incubation in 0.5 ml elution buffer at RT, the buffer was collected (E). Fractions were TCA precipitated, washed and dissolved in reducing SDS sample buffer at 95° C for 3 min. Proteins were resolved using SDS-PAGE (10% or 4–15% gradient gel, Bio-Rad, Hercules, CA) and immunoblotted to PVDF (Immobilon P, Millipore). Samples were visualized with chemiluminescence (ECL, Amersham or SuperSignal, Pierce) and recorded on autoradiography film. Each set of binding assays included HEDJ or WT-HA to validate assay conditions. The recovery of WT-HA or HEDJ in collected fractions was estimated for several of the columns by including a lane in the SDS-polyacrylamide gel for an aliquot of microsomes that had not been fractionated or processed.
Construction of HEDJ Mutants
Mutants were made using PCR mutagenesis. The wild type HA-tagged mutant (WT-HA) was made by using the pHED3 vector [8] with the forward primer referred to as GFP-1 (GGCCTCACAGGGCCGGGTGGGCTGG) and the reverse primer referred to as HEDJ-HA (GTATACAATGGACTGCAAGGATATGGCTATCCTTATGACGTGCCTGACTATGCCTCAGGCTAGCCG). The mutant (1, see nomenclature in Figure 1) containing the mutation of the first Cys in the sequence to a Ser was made by amplifying the 5′ portion with primers GFP-1 and Cys1R, CCGCATCTCTTGCCGACAATTTGACTTCCGTTTGCCAGG. The 3′ portion was amplified with the forward primer Cys1F, CCTGGCAAACGGAAGTCAA-ATTGTCGGCAAGAGATGCGG and the reverse primer HEDJ-HA. The full length, mutated cDNA was generated by combining the 5′ and 3′ PCR products and subjecting the mixture to PCR using flanking primers GFP-1 and HEDJ-HA. Similarly, mutant 2 was made as for mutant 1, except the mutant specific primers were Cys2R, CCGCATCTCTTGCCGTGAATTGCACTTCCGTTTGCC and Cys2F, GGCAAACGGAAGTG-CAATTCACGGCAAGAGATGCGG. For mutant 3, the primers were Cys 3R, GACATTAGGGCATTCGTCTGAGACCACCTCCTGGG and Cys3F, CCCAGGAGGTGGTCTCAGACGAATGCCCTAATGTC. Mutant 4 was made with the primers Cys4R, CACTAGTTTGACATTAGGTGATTCGTCGCAGACCACC and Cys4F, GGTGGTCTG CGACGAATCACCTAATGTCAAACTAGTG. The 1.2 mutant was made with 1.2R, GGTCCGCATCTCTTGCCGTGAATTTGACTTCCGTTTGCCAGGAGCC and 1.2F, GGCTCC TGGCAAACGGAAGTCAAATTCACGGCAAGAGATGCGGACC. The 3.4 mutant was made using primers 3.4R, CATTCACTAGTTTGACATTAGGTGATTCGTCTGAGACCACCTCCTGGGTC and 3.4F, GACCCAGGAGGTGGTCTCAGACGAATCACCTAATGTCAAACTAGTGAATG. The 1.3 double mutant was made using the single Cys 3 mutant as a template, GFP-1 and primer Cys1R for the 5′ half and primer Cys1F with HEDJ-HA for the 3′ half. The full-length construct was made using GFP-1 and HEDJ-HA. The other double mutants were made in the same way, using the appropriate single mutant as a template and the appropriate primers from the other single mutant. PCR products were inserted into the pcDNA3.1/V5-His TOPO TA vector (Invitrogen, Carlsbad, CA). To make the NoCys mutant, 1.2 and 3.4 mutants were cut with Apa I and ligated together. All inserts were sequenced to ensure fidelity.
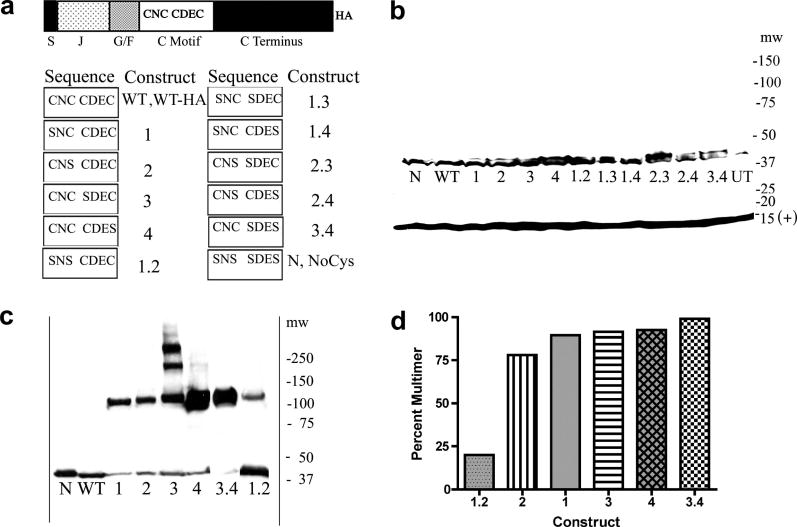
Fig. 1.
A. Domain structure of HEDJ, sequence of mutant constructs, nomenclature. Domains are labeled as follows: S, signal sequence; J, J domain; G/F, Gly/Phe rich; C motif, Cys-rich; C terminus, carboxyl-terminal domain; HA, hemagglutinin tag. The amino acids present in the Cys motifs of each mutant are listed in the columns marked Sequence and the corresponding names for each mutant are listed to the left, under the columns labeled Construct. B. WT-HA and mutant HA-tagged forms of HEDJ are expressed at similar levels. The proteins in TCA precipitated lysates from transfected COS-1 cells, were separated by using SDS-PAGE, 4–15%, with reducing conditions and immunoblotted with rabbit anti-HA antibody and rabbit anti-Sec 61β antibody (marked (+) at 14 kDa) which served as a loading control. Molecular weight markers (mw) are listed on the right. C. All the single Cys mutants and two double mutants, 3.4 and 1.2, form intermolecular disulfide-bridged multimers. Samples were processed as in B. except pellets were suspended in non-reducing SDS-PAGE sample buffer with 40 mM NEM and immunoblotting was done using mouse anti-HA antibody. The mobilities of WT and N are shown for comparison. D. Approximate quantification of the percent of the multimeric form present in each mutant in C.
Expression of Mutants
Transient transfections of the HA-tagged mutants were performed in COS-1 cells using Lipofectamine 2000. (Invitrogen) To characterize expression levels, 24 well plates of transfected cells were placed on ice, rinsed twice with cold PBS and lysed into 10% TCA. Pellets were collected, washed twice and dissolved in reducing SDS sample buffer. Proteins were separated using SDS-PAGE and immunoblotted with a rabbit antibody to HA or a mouse monoclonal antibody to HA. To determine which constructs formed intermolecular disulfide bonds, cells were processed as above, except pellets were dissolved in non-reducing SDS sample buffer containing 40 mM NEM.
