Abstract
Despite the vast number of studies on Parkinson’s disease (PD), its effective diagnosis and treatment remains unsatisfactory. Hence, the relentless search for an optimal cure continues. The emergence of neuroproteomics, with its sophisticated techniques and non-biased ability to quantify proteins, provides a methodology with which to study the changes in neurons that are associated with neurodegeneration. Neuroproteomics is an emerging tool to establish disease-associated protein profiles, while also generating a greater understanding as to how these proteins interact and undergo post-translational modifications. Furthermore, due to the advances made in bioinformatics, insight is created concerning their functional characteristics. In this review, we first summarize the most prominent proteomics techniques and then discuss the major advances in the fast-growing field of neuroproteomics in PD. Ultimately, it is hoped that the application of this technology will lead towards a presymptomatic diagnosis of PD, and the identification of risk factors and new therapeutic targets at which pharmacological intervention can be aimed.
Keywords: Alzheimer’s disease, Biomarker, Cerebrospinal fluid, Mass spectrometry, Neurodegenerative disease, Neuroproteomics, Parkinson’s disease
Introduction
This review discusses the potential offered in harnessing the technological advances made in proteomics. This technology can provide a clearer understanding of the pathogenesis and protective mechanisms against Parkinson’s disease (PD), by giving insight into the role played by the aggregation and deposition of proteins in human PD and in chemically induced models of the disease (Schulenborg et al. 2006). In addition, it can reveal the patterns of PD-specific cellular markers to provide for more effective treatment options and to allow the therapeutic regime to start earlier, when it is likely to have a more beneficial effect.
Despite the widespread, growing interest in the field, insight into the application of proteomics tools and technologies is only slowly penetrating the PD research community and relatively few studies have been performed in the PD context. The technology has grown rapidly as a subdiscipline of the life sciences to its current position, where it is increasingly applied as clinical proteomics, with the view to identify new biomarkers for diagnosis and to better understand the mechanisms, risks, state and progression of PD (Hanash 2004; Johnson et al. 2005).
Parkinson’s disease: neuropathology, genetics, clinical features and treatment
Parkinson’s disease is the second most common neurodegenerative disorder, affecting nearly 3% of the population over the age of 60 years (Gasser 2005). With the population of many countries aging, PD presents a major challenge to many national health care budgets. The nearly epidemic scale of the disease has led scientific research to identify its causes and improve the effectiveness of available treatment options, to ascertain PD research with a prominent place on many national research agendas.
PD manifests clinically as a dysfunctional involuntary motor system, with bradykinesia, tremor, postural instability and rigidity being the cardinal symptoms (Olanow and Tatton 1999; Zigmond and Burke 2002). These often co-present with non-motor related symptoms, including depression, dysarthria, sleep disturbances, dysphagia, sexual dysfunction and dementia (Truong et al. 2007; Miller et al. 2006). The central neuropathological hallmarks of PD are the progressive loss of dopamine (DA)-producing neurons in midbrain regions, particularly within the Substantia Nigra pars compacta (SNpc), and the presence of intraneuronal fibrillar cytoplasmic inclusions termed Lewy bodies (LB) and Lewy neurites (Nissbaum and Ellis 2003; Braak et al. 2003). The major proteinaceous constituent of these inclusions is α-synuclein (SNCA), but they also contain synphilin, neurofilament chains, ubiquitin, ubiquitin carboxy-terminal hydrolase L1 and tau (Spillantini et al. 1997; Goldman et al. 1983; Manetto et al. 1988; Goedert 2001). Many investigators have assigned a critical role to SNCA in the pathogenesis of PD (Dawson and Dawson 2003; Lee and Appel 2004).
Several environmental, occupational and life-style factors have been implicated in the onset and/or progression of PD. These include drinking rural well water and being exposed to welding and transition-series metals, such as manganese or iron (Racette et al. 2005; Gorell et al. 1998; Bloch et al. 2006). In addition, a possible inverse relationship has been established between cigarette smoking (Benedetti et al. 2000; Miksys and Tyndale 2006), caffeine consumption (Ross et al. 2000; Ascherio et al. 2001; Hernan et al. 2002), physical exercise (Chen et al. 2005a) and the risk of developing PD.
Evidence for an environmental, toxin-based cause of PD derives from an out-break in humans who had developed parkinsonian symptoms after having been exposed to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Other compounds, such as 6-hydroxydopamine (6-OHDA), rotenone and paraquat also reproduce several pathological features of PD, both in vitro and in animals, in which they cause the loss of dopaminergic neurons (Seniuk et al. 1990; Deumens et al. 2002) and formation of LBs (Forno et al. 1986; Betarbet et al. 2000; Manning-Boğ et al. 2002; Fornai et al. 2005). Rotenone is an inhibitor of the complex I of the mitochondrial respiratory chain, causing lowered cellular energy production, impairment of ATP-dependent cellular processes, and increased generation of free radicals (Betarbet et al. 2000; Fornai et al. 2005; Hardy et al. 2006; Kaur et al. 2003; Ascherio et al. 2006). Reduced complex I activity has also been found in the muscle and platelets of idiopathic PD patients (Schapira et al. 1990; Parker et al. 1989). In PD post-mortem brain tissue, elevated levels of lipid peroxidation, iron and protein nitration have been reported (Alam et al. 1997; Andersen 2004; Dexter et al. 1987; Yoritaka et al. 1996), further suggesting a mitochondrial dysfunction in PD. An inhibition of the mitochondrial respiratory chain was also found in other neurodegenerative diseases, including frontotemporal dementia (FTD), a pathological condition related to Alzheimer’s disease (AD) (David et al. 2005a), suggesting that this pathological process may underlie several progressive neurological disorders.
Cellular proteins accumulate in dopaminergic neurons of post-mortem PD brains (Conway et al. 2000; Lee 2008). These misfolded proteins include nitrated and phosphorylated SNCA, with the toxicity of SNCA thought to be induced by phosphorylation at Ser129 (Giasson et al. 2000; Conway et al. 2001; Matsuoka et al. 2001). The aggregates have been found in both cytoplasmic and extracellular LBs (Giasson et al. 2000; Fujiwara et al. 2002; Chen and Feany 2005). In familial cases, these insoluble protein aggregates can be caused by either of two dominantly inherited mutations in the SNCA protein (Ala/Thr substitution at residue 53 and Ala/Pro substitution at residue 30), resulting in early onset PD (Polymeropoulos et al. 1997; Krüger et al. 1998). In sporadic PD, SNCA aggregation may be caused due to the secondary effects of oxidative modifications or impaired degradation pathways, with the rotenone rat model suggesting that mitochondrial inhibition and free radical generation result in the production of SNCA inclusions (Schapira 2004). Pro-apoptotic patho-mechanisms have been proposed as an alternative disease pathomechanism (Hoglinger et al. 2003).
The importance of mitochondrial dysfunction as a central mechanism of PD pathogenesis is underscored by the finding that the proteins whose genes are mutated in familial PD, such as in the pink1-parkin pathway, regulate mitochondrial function (Clark et al. 2006; Park et al. 2006; Shen and Cookson 2004). While the vast majority of PD cases are sporadic, only a small subset is due to autosomal dominant or recessive genetic alterations, with several of these relating to the proteosomal system (Lee and Appel 2004). Mutations in at least five genes have been linked to PD, and these genetic variants typically manifest at a younger age than the more common sporadic forms (Klein and Lohmann-Hedrich 2007). Mutations have been found in genes encoding SNCA (PARK1), the E3 ligase parkin (PARK2), DJ-1 (PARK7), PTEN-induced kinase 1 (PINK1, also known as PARK6) and the leucine-rich repeat kinase 2 (LRRK2) (Bonifati et al. 2003; Kitada et al. 1998; Polymeropoulos et al. 1997; Giasson and Van Deerlin 2008). PARK1 is believed to be the most frequent gene underlying autosomal dominant familial PD (Trojanowski and Lee 1998; Polymeropoulos et al. 1997; Krüger et al. 1998).
Since PD is a complex disease, it is conceivable that either its etiology or its development may involve a multitude of proteins that participate in either establishing or aggregating pathogenesis. Thus far, a number of proteins have been associated with the pathophysiology underlying PD. The selective destruction of dopaminergic neurons, induced by the viral overexpression of SNCA in rats (Kirik et al. 2002), and overexpression of normal and mutant forms of human SNCA in Drosophila models (Feany and Bender 2000) strongly suggest that this small acidic protein may associate with elements of toxicity. However, a lack of consensus continues as to whether SNCA aggregation offers neuronal protection, perhaps by sequestering into inert bodies, thereby reducing the amount of diffuse toxic proteins available in the cell (Chen and Feany 2005), or whether protein overexpression fulfills a toxic cellular role (Cookson 2005).
