Abstract
We have studied relative efficacies of NR1 agonists glycine and d-cycloserine (DCS), and found efficacy to be dependent on the NR2 subunit. DCS shows partial agonism at NR1/NR2B but has higher relative efficacy than glycine at NR1/NR2C receptor. Molecular dynamics (MD) simulations of the NR1/NR2B and NR1/NR2C agonist binding domain dimer suggest only subtle differences in the interactions of DCS with NR1 binding site residues relative to glycine. The most pronounced differences were observed in the NR1/NR2C simulation between the orientation of helices F and G of the NR1 subunit. Interestingly, Helix F was previously proposed to influence receptor gating and to adopt an orientation depending on agonist efficacy. MD simulations and site-directed mutagenesis further suggest a role for residues at the agonist binding domain dimer interface in regulating DCS efficacy. To relate the structural rearrangements to receptor gating, we recorded single-channel currents from outside-out patches containing a single active NR1/NR2C receptor. DCS increased the mean open time and open probability of NR1/NR2C receptors compared with glycine. Maximum likelihood fitting of a gating model for NR1/NR2C receptor activation to the single-channel data suggests that DCS specifically accelerates the rate constant governing a fast gating step and reduces the closing rate. These changes appear to reflect a decreased activation energy for a pregating step and increased stability of the open states. We suggest that the higher efficacy of DCS at NR1/NR2C receptors involves structural rearrangements at the dimer interface and an effect on NR1/NR2C receptor pregating conformational changes.
Introduction
Glutamate, the major excitatory neurotransmitter in the mammalian CNS, activates three classes of ionotropic receptors classified on the basis of pharmacology and sequence homology (Dingledine et al., 1999; Erreger et al., 2004). Among these receptor subtypes, NMDA receptors are involved in key physiological processes such as synaptic plasticity and development. NMDA receptors are tetramers composed of two NR1 and two NR2 subunits, which bind glycine and glutamate, respectively. d-cycloserine (DCS) has previously been approved for use in humans for treatment of tuberculosis (Albouy et al., 1955; Boyd et al., 1955; Marshall, 2008), but more recently has become known as a NR1 agonist (Hood et al., 1989; McBain et al., 1989; Henderson et al., 1990) of the NMDA receptor with a number of intriguing neuroactive properties (Monahan et al., 1989; Flood et al., 1992; Schuster and Schmidt, 1992; Thompson et al., 1992). DCS exhibits clinical actions that appear relevant for adjunct exposure therapy for posttraumatic stress disorder and other anxiety disorders (Walker et al., 2002; Ressler et al., 2004; Davis et al., 2006). Interestingly, the efficacy of DCS at recombinant NMDA receptors is NR2 subunit-dependent (Sheinin et al., 2001). DCS acts as a partial agonist at NR1/NR2A, NR1/NR2B and NR1/NR2D receptors with lower relative efficacy than the endogenous agonists glycine or d-serine. In contrast, DCS has higher relative efficacy than glycine or d-serine at NR1/NR2C receptors (Sheinin et al., 2001).
NMDA receptor undergoes complex gating steps before the channel opens. Current models of NMDA receptor gating have been developed using partial agonists for NR1/NR2A and NR1/NR2B receptors. One of the gating schemes based on partial agonists suggests that the receptor may undergo NR1 and NR2 subunit-dependent gating steps before the channel opens (Banke and Traynelis, 2003; Erreger et al., 2005b). However, a recently proposed model suggests that partial agonists modify multiple gating steps and therefore mechanisms other than subunit-dependent gating may be involved (Kussius and Popescu, 2009). A common feature of these proposed models is that partial agonists reduce the efficiency of NMDA receptor gating. Since DCS has higher efficacy than glycine it is likely to increase the efficiency of gating of NR1/NR2C receptor compared with glycine. To understand the possible conformational changes induced by DCS that may account for its higher efficacy at the NR1/NR2C receptor we performed molecular dynamics (MD) simulations and single-channel studies. MD simulations of the DCS- and glycine-bound NR1/NR2B and NR1/NR2C ligand binding domain dimers suggest that the interaction of specific residues at multiple sites within the NR1:NR2 dimer interface is dependent on whether a partial or full agonist binds to the NR1 ligand binding pocket. Furthermore, maximum likelihood fitting of kinetic models to single-channel recordings suggest that a fast gating rate and closing rate are uniquely modified by DCS. These results highlight the importance of the dimer interface as a transducer of DCS efficacy at the NR1/NR2C receptor.
Materials and Methods
Two-electrode voltage-clamp recording from Xenopus laevis oocytes.
Isolation, injection, and recording from Xenopus oocytes was performed as previously described (Dravid et al., 2007). Briefly, female Xenopus laevis were anesthetized using 0.1–0.2% ethyl 3-aminobenzoate, and the ovaries were surgically removed. Stage V and VI oocytes were isolated after a 2 h incubation of the ovaries in 2 mg/ml collagenase at room temperature. Oocytes were injected within 24 h of isolation with 5 ng of NR1-1a subunit cRNA (hereafter NR1; GenBank accession number U11418) and 5–10 ng of NR2A, NR2B, NR2C or NR2D subunit cRNA (GenBank accession numbers D13211, U11419, M91563, or L31611, respectively) in a 50 nl volume, and incubated in Barth's solution at 18°C for 3–7 d; some oocytes were stored at 4°C after 3–5 d. RNA was synthesized in vitro according to manufacturer's recommendations (Ambion mMessage mMachine). Recording pipettes were filled with 0.3–3 m KCl. Current recordings were made using Warner OC725B two-electrode voltage-clamp amplifiers configured as recommended by the manufacturer. Oocytes were continuously perfused in a solution containing (in mm) 90 NaCl, 5 HEPES, 3 KCl, 0.5 BaCl2, pH 7.4, and recorded at a holding potential of −40 mV. All experiments were performed at room temperature (23°C). All laboratory practices and animal care were consistent with current National Institutes of Health guidelines and all experimental protocols were approved by the Creighton University or the Emory University Institutional Animal Care and Use Committee.
Mutagenesis.
Site-directed mutagenesis was accomplished by using the QuikChange kit (Stratagene) and verified by sequencing, as described previously (Low et al., 2000).
Voltage-clamp recording from HEK293 cells.
Whole-cell voltage-clamp recordings from transiently transfected HEK293 cells (ATCC-1573) were performed as previously described (Dravid et al., 2007). Briefly, HEK cells were plated on glass coverslips coated with poly-d-lysine and transiently transfected using the calcium phosphate method (Chen and Okayama, 1987) or Fugene transfection reagent (Roche Diagnostics) with NR1 (pCIneo vector), NR2C (pRK vector) and green fluorescent protein (GFP) at a ratio of 1:2:0.5 (0.5 μg of total DNA/well of 24 well plate). Steady-state current recordings from outside-out patches held under voltage clamp at a holding potential (VHOLD) of −80 mV were made as previously described (Dravid et al., 2008); the holding potential was not corrected for the liquid junction potential. Single-channel currents were recorded using an Axopatch 200B amplifier (Molecular Devices), filtered at 8 kHz (−3 dB, 8-pole Bessel), and digitized at 40 kHz with Axon Digidata board and pClamp9 or pClamp10 software. Whole-cell currents were filtered at 5 kHz and digitized at 20–25 kHz. The extracellular recording solution consisted of the following (in mm): 150 NaCl, 10 HEPES, 0.5 CaCl2, 3 KCl, with 1 mm glycine or 1 mm DCS and 1 mm glutamate used to activate NR1/NR2C receptors unless otherwise noted (pH 7.4, 23°C). This solution was supplemented with 0.01 mm EDTA to chelate trace amounts of contaminant divalent ions, such as Zn2+. The internal solution consisted of the following (in mm): 110 cesium gluconate, 30 CsCl2, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 Na2ATP, and 0.3 Na2GTP (pH 7.35 with CsOH). The extracellular recording solution and internal solution were identical for whole-cell and single-channel recordings. The agonist concentration for whole-cell and macroscopic jumps were 0.5–1 mm glycine, 0.5–1 mm DCS and 1 mm glutamate. A submaximal concentration of glutamate (5 μm) was used for macroscopic jumps for macroscopic fitting. Rapid solution exchange was achieved with a two-barrel theta glass pipette controlled by a piezoelectric translator (Burleigh); junction currents were used to estimate the speed of solution exchange after recordings. Exchange times for 10–90% solution at an open pipette tip were <1 ms, which should be representative of exchange time at outside-out patches.
Single-channel analysis.
Idealization was performed using the time course fitting method, which fits a filtered step response function to each transition between open and closed states (SCAN; www.ucl.ac.uk/Pharmacology/dcpr95.html). Fitted amplitudes were analyzed using EKDIST. Open and closed duration histograms were constructed from data analyzed with this method and fitted to the sum of Gaussian or exponential components using ChanneLab (all conductance levels were assumed to be equal for this analysis). Dwell-time histograms were generated with an imposed dead time of 50 μs for open events and 30 μs for shut events. Maximal interval likelihood fitting (MIL) (Qin et al., 1996) was performed on data idealized with SCAN with an imposed dead time of 30 μs using QUB software (www.qub.buffalo.edu); similar results were obtained when a dead time of 50 μs was imposed (data not shown). We used the approximation of Colquhoun and Hawkes (1990) to calculate the probability that only one active channel was present in the outside-out patches without any double openings when exposed to saturating concentration of agonists. This equation allows the prediction of the number of consecutive openings of a channel in a patch that contains 2 active channels without any double openings as a function of the open probability of the channel. E(r) is the mean number of consecutive openings observed in a run, and is related to Po2, the probability that a channel is open during an observed run of single openings that originates from two independent channels (Po2 is two times the channel open probability). That is, the mean number of consecutive openings in a run is related to Po2, the fraction of time for which a channel is open during the run, according to E(r) = (2/Po2) (1 − 0.5 Po2 − 0.75 Po22).
We used this equation to determine the probability that patches contain a single active channel; analysis was restricted to recordings for which p > 0.99 that the patch contains only one active channel.
Single-channel records were also idealized with a segmental k-means algorithm (Qin, 2004) using QUB software (www.qub.buffalo.edu) for comparison with SCAN. Although a similar increase in open period and Po in DCS compared with glycine were observed with QUB idealized data, the fitted shut time constants were found to differ somewhat between the two idealization methods (data not shown).
Molecular modeling and molecular dynamics simulations.