Modification of sulfhydryl groups with PEG-Mal
Methods for PEG-Mal conjugation, an alkylating agent that reacts with free SH groups, were modified from several sources [25–27]. Microsomes were prepared from each 100 mm plate of COS-1 cells and lysed into 120 μl denaturing solution (6M urea, 1% Triton X-100 and 150 mM Tris HCl, pH 7). In some of the microsomal preparations half of the samples (60 μl) received 40 mM NEM (N-ethylmaleimide). (Supplementary Figure B.) Samples were incubated at 30°C for 10 min, on ice for 50 min and were clarified by centrifugation at 12,000 × g at 4°C for 10 min. The NEM-treated samples were desalted using desalting spin columns (Pierce). Samples were divided into equal aliquots and were left untreated or were treated with 100 mM DTT, or 5 mM diamide, an oxidizing agent. Tubes were incubated for 30 min at RT, 100 mM DTT was re-added to the tubes that had previously received DTT and tubes were incubated for 10 min at 30°C. Samples were divided into aliquots, TCA precipitated and washed 3 times with acetone and 70% EtOH. Pellets were dissolved either in a deoxygenated solution of 6 M urea, 2 % SDS, 150 mM TrisHCl, pH 7.4 or this solution with 5 mM PEG-Mal added. Samples were incubated for 1 h at RT and 10 min at 30°C. 6X non-reducing gel buffer was added and samples were run by SDS-PAGE (7.5% or 4–15% gradient gels) and electroblotted to PVDF, developed with antibody to HEDJ or to HA, HRP conjugated secondary antibody and chemiluminescence.
Cell culture samples
Six well plates of confluent COS-1, HepG2 or HeLa cells were left untreated or were treated with 20 mM DTT (concentrations of DTT were lower than in the microsomal preparations because the cell morphology was altered with 100 mM DTT) added to the media and incubated for 20 min at 37°C. Plates of cells were chilled on ice, the media removed and cells washed once with cold PBS followed by 240 μl of 10% TCA per well. One third of this lysate was analyzed per condition. Pellets were washed 3 times and incubated with or without approximately 5 mM PEG-Mal that had been dissolved in the urea-SDS-Tris buffer as above and had been filtered over a desalting column (Pierce) to remove low molecular weight maleimides [28]. Samples were immunoblotted, as above.
Native Gel Electrophoresis
COS-1 cells were washed twice with DPBS and lysed with 50 mM Tris HCl pH 7.4, 150 mM NaCl, 10 mM EDTA, and 1% Triton X-100 [29]. The mixture was incubated on ice for 20 min and clarified by centrifugation for 10 min at 12,000 × g at 4°C. Supernatants were untreated or treated with 20 mM DTT or 5 mM diamide and incubated for 20 min at RT. NEM was added to a final concentration of 25 mM and samples were incubated for 5 min. 5X native gel buffer was added to each tube (100 mM TrisHCl, pH 6.8, 20% glycerol, bromophenol blue and 0.02% SDS). Proteins were separated using a 7.5% Tris-glycine gel (Bio-Rad) in Tris-glycine buffer at 60 V and electroblotted to PVDF (Millipore) following a 20 min equilibration in Tris-glycine-SDS running buffer. Blots were developed with the anti-HA antibody or the anti-HEDJ antibody, secondary antibody and chemiluminescence.
Quantification
Autoradiograms were scanned and bands were quantified by densitometry using the Image J program on a Macintosh computer (developed at the NIH,//rsb.info.nih.gov/nih-image). Symbols with an error bar represent mean percentage +SEM. Statistical significance was calculated by unpaired t test. Calculations were performed in Prism 4 for Macintosh (GraphPad software).
RESULTS
Constructs of HEDJ
Mutants of HEDJ were constructed and tagged with HA at the carboxyl terminus (Figure 1A). Four cysteines are present in the mature HEDJ, arranged in two motifs, C169NC171 and C193DEC196, as depicted in the WT-HA construct. Mutant constructs were made in which one or more cysteines were replaced with serines, in order to eliminate the possibility of one or both disulfide bridges. The nomenclature used to designate each construct is listed in Figure 1A. The Cys residues are numbered according to their occurrence in the linear sequence. For example, the construct with replacement of the first Cys in the linear sequence is 1, while the double mutant with replacement of both Cys 1 and 2 is referred to as 1.2. NoCys or N refers to the mutant with complete replacement of all four cysteines with serines.
The approximate size and expression level of the HA-tagged mutants were determined following transient transfections of the HA-tagged mutants in COS-1 cells. Cells were lysed into TCA and the pellets were dissolved in reducing SDS sample buffer, proteins were separated using SDS-PAGE and immunoblotted with rabbit antibody to HA. As shown in Figure 1B, all of the constructs were of the correct molecular mass and had approximately equal levels of expression. Because a small signal was sometimes seen from the untransfected lysate and several non-specific upper bands sometimes occurred in non-reducing gels with this antibody, in subsequent experiments a mouse monoclonal antibody to HA was used. Construct 2.3, which is missing the last two amino acids of the HA tag, reacted poorly to this antibody, so results were confirmed with the rabbit polyclonal antibody. The mouse monoclonal anti-HA antibody was characterized in an immunoprecipitation and western assay (data not shown.) It was found that the anti-HA antibody immunoprecipitated HA-tagged 3.4 and the antibody to the C-terminus of HEDJ recognized HA-tagged 3.4 in the immunoblot. (Similar results were seen with WT-HA and with 1.2 mutant HEDJ-HA.) Endogenous HEDJ was not immunoprecipitated by the anti-HA antibody.
To determine which constructs formed intermolecular disulfide bonds, samples were processed as in Figure 1B, except the pellets from the TCA lysis were dissolved in non-reducing SDS sample buffer containing 40 mM NEM. The constructs that had a significant amount of intermolecular disulfide bond formation are shown in Figure 1C. Figure 1D shows a quantification of the amount of each construct in intermolecular disulfide multimers. All of the single Cys mutants and two of the double Cys mutants, 3.4 and 1.2, formed intermolecular disulfide bridges resulting in multimers of approximately 120 kDa size, suggesting that the multimeric complexes are similar in structure. Only in mutant 3, did two higher molecular weight complexes form, indicating that the presence of Cys 3 in the other single Cys mutants, prevents the formation of additional complexes that have intermolecular disulfide bridges. The observation that free SH groups in the single Cys mutants allowed the formation of multimers that did not occur in the WT, suggested that the Cys residues in the WT are normally paired into intramolecular disulfide bridges. In the 3.4 mutant, remaining Cys residues 1 and 2 formed predominantly intermolecular disulfide-bridged multimers, which could indicate that 1 and 2 do not normally form an intramolecular disulfide bridge in HEDJ or that the formation of a disulfide bridge between 1 and 2 occurs if 3 and 4 are present in an intramolecular disulfide bridge. The 1.2 mutant was predominantly in the monomeric form, suggesting that the majority of its Cys residues 3 and 4 are present in intramolecular disulfide bridges. With longer exposure times, some of the other constructs showed a small amount of multimerization.
Evaluation of the Number of Disulfide Bridges in Wild Type and Mutant HEDJ
PEG-Mal binds to free sulfhydryl groups of reduced Cys side chains but does not react with Cys residues oxidized in disulfide bridges or with modified Cys residues [27]. The number of reduced or oxidized Cys residues in a protein can be estimated using SDS-PAGE, by comparing the relative mobility of untreated protein to protein that has been reacted with PEG-Mal (conjugation will retard the mobility) [26]. As controls, prior to reaction with PEG-Mal, proteins containing Cys residues may be oxidized with diamide to generate disulfide bridges or reduced with DTT to form sulfhydryl groups.