The mainstay pharmacological treatment for PD is the administration of the DA precursor 3,4-dihydroxy-L-phenylalanine, also known as levodopa (L-DOPA). After crossing the blood brain barrier, it is converted to DA thereby providing symptomatic relief. While dramatically improving bradykinesia and muscle tone, and decreasing tremor (Birkmayer and Hornykiewicz 1961), long-term use of L-DOPA leads to the onset and progressive worsening of motor fluctuations, often co-presenting with severe dyskinesia (Tinazzi et al. 2006). The non-motor symptoms of PD also fail to benefit from this particular form of pharmacotherapy (Wolters et al. 2000). Moreover, DA-producing cells continue to degenerate and L-DOPA becomes less effective when striatal DA falls below critical levels. Another treatment for patients suffering from advanced PD is deep brain stimulation of substructures of the basal ganglia (Limousin et al. 1998; The deep-brain stimulation for Parkinson study group 2001). Drawbacks of this approach are the high cost of electrodes, the need to replace batteries, and the time it takes for adjusting electric parameters (Tseng et al. 2007).
The effective treatment of PD is further compromised by the fact that it is often diagnosed only after symptoms appear. By then a substantial loss of striatal nerve perikarya has already occurred, often exceeding the critical (>80%) threshold level (Bernheimer et al. 1973), when the ideal window period for administering neuroprotective agents has surpassed.
At present, the tools to diagnose PD preclinically are not yet available to clinicians (Montgomery et al. 2000a, b). It is foreseen by many that the emerging field of neuroproteomics will allow for detecting new biomarkers with which to diagnose PD more accurately and earlier, thereby offering the possibility for early intervention (Zetterberg et al. 2008). This could allow the technological barriers imposed on previous studies to be overcome. By studying the unique expression pattern and level of PD-associated proteins, it is hoped that PD can be differentially diagnosed and insight gained into the mechanisms underlying this disorder. In addition, it may provide much-needed surrogate end-points with which to validate the clinical efficacy of novel treatment options (Michell et al. 2004).
A brief overview of proteomic techniques
The technology that is broadly described under the umbrella-term ‘proteomics’, derives from the term “proteome”, that refers to “the proteins expressed by a particular genome” (Wilkins et al. 1996), which is the large-scale analysis of many proteins simultaneously (David et al. 2005b; Hoerndli et al. 2005). While this could potentially include all the proteins encoded by the genome at any given time (i.e., the proteome), it more commonly refers to specific subsets of proteins found in specific regions or cell-type, under specific conditions (Pandey and Mann 2000; Righetti et al. 2004). Sensitive proteomics techniques have been developed to determine the structure, localization, biochemical activity, interactions (i.e., protein–protein, protein–lipid) and the cellular roles played by a multitude of proteins, with the results translated either to a natural physiological or to a pathological state of events (Onn and Mann 2005).
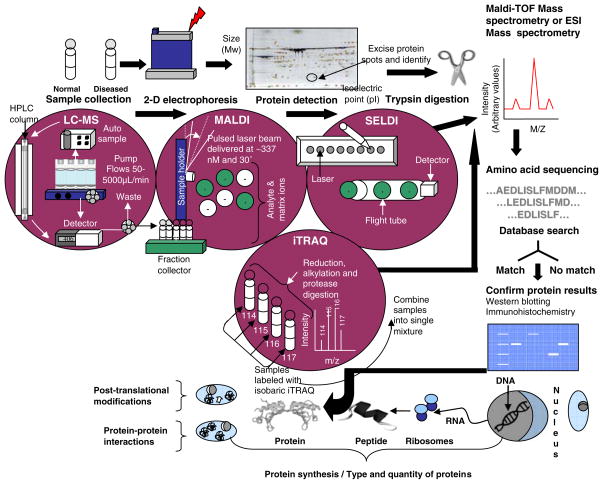
Substantial technological advances have been made in novel instrumentation, experimental strategies, and bioinformatics tools for analyzing the proteome (De Hoog and Mann 2004). However, despite the bewildering range of available techniques and analysis tools, some form of protein separation procedure, followed by techniques for peptide detection and identification are routinely required, with mass spectrometry (MS) remaining its key technology (Fig. 1). The speed of advancements in peptide-sequencing technology may allow quantifying most cellular proteins by high resolution MS (Lipton et al. 2002). To quantify proteins according to their degree of expression, MS technology is frequently applied in conjunction with two-dimensional polyacrylamide gel electrophoresis (2-D PAGE). This combined experimental approach allows for inter-sample comparisons relating to the disease state as well as between experimental subjects and normal controls (De Iuliis et al. 2005).
Fig. 1.
A flow-diagram illustrating the sequence of steps involved in sample analysis using SELDI, MALDI, LC-MS and iTRQ proteomic strategies. As illustrated, these methods provide information on the type and quantity of proteins, post-translational modifications and protein–protein interactions
There is an inherent risk of preferentially detecting proteins that are present in abnormally high concentration, thereby repressing the weaker signals by less abundant proteins (Fountoulakis 2001, 2004; Vercauteren et al. 2004; Wiederkehr 1991). To reduce the complexity of the protein sample, and thereby increase sample resolution, strategies have been developed to reduce levels of highly abundant (and generally less interesting) proteins, which may obscure detection of the less abundant ones. In blood-based proteomics this can be achieved by affinity separation of abundant proteins (such as albumin, fibrinogen, and transferrin) from either human or mouse plasma, using affinity-purified antibodies covalently conjugated to microbeads. Another practical and economically feasible approach is to first apply proteomic techniques to detect for markers of the disease in organs where protein concentrations are expectedly high, before searching for them in fluids where they are less abundant (Meyer and Stuhler 2007). The application of this approach to neurodegeneration is not trivial, as human brain tissue can be of bad quality and show high variability, for example, with regards to gender, age, and medication of patients. Nonetheless, one may be able to choose tissue from brain areas that are affected only relatively late in disease and thus spared from massive degeneration. For AD, this could be the brain stem as a brain area that is relatively preserved in late stage AD, but still displays the typical hallmark lesions of the disease. Alternatively, animal models of AD or PD may be employed to apply proteomics first to dissected brain areas and eventually body fluids such as plasma or CSF, which are likely to reflect, at least in parts, the deregulation of proteins in the brain (Götz et al. 2007). Subcellular proteomics provides another means for combining the direct analysis of the proteome of isolated organelles such as mitochondria. Methods for fractionating subcellular organelles include traditional techniques, such as centrifugation (e.g., high-speed sedimentation/density gradient) (Pasquali et al. 1997; Fialka et al. 1997; Tribl et al. 2006a, b), but also highly sensitive commercial fractionation kits for extracting nuclear, cytosolic, mitochondrial or membrane fractions (Huber et al. 2003; Heimlick and Cidlowski 2006; Chen et al. 2005b; Vila-Carriles et al. 2007). The limitations of the subfractionation approach is in its experimental variability and the fact that the fractions are rarely pure as, for example, in synaptosomal preparations, mitochondria are easily co-purified (J. Götz, unpublished observations). The subcellular fractionation and purification techniques can be effectively combined with gel-dependent methods, such as 2-D PAGE, or gel-independent ones.
In 2-D PAGE, proteins are first separated in the first dimension according to their charge (z) and molecular weight using isoelectric focusing (IEF) (Klose 1975; O’Farrell 1975; Görg et al. 1985) (Fig. 1). The proteins migrate through a thin gel-strip embedded with immobilized pH gradients (IPG). Migration terminates when the protein reaches the point where the net charge is neutral (i.e., the isoelectric point, or pI). Different IPG strips can be used: wide pH ranges (IPG 3–10) that cover many pH units, those that cover narrower ranges (IPG 4–7), or very narrow ones that only provide a single pH unit-width (IPG 5–6, or IPG 5.5–6.7) (Simpson 2003). Narrow pH ranges allow zooming in and give a better resolution of the protein spot pattern within a smaller window (Hoving et al. 2002). Proteins are then separated in the second dimension (orthogonal direction) using electrophoresis that takes place in acrylamide gels containing sodium dodecyl sulfate (SDS), where SDS imparts a net negative charge, allowing proteins to separate according to mass (m). Separation in these two dimensions allows for the resolution of multiple isoforms and variants of the same protein. A variety of chemical stains and fluorescent markers are available to visualize and detect the differences in protein staining intensities between samples (David et al. 2005a). Approximately 1,000–3,000 proteins can be visualized per stained gel, with the capacity to resolve up to 10,000 proteins in a single gel (Klose 1975; O’Farrell 1975). A comparison of different 2-D PAGE methods and their application to mouse brain has been done by several groups including Stühler and colleagues (2006).
Despite 2-D PAGE being a relatively old-fashioned biotechnological technique, from a proteomics perspective it remains popular since it allows for the use of high through-put screening in an easily executable and cost-effective manner. However, its main drawbacks are the limited dynamic range and often a failure of detecting membranous or low abundant proteins (Aebersold et al. 2000). In addition, it is often difficult to accurately compare two samples when the comparison is based on the degree of protein-expression alone, since protein spots often overlap or appear small and/or blurry, thereby increasing the risk of over- or undercounting them.