Starting from the crystal structure of the NR1/NR2A agonist binding domain dimer (PDB code 2A5T), models of the NR1/NR2B and NR1/NR2C agonist binding domain dimers were constructed using the comparative modeling package Prime (Schrödinger), similar to what we have previously described (Erreger et al., 2007). When aligned, the agonist binding domain of NR2A shows 86% sequence identity and 92% sequence similarity with NR2B, compared with 75% sequence identity and 85% sequence similarity with NR2C. The sequence of NR2A in our model was also modified to represent the wild-type polypeptide, which involved removal of a short linker peptide and addition of two unresolved residues in the crystal structure (pdb 2A5T). The NR1/NR2C and NR1/NR2B interfaces were subjected to multiple rounds of side chain optimization and energy minimization using Prime (Schrödinger) to alleviate any strain introduced by homology modeling. Analysis with PROCHECK (Laskowski et al., 1993) reveals little difference in the overall G factors between the crystal structure (0.34) and both the NR1/NR2B (0.28) and NR1/NR2C homology models (0.21); scores above −0.5, considered acceptable. The glycine and glutamate ligands were retained in their crystallographic positions before unrestrained molecular dynamics simulation in which both protein and ligand were allowed to move. Similarly the structure of DCS (net charge of zero) cocrystallized with the NR1 subunit was used for the DCS–receptor complex simulations as published by Furukawa et al. (2003). These complexes were then prepared for molecular dynamics (MD) simulation using Desmond (Bowers et al., 2006). Ligand and protein were treated using the OPLS2005 force field, and then solvated with a box volume of 1,414,255 Å3 (buffer distance of 10Å) containing 18,559 water molecules (SPC water model). The overall system was neutralized at pH 7.0 using an ion concentration of 0.15 NaCl. The protonation state of DCS as a zwitterion [net charge of zero, pKa 7.5 for exocyclic amine and 4.5 for the exocyclic oxygen (McBain et al., 1989) (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material)] was used as described by Furukawa et al. (2003), since it is supported in the OPLS2005 force field. This differs from previous MD simulations with DCS, which were modeled with a net charge of +1. The system was first relaxed using the Desmond relaxation model. The completed equilibration run was followed by a production run performed with NPT conditions using the Berendsen thermostat (300K and 1.103 bar) (Berendsen et al., 1984) and PME electrostatics (Essman et al., 1995) with a cutoff of 9Å. Time step calculations were performed every 2 fs. Average structures were prepared from the final 2 ns of simulation. Flexible loop regions of the D1 domains were excluded when performing domain alignments. The center of mass calculations of these structures were performed using the C-α carbons of the residue selection constituting the domains of interest. All figures from MD simulations were produced using VMD (Humphrey et al., 1996).
Statistics.
All values are given as mean ± SEM. We compared the differences of means using a t test; values p < 0.05 were considered significantly different. Concentration–response curves normalized to the response to maximally effective concentration of glycine were fitted by the Hill equation using Prism 4.0 (GraphPad Software Inc.), Response = Relative Efficacy/(1 + (EC50/[Concentration])N), where N is the Hill slope and EC50 is the concentration of agonist that produces a half-maximal response.
Results
d-Cycloserine activates NR1/NR2C receptors with higher relative efficacy than glycine
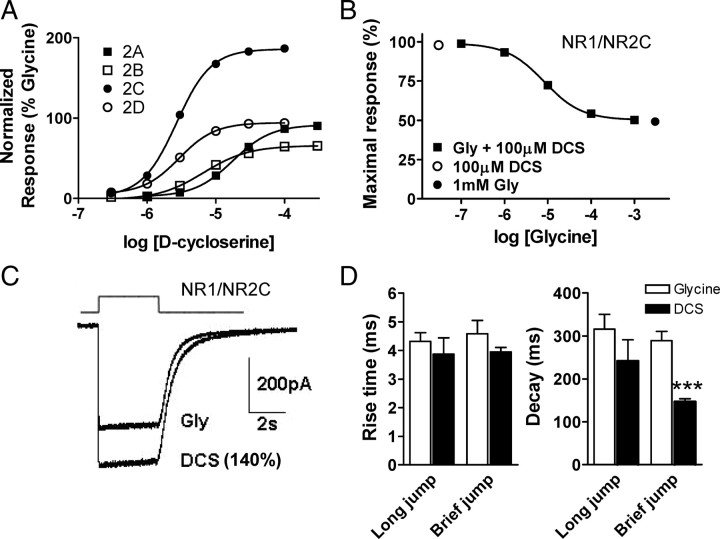
DCS is a partial agonist at NMDA receptors containing the NR2A, B or D subunits. However at NR1/NR2C receptors it shows higher relative efficacy than the endogenous agonist glycine (Sheinin et al., 2001). The underlying mechanism of differential efficacy of DCS is unknown. Sheinin et al. (2001) studied the efficacy of DCS at different NR1/NR2 subunit combinations in the presence of a maximal concentration of NMDA. NMDA is a partial agonist at the NR2 subunit, with relative efficacy that is itself dependent on the NR2 subunit (Erreger et al., 2007). We determined the relative efficacy of DCS at all NR1/NR2 subunit combinations in the presence of maximal concentration of the endogenous neurotransmitter glutamate (Fig. 1A). The EC50 and efficacy of DCS compared with glycine at all receptor combinations is presented in Table 1. The relative efficacy of DCS as a percentage of the response to a maximal concentration of glycine and glutamate was 90 ± 2 (n = 7), 65 ± 2 (n = 17), 190 ± 6 (n = 22) and 94 ± 2 (n = 8) at NR1/NR2A, NR1/NR2B, NR1/NR2C and NR1/NR2D receptors, respectively (Fig. 1A; Table 1). The supramaximal response of 100 μm DCS at NR1/NR2C receptors could be converted to maximal glycine response by coapplication of 100 μm DCS with increasing concentration of glycine (Fig. 1B). This apparent displacement of DCS by glycine confirms that the higher efficacy of DCS reflects its binding to the glycine site on NR1. The IC50 for glycine-mediated reduction of the maximal response by 100 μm DCS was 8 μm (n = 5, Fig. 1B).
Figure 1.
d-Cycloserine has higher relative efficacy than glycine at NR1/NR2C receptors. A, Concentration–response curves for DCS normalized to the response to a maximally effective concentration of glycine (100 μm glycine) at all NR1/NR2 subunit combinations determined by TEVC recordings from Xenopus oocytes (VHOLD, −40 mV). The relative efficacy and EC50 values are summarized in Table 1. All voltage-clamp recordings were performed with 100 μm glutamate as a coagonist. B, The higher relative efficacy of 100 μm DCS is reduced with increasing concentrations of glycine co-applied at the NR1/NR2C receptor. C, Whole-cell current responses to rapid agonist concentration jumps from HEK293 cells expressing NR1/NR2C receptors (VHOLD, −80 mV). Current responses were evoked by 1 mm glutamate and 1 mm glycine or 1 mm DCS. The relative amplitude of the peak response in the same cell for DCS relative to glycine is 140 ± 8% (n = 4). The top trace is open tip potential. D, Time course of macroscopic NR1/NR2C receptor current responses from outside-out patches. Agonist concentration jumps were performed from maximal concentration of glycine or DCS (0.5 mm) into 1 mm glutamate plus glycine or DCS. The long and brief jumps were for a duration of 2 s and 5–8 ms, respectively. Comparison was done using unpaired t test; ***p < 0.001.
Table 1.
EC50 and efficacy at wild-type and mutant NMDA receptors
| Receptor | Glycine EC50 (μm) | Fold change (mut/WT) | N | DCS EC50 (μm) | Fold change (mut/WT) | N | DCS efficacy (% glycine) | Fold change (mut/WT) | N |
|---|---|---|---|---|---|---|---|---|---|
| Wild-type receptors | |||||||||
| NR1/NR2A | 1.3 ± 0.1 | 9 | 19 ± 1.5 | 7 | 90 ± 2 | 7 | |||
| NR1/NR2B | 0.5 ± 0.1 | 11 | 8.2 ± 0.9 | 12 | 65 ± 2 | 17 | |||
| NR1/NR2C | 0.4 ± 0.02 | 21 | 3.3 ± 0.2 | 19 | 190 ± 6 | 22 | |||
| NR1/NR2D | 0.2 ± 0.03 | 8 | 2.9 ± 0.2 | 8 | 94 ± 2 | 8 | |||
| Binding site mutants | |||||||||
| NR1(R523A)/NR2B | 2853 ± 31 | 5700 | 4 | 129 ± 10 | 15 | 5 | 31 ± 2 | 0.5 | 6 |
| NR1(R523A)/NR2C | 1074 ± 49 | 2700 | 3 | 98 ± 1 | 30 | 6 | 254 ± 20 | 1.3 | 6 |
| NR1/NR2B(R693A) | 0.3 ± 0.01 | 1 | 6 | 9.2 ± 1.2 | 1 | 7 | 68 ± 3 | 1.0 | 12 |
| NR1/NR2C(R703A) | 0.4 ± 0.02 | 1 | 6 | 4.3 ± 0.2 | 1 | 4 | 190 ± 4 | 1.0 | 9 |
| Site II mutants | |||||||||
| NR1/NR2A(E798Q) | 3.0 ± 0.4 | 2 | 4 | 82 ± 4.8 | 4 | 4 | 90 ± 2 | 1.0 | 7 |
| NR1/NR2B(E790Q) | 0.5 ± 0.02 | 1 | 6 | 22 ± 1.3 | 3 | 4 | 78 ± 1 | 1.2 | 6 |
| NR1/NR2C(Q800E) | 0.7 ± 0.2 | 2 | 4 | 4.5 ± 0.4 | 1 | 8 | 130 ± 4 | 0.7 | 12 |
| NR1/NR2D(E791Q) | 0.3 ± 0.1 | 1 | 4 | 3.0 ± 0.5 | 1 | 4 | 110 ± 1 | 1.2 | 8 |
| NR1/NR2B(E790A) | 0.9 ± 0.03 | 2 | 5 | 12 ± 1.9 | 1 | 9 | 68 ± 3 | 1.0 | 13 |
| NR1/NR2C(Q800A) | 0.4 ± 0.03 | 1 | 4 | 5.8 ± 0.9 | 2 | 6 | 191 ± 7 | 1.0 | 8 |
| NR1(Y692A)/NR2A | 4.5 ± 0.1 | 3 | 3 | 110 ± 7.8 | 6 | 4 | 77 ± 1 | 0.9 | 7 |
| NR1(Y692A)/NR2B | 2.0 ± 0.2 | 4 | 3 | 33 ± 4.4 | 4 | 4 | 46 ± 1 | 0.7 | 7 |
| NR1(Y692A)/NR2C | 1.2 ± 0.1 | 3 | 3 | 9.5 ± 3.2 | 3 | 5 | 59 ± 2 | 0.3 | 8 |
| NR1(Y692A)/NR2D | 1.1 ± 0.1 | 5 | 5 | 19 ± 1.6 | 6 | 3 | 75 ± 1 | 0.8 | 8 |
| NR1(R755A)/NR2C | 0.9 ± 0.3 | 2 | 8 | 14.2 ± 2.2 | 4 | 9 | 100 ± 5 | 0.5 | 20 |
| Site III mutants | |||||||||
| NR1/NR2C(S707N,R710P) | 0.4 ± 0.03 | 1 | 6 | 3.9 ± 0.2 | 1 | 6 | 190 ± 7 | 1.0 | 12 |
| NR1(E781A)/NR2B | 1.7 ± 0.2 | 3 | 4 | 8.6 ± 3.0 | 1 | 4 | 10 ± 1 | 0.2 | 8 |
| NR1(E781A)/NR2C | 0.7 ± 0.1 | 2 | 6 | 7.9 ± 0.8 | 2 | 4 | 150 ± 10 | 0.8 | 8 |
| ATD deleted NR2C | |||||||||
| NR1/NR2C(ΔATD) | 2.5 ± 0.2 | 6 | 7 | 18 ± 0.8 | 5 | 8 | 72 ± 2 | 0.4 | 15 |
Individual concentration–effect curves for each oocyte were fitted with the Hill equation, as described in Materials and Methods; the fitted Hill slopes ranged between 0.7 and 1.7. Values are mean ± SEM.