The PEG-Mal method can also be modified to assess the number of disulfide bridges present in a protein, by including a pre-treatment with NEM [27]. NEM reacts only with free sulfhydryl groups, while Cys residues in disulfide bridges are protected from NEM. Following NEM pre-treatment, only the disulfide-bridged Cys residues will be reduced by DTT, generating free sulfhydryl groups. These can subsequently react with PEG-Mal, resulting in the lowered mobility of a protein in SDS-PAGE.
In order to validate our techniques, we reacted PEG-Mal with NoCys mutants (Figure 2A.), and with microsomes containing Sec61β, a protein with one Cys (Supplementary Figure, A). No shift in the relative mobility was seen in the case of NoCys indicating that the PEG-Mal reaction was specific for Cys, while Sec61β showed a mobility shift of approximately 15 kDa with no detectable unreacted Sec61β. Although the molecular weight of PEG-Mal is only 5 kDa, the mobility shift due to PEG-Mal conjugation has been shown to be ~15 kDa per PEG-Mal molecule [26].
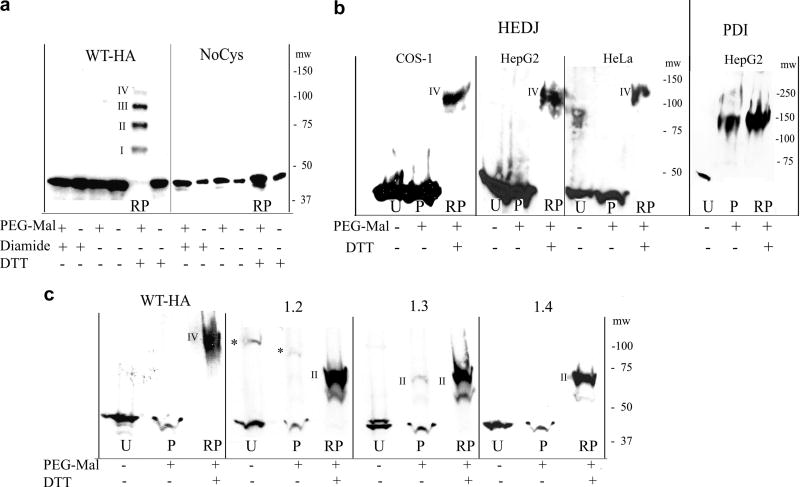
Fig. 2.
A. Microsomes, isolated from COS-1 cells that had been transfected with WT-HA or NoCys were reacted with 5 mM diamide, 100 mM DTT or untreated. They were TCA precipitated and treated with or without 5 mM PEG-Mal that had not been gel-filtered. Proteins were separated on a 7.5% non-reducing gel and immunoblotted with mouse anti-HA. In the lane labeled RP (DTT reduced + PEG-Mal), a ladder of bands approximately 15 kDa apart is seen, representing reduced, HA-tagged HEDJ conjugated to PEG-Mal on I, II, III or IV cysteines. The position of the molecular weight markers, mw, is shown on the right. B. HEDJ is predominantly oxidized in vivo. COS-1, HepG2 and HeLa cells were incubated with 20 mM DTT or untreated, TCA precipitated and the pellets processed as in A. except gel-filtered PEG-Mal was used and immunoblots were developed with an antibody to HEDJ or PDI. Lanes: U, untreated; P, untreated cells + PEG-Mal; RP, as in A (DTT reduced + PEG-Mal). The bands labeled IV in the RP lanes, correspond to band IV in Figure 2A based on their apparent molecular weights (see the molecular weight markers, mw on each panel). (For clarity, lane RP from the HepG2 cells, second panel, was taken from a shorter film exposure.) To validate the reaction conditions, an immunoblot of PDI from the processed HepG2 cells is shown in the right panel. C. WT-HA and several of the double mutants of HA-tagged HEDJ are predominantly oxidized in COS-1 cells. Cells were transfected with one of the constructs as shown, processed as in B., and the immunoblots developed with mouse anti-HA. Constructs conjugated to two PEG-Mal molecules are marked II. * denotes multimeric HA-tagged HEDJ
A calibration of the PEG-Mal reaction is shown in Figure 2A. Microsomes were isolated from COS-1 cells that had been transfected with WT-HA or NoCys and were treated as listed in the table below the panels. Proteins were resolved using non-reducing SDS-PAGE and immunoblots were developed with anti-HA antibodies. Results on WT-HA in microsomes were virtually the same regardless of pretreatment with NEM, an agent that blocks sulfhydryl groups (Supplementary Figure, B). DTT treatment alone did not change the mobility of WT-HA. However, a ladder of four bands of ~ 60, ~75, ~90 and ~ 110 kDa was seen when WT-HA, reduced by DTT, was reacted with PEG-Mal (Figure 2A, panel 1, lane RP.) This was consistent with the conjugation of one (I), two (II), three (III) or four (IV) PEG-Mal molecules per WT-HA molecule. A similar pattern was seen when endogenous HEDJ in COS-1 cells was analyzed under these conditions (Supplementary Figure, D). The fact that the total pool of modified HEDJ-HA did not shift to the position of band IV, indicated that not all four of the cysteines had reacted with PEG-Mal. This was most likely due to contaminating small molecular weight maleimides that competed with PEG-Mal for reaction with free sulfhydryl groups. [28]. In the experiments shown in Figures 2B and 2C, the PEG-Mal reagent was subject to gel filtration, to remove small molecular weight maleimides so that each reaction yielded only a single band [28].
In Figure 2A, only monomeric, 43 kDa WT-HA (HEDJ-HA) was present in the microsomes that had been pretreated with diamide prior to the addition of PEG-Mal. The absence of a reaction with PEG-Mal is expected as diamide reacts with thiol groups to form disulfide bridges. Similarly, untreated WT-HA also did not react with PEG-Mal indicating that all four Cys residues form disulfide bridges within the microsomes. These results are consistent with the results in intact cells, presented in Figure 2C, below.
Redox state of HEDJ in intact cells
We next examined the redox state of HEDJ in intact cells. In Figure 2B, COS-1, HepG2 and HeLa cell cultures were incubated in buffer alone, or treated with DTT. TCA was added to precipitate the proteins and the pellets from each condition were solubilized in the absence or the presence of gel-filtered PEG-Mal. Endogenous HEDJ from untreated COS-1, HepG2 or HeLa cells was predominantly oxidized and did not show a significant reaction with PEG-Mal as indicated by the similar mobility of HEDJ in lanes P (reacted with PEG-Mal) and lanes U (not reacted) (Figure 2B). Upon reduction with DTT, HEDJ from all three cell types was capable of reacting with PEG-Mal as judged by the lower mobility of modified HEDJ (lanes marked RP). The position of these bands is consistent with PEG-Mal modification on all four Cys residues and they are labeled with the Roman numeral IV, similarly to the notation of this band in Fig. 2A. In order to validate the experimental conditions, the HepG2 samples were immunoblotted for PDI (Figure 2B, right panel). PDI contained a substantial population of reduced Cys, as shown by its retarded mobility after PEG-Mal treatment, whether it had been pretreated with DTT or not (lanes RP and P, respectively). This result is consistent with previous reports showing that Cys residues are predominantly reduced in PDI taken from several mammalian cell lines [30, 31]. WT-HA-tagged HEDJ, expressed in COS-1 cells, was also tested using gel-filtered PEG-Mal (Figure 2C, first panel). The Cys residues were predominantly oxidized as evidenced by WT-HA having the same mobility in lanes P and U.