The technique known as ‘Differential In-Gel Electrophoresis’ (DIGE) by which the samples are labeled with different fluorescent dyes (such as Cy3 and Cy5) and then separated on the same gel has proved very useful in increasing the dynamic range and reducing technical variability (Wu 2006).
Liquid chromatography (LC) coupled to MS has proved to be an important alternative to 2-D gels (Fig. 1). Various LC techniques (such as affinity, size exclusion, reversed phase or charge chromatography) provide different means for separating complex samples (Wolters et al. 2001). The separated protein mixture is then fed directly into an electrospray ionisation (ESI) MS, using tubes of micro-capillary diameter which greatly reduces the amount of sample needed for analysis (Graves and Haystead 2002; Link et al. 1999; Opiteck et al. 1997; Washburn and Yates 2000). The LC-MS technique is particularly useful for membrane-associated proteins, phosphopeptides, enzymes such as kinases and phosphatases, and transcription factors that are difficult to detect through 2-D PAGE because of their low levels of abundance (Washburn et al. 2002). Of the various ionization methods developed for coupling, LC to MS-ESI is the most widely used technique that allows for the gentle ionization of large biomolecules (Jorge et al. 2007). An alternative technique is matrix-assisted laser desorption/ionization (MALDI), a “soft” ionization technique used for analyzing biomolecules and large organic molecules that tend to be too tender to be fragmented and ionized by other, more conventional means. Compared to MALDI, ESI takes longer to perform but has the advantage of easy coupling to separation techniques such as LC or HPLC. This allows for high-throughput and on-line analysis of peptide and protein mixtures.
An ionization method known as surface-enhanced laser desorption/ionisation (SELDI) makes use of the proteinchip array technology (Hutchens and Yip 1993). The “chip” consists of a thin strip of aluminium containing eight sample-loading areas. Depending on the nature of the chip surface, only specific types of proteins bind. In other words, the chip forms the target surface to which proteins and matrix are applied. Depending on the composition of matrix, particular types of proteins (acidic, basic, antigens, etc.) are then captured. For example, an IMAC30 ProteinChip Array that is activated by coating the surface with transition metals such as gallium allows for the detection of posttranslational modifications such as phosphorylation (Escher et al. 2007). This method is typically used in combination with time-of-flight MS (TOF-MS) and is similar to MALDI. The speed and ease of SELDI-TOF in principal allows for use in a clinical environment (Paweletz et al. 2001), but compared to other MS methods, SELDI does not allow for the identification of peptides or proteins but only detects signals.
Whereas some techniques have been developed with the aim of providing qualitative information on protein expression, a number of methods have been developed to specifically obtain quantitative information. One such method is the isotope coded affinity tag (ICAT) technique, where cysteine residues are differentially tagged with stable isotopes (Olsen et al. 2004). Proteins from two different samples (e.g., control and experiment) are labeled with either deuterium d0 (light) and d8 (heavy) tagging reagents, which have a mass difference of only eight mass units. This allows for mixing two samples prior to separating them on the same gel, thereby eliminating in-between-gel variation. A comparison of the peak intensities of these tags provides information on the differential expression of the proteins of interest under two different conditions. ICAT is particularly well suited for targeting membrane proteins, as it is compatible with strong detergents (Gygi et al. 1999; Moseley 2001). However, drawbacks of the ICAT method are that it requires the presence of a cysteine in the protein sequence and that it usually requires large initial sample sizes, in the order of 100 μg. Other labeling methods were recently developed to accommodate smaller sample sizes, such as labeling with 12C and 13C, or 16O and 18O (Zang et al. 2004).
Isobaric tagging for relative and absolute protein quantification (iTRAQ) is another quantitative protein identification approach (Fig. 1). It was the first reagent developed for LC-based differential proteome analysis. The isobaric tagging reagents consist of a reporter group, a balance group (to make up for the mass difference of the reporter group), and a peptide reactive group. The latter covalently links an iTRAQ tag with each lysine side chain and amino-terminal group of a peptide, allowing for labeling multiple peptides in a sample digest. The complex mixtures are then fractioned by chromatographic means and analyzed with LC/MS/MS (Olsen et al. 2004; Abdi et al. 2006). Together with isotope-coded protein label (ICPL) (Schmidt et al. 2005), iTRAQ has now more or less substituted ICAT. In biomarker research, however, label-free MS is increasingly applied, owing to the high costs of ICAT or iTRAQ labeling, since large numbers of samples need to be analyzed (Horvatovich et al. 2007).
Metal element chelated tags (MECT) uses eight uncommon earth metals that couples to peptides. It is cheaper than isotope-based methods, with the bicyclic anhydride diethylenetriamine-N,N,N′,N″,N″-pentacetic acid (DTPA) coupling covalently to primary amines of peptides, and the ligand then chelating to the rare earth metals Y and Tb. The tagged peptides are mixed, analyzed by LC-ESI-MS/MS, and peptides quantified by measuring the relative signal intensities for the Y and Tb tag pairs (Liu et al. 2006).
Strategies are constantly improved to functionally categorize the stream of proteins that are newly discovered through proteomics. One technique that was recently shown to be effective for establishing cofactor preferences for proteins is fingerprinting with saturation transfer difference (STD) NMR for detecting closely related cofactors. This approach was validated with both dehydrogenases and cyclic nucleotide-binding proteins (Yao and Sem 2005).
The increased use of proteomics in the field of neurodegeneration research is greatly assisted by the development of integrated technologies that apply high-resolution separation strategies to complex protein samples, combined with sophisticated computer-based identification methods. Ingenious novel hardware and software developments continuously lower detection limits, while widening the dynamic range, making profiling of highly complex protein mixtures for identification of PD biomarkers possible. Although the different search engines and databases (i.e., SwissProt, Mascot) vary in their sensitivity and accuracy for detecting protein sequences, eventually this should have little influence on experimental outcomes, due to the standardization in data collection and interpretation that is enforced by the proteomics community.
Protein markers for mitochondrial dysfunction and oxidative stress in PD
Mitochondrial dysfunction and oxidative stress have been implicated in PD (Schapira 1998; Betarbet et al. 2000; Hardy et al. 2003; Kaur et al. 2003; Lopes and Melov 2002; Beal 2003). Proteomics has proved to be a useful tool to examine the oxidative-stress induced changes in proteins (Butterfield and Castegna, 2003). A deficit of complex I of the respiratory chain has been shown by reduced activity of the enzyme NADH-ubiquinone reductase in neuronal tissue, such as the SN (Schapira et al. 1989; Dexter et al. 1994) and frontal cortex of PD patients (Parker et al. 2008), as well as in non-neuronal tissue such as blood platelets (Parker et al. 1989; Benecke et al. 1993; Haas et al. 1995; Swerdlow et al. 1996; Gu et al. 1998) and skeletal muscle (Bindoff et al. 1991; Shoffner et al. 1991). A proteomic analysis of PD and control SN revealed that neurofilament chains were less abundant in PD specimens, whereas peroxiredoxin II, mitochondrial complex III, and the ATP synthase D chain, among others, were significantly increased in PD samples compared to controls, indicating a compensatory upregulation in surviving SN neurons (Basso et al. 2004). A role for mitochondrial dysfunction in PD is further supported in animal models as administration of environmental toxins known to inhibit mitochondrial function caused dopaminergic cell loss (Betarbet et al. 2000; Di Monte et al. 2000) and increased production of ROS, another correlate of the clinical phenotype (Beal 2004).
Ubiquitin carboxy-terminal hydroxylase L-1 (UCHL1), also known as PGP9.5, is one of the most abundant neuronal proteins present in brain (Wilkinson et al. 1989). Studies have shown that it is extensively downregulated and structurally modified in the presence of oxidative stress (Castegna et al. 2002; Choi et al. 2004). In addition, mutating isoleucine 93 to methionine of UCHL1 causes autosomal dominant PD (Leroy et al. 1998). Furthermore, increased amounts of oxidatively modified UCH-L1 were found in sporadic PD cases compared to normal brain (Castegna et al. 2002; Choi et al. 2004; Butterfield et al. 2006). This deregulation has been implicated in the pathogenesis of PD since oxidative modification and the subsequent decrease in UCH-L1 enzymatic activity may affect normal neuronal function and plasticity (Setsuie and Wada 2007). Interestingly, increased expression of UCH-L1 has been found also in AD using MALDI MS (Sultana et al. 2007). Hence, the application of redox proteomics may be an invaluable tool to help ascertain the role played by oxidized UCH-L1 and other proteins in the pathogenesis of PD and possibly, AD.
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine is a potent mitochondrial toxin that causes dopaminergic degeneration and LB formation in nigral neurons. When mice were chronically treated with MPTP, compared to control animals, more than 300 mitochondrial-related proteins were identified, of which more than 100 displayed a differential expression pattern (Jin et al. 2005). A category analysis revealed their involvement in signal transduction, the ubiquitin-protease system, oxidative stress and mitochondrial function, all of which has been implicated in the pathogenesis of PD (Dauer and Przedborski 2003; Fiskum et al. 2003; Rego and Oliveira 2003). Of the more than 300 proteins detected, DJ-1 was validated by Western blotting.