We subsequently evaluated the efficacy of rapidly applied DCS relative to glycine for activation of NR1/NR2C receptors in a mammalian expression system. The whole-cell peak response from HEK293 cells to 1 mm DCS was found to be 140 ± 8% of that observed with 1 mm glycine (n = 4), with 1 mm glutamate as the coagonist with both glycine and DCS (Fig. 1C). The time course of the NR1/NR2C current responses to rapid application of DCS or glycine was measured from macroscopic responses of NR1/NR2C in outside-out patches. The rise time and glutamate deactivation time constants of the macroscopic currents in response to a short 5–8 ms application of maximal concentrations of glutamate in the continuous presence of DCS were 3.9 ± 0.2 ms and 147 ± 7 ms, respectively (n = 4). Rise time and glutamate deactivation time course of the current responses to a 1 s concentration jump from DCS into glutamate plus DCS were 3.8 ± 0.7 ms and 242 ± 50 ms, respectively (n = 4). The rise time in response to DCS/glutamate is similar to that described previously for glycine/glutamate activation of NR1/NR2C receptors (3.9 ms, Dravid et al., 2008). The deactivation time course observed following removal of glutamate is modestly faster in DCS- compared with glycine-bound receptors (320–420 ms; Dravid et al., 2008). We measured the EC50 of glutamate in the presence of saturating concentration of glycine or DCS (100 μm) in oocytes expressing NR1/NR2B or NR1/NR2C receptors. The EC50 of glutamate at NR1/NR2B receptors was 1.19 ± 0.04 μm in saturating glycine and 1.52 ± 0.08 μm in saturating DCS (n = 8 for both, p < 0.01, unpaired t test). The EC50 for glutamate at the NR1/NR2C receptors was 0.42 ± 0.04 μm (glycine) and 0.47 ± 0.06 μm (DCS, n = 8 for both, p = 0.5). A slightly higher glutamate EC50 in the presence of DCS compared with glutamate would be consistent with the faster deactivation rate following glutamate removal in the presence of DCS (Fig. 1). The lack of detectable difference at NR1/NR2C receptors may be due to higher affinity of glutamate and perhaps the presence of contaminating glycine in the recording solution.
Molecular dynamics simulations of the NR1/NR2C andNR1/NR2B ligand-binding domain dimers
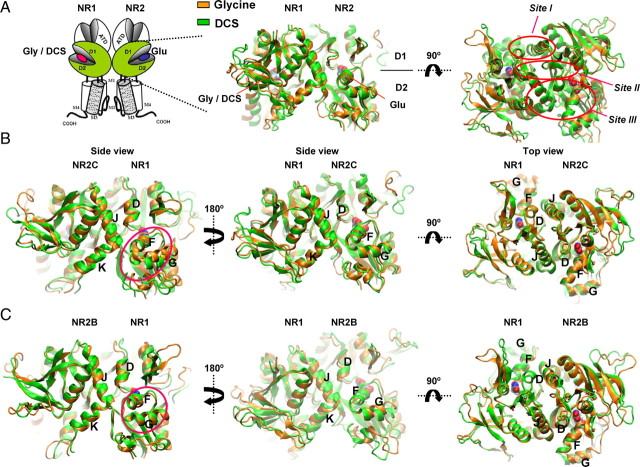
We performed molecular dynamics simulations at 300K on the hydrated homology models of the ligand binding domain (LBD) dimers of NR1/NR2C and NR1/NR2B built from the NR1/NR2A agonist binding domain structures (Furukawa et al., 2005) (see Materials and Methods) with glutamate in the NR2 binding pocket and glycine or DCS in the NR1 binding pocket. The goal of these simulations was to explore the conformational rearrangements within the ligand binding domain that might contribute to the differential relative efficacy of DCS (Fig. 2A). These two subunits (NR2B and NR2C) were selected because they show the largest difference in the relative efficacy of DCS. After 10 ns of simulation, the dimer retained the three interacting sites between protomers described by Furukawa et al. (2005) (Fig. 2A, Sites I, II, III). An alignment of the average structures over the last 2 ns based on the protein backbone for the glycine and DCS molecular dynamics simulations gave RMSD values of 2.2Å and 2.4Å for NR1/NR2C and NR1/NR2B, respectively. The most pronounced differences were observed in the NR1/NR2C simulation between the orientation of helix F and G of the NR1 subunit along with modest differences in helix F and K of the NR2 subunit (Fig. 2B). Furthermore differences in orientation of helix F and G of NR1 were also observed along with helix K of the NR2 subunit in the NR1/NR2B simulations (Fig. 2C).
Figure 2.
Molecular dynamics simulations of the NR1/NR2B and NR1/NR2C ligand binding domain dimer. A, Schematic showing the NMDA receptor subunit arrangement with the ligand binding domains of NR1 and NR2 subunit shown in green. Homology models of NR1/NR2B and NR1/NR2C were based on the crystal structure of NR1/NR2A (Furukawa et al., 2005); molecular dynamics simulations were run on the hydrated protein with glycine (orange) or DCS (green) docked into the NR1 pocket and glutamate in the NR2 pocket. Dimer interaction sites Site I-III are indicated by red circles. B, Molecular dynamics simulations of NR1/NR2C ligand binding domain dimer. The difference in orientation of helix F and G of NR1 is evident (circled in side view). In addition, differences in arrangement of helices F and K of NR2C can be seen. C, Molecular dynamics simulations of NR1/NR2B ligand binding domain dimer. The difference in orientation of helices F and G of NR1 is evident (circled in side view).
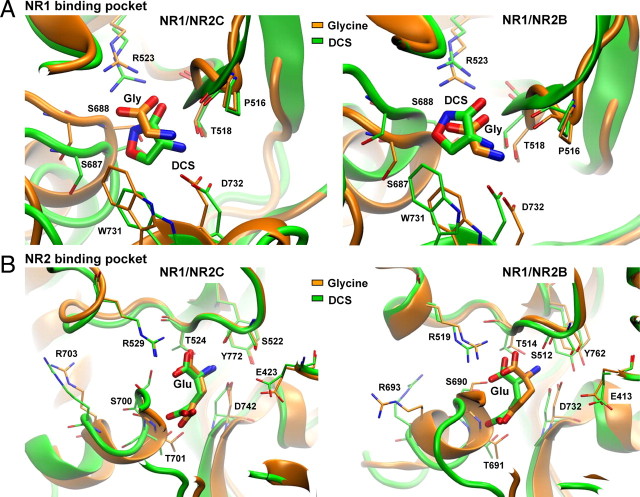
We examined in detail the orientation of glycine and DCS in the NR1 agonist binding pocket following MD simulations, which suggests that these ligands largely retain their expected positions from crystallographic data (Furukawa and Gouaux, 2003; Furukawa et al., 2005). Similar to the crystal structure and previous MD simulations, glycine interacts with Arg523 and Thr518 in the D1 and Asp732 and Ser688 in D2. The hydrogen bonds formed between the amino groups of Gly and DCS and the backbone carbonyl of Pro516 after 10 ns of MD for both the NR1/NR2B and NR1/NR2C dimers are consistent with the interaction observed in the monomeric NR1 crystal structure (Furukawa and Gouaux, 2003) (Fig. 3A). Consistent with data from both crystal structures of monomeric NR1 and dimeric NR1/NR2A, the α-carboxy group of glycine interacts with the guanidinium group of NR1 Arg523 through a salt-bridge in both the NR1/NR2C and the NR1/NR2B dimers throughout the MD simulations. In the presence of DCS this salt-bridge is replaced by a similar interaction between the negatively charged exocyclic oxygen and the endocyclic nitrogen with Arg523, which persists throughout the simulation (Fig. 3A). Furthermore it was observed that mutation of Arg523 to Ala dramatically increases the EC50 for both glycine and DCS, as expected for a directly interacting residue (Table 1). However, the EC50 of DCS is lower than glycine at NR1(R523A)/NR2B and NR1(R523A)/NR2C. The reduced effect of NR1(R523A) for DCS might be explained by additional hydrogen bond formation that is observed during the MD simulation between both the exo- and endocyclic oxygens of DCS with Thr518 and Ser688, respectively. These interactions should stabilize DCS in the glycine binding pocket, but still allow a response upon binding. We also studied the glutamate interactions within the NR2 binding pocket for the NR1-bound glycine and NR1-bound DCS MD simulations. All glutamate interactions within glycine- or DCS-bound NR1/NR2 simulations were the same, except for a few relatively minor differences involving interactions of the ligand amino group with Ser512/522 and Glu413/423 of NR2B/C.
Figure 3.
Comparison of glycine, DCS, and glutamate binding, with models aligned by D1 domains of NR1 and NR2. A, Molecular dynamics simulation with glycine (orange) or DCS (green) in the binding pocket for NR1/NR2B and NR1/NR2C receptors. The α-carboxy group of glycine interacts with the guanidinium group of Arg523 through a salt-bridge in both NR1/NR2B and NR1/NR2C MD simulations. B, Molecular dynamics simulations show the interactions of glutamate within the binding pocket with either glycine (orange) or DCS (green) in the NR1 binding pocket.
Conformational changes within the NR1 and NR2 ligand-binding domains induced by d-cycloserine
The efficacy of glycine site agonists depend on the NR2 subtype, which suggests that intersubunit communication must occur. The most apparent site for such long-range intraprotein conformational changes is the dimer interface of the NR1 and NR2 agonist binding domains, where the residues of NR1 domain may interact differentially with the four NR2 subunits. Our working hypothesis is that differential dimer interface interactions between NR1 and NR2C ligand binding domains are responsible for the high relative efficacy of DCS on NR1/NR2C. Differences in the intersubunit contacts between NR1 and NR2A compared with NR1 and NR2D (particularly at D2) have been suggested to influence the glycine potencies at these two receptors (Chen et al., 2008). We therefore examined the MD simulations for apparent differences at the dimer interface that could explain the differential agonist efficacy of DCS at NR1/NR2C and NR1/NR2B receptors. We first examined Site I (Fig. 2A) (Furukawa et al., 2005) of the dimer interface formed by helix D of NR1 and helix J of NR2. No apparent differences in the orientation of helix D of NR1 and helix J of NR2C were observed between glycine- and DCS-bound NR1 (Fig. 2B). Similarly no difference in the orientation of NR1 helix D and NR2B helix J was observed for glycine and DCS, although in the DCS simulation they appear to orient at a somewhat greater angle than in glycine simulation (Fig. 2C). Hydrophobic and polar residues at Site I are conserved among all the NR2 subunits and are therefore unlikely to lead to differential relative efficacy of DCS (Furukawa et al., 2005). As expected, molecular dynamics simulations did not identify residues at Site I that could account for the differences in relative efficacy of DCS compared with glycine.
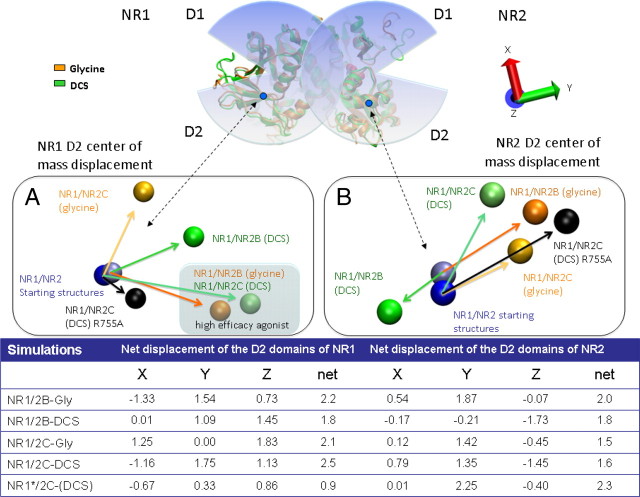
Helix F has been suggested to play an important role in agonist binding, receptor gating, and agonist efficacy (McFeeters and Oswald, 2002; Erreger et al., 2005b; Furukawa et al., 2005; Inanobe et al., 2005). In crystal structures of isolated NR1 glycine- or DCS-bound agonist binding domain, helix F does not exhibit any difference in orientation (Inanobe et al., 2005). Interestingly, different orientations of helix F and G in NR1 develop between glycine and DCS in simulations of the NR1/NR2 heterodimer (Fig. 2B,C). Similar displacements for NR1/NR2B and NR1/NR2C were observed for both the most and least efficacious agonists. That is, the relative positions of NR1 helices F and G of the DCS (NR1/NR2C) and glycine (NR1/NR2B) simulation (RMSD of 1.1Å) were found to be similar, as were the corresponding NR1 helices of the DCS (NR1/NR2B) and glycine (NR1/NR2C) simulations (RMSD of 1.8 Å). The largest deviation was observed between NR1 helices F and G of NR1/NR2C simulated with DCS and glycine, compared with the starting structure (aligned by NR1-D1, NR2-D1). The movement of these NR1 helices correlated with movement of the center of mass of the NR1-D2 domain from the starting configuration (Fig. 4A). The most efficacious agonists of the NR1/NR2B and NR1/NR2C simulations showed similar domain movement compared with the starting configuration of the simulation (Fig. 4A, see shaded region), perhaps explaining the similar movement/orientation of helices F and G. We therefore postulate that the importance of these helices in different agonist efficacies is related to NR1-D2 domain movement. No clear conclusion regarding agonist dependency could be drawn from the domain movements observed during the MD simulations of the NR2 subunit, however it was noted that the higher efficacy agonists simulations grouped together (Fig. 4B).