Overall, the results indicate that the majority of the Cys residues in HEDJ do not exist as free sulfhydryls within the ER. This compartment itself is relatively oxidized with a much lower ratio of reduced to oxidized glutathione compared to the cytoplasm [30–32]. However, not all Cys residues of other ER resident proteins are oxidized, as shown by the presence of free sulfhydryl groups in untreated PDI (Figure 2B, right panel).
The results presented thus far indicate that each of the cysteine residues in HEDJ is oxidized under normal conditions in the ER. It is not clear if the native disulfide bridges within HEDJ are formed by pairing of the two Cys residues within each motif or pairing between Cys residues in the two different motifs. COS-1 cells expressing mutant constructs 1.2, 1.3 or 1.4 were treated with PEG-Mal to determine if disulfide bridges were present. The results with construct 1.2 are demonstrated in the second panel of Figure 2C. In this mutant, the remaining Cys residues 3 and 4 are in the CXXC motif. The majority of the untreated 1.2 construct did not react with PEG-Mal, (lane P), indicating that Cys residues 3 and 4 can form a disulfide bridge in this mutant. Approximately ~20% of this construct forms an intermolecular disulfide-bridged multimer that would not be available for reaction with PEG-Mal (* marks the position in Figure 2C, panel 1.2. See also Figure 1C and Supplementary Figure B, 1.2 panels). Nevertheless, the monomeric form of mutant 1.2 must be predominantly in the intramolecular disulfide-bonded form, given the small amount of PEG-Mal reactivity overall. Consistent with this, upon reduction with DTT, construct 1.2 had a mobility shift corresponding to band II in part A, indicating that the two cysteine residues were reduced and capable of reacting with PEG-Mal. In the third panel, untreated construct 1.3 showed a slight amount of PEG-Mal modification in lane P, but most of the Cys 2 and Cys 4 residues formed disulfide bridges, indicating that Cys 2 in the CXC motif could pair with Cys 4 in the CXXC motif. Construct 1.4, (final panel) had no reactivity with PEG-Mal showing that Cys residues 2 and 3 formed a disulfide bridge. These data show that in each of the double mutants 1.2, 1.3 and 1.4, a disulfide bridge was formed regardless of which of the two Cys residues remained. (Supplementary Figure C using unfiltered PEG-Mal on microsomes provides some indication of the degree of oxidation of mutants 2.3, and 2.4 in comparison to mutants 1.3 and 1.4.) These results suggest that oxidization of the HEDJ cysteine residues is highly favored, but do not alone allow prediction of the likely partner for each Cys residue in the native protein.
Comparison of the mobility of WT and mutant HEDJ in native gels
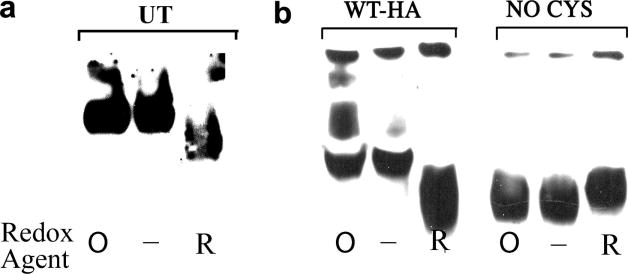
In native gel electrophoresis, the mobility of proteins is a complex function of their charge, size and shape. Redox conditions may influence the overall conformation of a protein, as shown with a number of protocols for native gel electrophoresis of proteins such as HSF-1 and PDI family members [30, 33–35]. Under our native gel conditions, untreated and diamide-treated HEDJ had equivalent mobilities, while DTT-treated HEDJ had a faster mobility (Figure 3A). Native gel electrophoresis of WT-HA demonstrated a similar mobility for diamide treated (O) and untreated (−) samples whereas DTT treatment resulted in a faster mobility of the wild-type protein (Figure 3B). These results indicate that the reduced and oxidized forms of HEDJ can be distinguished by native gel electrophoresis and demonstrate that WT-HA or endogenous HEDJ migrates with the same mobility as the diamide treated, fully oxidized protein. The NoCys mutant form of HEDJ showed no change in mobility with differing redox states and migrated similarly to reduced WT-HA, consistent with a protein that has an altered conformation as a result of the absence of disulfide bond formation.
Fig. 3.
The native conformation of wild type HEDJ is similar to oxidized, diamide treated HEDJ and unlike reduced or Cys mutant HEDJ. A. Lysates of untransfected COS-1 cells (UT) were not treated (−) or were incubated with 5 mM diamide (O) or 20 mM DTT (R). Following incubation, 25 mM NEM was added to each sample to block sulfhydryl groups. Native gel sample buffer was added, samples were separated on a 7.5% Tris-glycine gel and immunoblotted using anti-HEDJ. B. Lysates from COS-1 cells that had been transfected with HEDJ-HA tagged constructs were treated and analyzed as in A. using mouse anti-HA antibody.
Functional studies: binding of chaperones to immobilized substrates and effect of ATP
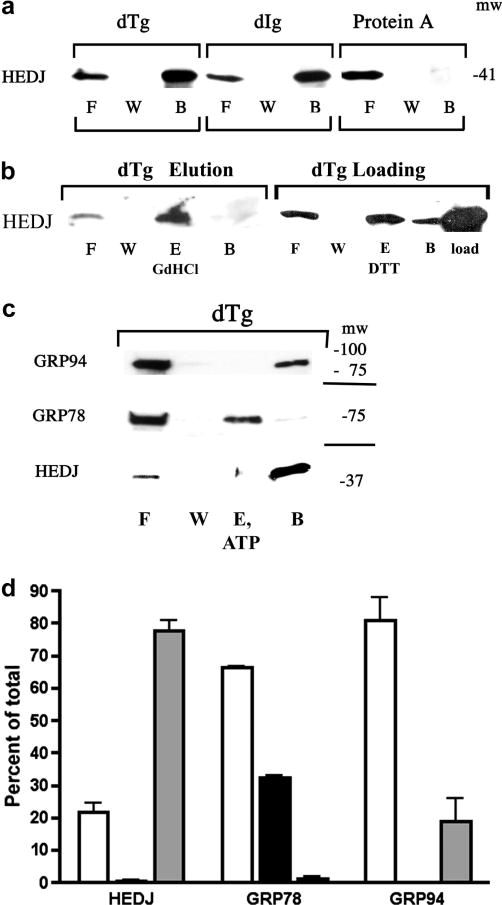
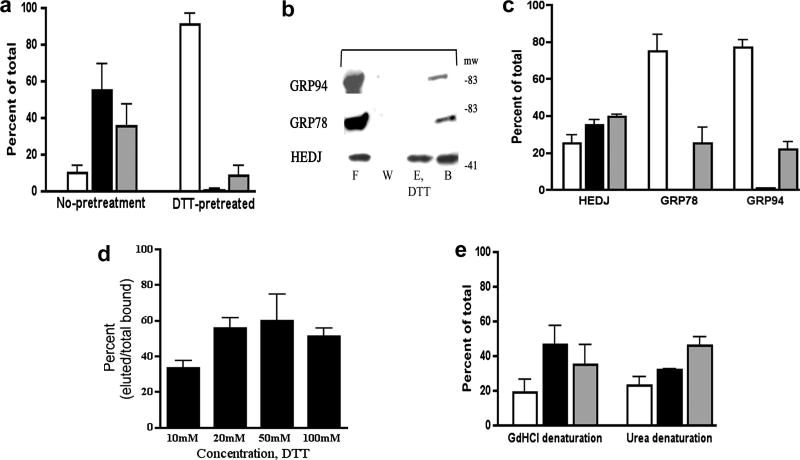
Previous studies have shown that several cytoplasmic Type I and Type II, DnaJ/Hsp40 proteins can bind to unfolded client proteins independently of Hsp70 and target them to Hsp70 chaperones [16]. Few functional studies have been done on mammalian ER homologues of DnaJ/Hsp40 proteins such as HEDJ [20, 22, 36]. HEDJ and GRP78 have been identified as part of a network of chaperones that bind to denatured proteins [21]. Recently, evidence that HEDJ may bind to unfolded client proteins independently of GRP78 has been obtained in vivo [20]. In our study, a cell-free system was used to examine the ability of HEDJ to bind denatured substrates and determine the contribution of ATP, redox state and the role of the Cys-rich domain in substrate interactions. An affinity chromatography system was used that was similar to one employed to assess the ability of ER chaperones (GRP78, GRP94, calreticulin and members of the PDI family) to bind to immobilized, denatured proteins and to be eluted with ATP [24]. We assessed the binding of native glycosylated HEDJ to immobilized, denatured thyroglobulin (dTg) and immunoglobulin (dIg). As shown Figure 4A, a substantial amount of HEDJ from a microsomal lysate bound to immobilized dTg or dIg (~69% and 73%, respectively), while there was no significant binding to native protein A agarose that was used as a negative control. Bound HEDJ could be completely eluted from dTg with GdHCl, a chaotropic agent that denatures proteins, in part by its effects on hydrophobic interactions (Figure 4B). In some cases, the recovery of protein from the dTg and dIg columns was estimated by including a lane of microsomal protein equivalent to the amount of protein originally applied to the column. An example is shown in Figure 4B, dTg Loading. (See Figure 5 for details of the DTT elution.) We estimate that the average recovery of protein was 79 % + 19 %, n=5. GRP78 and GRP94 also bound to dTg (Figures 4C, 4D, 5B, 5C) and non-specific binding to protein A agarose was low (data not shown). Whereas GRP78 bound to dTg was completely eluted with 2 mM ATP, the binding of HEDJ and GRP94 were not affected by ATP addition (Figures 4C and 4D). These results indicate that HEDJ is capable of binding to denatured substrates in an ATP-independent fashion and demonstrate that binding is maintained in the absence of GRP78.
Fig. 4.
HEDJ can bind to immobilized dTg or dIg directly. A. Immobilized dTg or dIg or native protein A agarose beads were incubated with a microsomal lysate. The initial washes and flow through (F) and the final wash (W) were collected and concentrated. Material bound to the beads was released by boiling in SDS gel sample buffer (B). The proteins in all fractions were separated by using a 4–15% SDS-PAGE and immunoblotted with anti-HEDJ. B. In both immunoblots, binding assays to dTg were performed as in A. In the immunoblot labeled dTg Elution, an elution step (E, GdHCl) was included after washing the beads. The second blot, labeled dTg Loading, included a lane of unfractionated, unprocessed microsomes adjacent to the processed fractions in order to estimate total recovery. (See Figure 5 for explanation of the DTT elution shown in lane E.) C. GRP78 can be eluted from dTg using ATP while HEDJ and GRP94 cannot. The binding assay to dTg was performed as in B., using 2 mM ATP in the elution buffer. D. Quantification of C. For each assay, the total amount of recovered protein (density of the signals) was quantified and the percent of the total present in each fraction was determined. The value of each fraction is given as the mean +SEM. Open columns represent F, black columns represent E, and grey columns represent B. HEDJ (n=3); GRP78 (n=3); GRP94 (n=2). The percent of the total GRP78 that was recovered in the elution fraction was significantly different from the percent of HEDJ or GRP94 in the elution fraction. (p<0.0001
Fig. 5.
A. Pretreatment of HEDJ with reducing agent prevents initial binding to dTg. Lysed microsomes were pre-incubated in binding buffer with or without 50 mM DTT, prior to incubation with immobilized dTg. DTT pretreated refers to samples that were pretreated, washed and eluted with 50 mM DTT. No pretreatment indicates that 50 mM DTT was included only in the elution buffer. Open columns, F; black, E; gray, B. There was significantly more HEDJ in the flow-through and wash fraction (F) of samples pretreated with DTT as compared to the control. (p< 0.02, n=3, t test) B. HEDJ can be partially eluted from dTg with DTT while binding by GRP78 and GRP94 is not affected. Immobilized dTg, denatured with urea and 2-mercaptoethanol, was processed as in Figure 4A. The elution buffer contained 100 mM DTT. C. Quantification of chaperone binding and DTT elution experiments. The amount of HEDJ in the eluted fraction (E, DTT) was significantly higher in the case of HEDJ as compared with GRP78 or GRP94. (p<0.05, unpaired t test, n=2). D. The effect of DTT concentration on the fraction of HEDJ eluted from dTg. Experiments were done as in B. except 10, 20, 50, or 100 mM DTT was used for the elution step. The proportion of HEDJ eluted with 20, 50 or 100 mM DTT was significantly higher than that eluted with 10 mM DTT, while there was no significant difference in the range of 20–100 mM DTT. (p<0.05) n=2, 10 mM; n=3 for 20, 50, 100 mM (Data from C. was also used in D.) E. Binding and elution of HEDJ from dTg does not depend upon free sulfhydryl groups in dTg. GdHCl denaturation included IA treatment that blocked sulfhydryl groups, making them unavailable for binding to HEDJ, whereas urea denaturation did not include IA. n=4 for each denaturant Values for F, E and B were not significantly different.
Redox conditions affect the binding of HEDJ to client proteins
Although ATP did not affect the binding of HEDJ to dTg, reducing conditions prevented the binding. Microsomal lysates were pretreated with 50 mM DTT to reduce HEDJ and DTT was included throughout the binding assay. Fractions were compared to those of untreated HEDJ. The binding to dTg was markedly diminished for reduced HEDJ, with more than 90% of it found in the flow-through fraction as compared to only 10% of untreated HEDJ (Figure 5A). By using a protocol that incorporated reducing conditions prior to binding to dTg and throughout the assay, we were successful in demonstrating that completely reduced HEDJ was unable to bind to immobilized dTg.
We next examined the effect of reducing conditions on the binding of GRP78, GRP94 and HEDJ to immobilized dTg. HEDJ, GRP78 and GRP94 were bound to immobilized dTg and subjected to an elution step performed under reducing conditions. While the binding of GRP78 and GRP94 were not affected by highly reducing conditions of 100 mM DTT, approximately half of the HEDJ bound to dTg was released under these conditions (Figures 5B and 5C). The elution of HEDJ from dTg was determined as a function of the concentration of DTT, with 20 mM DTT or higher eluting a similar amount of HEDJ (Figure 5D).