As discussed before, genetic mutations have been identified in a small subset of cases, mainly in families with early onset familial PD. DJ-1 is one of the genes implicated in disease pathogenesis (Bonifati et al. 2003). DJ-1 has also been identified as a possible marker in plasma for the progression of sporadic PD (Waragai et al. 2007). Gene-based animal models indicate that DJ-1-associated disease onset and progression may operate through the ubiquitin-proteasomal pathway, which is a molecular pathway common to all genetic variants of PD (Dauer and Przedborski 2003; von Bohlen und Halbach et al. 2004).
In its mutant form, DJ-1 confers a reduced protective function against endoplasmic reticulum stress. Two recent reports (Taira et al. 2004; Yokota et al. 2003) show that DJ-1 has a repressive effect on cellular toxicity induced by overexpression of Pael-R, a newly identified substrate for parkin that is localized within the core of LBs (Yokota et al. 2003). Pael-R has also been assigned a distinct role in LB formation (Murakami et al. 2004).
DJ-1 seems to antagonize many of the deleterious effects of SNCA, including its aggregation (Shendelman et al. 2004; Zhou and Freed 2005; Zhou et al. 2006). Therefore, although the exact physiological function of DJ-1 remains elusive, increasing evidence suggests that it may confer protection, for instance as an anti-oxidative protein. In this regard, DJ-1 has been identified as a hydroperoxide-responsive protein that increases its acidity following exposure to oxidative stress (Mitsumoto and Nakagawa 2001; Mitsumoto et al. 2001), thereby revealing the presence of oxidative stress (Kinumi et al. 2004; Taira et al. 2004).
In addition to this role, many lines of evidence argue that DJ-1 modulates the function of SNCA and vice versa (Zhou et al. 2004; Jin et al. 2005). Robust proteomics, consisting of stable isotope labeling by amino acids in cell culture (SILAC) was used on dopaminergic neurons exposed to the mitochondrial toxin rotenone (Jin et al. 2007). Compared to controls, 324 proteins were associated with SNCA and 306 with DJ-1. Although no direct interaction has been observed between DJ-1 and SNCA, 144 proteins mutually associated with DJ-1 and SNCA, displaying significant changes in terms of relative abundance. Further validation was performed on a subset of these, revealing that the docking proteins mortalin (mthsp70/GRP75), nucleolin, grp94, calnexin and clathrin associate with both SNCA and DJ-1. The study confirmed the absence of a direct interaction between SNCA and DJ-1 and reported the identification of five novel proteins that associate with both SNCA and DJ-1. It is anticipated that future work will shed light on the functional interaction between these two critical proteins and the mechanism by which they participate in cellular functions and PD pathogenesis.
Mitochondria-enriched fractions obtained from the SN of mice chronically treated with MPTP have been subjected to ICAT and compared with controls (Jin et al. 2005). The animals received a 5-week treatment regime together with an adjuvant, probenecid (prob) that caused selective nigrostriatal neurodegeneration and the formation of LB-like cytoplasmic inclusions in surviving nigral neurons. After identifying more than 300 proteins, of which >100 displayed changes in relative abundance between the MPTP/prob-treated mice and controls, the high throughput proteomic analysis was validated by Western blotting. This demonstrated that one of the identified proteins, DJ-1, co-localized with SNCA in dopaminergic neurons as well as in cytoplasmic inclusions in the mice that had been treated with MPTP/prob. By using this particular proteomics approach, the study successfully overcame a limitation usually imposed when using pooled samples that prevents the results from telling whether the DJ-1 increase results from a single mouse or from multiple mice. By conducting Western blot analyses in separate animals a DJ-1 increase was demonstrated in the majority of subjects.
Jin et al. (2007) created a quantitative profile of mitochondrial proteins harvested from PD patients compared to normal age-matched controls. In addition, they used a cellular model of PD, consisting of rotenone-treated dopaminergic cells. In this study, a ‘shotgun proteomic’ approach was used called multidimensional protein identification technology (MudPIT), that entails the direct analysis of complex samples for rapidly producing a global profile of the protein complement (Link et al. 1999; Wu and MacCoss 2002). To quantify the proteins, ICAT was combined with SILAC (Jin et al. 2007). Among the differentially expressed proteins was mortalin, a mitochondrial stress protein that was substantially decreased in both PD brain and the cellular PD model. The study further showed that several mortalin-binding proteins might participate in rotenone-mediated toxicity and that by overexpressing and/or silencing mortalin expression, PD-related pathology was significantly affected. Together, the studies suggest that mortalin modulates PD development via pathways involving mitochondrial and proteasomal functions as well as oxidative stress (Jin et al. 2007).
By integrating proteomics technologies (2-D gel electrophoresis and MALDI-TOF), Lee et al. (2003) gathered evidence that expression levels of calreticulin, a high-capacity calcium buffering protein located in MN9D dopaminergic cells may correlate with the degree of cell death following 6-OHDA treatment. This implies that calreticulin may either directly mediate 6-OHDA-induced cell death or, alternatively, it may participate in inducing a general cellular response prior to apoptosis.
Calreticulin further functions as a chaperone by binding to oligosaccharide moieties on proteins in addition to retaining proteins in the ER (Spiro et al. 1996). Its other assigned roles have been controversial, such as promoting cell death when over-expressed (Arnaudeau et al. 2002; Nakamura et al. 2000), as other studies suggest that it might be involved in preventing oxidative stress-induced cell death (Liu et al. 2001; Nunez et al. 2001). Although the direct mechanisms through which calreticulin might induce dopaminergic neuronal death remain to be established, proteomics suggests that this protein may be a marker of oxidative stress (Lee et al. 2003).
Parkin is another marker for oxidative stress and mitochondrial dysfunction in PD. Periquet et al. (2005) generated parkin knock-out mice and searched for possible changes in the brain proteome using 2-D-DIGE. The study indicated compensatory mechanisms that protect dopaminergic neurons from ROS-induced death. This phenomenon was ascribed to 87 proteins that differed in relative abundance compared to wild-type brains. A high proportion of the altered proteins related to energy metabolism, detoxification, stress-related chaperones and components of the ubiquitin-proteasome pathway. In a related study, parkin knock-out mice were analyzed by 2D-PAGE followed by MS (Palacino et al. 2004). This revealed a decreased abundance of a number of proteins involved in mitochondrial function or oxidative stress that could be confirmed functionally. The mice also showed decreased serum anti-oxidant capacity and increased protein and lipid peroxidation. Together, this identifies an essential role for parkin in the regulation of mitochondrial function. Interestingly, in the mice mitochondrial dysfunction and oxidative damage was found in the absence of nigral degeneration (Palacino et al. 2004). This is supported by studies in parkin-deficient flies (Greene et al. 2003) and reminiscent of P301L tau mutant mice that model aspects of AD and also show mitochondrial dysfunction and oxidative damage, in the absence of overt cell loss (David et al. 2005a; David et al. 2006; Götz et al. 2008).
Another protein found in an oxidatively modified state in PD brain tissue is superoxide dismutase 1 (SOD1) that was identified by MALDI-TOF MS in a cell-culture system in a screen for changes in mitochondrial protein expression (Wood-Allum et al. 2006). In PD, not only is residue Cys-146 of SOD oxidized to cysteic acid, but SOD1 is also carbonylated (Choi et al. 2005). The nature of these modifications are still unclear, and it is hoped that proteomics will provide an explanation for the accumulation and posttranslational modifications of SOD1 in the PD brain (Bandopadhyay et al. 2004).
α-Synuclein (SNCA) and its role in PD
Alpha-synuclein (SNCA) is a presynaptic protein associated with PD pathogenesis. It appears in both a soluble and membrane-bound form (Zhu et al. 2006), and comprises the central filamentous component of LBs. Under physiological conditions, it forms a protein complex in the cytosol. As interacting proteins in this complex, 250 candidate proteins have been identified by Zhou and coworkers (2004). However, DJ-1 was not among them. It is important to note, that a ‘negative’ proteomic result does not necessarily entail that the protein of interest is completely absent within the protein complex (Corthals et al. 2000), especially when immunohistochemistry has been applied. For example, Neumann et al. (2004) and Jin et al. (2007) convincingly demonstrated that DJ-1 is present within the halo part of LBs that is known to contain insoluble SNCA.