Figure 4.
A representation of the net displacement observed for the center of mass of the D2 domains for both the NR1 and NR2 subunit at the conclusion of the simulations. To determine the displacement of the D2 domains, the average structure of the last 2 ns of the 10 ns Molecular dynamics simulations were aligned to the starting structures based on the D1 domain of both the NR1 and NR2 subunit. A, The spheres are representative of the center of mass of the D2 domain of the NR1 subunit. The sphere in light blue represents the center of mass of the starting structure of NR1/NR2B and the dark blue that of NR1/NR2C; the starting structure reflects the position of D2 in the NR1/NR2A crystal structure from which NR1/NR2B and NR1/NR2C homology models were built. The NR1 simulations with Gly are represented in orange and that of DCS in green. The black spheres are representative of the center of mass displacement of the D2 domains of a NR1(R755A)/NR2C mutant, simulated in the presence of DCS and Glu. The shaded area highlights the similar displacement at the end of the simulation for the most efficacious agonist for both NR1/NR2B (glycine) and NR1/NR2C (DCS). B, The spheres are representative of the center of mass of the D2 domain of the NR2 subunit. The same color scheme as in A. The net displacement on the x-, y-, and z-axis are presented in the table below. The orientation of the axis can be found in the top right-hand corner.
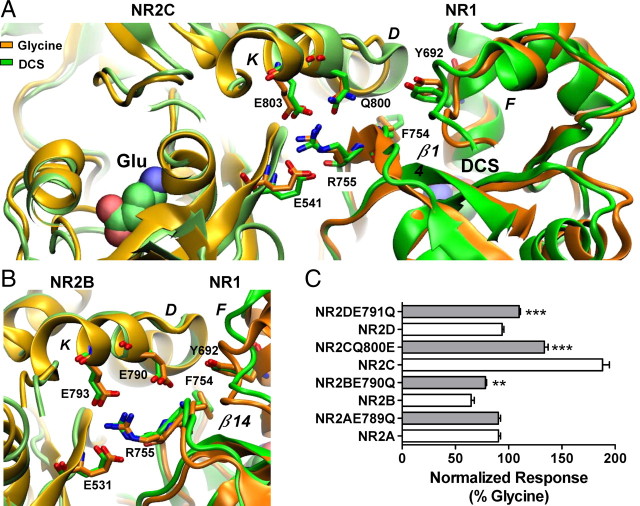
We identified several interactions within Site II between NR2B-Glu531, NR2B-Glu793 (Glu541, Glu803 of NR2C) and NR1-Arg755 that may play an important role in correlating movement between the D2 domains of the NR1/NR2 complex (Fig. 5A,B). To investigate this hypothesis, the effect of DCS was tested on NR1(R755A)/NR2C. The relative efficacy of DCS was reduced from 190% to 100% of the maximal glycine response (Table 1). The NR1(R755A)/NR2C mutant was simulated with DCS and glutamate docked in the agonist binding sites. The center of mass displacement of the NR1-D2 domain was found to behave similarly, with a reduced magnitude compared with the wild-type (Fig. 4A). Similarly the NR2-D2 domain showed the same movement as the wild-type but with a reduced magnitude (Fig. 4B). The data supports the idea that NR1-Arg755 participates in the transfer of actions that DCS has on the NR1 subunit to the NR2 subunit. It is further thought that NR1-Arg755 mediates the transfer of the DCS effects by coordinating the D2 domain movements of both the NR1 and NR2 subunits with respect to each other. The mediation takes place by the interaction of NR1-Arg755 with both NR2C-Glu803 and NR2C-Glu541 (corresponding to NR2B-Glu793 and NR2B-Glu531; Fig. 5).
Figure 5.
Residues at the dimer interface play a role in the control of the relative efficacy of DCS. A, Hydrogen bond formation between residues Arg755, Glu803 and Glu541 illustrating the dimer interface interaction, which correlates domain movement between the D2 domains of NR1 and NR2. Left, The NR2C subunit is colored lime and light orange for the DCS and glycine, respectively. Right, The NR1 subunit is colored green and orange with Arg755 protruding from the NR1–D2 domain interacting with Glu803 and Glu541 from the NR2C-D2 domain. The orientation change Phe754 undergoes between the DCS and glycine simulations can be observed on β-strand 14. Gln800 is shown protruding from helix K, no hydrogen bond formation with Tyr692 and Phe754 was observed. B, Hydrogen bond formation between residues Arg755, Glu793 and Glu531 illustrating the dimer interface interaction, which correlates movement between the D2 domains of NR1 and NR2. Left, The NR2B subunit is colored lime and light orange for the DCS and glycine, respectively. Right, The NR1 subunit is colored green (DCS) and orange (glycine) with Arg755 protruding from the NR1–D2 domain interacting with Glu793 and Glu531 from the NR2B-D2 domain. Glu790 protrudes from helix K to form hydrogen bonds with Tyr692 and the Phe754. C, Summary of changes in relative efficacy; efficacy between wild type and respective mutant receptors was compared by unpaired t test; **p < 0.01, ***p < 0.001. The fitted EC50 values are presented in Table 1.
Furthermore, stabilizing interactions involving NR2B-Glu790 and NR2C-Gln800 (Fig. 5A,B) could also partially explain the extent of domain D2 displacement; residues corresponding to NR2B-Glu790 are conserved in all NR2 subtypes except NR2C. NR2B-Glu790 initially forms hydrogen bonds with NR1-Tyr692 at the start of the simulations, but this interaction is lost after ∼1.5 ns or absent in NR1/NR2C simulations. However, in the glycine-bound NR1/NR2B simulation, Glu790 forms hydrogen bonds with the amino group of Phe754 for 60% of the time and the guanidinium moiety of Arg755 via a water molecule (Fig. 5B). In contrast to the NR1/NR2B simulation, the NR1/NR2C simulation suggested that NR2C-Gln800 forms interactions similar to those in NR2B-Glu790 but only for 7% of the time. These interactions were absent from DCS-bound simulations, which highlights the potential importance of NR2B-Glu790 and NR2C-Gln800 in understanding the relative efficacy of NR1 agonists such as DCS. By altering the protein-protein interactions between NR1 and NR2 subunits, these residues may influence changes between the interacting domains contributing to differential DCS efficacy at NR1/NR2C.
In an attempt to test whether NR2C-Q800 uniquely influences relative efficacy compared with glutamate at the corresponding position at NR2A, NR2B, and NR2D, we tested the functional consequence of changing Gln to Glu, and vice versa. The relative efficacy of DCS at NR1/NR2C(Q800E) was reduced from 188% to 134 ± 4% of the maximal glycine response (n = 12, p < 0.001, unpaired t test, Fig. 5C), with no significant change in the EC50 of glycine and DCS (Table 1), consistent with the working hypothesis. The effect of the reverse mutations NR1/NR2B(E790Q) and NR1/NR2D(E791Q) receptors showed a modest increase in relative efficacy of DCS compared with wild type receptors (Table 1). These data suggest that NR2C-Gln800 and NR2B-Glu790 partially contribute to the differential efficacy of DCS on NR1/NR2B and NR1/NR2C. Mutation of either NR2C-Gln800 or NR2B-Glu790 to Ala showed no effect on the relative efficacy of DCS and a <2-fold increase in EC50 of glycine or DCS (Table 1), suggesting that the hydrogen bonding within this region of the dimer interface is important in these interactions.
We additionally investigated the role of NR1 Tyr692 by mutating it to Ala, which reduced the relative efficacy of DCS activation of NR1(Y692A)/NR2C to 59 ± 2% (n = 8, Fig. 5C) of glycine. This level of relative efficacy was similar to that for other NR2 subunits (relative efficacy 46–77%; Table 1). NR1(Y692A) also showed an ∼3- to 5-fold increase in DCS and glycine EC50 values for various NMDA receptor combinations (Table 1). The effects of NR1(Y692A) could be explained through the significant change this mutation imparts to the surrounding hydrophobic environment, which consequently disrupts the interactions at Site I.
We also examined the dimer interface at Site II formed by the β-strands 10 and 14 in NR1 and β-strands 10 and 14 in NR2 (Furukawa et al., 2005). In the NR1 crystal structure with partial agonists ACPC and ACBC, the residues of the NR1 β-strand 14 show differences in conformation compared with the NR1 crystal structure bound by glycine (Inanobe et al., 2005). In the NR1/NR2B simulations with glycine and DCS, no clear trend in conformational differences was observed in NR1 β-strand 14 that could be correlated with agonist efficacy. However, in the NR1/NR2C simulations, a rotation of almost 90 degrees was observed for the phenyl ring of NR1-Phe754 during the DCS simulation with respect to that observed in the simulation of glycine-bound NR1/NR2C (Fig. 5A). It appears that DCS allows for a slight expansion of the glycine binding cavity, which together with the absence of a hydrogen bond with the backbone amino group of NR1-Phe754 (with NR2C-Gln800) allows for the phenyl group reorientation throughout the simulation. The lack of movement of NR1-Phe754 within NR1/NR2B appears to be due to the hydrogen bond formation between NR2B-Glu790 and the amino group of NR1-Phe754. Although no conclusive correlation for agonist efficacy could be drawn from this observation, it is consistent with the hypothesis that NR2C-Gln800 plays a role in agonist efficacy due to the minimal interactions with NR1-Phe754.