It is interesting to note that pretreatment of microsomal extracts with reducing agent completely abolishes HEDJ interaction with substrate, whereas once bound to dTg under non-reducing conditions, only approximately half of HEDJ is released, even under highly reducing conditions. These observations suggest that the redox state of HEDJ is important for the initial interaction with substrate, whereas bound HEDJ may form substrate interactions that are less dependent upon on redox state.
Binding studies in Figure 5 were performed with dTg that had been denatured with urea and 2-mercaptoethanol and with dTg containing blocked sulfhydryl groups (denatured with GdHCl and DTT, followed by iodoacetamide, which forms carboxamidomethylated sulfhydryl groups). The fraction of HEDJ bound to dTg and its partial elution with 20, 50 or 100 mM DTT were not significantly different following either denaturation method (Figure 5E). Carboxamidomethylation of sulfhydryl groups by iodoacetamide indicated that intermolecular disulfide bridges between dTg and HEDJ were not needed for binding.
Contribution of the Cys-rich domain to substrate interactions
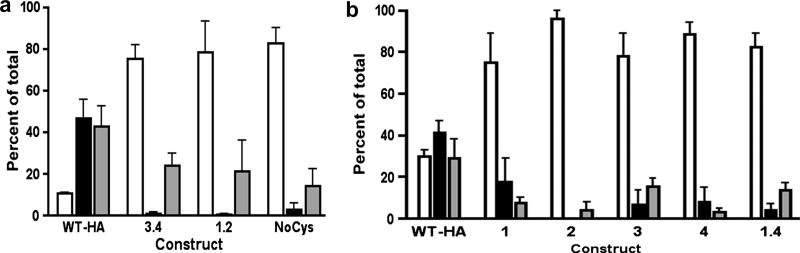
The substrate binding activity of WT-HA and mutant HA-tagged forms of HEDJ was examined. Assays were performed as in Figure 5, and the elution was performed with 20 mM DTT since higher concentrations did not appreciably increase the amount of bound HEDJ that could be eluted. These results demonstrated that WT-HA exhibited a binding pattern to dTg that was very similar to that of endogenous HEDJ, in that it bound to dTg efficiently and a considerable portion of bound WT-HA was eluted by 20 mM DTT. In contrast, all of the HA-tagged cysteine-mutant forms of HEDJ bound the denatured substrate very poorly. Figure 6A shows a comparison of the binding of 1.2, 3.4 and NoCys to that of WT-HA. While ~10% of the WT-HA was found in the flow through, ~ 80% of each mutant was found in the non-binding flow-through fraction. As with endogenous HEDJ, approximately half of the WT-HA protein that bound to dTg could be eluted with 20 mM DTT (fraction E, DTT) while none of the 3.4, 1.2 and NoCys mutants could be eluted, suggesting that an intact Cys-rich domain is required for substrate interaction. A second series of assays compared the binding to dTg of single Cys mutants 1, 2, 3 and 4, the double mutant 1.4 and the WT-HA. The results of this comparison are quantified in Figure 6B and show that the mutants bind poorly compared to the WT-HA. A binding assay using the double mutants 1.3, 2.3 and 2.4 also showed poor binding (data not shown). These mutant constructs variously form intermolecular disulfide bridges and are multimers or they have only one or no intramolecular disulfide bridges, as demonstrated previously. We conclude that any substitution of Cys with Ser results in mutants that bind poorly to immobilized substrate, regardless of the possession of an intermolecular or intramolecular disulfide bond.
Fig. 6.
Cys mutants of HEDJ bind poorly to denatured protein. A. Immobilized dTg was incubated with lysed microsomes isolated from cells expressing wild type (WT-HA) or mutant (NoCys, 3.4, 1.2) HA-tagged HEDJ. The elution buffer contained 20 mM DTT. Proteins were separated by using a reducing 10% SDS-PAGE, immunoblotted using mouse anti-HA and quantified. Column legend: Open =F; black= E; gray = B. The differences between the amount of WT-HA and the mutants in (F) and (E) are statistically significant. (p<0.05, n=3) The proportion of WT-HA that remained bound to dTg (B) was significantly higher than that of the NoCys mutant (p<0.05, n=3). B. Quantification of the binding of single Cys mutants and the double mutant 1.4 to dTg. Assays were performed and fractions labeled as in A. There was a statistically significant difference in the proportion of recovered WT-HA that bound to the column as compared to each mutant. (P<0.05, one tailed P value) n=2.
DISCUSSION
The ER-localized chaperone HEDJ is homologous to other type I Hsp40 chaperones with conserved J and Gly/Phe regions. However, the Cys-rich domain differs considerably in sequence and arrangement[8, 36]. Instead of the typical 4 motifs of CXXCXG, the Cys-rich region of HEDJ consists of a CXC and a CXXC motif, more closely resembling regions found in thioredoxin and PDI family members. Whereas the Cys-rich region in type I proteins contributes to their chaperone activities, both stabilizing the binding pocket and affecting the transfer of substrate to the Hsp70 partner[18], the Cys-rich regions of thioredoxin/PDI family members serve predominantly enzymatic roles as part of the active site involved in oxidoreductase activity. Consequently, we were interested in determining the role of the Cys-rich domain of HEDJ in substrate interactions.
We demonstrated that the Cys-rich domain in HEDJ is predominantly in the oxidized form with two intramolecular disulfide bonds. Reduction of the disulfide bonds within the Cys-rich region altered the protein’s conformation, as judged by migration in native gel electrophoresis. An in vitro assay demonstrated that a fully intact Cys-rich domain is required for interaction of HEDJ with client proteins. Taken together, these data suggest that the Cys-rich domain consists of two disulfide bonds that stabilize the protein in a binding-competent structure.
The redox state of HEDJ was investigated using the PEG-Mal reagent to react with sulfhydryl groups, similarly to previous studies on PDI family members [26, 30]. Our results indicate that the bulk of HEDJ exists in an oxidized state and the Cys residues form intramolecular disulfide bridges (Figures 1–3). Several arrangements of the Cys motifs are possible. Cys residues that are nearest each other in the linear sequence might pair: Cys 1 with Cys 2 and Cys 3 with Cys 4. Cys 1 and Cys 2, separated by only one amino acid, would form a strained disulfide bond, which occurs relatively rarely [37]. Cys residues, in separate motifs and farther apart in the linear sequence, could be paired if the tertiary structure of the protein had a loop(s) with 1 and 2 on one branch and 3 and 4 on the other branch. Neither model can be favored, because non-native disulfide bridges formed in a number of our mutants. All mutants showed impaired binding to denatured client protein. Previous studies have shown that mutation of Cys residues in some proteins impairs folding and function and some mutants can form non-native disulfide bridges [38, 39]. Under our experimental conditions, HEDJ required both the native disulfide bridges for proper folding and function.
Our results with PEG-Mal indicate that Zn coordination is unlikely in HEDJ. In general, reduced Cys residues that coordinate Zn are able to react with alkylating agents. For example, reduced Hsp33 coordinates Zn through its Cys residues, which can be modified by IA [40] Similarly, untreated HEDJ should have reacted with PEG-Mal if it had Cys residues that were reduced, even if the residues bound Zn. Nonetheless, we cannot rule out the possibility that the conformation of HEDJ could be influenced by Zn binding under some conditions, as we did not purify the protein and directly assay for the presence of Zn.