In an effort to identify proteins that are associated with SNCA and may contribute towards its aggregation, Zhou et al. (2004) exposed MES rat cells to rotenone. Duplicate 2-D gels were run with one gel probed with an anti-SNCA antibody, while the other was stained with Coomassie Blue. The stained gel sections that corresponded to SNCA-immunoreactive bands were cut, trypsin-digested and analyzed with LC-MS/MS spectrometry. The study identified more than 250 proteins associated with SNCA. Under conditions where rotenone was cytotoxic and induced the formation of SNCA-immunoreactive cytoplasmic inclusions, >51 proteins displayed significant differences in terms of relative abundance. This list included heat shock proteins (HSP) 70 and 90 which, when overexpressed in MES cells not only protected cells from rotenone-mediated cytotoxicity, but also decreased SNCA aggregation. From this, it can be concluded that the protection afforded by hsp70 transfection may relate to suppression by rotenone-induced oxidative stress (Zhou et al. 2004). This putative protective role of hsp70 was further supported in subsequent in vivo studies carried out by Klucken et al. (2004). Other neuronal proteins that may couple to SNCA in a similar fashion include parkin (Paciello et al. 2006) and the microtubule-associated protein tau (Jensen et al. 1999), the principal component of the neurofibrillary tangles in AD (Chen et al. 2004).
A ‘shotgun proteomic’ experimental approach was applied to A30P SNCA transgenic Drosophila and age-matched controls (Xun et al. 2007). By conducting the analysis at seven different time-points across the organism’s adult lifespan, disease-associated differences appeared to change substantially as the flies aged. Interestingly, the majority of the perturbed protein levels only existed over a narrow distribution of ages indicating the necessity to take multiple time-points for analysis.
Role of the cytoskeleton and small peptides in PD
Axonal transport and in particular the integrity of microtubules is essential for neuronal function. Not surprisingly, impaired axonal transport has been implicated in a range of neurodegenerative disorders (Götz et al. 2004). Among the proteins with a causal role in AD is tau, a microtubule-associated protein, that similar to SNCA, is found in an unfolded state when in solution. Tau is mainly a neuronal protein. However, it has been found also, although at low levels, in astrocytes and oligodendrocytes (Chen et al. 2004; Götz et al. 2001; Davidsson and Sjögren 2005). Tau’s cardinal functions in neurons are to regulate the assembly and stabilization of microtubules that determine cellular morphology and serve as tracks for vesicles, axonal proteins and mitochondria (Shahani and Brandt 2002). Under physiological conditions, tau is transported along axons at a rate compatible with a slow transportation mechanism (Mercken et al. 1995; Utton et al. 2002; Zhang et al. 2004). Under pathologic conditions such as AD and frontotemporal dementia (FTD), tau aggregates and form filaments, which fill up the entire soma of a degenerating neuron. Here, a failure of axonal transport has been implicated as a possible mechanism underlying tau accumulation (Praprotnik et al. 1996; Flament-Durand and Couck 1979; Richard et al. 1989; Bendiske et al. 2002; Dai et al. 2002; Götz et al. 2006).
There is increasing evidence for an overlap between PD and AD, not only in clinical terms (Kurosinski et al. 2002), but also biochemically and histopathologically (Hoerndli et al. 2005). For example, LBs are not only present in PD brain, but also abundant in Dementia with Lewy bodies (DLB) and in the LB variant of AD (Hansen et al. 1990). Specifically, LBs have been detected in the amygdala of more than half of all familial AD cases, and some LBs colocalized with tau-positive neurofibrillary tangles (NFTs) (Lippa et al. 1998). Biochemically, many of the functional categories which are deregulated in PD are also deregulated in AD (Hoerndli et al. 2005).
Another cytoskeletal protein with a role in neurodegeneration is β-actin, found abundantly within neurons, astrocytes, and blood vessels in the CNS, and concentrated particularly along the periphery of the neuronal perikaryon (De Iuliis et al. 2005). This preferred location places it in an ideal position to integrate incoming signals and produce the mechanical tension necessary for shaping the perikaryal surface (Pannese et al. 1996). A recent study found that specific protein carbonyl levels in β-actin are significantly higher in postmortem human AD brain samples than in controls (Aksenov et al. 2001). Also, when a ‘hemiparkinson’ rat model of PD was obtained by unilaterally injecting the neurotoxin 6-OHDA, De Iuliis et al. (2005) found increased levels of β-actin in the ipsilateral striatum, compared to the contralateral side (Lee et al. 2003). Previous studies described β-actin to be in an oxidized state in AD again suggesting common patho-mechanisms for the two diseases.
Much of the signaling in brain is mediated by small peptides. The study of endogenous peptides, termed peptidomics; however, is complicated by the presence of protein fragments produced post-mortem during conventional sample handling. This can be partially prevented by rapidly heat-denaturing brain tissue before analysis (Svensson et al. 2007). This approach has been applied to the MPTP model for PD, by which a polypeptide termed PEP-19 (Peptide 19) was found to be significantly decreased in the striatum of treated animals compared to controls (Svensson et al. 2007). PEP 19 is a 7.6-kDa polypeptide that binds calmodulin and prevents calcium-calmodulin-dependent signaling (Ichikawa and Sugimoto 2005). As a neuron-specific calmodulin-binding polypeptide, it is believed to play a pivotal role within the second messenger system that allows for the transduction of signals within cells. Elevated levels have been found in selective neuronal populations, such as the granule cells of the hippocampus and the Purkinje cells of the cerebellum, implying resistance towards degeneration (Johanson et al. 2000). In contrast, Skold et al. (2006) recently demonstrated by in situ hybridization analysis, that PEP-19 mRNA levels are significantly decreased in the striatum, following MPTP administration in mice. Disease-specific differences have been found in an earlier study (Utal et al. 1998) that reported that although PEP-19 immunoreactivity appeared to be significantly reduced in AD and Huntington’s disease brain, no apparent alterations were detected in PD brain. Thus, the exact nature of PEP-19 in PD remains controversial.
From post mortem tissue to body fluids as source of proteins
Proteome searches are frequently conducted within postmortem brain tissue of a diseased state. This approach entails its own set of difficulties, including the potential confounds caused by additional age-related diseases that the usually aged patient may have suffered from, the time between death and autopsy that could have resulted in protein changes unrelated to the disease, how rapidly death ensued, the time of death and the duration of the coma that preceded death. In addition, the pharmacological treatment, that is, the medication, may result in deranged protein levels, that may be mistaken for being disease-related. It is essential to consider the homogeneity of all these parameters in both the control and the patient group (Riederer and Wuketich 1976). However, in spite of these difficulties several groups have proteomically probed brain tissue for parameters that may assist in predicting or diagnosing the disease, monitor its progression, or be used to follow-up therapy outcomes (Hoerndli et al. 2005).
In addition to post mortem tissue, body fluids such as cerebrospinal fluid (CSF), serum, blood or urine are increasingly collected from either animal models or humans and subjected to proteomics analysis. Obviously, animal models do not reproduce all aspects of disease, especially as far as the clinical features and the spatiotemporal distribution of brain lesions is concerned, but at a single cell level and biochemically, they have proven to be a very powerful tool (Götz et al. 2004).
Although there is currently no blood test available with which to confirm the diagnosis or monitor PD, the accessibility of serum, as a component of blood, that is more readily available than any other body-fluid, offers potential exploitation using proteomics technology. Sophisticated techniques, including MS are used to identify rare proteins in blood serum and CSF to identify biomarkers that represent unique proteomes. At present, 34 blood serum protein biomarkers are used for diagnosing neurodegenerative diseases (Sheta et al. 2006).
In particular, CSF has been used as a medium from which to launch a proteome-based search strategy for detecting disease biomarkers, since the CSF reflects the state of proteins present in the brain under healthy and diseased conditions (Hühmer et al. 2006). As a complex mixture, consisting of proteins, peptides, proteolytic fragments, and antibodies, CSF provides an excellent repository of pathologic information concerning the CNS (Romeo et al. 2005).
In one study, quantitative proteomics has been applied successfully to CSF samples from patients with PD at different stages as well as with AD and compared to normal controls (Pan et al. 2008). The frequently encountered problem was addressed that while biomarkers are generally validated by enzyme-linked immunosorbent assays (ELI-SAs), often there are no specific antibodies available to set up these assays. Hence Pan et al. (2008) used a quantitative LC-MALDI TOF/TOF approach by spiking CSF samples with isotope-labeled peptides derived from a total of 14 proteins that had been previously identified as being deregulated in disease. Importantly, they were able to identify and quantify the peptides in CSF without prior depletion of abundant proteins (Pan et al. 2008). It should be mentioned that for quantitative proteomics of body fluids, label-free profiling has been successfully used by applying a microfluidics-based chip-LC-MS system, a method that has gained increased attraction for reasons of cost (Horvatovich et al. 2007). Sophisticated softwares have been developed to assist in the quantification of label-free profiling, including the open software tool, SuperHirn (Mueller et al. 2007).