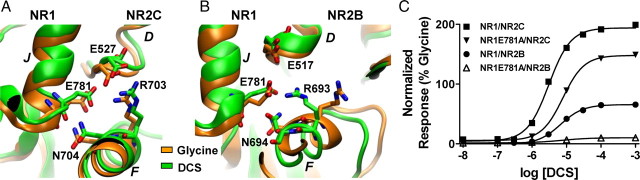
The residues in helix D of NR2 subtypes are highly conserved, including the hydrophobic residues forming Site III (Furukawa et al., 2005). The NR1 helix J residue Glu781 has been shown to be a key residue at the dimer interface. Mutation of NR1-Glu781 can modify allosteric inhibition via the amino terminal domain (ATD) of NMDA receptors (Gielen et al., 2008). We studied the functional effects of the polar interactions formed by NR1 Glu781. In the NR1/NR2A crystal structure as well as the MD simulation, NR1-Glu781 forms several polar contacts with residues on helix D and helix F of the NR2A subunit. In both NR1/NR2B and NR1/NR2C simulations, similar polar interactions are present between the NR1-Glu781 and both helices F and D of the NR2 subunit. NR1-Glu781 forms a hydrogen bond with the amino group of NR2B-Glu517 in helix D and NR2C-Glu527 in helix D, stabilizing the dimer interface between NR1 and NR2. NR1-Glu781 also shares a hydrogen bond with helix F NR2B-Asn694 and NR2C-Asn704 in all the simulations using either its backbone carbonyl group and/or its γ-carboxyl group. In addition, NR1-Glu781 formed hydrogen bonds between its γ-carboxyl group and helix F NR2B-Arg693 or NR2C-Arg703 in all except the glycine-bound NR/NR2B simulation. In the glycine-bound NR1/NR2B simulation the NR1-Glu781 instead interacts with NR1-Lys778, and the NR2B-Arg693 interacts with NR2B-Glu518 (data not shown; Fig. 6). These data suggest that NR1-Glu781 plays an integral role in dimer stabilization between the NR1 and NR2 subunits. To evaluate the effect of dimer stabilization by NR1-Glu781 on DCS and glycine efficacy, we studied the functional consequence of mutating NR1-Glu781 to Ala. This mutation significantly reduced the DCS relative efficacy to 150 ± 10% of glycine at NR1(E781A)/NR2C and to 10 ± 1% of glycine at NR1(E781A)/NR2B (Fig. 6C, n = 8), with a modest 2- to 3-fold change in the EC50 values of DCS or glycine. These results further highlight the importance of residues at the dimer interface in controlling the relative efficacy of NR1 agonists.
Figure 6.
Structural rearrangements at dimer interface involving helix J of NR1 and both helices F and D of NR2. A, Molecular dynamics simulations showed polar interactions between NR1-Glu781 and both helices D and F of NR2C. The γ-carboxyl group of NR1-Glu781 interacts with NR2C-Arg703 whereas its backbone carbonyl group interacts with NR2C-Asn704 in both the DCS and Gly simulations. In addition Arg703 of helix F also interacts with NR2C-Glu527 of helix D in both simulations. B, In the NR2B simulations the backbone carbonyl group of NR1-Glu781 interacts with the NR2B-Asn694 of helix F in both simulations. The γ-carboxyl group of NR1-Glu781 interacts with NR2B-Arg693 in the DCS simulation, which in turn interacts with NR2B-Glu517 of helix D. The latter was however not observed in the glycine simulation. C, Mutation of NR1-Glu781 to Ala reduced the relative efficacy of DCS. The DCS relative efficacy was 149 ± 10% and 10 ± 1% at NR1(E781A)/NR2C and NR1(E781A)/NR2B, respectively (n = 8). There was no significant change in EC50 for DCS or glycine activation of NR1(E781A)/NR2C or NR1(E781A)/NR2B (see Table 1).
Recent studies have shown a role of ATD of the NR2 subunit in regulating the open probability of the receptor (Gielen et al., 2009; Yuan et al., 2009). We thus tested the effect of deletion of the NR2C ATD on agonist EC50 and efficacy. The EC50 for both glycine and DCS were 5- to 6-fold higher at NR1/NR2C(ΔATD) receptors (Table 1) (see also Yuan et al., 2009). Moreover the relative efficacy of DCS was reduced to 72 ± 2% of glycine at NR1/NR2C(ΔATD) receptors. These data suggest that the presence of ATD is required for DCS to exhibit a higher relative efficacy than glycine at NR1/NR2C. This result is also consistent with the idea that multiple factors are responsible for the differences in the DCS and glycine relative efficacy. That is, multiple and complementary structural determinants of relative efficacy of NR1 agonists appear to exist.
d-Cycloserine increases the apparent mean open time and open probability of NR1/NR2C receptors
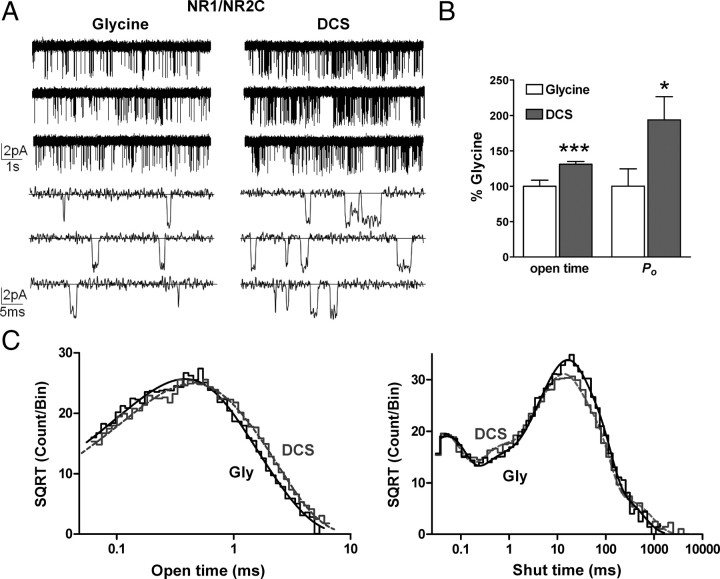
To examine the mechanism underlying the enhanced function of DCS-bound NR1/NR2C compared with glycine-bound NR1/NR2C, we recorded single-channel currents from outside-out patches containing one active NR1/NR2C receptor. Single-channel currents were evoked by steady-state application of a maximally effective concentration of glycine (1 mm) and glutamate (1 mm), or DCS (1 mm) and glutamate (1 mm) (see Materials and Methods). Only patches in which we were able to obtain paired recordings of both DCS and glycine were analyzed (Fig. 7A). We observed only one active channel (see Materials and Methods) in 6 patches. Using a previously described procedure (Dravid et al., 2008) (see Materials and Methods), which is based on the approximation of Colquhoun and Hawkes (1990), we estimated these patches to contain one active channel with at least 99% certainty. We used time course fitting to measure the two NR1/NR2C conductance states observed in the presence of 0.5 mm extracellular Ca2+. The subconductance levels and associated areas were unchanged in the presence of DCS compared with glycine, and were 43 ± 3 pS (68 ± 5%), 32 ± 3 pS (33 ± 5%) for glycine and 45 ± 5 pS (72 ± 8%), 28 ± 3 pS (28 ± 5%) for DCS (reversal potential was assumed to be 0 mV; n = 3; p > 0.05, paired t test).
Figure 7.
DCS increases the mean open time and open probability of NR1/NR2C receptors. A, Steady-state recordings of NR1/NR2C unitary currents from an outside-out patch that contained one active channel in the presence of maximally effective concentration of glutamate and glycine or DCS (1 mm glutamate, 1 mm glycine or 1 mm DCS; VHOLD −80 mV, digitized at 40 kHz, filtered at 5–8 kHz, −3 dB) under two different time scales showing the increase in open probability and mean open time of the channel. B, Paired recordings in glycine and DCS in which we observed only one active channel were analyzed by time course fitting analysis using SCAN software (see Materials and Methods). The mean open time of NR1/NR2C receptors was increased by 131 ± 4% in the presence of DCS (n = 6, ***p < 0.001, paired t test). The open probability was increased by 194 ± 33% in the presence of DCS (n = 6, *p < 0.05, paired t test). C, The composite open time histogram from 6 patches was fitted by two exponential components [glycine: n = 6 patches, 11,275 open periods, τ1 = 0.32 ms (64%), τ2 = 0.68 ms (36%) and DCS: n = 6 patches, 16,662 open periods, τ1 = 0.32 ms (49%), τ2 = 0.76 ms (51%)]. The composite shut time histogram from 6 patches was fitted by five exponential components [glycine: n = 6 patches, 11,272 closed periods, τ1 = 0.042 ms (18%), τ2 = 0.36 ms (7%), τ3 = 9.1 (29%), τ4 = 32 ms (43%) and τ5 = 138 ms (3%) and DCS: n = 6 patches, 16,657 closed periods, τ1 = 0.037 ms (19%), τ2 = 0.48 ms (10%), τ3 = 9.5 ms (36%), τ4 = 36 ms (32%) and τ5 = 248 ms (3%)].
The mean duration of apparent open periods of NR1/NR2C receptors increased from 0.46 ± 0.04 ms in the presence of glycine to 0.61 ± 0.06 ms in the presence of DCS, which reflected a mean increase to 132% of control (n = 6, p < 0.001, paired t test; Fig. 7B). In addition, the open probability (Po) obtained from patches containing a single active channel increased from 0.018 ± 0.004 in the presence of glycine to 0.029 ± 0.006 in the presence of DCS, which corresponded to a mean increase to 163% of control (n = 6, p < 0.05, paired t test; Fig. 7B). DCS does not detectably change the mean open time of NR1/NR2A or NR1/NR2B receptors (Banke and Traynelis, 2003; Kussius and Popescu, 2009), suggesting somewhat different mechanisms of activation between NR2C and NR2A or NR2B. The composite open time histogram from 6 patches was fitted by the sum of two exponential components (Fig. 7C). Time constants and areas for glycine-activated NR1/NR2C were τ1 = 0.32 ms (64%), τ2 = 0.68 ms (36%) (n = 6 patches, 11,275 open periods). Compared with glycine, DCS-activated NR1/NR2C channels showed almost identical fitted open time constants with different relative areas, being 0.32 ms (49%), τ2 = 0.76 ms (51%) (n = 6 patches, 16,662 open periods). These data suggest that the increase in mean open time by DCS appears to be due to increase in the area of the longer time constant.
Analyses of recordings from outside-out patches that contain one active channel allow unambiguous determination of all intra-activation shut durations. The shut duration histograms constructed for NR1/NR2C receptors were fitted individually from 6 patches with >1000 closed events under both glycine and DCS. The shut duration histogram could be fitted by the sum of five exponential functions (Fig. 7C) with the average time constants ± SEM (in ms) and areas as follows: Glycine, τ1 = 0.04 ± 0.005 (16%), τ2 = 0.48 ± 0.06 (6%), τ3 = 12.1 ± 2 (37%), τ4 = 43 ± 7 (38%) and τ5 = 267 ± 130 (3%) (n = 6 patches, 11,272 closed periods); DCS, τ1 = 0.04 ± 0.005 (19%), τ2 = 0.55 ± 0.16 (10%), τ3 = 8.5 ± 1.2 (30%), τ4 = 34 ± 4 (36%) and τ5 = 220 ± 79 (5%) (n = 6 patches, 16,657 closed periods). We did not observe a significant shift in any of the time constants within this dataset, although it is possible that changes in τ3 and τ4 may have been significant if we were able to record more single-channel openings.