A number of studies have shown that DnaJ and Ydj1, Hsp40/DnaJ type I proteins, can bind to proteins independently of Hsp70 chaperones [41–43]. These proteins bind to a core of aromatic and large aliphatic hydrophobic residues in the client protein [43, 44]. The client protein may then be presented to DnaK/Hsp70. Interactions between DnaJ/Hsp40 and DnaK/Hsp70 encompass regions within the ATPase domain of Hsp70 and part of the J domain of Hsp40, including a highly conserved His-Pro-Asp motif. DnaJ/Hsp40 proteins stimulate the hydrolysis of ATP by the Hsp70 protein. Ternary complexes of DnaJ/Hsp40, DnaK/Hsp70 and client protein form [42, 45]. DnaJ/Hsp 40 leaves the complex and the client protein is bound by DnaK/Hsp70 [16, 42, 45]. HEDJ and GRP78 have been shown to bind to each other in the presence of ATP and the J domain of HEDJ has been shown to stimulate the ATPase activity of GRP78 [8, 20, 36]. In a recent study, a construct of HEDJ with the His-Pro-Asp motif altered to Gln-Pro-Asp did not bind GRP78 yet still bound client proteins, presumably through a direct interaction [20].
In order to facilitate experiments examining a direct interaction between HEDJ and substrate, we developed an in vitro binding assay, similar to that used to elucidate the conditions for interaction of GRP78 with substrate [24]. Using this assay, we were able to demonstrate binding of HEDJ to immobilized, denatured substrate proteins. In our microsomal extracts, we could not initially exclude the possibility that HEDJ interaction with substrate proteins was indirect and mediated through interaction with other cofactors or chaperones, particularly GRP78. By altering ATP and redox conditions, we were able to differentiate the binding of HEDJ to client protein from the binding of GRP78 and GRP94 to client protein. Unlike GRP78, the binding of DnaJ and other Hsp40 homologues to client proteins is not directly affected by ATP [41]. In our study, ATP did not elute HEDJ from a complex with denatured client protein, whereas bound GRP78 was released from client protein in the presence of ATP, as expected. With respect to other chaperone molecules, we found a small amount of binding by GRP94 to immobilized, denatured protein and like HEDJ, this chaperone was not eluted following addition of ATP, consistent with the ATP-independent binding of this chaperone to target molecules [46]. However, we were able to differentiate binding of HEDJ and GRP94 by altering redox conditions. Reducing conditions did not result in the elution of GRP78 or GRP94 but did elute a significant fraction of bound HEDJ.
Although the Cys-rich domains are not similar, HEDJ has homology to Scj1p, a yeast type I, DnaJ/Hsp40 protein localized to the ER and containing the typical Zn finger-like motifs [36] and also has similarity to the type II proteins [20]. Crystallographic and mutational studies demonstrate that type I and type II proteins dimerize in the carboxyl terminal domain for efficient binding to client proteins [47–50]. Another region, the hydrophobic binding pocket, forms contacts with the client protein [17, 48]. In the monomer form, the binding region of Ydj1, a cytosolic type I protein from yeast that is related to DnaJ, has an L shape with the binding pocket on the long arm of the L and a Cys-rich domain consisting of two zinc finger motifs that form the ninety-degree turn and the short arm of the L. Although the hydrophobic binding pocket does not include the adjacent Cys-rich region, disrupting the structure of zinc finger I could destabilize the binding domain [18]. Mutants of the type I protein DnaJ, containing substitutions for all four Cys in zinc finger I, were unable to bind independently to substrates [19]. Oxidation of the cysteines and release of zinc in the DnaJ homologue Hdj2, resulted in its inactivation [51].
Given its homology to Hp40/DnaJ proteins except for the Cys motifs, it is likely that HEDJ does dimerize in its carboxyl terminal region and bind to client proteins through a substrate-binding pocket adjacent to its Cys-rich region. The atypical Cys-rich domain in HEDJ is much shorter in length than the type I domain and the Cys residues are oxidized. Reduced HEDJ was unable to bind to substrate and had an altered conformation from oxidized HEDJ. Possibly dimerization was affected by the misfolding of reduced HEDJ and this lowered the overall affinity for substrate. Complete replacement of Cys residues with Ser or single or double Cys to Ser mutations markedly impaired the binding of HEDJ to immobilized substrate and resulted in a non-native conformation. These experiments, under cell-free conditions, demonstrate the crucial contribution of the Cys-rich domain and its oxidation state in facilitating HEDJ binding to substrate and may indicate that the Cys-rich motifs are adjacent to the binding domain.
All of our binding assays were performed under cell-free conditions and used DTT as an analytical tool with levels that should have ensured reduction of the Cys residues in HEDJ. Recently, an in vivo study demonstrated, under reducing conditions, the association of a considerable amount of unfolded immunoglobulin light chain with GRP78 and detectable binding of the light chain to HEDJ/ERdj3 [20]. It is notable that, under our conditions, abrogation of HEDJ binding in vitro required relatively high DTT concentrations, suggesting that the oxidized form is highly favored. It is possible that conditions used in the previous study in whole cells allowed reduction of the light chain but did not completely reduce HEDJ. Under some conditions, as detailed above, we have seen that a fraction of HEDJ continues to bind to substrate in the presence of reducing agents, but we have not yet studied HEDJ binding to client proteins within the cell under conditions that can be demonstrated to completely reduce HEDJ.
Clearly the Cys-rich motifs of HEDJ are important for its chaperone functions, but it is not clear if redox conditions can play a regulatory role in its chaperone activities. Several proteins with a CXXC motif(s) have been demonstrated to have redox-regulated chaperone activities. Hsp33 assumes an inactive conformation under the normal reducing conditions of the cytoplasm and an active conformation under conditions of oxidative stress [52]. PDI interacts with cholera toxin A1 chain in a redox-regulated manner, binding when reduced and releasing toxin when oxidized by Ero1 [53, 54]. It has also been suggested that the chaperone activity of PDI is redox regulated in its interaction with the ERAD substrate BACE457 [55]. However, changes in redox conditions do not alter the ability of PDI to bind to the C propeptide of procollagen or to act as a subunit for prolyl-4-hydroxylase [29].
In summary, we have demonstrated that the Cys-rich domain in HEDJ differs in sequence, arrangement and oxidation state from those in the typical type I Cys-rich proteins but functionally, both proteins require regions within the Cys-rich domain for several chaperone activities. Specifically for HEDJ, in vitro, this domain contributes structural information that is crucial for substrate interactions. In the cytoplasmic chaperones, cysteines are in the reduced state and provide a structural scaffold by coordinating zinc but in HEDJ, the formation of disulfide bonds among the cysteine containing motifs appears to provide the same structural stability to the substrate-binding domain. It is notable that one of the cysteine motifs in HEDJ is similar to that of thioredoxin and PDI family members, in which CXXC motifs are essential for catalytic oxidoreductase activity. As binding of HEDJ occurred to substrates that had no free sulfhydryl groups or available disulfide bonds, there could be no direct interaction of the Cys-rich domain with Cys residues in client proteins, such as occurs during oxidoreductase activity. The possibility that the HEDJ Cys-rich domain also possesses oxidoreductase activity and the capacity for redox-exchange reactions has not yet been examined. In this regard, in vitro studies have demonstrated that regions within the DnaJ Cys-rich domain contribute to chaperone activities [19] and oxidoreductase activities [56], suggesting that the Cys-rich domain of HEDJ may likewise have dual functions.