Several strategies exist for characterizing lipid-metabolizing proteins in human CSF. These proteins are potentially therapeutic targets due to their ability to transport lipids required for neural growth or to convert these into molecules that control brain physiology (Fonteh et al. 2006). By combining lipid analysis with proteomics, the existing knowledge of disease pathology may be enhanced and the likelihood increased for discovering distinct markers and biochemical mechanisms of disease. In an effort to profile tau in CSF as a reflection of the degree of neuronal degeneration and damage sustained, Blennow et al. (1995) combined a pre-fractionation step consisting of liquid phase isoelectric focusing (LP-IEF) with immunoblotting. This revealed that both phosphorylated and unphosphorylated forms of tau are present in CSF and that tau appeared both in truncated and full-length versions. For a comprehensive review of the posttranslational modifications of tau, the reader is referred to Chen et al. (2004).
Quantitative 2-D gel electrophoresis has been used to analyze serum proteins derived from 422 patients suffering from different neurodegenerative diseases and compared with normal controls, in an effort to identify potential biomarkers (Goldknopf et al. 2006). Differential protein spots were found for a total of 34 serum proteins between amyotrophic lateral sclerosis (ALS), PD and related disorders, with nine relating to the complement system. Components of complement C3, including C3c and C3dg as well as complement factor H showed a marked increase in both disease-types compared to controls. In addition, the results indicated elevated levels for full-length factor B for PD, but not the control group. This reported elevation of factors H and B contradicts recent work proclaiming that they are significantly reduced in PD CSF (Finehout et al. 2005). In this study, it is possible that elevations of fragment Bb arose from elevated levels of factor H, which has been found to induce dissociation of C3 convertase (C3bBb(Mg2+)) (Hourcade et al. 2002), thereby liberating factor Bb and interrupting the alternative pathway of the complement cascade (Goldknopf et al. 2006). The study generated a list of potentially useful biomarkers for further exploitation in future studies. In addition, the identified proteins provide evidence for an involvement of neuroinflammatory processes in the pathogenesis of ALS and PD. The study indicates further that a comparison of the expression level of individual protein isoforms in conjunction with a measurement of the total protein expression level may be useful for investigating a protein as a possible biomarker.
The results on the complement cascade add strength to existing evidence suggesting that inflammatory processes involving complement activation may play a definitive role in the onset of PD. It also complements work that provides evidence for neuronal injury resulting from activated microglial release of free radicals in PD (He et al. 2002). This finding provides a potential avenue for developing treatment targets and for monitoring their clinical application. It further shows that two different neurodegenerative diseases can differentially express members of the complement system. In another study performed by Sjögren et al. (2001) it was found that the CSF concentrations of tau and phosphotau are increased in around two-thirds of probable AD cases while revealing normal levels in PD and in controls. The insight gained from such studies may potentially provide valuable information regarding their respective pathological mechanisms and identify markers that are shared between diseases and others which are disease-specific. In these endeavors, it is worth mentioning that the analysis of post-translational modifications remains a technological challenge, but various generic strategies have recently emerged to aid this strategy (Reinders et al. 2004; Fountoulakis and Kossida 2006).
Conclusions and future directions
It is evident that our present knowledge of PD pathogenesis and potential treatment strategies is inadequate. A growing body of evidence indicates that the accumulation of altered proteins and impaired protein clearance may be a common pathomechanism in both familial and sporadic PD. To fully understand the role played by abnormal protein aggregates in PD, it is imperative that additional proteins with a role in disease are identified and that their disease-related roles are defined. Recent advances in proteomics indicate that this technology may equip us with methodologies with which to study PD in a systematic manner that has not been possible to this extent in the past, by offering the opportunity to explore the proteome at all levels, ranging from fundamental neuroscience to clinical trials. However, to utilize neuroproteomics to its full potential, it is imperative that standard operating protocols are developed that allow these techniques to be applied reproducibly in a clinical setting.
The systems-based approach offered by proteomics not only allows for identifying multiple key proteins and signaling cascades, but also shows how these proteins interact with each other. It is anticipated that such data may eventually improve our understanding of the disease at a biochemical level and help identify novel biomarkers to assist in making an early, presymptomatic diagnosis of PD. In addition, it is hoped that the application of this technology will lead to the identification of new targets for more effective therapeutic intervention. The clinico-pathological profiles of AD and PD seem to overlap, or at least synergise to a certain extent (Kurosinski et al. 2002). Information is increasingly emerging to support such a postulation and therefore, the possibility is increasingly upheld that drugs that target the blocking of SNCA or tau may therapeutically benefit a much broader spectrum of neurodegenerative disorders.
Since PD likely has a multifactorial etiology, proteomics provides the possibility for characterizing inter-patient variance. The lessons learned from this may eventually assist in developing personalized therapies, resting on the basis of individual patterns of protein expression (Jain 2004). With this in mind, the future development of protein markers with which to identify not only the motor-related, but also the non-motor manifestations of Parkinson’s disease, including cognitive dysfunction, autonomic dysfunction and speech disturbance, will contribute greatly towards a more global perspective of the disease.
The greatest challenge facing the optimal utilization of this technology lies in detecting and quantifying low-abundant and hydrophobic proteins. In addition, the accurate detection of post-translational modifications, their origin and the role they play in PD should also be made a priority. However, it is anticipated that protein chips and miniature separation systems will play a significant role in overcoming these limitations.
Acknowledgments
IP is supported by an International Brian Research Organization (IBRO) Fellowship and wishes to thank IBRO for the generous financial support of her work. This work was also supported by an NIH Fogarty International Centre Research Grant (R21DA018087 to Michael Zigmond) and the Medical Research Council (MRC) of South Africa. The financial assistance of the National Research Foundation (NRF) of South Africa towards this work is also acknowledged. JG is a Medical Foundation Fellow. JG is supported by the University of Sydney, the National Health & Medical Research Council (NHMRC), the Australian Research Council (ARC), the New South Wales Government through the Ministry for Science and Medical Research (BioFirst Program), the Nerve Research Foundation, the Medical Foundation (University of Sydney) and the Judith Jane Mason & Harold Stannett Williams Memorial Foundation.
Abbreviations
- SNCA
Alpha-synuclein
- AD
Alzheimer’s disease
- SDS
Dodecyl sulphate
- DA
Dopamine
- ESI
Electrospray ionization
- L-DOPA
Levodopa
- LB
Lewy bodies
- MS
Mass spectrometry
- MPTP
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- 6-OHDA
Parkinson’s disease
- PD
6-Hydroxydopamine
- SNpc
Substantia Nigra pars compacta
References
- Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, Nixon R, Nutt J, Chung K, Zabetian C, Samii A, Lin M, Hattan S, Pan C, Wang Y, Jin J, Zhu D, Li GJ, Liu Y, Waichunas D, Montine TJ, Zhang J. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Rist B, Gygi SP. Quantitative proteome analysis: methods and applications. Ann NY Acad Sci. 2000;919:33–47. doi: 10.1111/j.1749-6632.2000.tb06865.x. [DOI] [PubMed] [Google Scholar]
- Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Alam Z, Daniel S, Lees A, Marsden D, Jenner P, Halliwell P. A generalized increase in protein carbonyls in the brain in Parkinson’s but not incidental Lewy body disease. J Neurochem. 1997;69:1326–1329. doi: 10.1046/j.1471-4159.1997.69031326.x. [DOI] [PubMed] [Google Scholar]
- Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10:S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- Arnaudeau S, Frieden M, Nakamura K, Castelbou C, Michalak M, Demaurex N. Calreticulin differentially modulates calcium uptake and release in the endoplasmic reticulum and mitochondria. J Biol Chem. 2002;277:46696–46705. doi: 10.1074/jbc.M202395200. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Zhang SM, Hernan MA, Kawachi I, Colditz GA, Speizer FE, Willett WC. Prospective study of caffeine consumption and risk of Parkinson’s disease in men and women. Ann Neurol. 2001;50:56–63. doi: 10.1002/ana.1052. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Chen H, Weisskopf MG, O’Reilly E, McCullough ML, Calle EE, Schwarzschild MA, Thun MJ. Pesticide exposure and risk for Parkinson’s disease. Ann Neurol. 2006;60:187–203. doi: 10.1002/ana.20904. [DOI] [PubMed] [Google Scholar]
- Bandopadhyay R, Kingsbury AE, Cookson MR, Reid AR, Evans IM, Hope AD, Pittman AM, Lashley T, Canet-Aviles R, Miller DW, McLendon C, Strand C, Leonard AJ, Abou-Sleiman PM, Healy DG, Ariga H, Wood NW, de Silva R, Revesz T, Hardy JA, Lees AJ. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain. 2004;127:420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- Basso M, Giraudo S, Corpillo D, Bergamasco B, Lopiano L, Fasano M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria, oxidative damage, and inflammation in Parkinson’s disease. Ann NY Acad Sci. 2003;991:120–131. doi: 10.1111/j.1749-6632.2003.tb07470.x. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondrial dysfunction and oxidative damage in Alzheimer’s and Parkinson’s diseases and coenzyme Q10 as a potential treatment. J Bioenerg Biomembr. 2004;36:381–386. doi: 10.1023/B:JOBB.0000041772.74810.92. [DOI] [PubMed] [Google Scholar]
- Benecke R, Strümper P, Weiss H. Electron transfer complexes I and IV of platelets are abnormal in Parkinson’s disease but normal in Parkinson-plus syndromes. Brain. 1993;116:1451–1463. doi: 10.1093/brain/116.6.1451. [DOI] [PubMed] [Google Scholar]
- Bendiske J, Caba E, Brown QB, Bahr BA. Intracellular deposition, microtubule destabilization, and transport failure: an “early” pathogenic cascade leading to synaptic decline. J Neuropathol Exp Neurol. 2002;61:640–650. doi: 10.1093/jnen/61.7.640. [DOI] [PubMed] [Google Scholar]
- Benedetti MD, Bower JH, Maraganore DM, McDonnell SK, Peterson BJ, Ahlskog JE, Schaid DJ, Rocca WA. Smoking, alcohol, and coffee consumption preceding Parkinson’s disease. Neurology. 2000;55:1350–1358. doi: 10.1212/wnl.55.9.1350. [DOI] [PubMed] [Google Scholar]
- Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci. 1973;20:415–455. doi: 10.1016/0022-510x(73)90175-5. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Bindoff LA, Birch-Machin M, Cartilidge NE, Parker WD, Turnbull DM. Respiratory chain abnormalities in skeletal muscle from patients with Parkinson’s disease. J Neurol Sci. 1991;104:203–208. doi: 10.1016/0022-510x(91)90311-t. [DOI] [PubMed] [Google Scholar]
- Birkmayer W, Hornykiewicz O. The effect of L-3,4-dihydroxyphenylalanine (L-DOPA) on akinesia in Parkinsonism. Wiener Klin Wochenschr 1998. 1961;73:787–788. [PubMed] [Google Scholar]; Parkinsonism Relat Disord. 4:59–60. (English translation) [Google Scholar]
- Blennow K, Wallin A, Agren H, Spenger C, Siegfried J, Vanmechelen E. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol Chem Neuropathol. 1995;26:231–245. doi: 10.1007/BF02815140. [DOI] [PubMed] [Google Scholar]
- Bloch F, Houetto JL, Tezenas du Montcel S, Bonneville F, Etchepare F, Welter ML, Rivaud-Pechoux S, Hahn-Barma V, Maisonobe T, Behar C, Lazennec JY, Kurys E, Arnulf I, Bonnet AM, Agid Y. Parkinson’s disease with camptocormia. J Neurol Neurosurg Psychiatry. 2006;77:1223–1228. doi: 10.1136/jnnp.2006.087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Castegna A. Proteomic analysis of oxidatively modified proteins in Alzheimer’s disease brain: insights into neurodegeneration. Amino Acids. 2003;25:419–425. [PubMed] [Google Scholar]
- Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer’s disease: an initial assessment. J Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part II: Dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- Chen F, David D, Ferrari A, Götz J. Posttranslational modifications of tau: role in human tauopathies and modeling in transgenic animals. Curr Drug Targets. 2004;5:503–515. doi: 10.2174/1389450043345236. [DOI] [PubMed] [Google Scholar]
- Chen L, Feany MB. Alpha-synuclein phospohorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson’s disease. Nat Neurosci. 2005;8:657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang SM, Schwarzschild MA, Hernan MA, Ascherio A. Physical activity and the risk of Parkinson disease. Neurology. 2005a;64:664–669. doi: 10.1212/01.WNL.0000151960.28687.93. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Yu H, Wang F, Xu W. The cross talk between protein kinase A- and RhoA-mediated signaling in cancer cells. Exp Biol Med. 2005b;230:731–741. doi: 10.1177/153537020523001006. [DOI] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- Choi J, Rees HD, Weintraub ST, Levey AI, Chin LS, Li L. Oxidative modifications and aggregation of Cu, Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J Biol Chem. 2005;280:11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- Clark I, Dodson M, Jiang C, Cao J, Huh J, Seol J, Yoo S, Hay B, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Rochet JC, Bieganski RM, Lansbury PT. Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- Cookson MR. The biochemistry of Parkinson’s disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- Corthals GL, Wasinger VC, Hochstrasser DF, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Dai J, Buijs RM, Kamphorst W, Swaab DF. Impaired axonal transport of cortical neurons in Alzheimer’s disease is associated with neuropathological changes. Brain Res. 2002;948:138–144. doi: 10.1016/s0006-8993(02)03152-9. [DOI] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- David DC, Hauptmann S, Scherping I, Schuessels K, Keil U, Rizzu P, Ravid R, Dröse S, Brandt U, Muller WE, Eckert A, Götz J. Proteomic and functional analysis reveal a mitochondrial dysfunction in P301L Tau transgenic mice. J Biol Chem. 2005a;280:23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- David DC, Hoerndli F, Götz J. Functional genomics meets neurodegenerative disorders. Part 1: Transcriptomic and proteomic technology. Progress Neurobiol. 2005b;76:153–168. doi: 10.1016/j.pneurobio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- David DC, Ittner LM, Gehrig P, Nergenau D, Shepherd C, Halliday G, Gotz J. β-Amyloid treatment of two complementary P301L tau-expressing Alzheimer’s disease models reveals similar deregulated cellular processes. Proteomics. 2006;6:6566–6577. doi: 10.1002/pmic.200600634. [DOI] [PubMed] [Google Scholar]
- Davidsson P, Sjögren M. The use of proteomics in biomarker discovery in neurodegenerative diseases. Disease Markers. 2005;18:1–12. doi: 10.1155/2005/848676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- De Hoog CL, Mann M. Proteomics. Annu Rev Genomics Hum Genet. 2004;5:267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- De Iuliis A, Grigoletto J, Recchia A, Giusti P, Arslan P. A proteomic approach in the study of an animal model of Parkinson’s disease. Clin Chim Acta. 2005;357:202–209. doi: 10.1016/j.cccn.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Deumens R, Blokland A, Prickaerts J. Parkinson’s disease in rats: an evaluation of 6-OHDA lesions of the nigrostriatal pathway. Exp Neurol. 2002;175:303–317. doi: 10.1006/exnr.2002.7891. [DOI] [PubMed] [Google Scholar]
- Dexter D, Wells F, Agid F, Agid Y, Lees AJ, Jenner P, Marsden CD. Increased nigral iron content in postmortem parkisonian brain. Lancet. 1987;8569:1219–1220. doi: 10.1016/s0140-6736(87)91361-4. [DOI] [PubMed] [Google Scholar]
- Dexter D, Sian J, Rose S, Hindmarsh JG, Mann VM, Cooper JM, Wells FR, Daniel SE, Lees AJ, Schapira AH, et al. Indices of oxidative stress and mitochondrial function in individuals with incidental Lewy body disease. Ann Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- Di Monte DA, McCormack A, Petzinger G, Janson AM, Quik M, Langston WJ. Relationship among nigrostriatal denervation, parkinsonism, and dyskinesias in the MPTP primate model. Mov Disord. 2000;15:459–466. doi: 10.1002/1531-8257(200005)15:3<459::AID-MDS1006>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Escher N, Kaatz M, Melle M, Hipler C, Ziemer M, Driesch D, Wollina U, von Eggeling F. Posttranslational modifications of transthyretin are serum markers in patients with mycosis fungiodes. Neoplasia. 2007;9:254–259. doi: 10.1593/neo.06805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Fialka I, Pasquali C, Lottspeich F, Ahorn H, Huber LA. Subcellular fractionation of polarized epithelial cells and identification of organelle-specific proteins by two-dimensional gel electrophoresis. Electrophoresis. 1997;18:2582–2590. doi: 10.1002/elps.1150181414. [DOI] [PubMed] [Google Scholar]
- Finehout EJ, Franck Z, Lee KH. Complement protein isoforms in CSF as possible biomarkers for neurodegenerative disease. Dis Markers. 2005;21:93–101. doi: 10.1155/2005/806573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskum G, Starkov A, Polster BM, Chinopoulos C. Mitochondrial mechanisms of neural cell death and neuroprotective interventions in Parkinson’s disease. Ann NY Acad Sci. 2003;99:111–119. doi: 10.1111/j.1749-6632.2003.tb07469.x. [DOI] [PubMed] [Google Scholar]