d-Cycloserine accelerates rate constants governing a pregating step
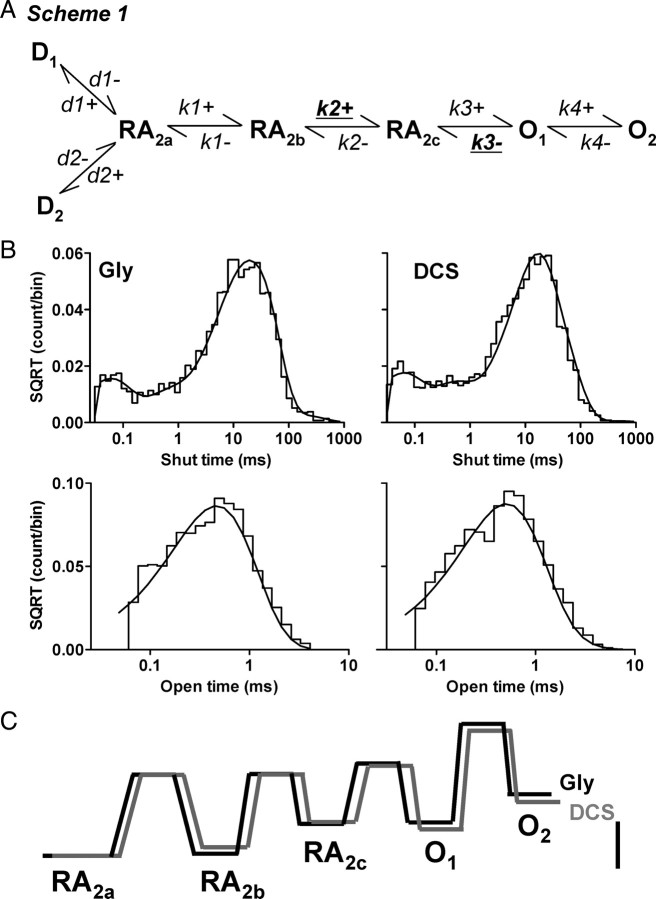
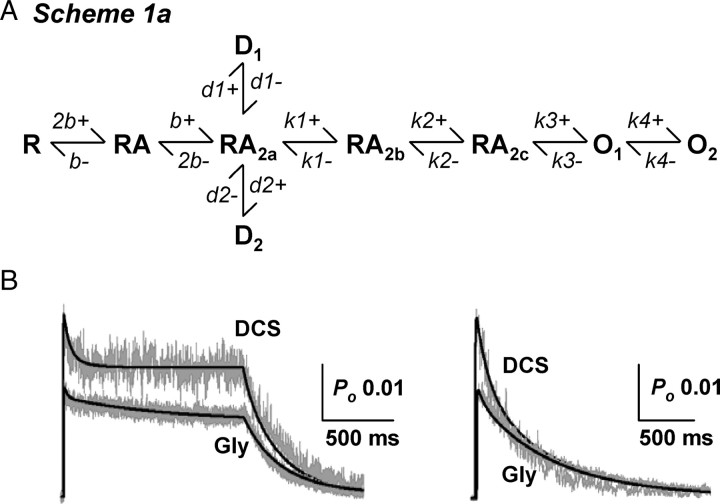
Fitting of conceptual models to single-channel records provides an opportunity to test specific hypotheses about channel gating. A number of recent studies have presented conceptual models for the activation of NR1/NR2A, NR1/NR2B and NR1/NR2C receptors that can accurately describe the single-channel and macroscopic properties of these receptors (Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Popescu et al., 2004; Auerbach and Zhou, 2005; Erreger et al., 2005a,b; Schorge et al., 2005; Dravid et al., 2008). To understand the gating mechanism of DCS at NR1/NR2C receptors, we fitted a previously published model of NR1/NR2C activation (Dravid et al., 2008, their Scheme 1) (Fig. 8A) to the idealized sequence of single-channel open and closed times using the maximum interval likelihood (MIL) method (see Materials and Methods). Scheme 1 is a linear model previously shown to account for both single-channel and macroscopic properties of NR1/NR2C receptors (Dravid et al., 2008). Because the recordings were performed in the continuous presence of a saturating concentration of both glutamate and glycine, no explicit binding steps are included and full occupancy of ligand binding sites was assumed. Two desensitized states are included in the model to account for observed macroscopic desensitization and longer shut times (Dravid et al., 2008). The quality of fit as estimated by the log(likelihood) is insensitive to whether the desensitized states are connected to either of the shut gating states (data not shown). We fitted Scheme 1 to the sequence of channel openings in response to a steady-state application of maximally effective concentration of glutamate plus glycine or DCS in the same patch; open and closed durations were determined using time course fitting. Maximum interval likelihood (MIL) fitting of kinetic models to idealized data was performed using QUB software, which takes into account missed events caused by limited time resolution of the recording system (Qin et al., 1996). We found that our working model fit the data from each patch well, with modest variability between rate constants among patches. Figure 8B compares the predicted probability density function to the observed shut time and open time distribution for one patch. The average log likelihood (a measure of the goodness of the fit of the model to the data) values were similar for glycine and DCS (log likelihood per event: glycine, 4.91 ± 0.04; DCS, 4.86 ± 0.05, n = 6). Table 2 compares the mean (±SEM) rate constants for activations by glycine or DCS fitted to Scheme 1. The fitted rate constants show that the forward rate of the fast gating step (k2+) is significantly altered by DCS. The forward rate k2+ increased from 860 ± 160 s−1 in glycine to 1395 ± 215 s−1 in DCS (n = 6, p < 0.05, paired t test). Consistent with our observations that DCS prolonged the mean open time, DCS significantly slowed the closing rate (k3−) of the NR1/NR2C receptors compared with glycine; the closing rate k3− decreased from 2630 ± 270 s−1 in glycine to 2150 ± 210 s−1 in DCS (n = 6, p < 0.05, paired t test; Table 2).
Figure 8.
DCS augments a fast gating step in NR1/NR2C activation. A, A linear model of NR1/NR2C receptor activation is shown (Dravid et al., 2008); all ligand binding steps are omitted, and all states are assumed to be liganded. The forward rate (k2+) was significantly augmented by DCS whereas the closing rate k3− was significantly slowed by DCS. B, MIL fit of single-channel data (idealized using SCAN) with Scheme 1 is shown. The representative recording from one patch contained a total of 2917 and 5128 open durations; 2916 and 5127 shut durations; an open probability of 0.021 and 0.026 and apparent mean open time of 0.46 ms and 0.52 ms in glycine and DCS, respectively (imposed resolution of 50 μs for open period and 30 μs for shut period). The rate constant k2+ for this particular patch was 1386 s−1 in glycine and 2150 s−1 in DCS and rate constant k3− was 2614 s−1 in glycine and 2388 s−1 in DCS. C, Free-energy relationship plot normalized to RA2a state was obtained using QUB. The desensitized states were excluded. Scale bar is 20 kJ mol−1. DCS reduced the height of the second activation barrier (RA2b to RA2c).
Table 2.
Maximum interval likelihood fitting of the sequence of open and closed durations
| Rates | Experimental |
Scheme I |
||
|---|---|---|---|---|
| Glycine | DCS | Glycine | DCS | |
| k1+ | 470 ± 85 | 420 ± 80 | ||
| k1− | 890 ± 265 | 1270 ± 340 | ||
| k2+ | 860 ± 160 | 1395 ± 215* | ||
| k2− | 15,630 ± 1700 | 17,510 ± 2135 | ||
| k3+ | 3735 ± 590 | 4795 ± 815 | ||
| k3− | 2630 ± 270 | 2150 ± 210* | ||
| k4+ | 60 ± 15 | 75 ± 35 | ||
| k4− | 1310 ± 225 | 1070 ± 130 | ||
| d1+ | 0.8 ± 0.3 | 1.2 ± 0.4 | ||
| d1− | 3.8 ± 1.0 | 4.2 ± 1.0 | ||
| d2+ | 17 ± 5 | 25 ± 8 | ||
| d2− | 33 ± 8 | 38 ± 7 | ||
| LL/event | 4.91 ± 0.03 | 4.86 ± 0.05 | ||
| Po | 0.019 | 0.028 | 0.018 | 0.029 |
| MOT | 0.46 | 0.57 | 0.46 | 0.61 |
Idealized current single-channel records were fitted to Scheme 1 as described in Figure 8. All rates have units of s−1. Data are mean ± SEM from six patches, each containing one active channel with a total of 11,275 and 16,662 open periods and 11,272 and 16,657 closed periods for glycine and DCS, respectively. The rates were compared by paired t test. *p < 0.05. MOT, mean open time.
It should be noted that the rate constants we obtained by MIL fitting in the current study are different from those previously described by Dravid et al. (2008). This difference reflects the use of time course fitting to idealize the data record in the current study compared with QUB used previously. Evaluation of this same dataset with QUB idealization and MIL fitting yields similar rate constants to those described by Dravid et al. (2008) (supplemental Table 1, available at www.jneurosci.org as supplemental material). We used time course fitting for this study in which we sought to evaluate the effect of DCS compared with glycine on mean open time, which is exceptionally brief for both conductance levels of NR1/NR2C receptors.
To test the ability of gating models in Table 2 to correctly predict the single-channel data obtained from glycine- and DCS-activated NR1/NR2C receptors, we performed Monte Carlo simulations and measured the properties of the simulated channel responses. The gating models correctly predicted the increase in the apparent mean open time and Po by DCS (Table 2). Free-energy plot of the linear model without the desensitized state suggested that the DCS reduces the height of the second activation barrier, which leads to an increase in the occupancy of the open states (Fig. 8C).
The ability of Scheme 1 to reproduce the macroscopic currents in response to glutamate in the presence of glycine or DCS was also tested. The following normalized macroscopic current responses were simultaneously fitted with Scheme 1a (Fig. 9) using a nonlinear least-squares algorithm: (1) a long 2 s jump into 1 mm glutamate + 0.5–1 mm glycine or DCS, (2) a short 5–8 ms jump into 1 mm glutamate + 0.5–1 mm glycine or DCS, and (3) a long 2 s jump into 5 μm glutamate + 0.5–1 mm glycine. The binding rates and desensitization rates were allowed to vary. The fitted rate constants are presented in Table 3, and the quality of fit is shown in Figure 9. Scheme 1a was able to predict the low open probability, modest degree of desensitization, deactivation kinetics as well as concentration dependence of amplitude and rise time for both glycine and DCS (Table 3). In addition, the macroscopic model was able to predict the higher Po for DCS. The desensitization rates obtained by macroscopic fitting were similar (1- to 3-fold different) to those obtained by MIL fit to single-channel data (Tables 2 and 3). However, the fitted rise time (2.3–2.4 ms) was modestly faster than the experimental rise time for both glycine and DCS (3.1–4.9 ms), and there was minor divergence in predicted and expected deactivation for glycine. Nevertheless, fitting of gating schemes to single-channel and macroscopic currents and simulations suggests that the higher open probability of DCS can be explained primarily by the higher forward rate constant (k2+) in the presence of DCS compared with glycine. Furthermore, the binding and unbinding rates obtained by macroscopic fitting are similar to those described in the previous study (Dravid et al., 2008), however the fast desensitization rates are different. This difference may be explained by the faster pregating steps in the current study when single-channel records were idealized using time course analysis.
Figure 9.
Least-squares fitting of NR1/NR2C activation model to macroscopic currents. A, Scheme 1a is an extension of Scheme 1 with explicit glutamate binding steps. B, The long and brief current responses were fitted simultaneously for each agonist as described in Materials and Methods (see also Erreger et al., 2005a). Only the binding, unbinding and desensitization rates were allowed to vary. The fitted rate constants are presented in Table 3. Scheme 1a was able to describe the main features of the macroscopic current response.
Table 3.
Fitting of macroscopic NR1/NR2C current–response time course
| Rates | Experimental |
Scheme Ia |
||
|---|---|---|---|---|
| Glycine | DCS | Glycine | DCS | |
| b+ | 0.98 | 0.56 | ||
| b− | 2.1 | 3.4 | ||
| d1+ | 0.4 | 1.4 | ||
| d1− | 0.7 | 5.7 | ||
| d2+ | 5.1 | 6.7 | ||
| d2− | 26 | 16 | ||
| Residual | 1.30 × 10−3 | 2.67 × 10−3 | ||
| Kd (μm) | 2.2 | 6 | ||
| EC50 in oocytes (μm) | 0.42 | 0.47 | ||
| Peak Po | 0.027 | 0.042 | ||
| Steady-state Po | 0.019 | 0.028 | 0.018 | 0.030 |
| Rise time | 3.7–4.1 | 3.1–4.9 | 2.3 | 2.4 |
| Deactivation | 240–340 | 130–160 | 390 | 245 |
| Desensitization (τ1) | 59 | 32 | 34 | 15 |
| Desensitization (τ2) | 720 | 540 | 1138 | 130 |
Normalized current response to fast agonist jumps were fitted by least sums of square algorithm to Scheme 1a as described in Figure 9. The gating rates were fixed to those derived by MIL fits (Table 2); the desensitization, binding, and unbinding rates were set as free parameters. The agonist binding rates are in μm−1s−1 and all other rates have units of s−1. The rise time, deactivation, and desensitization time constants are in milliseconds. Normalized residual sum of squares is indicated as Residual.