Supplementary Material
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PEG-Mal, methoxy polyethylene glycol maleimide; NEM, N-ethyl maleimide; DTT, dithiothreitol; IA, iodoacetamide; dTg, denatured thyroglobulin; GdHCl, guanidine hydrochloride; dIg, denatured immunoglobulin; Cys-rich, amino acid motif with cysteine or cystine residues
The authors appreciate the work of Corinne Garofalo for construction of two of the mutants and Malcolm DeBaun for his help in constructing one of the mutants. We would like to thank Joe St. Geme and his laboratory members for their helpful suggestions. This work was supported by the March of Dimes Foundation (#1-FY01-178), and the NIAID (RO1AI47900). D.H. is the recipient of an Investigator in Microbial Pathogenesis Award from the Burroughs Wellcome Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brodsky JL, Goeckeler J, Schekman R. Proc Natl Acad Sci. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gething MJ. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 3.Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y. Proc Natl Acad Sci. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 5.Brightman SE, Blatch GL, Zetter BR. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a. [DOI] [PubMed] [Google Scholar]
- 6.Skowronek MH, Rotter M, Haas IG. Biol Chem. 1999;380:1133–1138. doi: 10.1515/BC.1999.142. [DOI] [PubMed] [Google Scholar]
- 7.Bies C, Guth S, Janoschek K, Nastainczyk W, Volkmer J, Zimmermann R. Biol Chem. 1999;380:1175–1182. doi: 10.1515/BC.1999.149. [DOI] [PubMed] [Google Scholar]
- 8.Yu M, Haslam RH, Haslam DB. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y, Meunier L, Hendershot LM. J Biol Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- 10.Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, Leinonen S, Huikko MP, Gustafsson JA, Sitia R, Spyrou G. J Biol Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 11.Hosoda A, Kimata Y, Tsuru A, Kohno K. J Biol Chem. 2003;278:2669–2676. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsuka K, Hata M. Cell Stress Chaperon. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier M, Rhee H, Elguindi EC, Blond SY. J Biol Chem. 2000;275:19620–19627. doi: 10.1074/jbc.M001333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, Haas IG, Heim N, Nastainczyk W, Volkmer J, Zimmermann R. Proc Natl Acad Sci. 2000;97:7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheetham ME, Caplan AJ. Cell Stress Chaperon. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan CY, Lee S, Cyr DM. Cell Stress Chaperon. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Qian X, Sha B. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. J Biol Chem. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 19.Linke K, Wolfram T, Bussemer J, Jakob U. J Biol Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Hendershot LM. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meunier L, Usherwood YK, Chung KT, Hendershot LM. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M, Haslam DB. Infect & Immun. 2005;73:2524–2532. doi: 10.1128/IAI.73.4.2524-2532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machamer CE, Doms RW, Bole DG, Helenius A, Rose JK. J Biol Chem. 1990;265:6879–6883. [PubMed] [Google Scholar]
- 24.Nigam SK, Goldberg AL, Ho S, Rohde MF, Bush KT, Sherman M. J Biol Chem. 1994;269:1744–1749. [PubMed] [Google Scholar]
- 25.Tyagarajan K, Pretzer E, Wiktorowicz JE. Electrophoresis. 2003;24:2348–2358. doi: 10.1002/elps.200305478. [DOI] [PubMed] [Google Scholar]
- 26.Schwaller M, Wilkinson B, Gilbert HF. J Biol Chem. 2003;278:7154–7159. doi: 10.1074/jbc.M211036200. [DOI] [PubMed] [Google Scholar]
- 27.Wu HH, Thomas JA, Momand J. Biochem J. 2000;351:87–93. doi: 10.1042/0264-6021:3510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao R, Wilkinson B, Solovyov A, Winther JR, Holmgren A, Lundstrom-Ljung J, Gilbert HF. J Biol Chem. 2004;279:49780–49786. doi: 10.1074/jbc.M409210200. [DOI] [PubMed] [Google Scholar]
- 29.Lumb RA, Bulleid NJ. EMBO J. 2002;21:6763–6770. doi: 10.1093/emboj/cdf685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molteni SN, Fassio A, Ciriolo MR, Filomeni G, Pasqualetto E, Fagioli C, Sitia R. J Biol Chem. 2004;279:32667–32673. doi: 10.1074/jbc.M404992200. [DOI] [PubMed] [Google Scholar]
- 31.Jessop CE, Bulleid NJ. J Biol Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 32.Hwang C, Sinskey AJ, Lodish HF. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 33.Manalo DJ, Liu AY. J Biol Chem. 2001;276:23554–23561. doi: 10.1074/jbc.M011300200. [DOI] [PubMed] [Google Scholar]
- 34.Watson WH, Pohl J, Montfort WR, Stuchlik O, Reed MS, Powis G, Jones DP. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 35.Zapun A, Bardwell JC, Creighton TE. Biochemistry. 1993;32:5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 36.Bies C, Blum R, Dudek J, Nastainczyk W, Oberhauser S, Jung M, Zimmermann R. Biol Chem. 2004;385:389–395. doi: 10.1515/BC.2004.043. [DOI] [PubMed] [Google Scholar]
- 37.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. Nat Struct Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 38.Allen S, Lu H, Thornton D, Tokatlidis K. J Biol Chem. 2003;278:38505–38513. doi: 10.1074/jbc.M306027200. [DOI] [PubMed] [Google Scholar]
- 39.Wendland M, von Figura K, Pohlmann R. J Biol Chem. 1991;266:7132–7136. [PubMed] [Google Scholar]
- 40.Barbirz S, Jakob U, Glocker MO. J Biol Chem. 2000;275:18759–18766. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- 41.Wawrzynow A, Zylicz M. J Biol Chem. 1995;270:19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- 42.Han W, Christen P. J Biol Chem. 2003;278:19038–19043. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- 43.Fan CY, Lee S, Ren HY, Cyr DM. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudiger S, Schneider-Mergener J, Bukau B. EMBO Journal. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wawrzynow A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M. J Biol Chem. 1995;270:19307–19311. doi: 10.1074/jbc.270.33.19307. [DOI] [PubMed] [Google Scholar]
- 46.Rosser MF, Trotta BM, Marshall MR, Berwin B, Nicchitta CV. Biochemistry. 2004;43:8835–8845. doi: 10.1021/bi049539q. [DOI] [PubMed] [Google Scholar]
- 47.Borges JC, Fischer H, Craievich AF, Ramos CH. J Biol Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 48.Sha B, Lee S, Cyr DM. Structure. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 49.Shi YY, Hong XG, Wang CC. J Biol Chem. 2005;280:22761–22768. doi: 10.1074/jbc.M503643200. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Li J, Jin Z, Fu Z, Sha B. J Mol Biol. 2005;346:1005–1011. doi: 10.1016/j.jmb.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 51.Choi HI, Lee SP, Kim KS, Hwang CY, Lee YR, Chae SK, Kim YS, Chae HZ, Kwon KS. Free Radical Biol Med. 2006;40:651–659. doi: 10.1016/j.freeradbiomed.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 52.Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. J Biol Chem. 2004;279:20529–20538. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- 53.Tsai B, Rapoport TA. J Cell Biol. 2002;159:207–216. doi: 10.1083/jcb.200207120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 55.Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi YY, Tang W, Hao SF, Wang CC. Biochemistry. 2005;44:1683–1689. doi: 10.1021/bi0480943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.