Discussion
In this study we used single-channel recordings, site-directed mutagenesis, and molecular dynamics simulations to examine the structure–function relationship that accounts for the higher relative efficacy of DCS compared with glycine at the NR1/NR2C receptors. We predict that intersubunit interactions at the ligand binding domain dimer interface allow the NR1 agonist DCS to influence efficacy in a manner dependent on the NR2-subunit, a finding consistent with the allosteric nature of glutamate receptor function. Molecular dynamics simulations and site-directed mutagenesis experiments together identified a series of residues at the dimer interface that are candidates to control the NR2C-specific enhanced efficacy of DCS relative to glycine. These results suggest that long-range intra- and interprotein conformational changes are orchestrated by DCS binding to NR1. Moreover, single-channel recordings from one active NR1/NR2C channel show that DCS increases the apparent mean open time and open probability of the channel, and accelerates a kinetically distinct pregating step. These results suggest that DCS-bound receptors exhibit a decreased activation energy for opening and a more stable open state relative to glycine-bound receptors.
Structural determinants of DCS action
Computational methods were used to build and evaluate potential structures of the NR1/NR2B and NR1/NR2C ligand binding domain dimers in isolation. As observed in the crystal structure of monomeric NR1 and previous molecular dynamics simulations, the general conformation of the NR1 pocket housing glycine or DCS was similar (Furukawa and Gouaux, 2003; Kaye et al., 2006). The mechanism of partial agonism at the NR1 subunit has been explored in mutagenesis studies (Kuryatov et al., 1994; Hirai et al., 1996; Williams et al., 1996; Wood et al., 1999; Kalbaugh et al., 2004) and a crystallographic study of the isolated NR1 ligand binding domain occupied by both the full agonists glycine, d-serine and by the partial agonists d-cycloserine, ACPC, ACBC and cycloleucine (Inanobe et al., 2005). Although the degree of domain closure was found to be similar for full and partial agonists, partial agonists were not able to induce all of the conformational changes within the monomeric ligand binding domain as elicited by the full agonist glycine (Inanobe et al., 2005). We identified several agonist- and subunit-dependent differences in the molecular dynamics simulations of dimeric ligand binding domain that might account for the lower efficacy of DCS at NR1/NR2B and the higher efficacy at NR1/NR2C. For example, multicomponent interactions formed by Tyr692 and NR1-Phe754 with NR2B-Glu790 and NR2C-Gln800 showed agonist dependence. Moreover these interactions were partly conserved at other NR2 subtypes, since mutation of NR2D-Glu791 to Gln (as in NR2C) increased DCS relative efficacy to be greater than glycine. We also identified interactions between residues Arg755 (NR1), and Glu531/Glu793 of NR2B or Glu541/Glu803 of the NR2C, which appear to coordinate movement between the D2 domains of the NR1 and NR2 subunits.
The computational limitations that arise when simulating only a portion of the much larger NMDA receptor complex create important caveats to the interpretation of these data. To address these caveats, a subset of in silico results were used to design functional experimentation using mutagenesis to probe perturbations at residues predicted to change in an agonist-dependent manner. We found that no single residue is responsible for the subunit-specific differences in the agonist efficacy for DCS. Rather, mutagenesis experiments showed that multiple residues within the dimer interface control agonist efficacy, suggesting that the intersubunit interface may determine the efficacy of NR1 agonists. In addition, helices F and G of NR1 subunit and helices F and K of NR2C subunit in the NR1/NR2C dimer show different orientations for DCS binding compared with glycine. These simulations showing specific NR2 subunit-dependent changes are consistent with electrophysiological data showing that the identity of the NR1 agonist can differentially influence the function of the tetrameric receptor depending on the NR2 subunit. This is consistent with the idea that NMDA receptors are allosteric proteins (Benveniste et al., 1990; Vyklický et al., 1990; Benveniste and Mayer, 1991; Kemp and Priestley, 1991; Priestley and Kemp, 1994; Regalado et al., 2001) comprised of multiple semiautonomous interacting domains (Sobolevsky et al., 2009).
d-Cycloserine and gating of NMDA receptor
Upon agonist association with the ligand binding domain, the NMDA receptor undergoes a series of kinetically distinct conformational changes that precede concerted and rapid opening of the conduction pore most likely due to rotation and movement of the M3 transmembrane helix (Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Popescu et al., 2004; Auerbach and Zhou, 2005; Erreger et al., 2005a,b; Schorge et al., 2005; Sobolevsky et al., 2009). Partial agonists interacting with either the NR1 or NR2 agonist binding domains of the NMDA receptor have been shown to reduce the efficiency of gating by slowing forward rate constants describing steps that precede rapid pore opening (Banke and Traynelis, 2003; Erreger et al., 2005b; Kussius and Popescu, 2009). Our single-channel data support these findings, and mutagenesis and molecular dynamics studies identify specific coordinated protein interactions between NR1 and NR2C subunits that may underlie the unique effects of DCS at NR2C-containing NMDA receptors function. The mutagenesis experiments in particular provide support for molecular dynamics simulations that identify, for example, Gln800 as a key residue at the interface of NR1 and NR2C.
Our working hypothesis is that the kinetically distinct pregating steps likely represent the rate-limiting conformational changes within portions of protein that control opening of the M3 gate (Sobolevsky et al., 2009). Based on the molecular dynamics simulations with DCS and previous simulations with a NR2A subunit partial agonist (Erreger et al., 2005b), we hypothesize that brief kinetically distinguishable closed states in the single-channel record primarily may reflect rearrangements that involve helix F and the S2-M1/M3 linkers. The pre-M1 helix identified in the recent GluR2 structure is an excellent candidate for an element within the protein that may be required to move before opening of the M3 gate (Sobolevsky et al., 2009). That is, movement of pre-M1 helix within each subunit following closing of the ligand binding domain after agonist binding could represent a rate-limiting step.
The molecular dynamics simulations of the ligand binding domain dimer suggest agonist-dependent long-range interactions from NR1 to NR2 occur, which could impact NR2-gating steps, and vice versa. This allosteric interaction between NR1 and NR2 subunits could explain the changes in multiple gating rates observed by some partial agonists (Kussius and Popescu, 2009). In the case of DCS binding to NR1/NR2C receptors, maximum interval likelihood fitting of models to the single-channel data showed that the forward pregating rate constant k2+ is significantly accelerated by DCS. However interpretations from single-channel analysis have limitations due to the variability among patches, and it is likely that with more data other rates (such as k1−) may also be different between DCS and glycine, which could suggest that although the main effect of DCS is on NR1 conformational changes, through the dimer interactions it may also influence agonist-induced conformational changes in NR2. MD simulations show that DCS leads to more pronounced change in orientation of the helix F of NR1 subunit compared with the helix F of NR2C, and it is possible that this rearrangement of NR1 helix F favors activation of the receptor. More structural information and complimentary functional analysis will be required to fully understand the mechanism of NMDA receptor gating.
Clinical relevance of these findings
Exposure-based psychotherapy for human anxiety disorders is conceptually similar to extinction training in animals. Recently it was found that DCS facilitates extinction of conditioned fear in rats (Walker et al., 2002). This observation initiated several animal and human translational studies testing the complimentary effect of DCS for psychotherapy in other anxiety disorders (Walker et al., 2002; Ledgerwood et al., 2003, 2005; Ressler et al., 2004; Parnas et al., 2005; Hofmann et al., 2006; Lee et al., 2006; Woods and Bouton, 2006; Kushner et al., 2007; Storch et al., 2007; Weber et al., 2007; Guastella et al., 2008; Wilhelm et al., 2008). It is generally believed that the facilitatory effect of DCS on extinction learning is due to augmentation of NMDA transmission. This could occur in a manner independent of the NR2 subunit if NMDA receptors at key synapses are not saturated with glycine, and administration of DCS enhances NMDA receptor function by increasing agonist occupancy at the NR1 ligand binding site. In contrast, results here show that DCS efficacy is NR2 subunit-dependent with DCS being less efficacious than endogenous glycine or d-serine at NR1/NR2B receptors (see also Sheinin et al., 2001). Thus, DCS will not augment NMDA neurotransmission at a predominantly NR1/NR2A, NR1/NR2B, or NR1/NR2D synapses if endogenous glycine and d-serine are at levels sufficient to maximally activate the receptor. However, DCS could selectively augment NMDA neurotransmission at synaptic NR2C-containing NMDA receptors regardless of the endogenous level of glycine or d-serine. We thus propose that the effects of DCS in facilitation of fear extinction are mediated by its action on NR2C-containing NMDA receptors through actions at the NR1/NR2C dimer interface. This is a novel hypothesis that will require additional testing.
Footnotes
This work was supported by the National Institutes of Health–National Institute of Neurological Disorders and Stroke (Grant NS036654; S.F.T.), the Health Future Foundation (S.M.D.), and the American Epilepsy Foundation (S.M.D.). We thank Drs. Kasper B. Hansen and Mark Mayer for critical comments on the manuscript. We thank Dr. Pierre Paoletti for sharing NR1 E781A mutant.
References
- Albouy R, Eloy P, Pestel M, Ravina A, Rey M. First clinical results obtained with cycloserine in the treatment of human tuberculosis. Antibiot Annu. 1955;3:148–152. [PubMed] [Google Scholar]
- Auerbach A, Zhou Y. Gating reaction mechanisms for NMDA receptor channels. J Neurosci. 2005;25:7914–7923. doi: 10.1523/JNEUROSCI.1471-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6:144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- Banke TG, Dravid SM, Traynelis SF. Protons trap NR1/NR2B NMDA receptors in a nonconducting state. J Neurosci. 2005;25:42–51. doi: 10.1523/JNEUROSCI.3154-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML. Kinetic analysis of antagonist action at N-methyl-d-aspartic acid receptros: two binding sites each for glutamate and glycine. Biophys J. 1991;59:560–573. doi: 10.1016/S0006-3495(91)82272-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Clements J, Vyklický L, Jr, Mayer ML. A kinetic analysis of the modulation of N-methyl-d-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J Physiol. 1990;428:333–357. doi: 10.1113/jphysiol.1990.sp018215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossváry I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (SC06); November 11–17, 2006; Tampa, Florida. 2006. [Google Scholar]
- Boyd LJ, Epstein IG, Nair KG. The treatment of human tuberculosis with cycloserine: a year's progress. Antibiot Annu. 1955;3:141–147. [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PE, Geballe MT, Katz E, Erreger K, Livesey MR, O'Toole KK, Le P, Lee CJ, Snyder JP, Traynelis SF, Wyllie DJ. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol. 2008;586:227–245. doi: 10.1113/jphysiol.2007.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D, Hawkes AG. Stochastic properties of ion channel openings and bursts in a membrane patch that contains two channels: evidence concerning the number of channels present when a record containing only single openings is observed. Proc R Soc Lond B Biol Sci. 1990;240:453–477. doi: 10.1098/rspb.1990.0048. [DOI] [PubMed] [Google Scholar]
- Davis M, Ressler K, Rothbaum BO, Richardson R. Effects of d-cycloserine on extinction: translation from preclinical to clinical work. Biol Psychiatry. 2006;60:369–375. doi: 10.1016/j.biopsych.2006.03.084. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Dravid SM, Erreger K, Yuan H, Nicholson K, Le P, Lyuboslavsky P, Almonte A, Murray E, Mosely C, Barber J, French A, Balster R, Murray TF, Traynelis SF. Subunit-specific mechanisms and proton sensitivity of NMDA receptor channel block. J Physiol. 2007;581:107–128. doi: 10.1113/jphysiol.2006.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. J Physiol. 2008;586:4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Chen PE, Wyllie DJ, Traynelis SF. Glutamate receptor gating. Crit Rev Neurobiol. 2004;16:187–224. doi: 10.1615/critrevneurobiol.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005a;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF. Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci. 2005b;25:7858–7866. doi: 10.1523/JNEUROSCI.1613-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Kristensen A, Chen PE, Hansen KB, Lee CJ, Yuan H, Le P, Lyuboslavsky PN, Micale N, Jørgensen L, Clausen RP, Wyllie DJ, Snyder JP, Traynelis SF. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-d-aspartate glutamate receptors. Mol Pharmacol. 2007;72:907–920. doi: 10.1124/mol.107.037333. [DOI] [PubMed] [Google Scholar]
- Essman U, Perera L, Berkowitz ML, Darden HLT, Pedersene LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8592. [Google Scholar]
- Flood JF, Morley JE, Lanthorn TH. Effect on memory processing by d-cycloserine, an agonist of the NMDA/glycine receptor. Eur J Pharmacol. 1992;221:249–254. doi: 10.1016/0014-2999(92)90709-d. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastella AJ, Richardson R, Lovibond PF, Rapee RM, Gaston JE, Mitchell P, Dadds MR. A randomized controlled trial of d-cycloserine enhancement of exposure therapy for social anxiety disorder. Biol Psychiatry. 2008;63:544–549. doi: 10.1016/j.biopsych.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Henderson G, Johnson JW, Ascher P. Competitive antagonists and partial agonists at the glycine modulatory site of the mouse N-methyl-d-aspartate receptor. J Physiol. 1990;430:189–212. doi: 10.1113/jphysiol.1990.sp018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Kirsch J, Laube B, Betz H, Kuhse J. The glycine binding site of the N-methyl-d-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3–M4 loop region. Proc Natl Acad Sci U S A. 1996;93:6031–6036. doi: 10.1073/pnas.93.12.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann SG, Pollack MH, Otto MW. Augmentation treatment of psychotherapy for anxiety disorders with d-cycloserine. CNS Drug Rev. 2006;12:208–217. doi: 10.1111/j.1527-3458.2006.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood WF, Compton RP, Monahan JB. D-cycloserine: a ligand for the N-methyl-d-aspartate coupled glycine receptor has partial agonist characteristics. Neurosci Lett. 1989;98:91–95. doi: 10.1016/0304-3940(89)90379-0. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Kalbaugh TL, VanDongen HM, VanDongen AM. Ligand-binding residues integrate affinity and efficacy in the NMDA receptor. Mol Pharmacol. 2004;66:209–219. doi: 10.1124/mol.66.2.209. [DOI] [PubMed] [Google Scholar]
- Kaye SL, Sansom MS, Biggin PC. Molecular dynamics simulations of the ligand-binding domain of an N-methyl-d-aspartate receptor. J Biol Chem. 2006;281:12736–12742. doi: 10.1074/jbc.M512728200. [DOI] [PubMed] [Google Scholar]
- Kelley JB, Anderson KL, Itzhak Y. Long-term memory of cocaine-associated context: disruption and reinstatement. Neuroreport. 2007;18:777–780. doi: 10.1097/WNR.0b013e3280c1e2e7. [DOI] [PubMed] [Google Scholar]
- Kemp JA, Priestley T. Effects of (+)-HA-966 and 7-chlorokynurenic acid on the kinetics of N-methyl-d-aspartate receptor agonist responses in rat cultured cortical neurons. Mol Pharmacol. 1991;39:666–670. [PubMed] [Google Scholar]
- Kuryatov A, Laube B, Betz H, Kuhse J. Mutational analysis of the glycine-binding site of the NMDA receptor: structural similarity with bacterial amino acid-binding proteins. Neuron. 1994;12:1291–1300. doi: 10.1016/0896-6273(94)90445-6. [DOI] [PubMed] [Google Scholar]
- Kushner MG, Kim SW, Donahue C, Thuras P, Adson D, Kotlyar M, McCabe J, Peterson J, Foa EB. D-cycloserine augmented exposure therapy for obsessive-compulsive disorder. Biol Psychiatry. 2007;62:835–838. doi: 10.1016/j.biopsych.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK. Kinetic basis of partial agonism at NMDA receptors. Nat Neurosci. 2009;12:1114–1120. doi: 10.1038/nn.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Ledgerwood L, Richardson R, Cranney J. Effects of d-cycloserine on extinction of conditioned freezing. Behav Neurosci. 2003;117:341–349. doi: 10.1037/0735-7044.117.2.341. [DOI] [PubMed] [Google Scholar]
- Ledgerwood L, Richardson R, Cranney J. D-cycloserine facilitates extinction of learned fear: effects on reacquisition and generalized extinction. Biol Psychiatry. 2005;57:841–847. doi: 10.1016/j.biopsych.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Lee JL, Milton AL, Everitt BJ. Reconsolidation and extinction of conditioned fear: inhibition and potentiation. J Neurosci. 2006;26:10051–10056. doi: 10.1523/JNEUROSCI.2466-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-d-aspartate NR1/NR2A receptors. Proc Natl Acad Sci U S A. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E. Anti-TB drugs: and then there were none. Science. 2008;321:364. doi: 10.1126/science.321.5887.364. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Armstrong N. Structure and function of glutamate receptor ion channels. Annu Rev Physiol. 2004;66:161–181. doi: 10.1146/annurev.physiol.66.050802.084104. [DOI] [PubMed] [Google Scholar]
- McBain CJ, Kleckner NW, Wyrick S, Dingledine R. Structural requirements for activation of the glycine coagonist site of N-methyl-d-aspartate receptors expressed in Xenopus oocytes. Mol Pharmacol. 1989;36:556–565. [PubMed] [Google Scholar]
- McFeeters RL, Oswald RE. Structural mobility of the extracellular ligand-binding core of an ionotropic glutamate receptor. Analysis of NMR relaxation dynamics. Biochemistry. 2002;41:10472–10481. doi: 10.1021/bi026010p. [DOI] [PubMed] [Google Scholar]
- Monahan JB, Handelmann GE, Hood WF, Cordi AA. D-cycloserine, a positive modulator of the N-methyl-d-aspartate receptor, enhances performance of learning tasks in rats. Pharmacol Biochem Behav. 1989;34:649–653. doi: 10.1016/0091-3057(89)90571-6. [DOI] [PubMed] [Google Scholar]
- Parnas AS, Weber M, Richardson R. Effects of multiple exposures to d-cycloserine on extinction of conditioned fear in rats. Neurobiol Learn Mem. 2005;83:224–231. doi: 10.1016/j.nlm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6:476–483. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- Priestley T, Kemp JA. Kinetic study of the interactions between the glutamate and glycine recognition sites on the N-methyl-d-aspartic acid receptor complex. Mol Pharmacol. 1994;46:1191–1196. [PubMed] [Google Scholar]
- Qin F. Restoration of single channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Auerbach A, Sachs F. Estimating single channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado MP, Villarroel A, Lerma J. Intersubunit cooperativity in the NMDA receptor. Neuron. 2001;32:1085–1096. doi: 10.1016/s0896-6273(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Rothbaum BO, Tannenbaum L, Anderson P, Graap K, Zimand E, Hodges L, Davis M. Cognitive enhancers as adjuncts to psychotherapy: use of d-cycloserine in phobic individuals to facilitate extinction of fear. Arch Gen Psychiatry. 2004;61:1136–1144. doi: 10.1001/archpsyc.61.11.1136. [DOI] [PubMed] [Google Scholar]
- Schorge S, Elenes S, Colquhoun D. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol. 2005;569:395–418. doi: 10.1113/jphysiol.2005.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster GM, Schmidt WJ. D-cycloserine reverses the working memory impairment of hippocampal-lesioned rats in a spatial learning task. Eur J Pharmacol. 1992;224:97–98. doi: 10.1016/0014-2999(92)94825-g. [DOI] [PubMed] [Google Scholar]
- Sheinin A, Shavit S, Benveniste M. Subunit specificity and mechanism of action of NMDA partial agonist d-cycloserine. Neuropharmacology. 2001;41:151–158. doi: 10.1016/s0028-3908(01)00073-9. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch EA, Merlo LJ, Bengtson M, Murphy TK, Lewis MH, Yang MC, Jacob ML, Larson M, Hirsh A, Fernandez M, Geffken GR, Goodman WK. D-cycloserine does not enhance exposure-response prevention therapy in obsessive-compulsive disorder. Int Clin Psychopharmacol. 2007;22:230–237. doi: 10.1097/YIC.0b013e32819f8480. [DOI] [PubMed] [Google Scholar]
- Thompson LT, Moskal JR, Disterhoft JF. Hippocampus-dependent learning facilitated by a monoclonal antibody or d-cycloserine. Nature. 1992;359:638–641. doi: 10.1038/359638a0. [DOI] [PubMed] [Google Scholar]
- Vyklický L, Jr, Benveniste M, Mayer ML. Modulation of N-methyl-d-aspartic acid receptor desensitization by glycine in mouse cultured hippocampal neurones. J Physiol. 1990;428:313–331. doi: 10.1113/jphysiol.1990.sp018214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Ressler KJ, Lu KT, Davis M. Facilitation of conditioned fear extinction by systemic administration or intra-amygdala infusions of d-cycloserine as assessed with fear-potentiated startle in rats. J Neurosci. 2002;22:2343–2351. doi: 10.1523/JNEUROSCI.22-06-02343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hart J, Richardson R. Effects of d-cycloserine on extinction of learned fear to an olfactory cue. Neurobiol Learn Mem. 2007;87:476–482. doi: 10.1016/j.nlm.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Wilhelm S, Buhlmann U, Tolin DF, Meunier SA, Pearlson GD, Reese HE, Cannistraro P, Jenike MA, Rauch SL. Augmentation of behavior therapy with d-cycloserine for obsessive-compulsive disorder. Am J Psychiatry. 2008;165:335–341. doi: 10.1176/appi.ajp.2007.07050776. quiz 409. [DOI] [PubMed] [Google Scholar]
- Williams K, Chao J, Kashiwagi K, Masuko T, Igarashi K. Activation of N-methyl-d-aspartate receptors by glycine: role of an aspartate residue in the M3–M4 loop of the NR1 subunit. Mol Pharmacol. 1996;50:701–708. [PubMed] [Google Scholar]
- Wood MW, VanDongen HM, VanDongen AM. A mutation in the glycine binding pocket of the N-methyl-d-aspartate receptor NR1 subunit alters agonist efficacy. Brain Res Mol Brain Res. 1999;73:189–192. doi: 10.1016/s0169-328x(99)00247-8. [DOI] [PubMed] [Google Scholar]
- Woods AM, Bouton ME. D-cycloserine facilitates extinction but does not eliminate renewal of the conditioned emotional response. Behav Neurosci. 2006;120:1159–1162. doi: 10.1037/0735-7044.120.5.1159. [DOI] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of N-methyl-d-aspartate receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]