- Flament-Durand J, Couck AM. Spongiform alterations in brain biopsies of presenile dementia. Acta Neuropathol (Berl) 1979;46:159–162. doi: 10.1007/BF00684819. [DOI] [PubMed] [Google Scholar]
- Fonteh AN, Harrington RJ, Huhmer AF, Biringer RG, Riggings JN, Harrington MG. Identification of disease markers in human cerebrospinal fluid using lipidomic and proteomic methods. Dis Markers. 2006;22:39–64. doi: 10.1155/2006/202938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornai F, Schluter OM, Lenzi P, Ruffoli R, Ferrucci M, Lazzeri G, Busceti CL, Pontarelli F, Battaglia G, Pellegrini A, Nicoletti F, Ruggieri S, Paparelli A, Südhof TC. Parkinson-like syndrome induced by continuous MPTP infusion: Convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci USA. 2005;102:3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forno LS, Langston JW, DeLanney LE, Irwin I, Ricaurte GA. Locus coeruleus lesions and eosinophilic inclusions in MPTP-treated monkeys. Ann Neurol. 1986;20:449–455. doi: 10.1002/ana.410200403. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M. Proteomics: current technologies and applications in neurological disorders and toxicology. Amino Acids. 2001;21:363–381. doi: 10.1007/s007260170002. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M. Application of proteomics technologies in the investigation of the brain. Mass Spectrom Rev. 2004;23:231–258. doi: 10.1002/mas.10075. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Kossida S. Proteomics-driven progress in neurodegeneration research. Electrophoresis. 2006;27:1556–1573. doi: 10.1002/elps.200500738. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, Iwatsubo T. Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- Gasser T. Genetics of Parkinson’s disease. Curr Opin Neurol. 2005;18:363–369. doi: 10.1097/01.wco.0000170951.08924.3d. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;294:1346–1349. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Van Deerlin VM. Mutations in LRRK2 as a cause of Parkinson’s disease. Neurosignals. 2008;16:99–105. doi: 10.1159/000109764. [DOI] [PubMed] [Google Scholar]
- Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Sheta EA, Bryson J, Folsom B, Wilson C, Duty J, Yen AA, Appel SH. Complement C3c and related protein biomarkers in amyotrophic lateral sclerosis and Parkinson’s disease. Biochem Biophys Res Commun. 2006;342:1034–1039. doi: 10.1016/j.bbrc.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Goldman JE, Yen SH, Chiu FC, Peress NS. Lewy bodies of Parkinson’s disease contain neurofilament antigens. Science. 1983;221:1082–1084. doi: 10.1126/science.6308771. [DOI] [PubMed] [Google Scholar]
- Gorell JM, Johnson CC, Rybicki BA, Peterson EL, Richardson RJ. The risk of Parkinson’s disease with exposure to pesticides, farming, well water, and rural living. Neurology. 1998;50:1346–1350. doi: 10.1212/wnl.50.5.1346. [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Günther S, Weser J. Improved horizontal two-dimensional electrophoresis with hybrid isoelectric focusing in immobilized pH gradients in the first dimension and laying-on transfer to the second dimension. Electrophoresis. 1985;12:653–658. [Google Scholar]
- Götz J, Tolnay M, Barmettler R, Ferrari A, Bürki K, Goedert M, Probst A, Nitsch RM. Human tau transgenic mice. Towards an animal model for neuro- and glialfibrillary lesion formation. Adv Exp Med Biol. 2001;487:71–83. [PubMed] [Google Scholar]
- Götz J, Streffer JR, David D, Schild A, Hoerndli F, Pennanen L, Kurosinski P, Chen F. Transgenic animal models of Alzheimer’s disease and related disorders: Histopathology, behavior and therapy. Mol Psychiatry. 2004;9:664–683. doi: 10.1038/sj.mp.4001508. [DOI] [PubMed] [Google Scholar]
- Götz J, Ittner LM, Kins S. Do axonal defects in tau and amyloid precursor protein transgenic animals model axonopathy in Alzheimer’s disease? J Neurochem. 2006;98:993–1006. doi: 10.1111/j.1471-4159.2006.03955.x. [DOI] [PubMed] [Google Scholar]
- Götz J, Deters N, Doldissen A, Bokhari L, Ke Y, Wiesner A, Schonrock N, Ittner LM. A decade of tau transgenic animal models and beyond. Brain Pathol. 2007;17:91–103. doi: 10.1111/j.1750-3639.2007.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz J, David D, Hoerndli F, Ke YD, Schonrock N, Wiesner A, Fath T, Bokhari L, Lim YA, Deters N, Ittner LM. Functional genomics dissects pathomechanisms in tauopathies: mitosis failure and unfolded protein response. Neurodegener Dis. 2008;5:179–181. doi: 10.1159/000113696. [DOI] [PubMed] [Google Scholar]
- Graves PR, Haystead TA. Molecular biologist’s guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39–63. doi: 10.1128/MMBR.66.1.39-63.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Cooper JM, Taanman JW, Schapira AHV. Mitochondrial DNA transmission of the mitochondrial defect in Parkinson’s disease. Ann Neurol. 1998;44:177–186. doi: 10.1002/ana.410440207. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Haas RH, Nasirian F, Nakano K, Ward D, Pay M, Hill R, Shults CW. Low platelet mitochondrial complex I and complex II/III activity in early untreated Parkinson’s disease. Ann Neurol. 1995;37:714–722. doi: 10.1002/ana.410370604. [DOI] [PubMed] [Google Scholar]
- Hanash S. HUPO initiatives relevant to clinical proteomics. Mol Cell Proteomics. 2004;3:298–301. doi: 10.1074/mcp.R400004-MCP200. [DOI] [PubMed] [Google Scholar]
- Hardy J, Cookson MR, Singleton A. Genes and parkinsonism. Lancet Neurol. 2003;2:221–228. doi: 10.1016/s1474-4422(03)00350-8. [DOI] [PubMed] [Google Scholar]
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson’s disease and parkinsonism. Ann Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- Hansen L, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. The Lewy body variant of Alzheimer’s disease: a clinical and pathologic entity. Neurology. 1990;40:1–8. doi: 10.1212/wnl.40.1.1. [DOI] [PubMed] [Google Scholar]
- He Y, Le WD, Appel SH. Role of fcgamma receptors in nigral cell injury induced by Parkinson’s disease immunoglobulin injection into mouse substantia niga. Exp Neurol. 2002;176:322–327. doi: 10.1006/exnr.2002.7946. [DOI] [PubMed] [Google Scholar]
- Heimlick G, Cidlowski JA. Selective role of intracellular chloride in the regulation of the intrinsic but not extrinsic pathway of apoptosis in Jurkat T-cells. J Biol Chem. 2006;281:2232–2241. doi: 10.1074/jbc.M507367200. [DOI] [PubMed] [Google Scholar]
- Hernan MA, Takkouche B, Caamano-Isorna F, Gestal-Otero JJ. A meta-analysis of coffee drinking, cigarette smoking, and the risk of Parkinson’s disease. Ann Neurol. 2002;52:276–284. doi: 10.1002/ana.10277. [DOI] [PubMed] [Google Scholar]
- Hoving S, Gerrits B, Voshol H, Müller D, Roberts RC, van Oostrum J. Preparative two-dimensional gel electrophoresis at alkaline pH using narrow range immobilized pH gradients. Proteomics. 2002;2:127–134. doi: 10.1002/1615-9861(200202)2:2<127::aid-prot127>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Hutchens TW, Yip TT. New desorption strategies for the mass spectrometric analysis of macromolecules. Rapid Commun Mass Spectrom. 1993;7:576–580. [Google Scholar]
- Hoerndli F, David DC, Götz J. Functional Genomics meets neurodegenerative disorders. Part II: application and data integration. Prog Neurobiol. 2005;76:169–188. doi: 10.1016/j.pneurobio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Hoglinger GU, Carrard G, Michel PO, Medja F, Lombes A, Ruberg M, Friguet B, Hirsch EC. Dysfunction of mitochondrial complex I and the proteasome: interactions between two biochemical deficits in a cellular model of Parkinson’s disease. J Neurochem. 2003;86:1297–1307. doi: 10.1046/j.1471-4159.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- Horvatovich P, Govorukhina NI, Reijmers TH, van der Zee AG, Suits F, Bischoff R. Chip-LC-MS for label-free profiling of human serum. Electrophoresis. 2007;28:4493–4505. doi: 10.1002/elps.200600719. [DOI] [PubMed] [Google Scholar]
- Hourcade DE, Mitchell L, Kuttner-Kondo LA, Atkinson JP, Medof ME. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J Biol Chem. 2002;277:1107–1112. doi: 10.1074/jbc.M109322200. [DOI] [PubMed] [Google Scholar]
- Huber LA, Pfaller K, Vietor K. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ Res. 2003;92:962–968. doi: 10.1161/01.RES.0000071748.48338.25. [DOI] [PubMed] [Google Scholar]
- Hühmer AF, Biringer RG, Amato H, Fonteh AN, Harrington MG. Protein analysis in human cerebrospinal fluid: physiological aspects, current progress and future challenges. Disease Markers. 2006;22:2–26. doi: 10.1155/2006/158797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H, Sugimoto T. Peptide 19 in the rat vagal and glossopharyngeal sensory ganglia. Brain Res. 2005;1038:107–112. doi: 10.1016/j.brainres.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Jain KK. Role of pharmacoproteomics in the development of personalized medicine. Pharmacogenomics. 2004;5:331–336. doi: 10.1517/phgs.5.3.331.29830. [DOI] [PubMed] [Google Scholar]
- Jensen PH, Hager H, Nielsen MS, Højrup P, Gliemaann J, Jakes R. α-Synuclein binds to tau and stimulates the protein kinase A-catalyzed tau phosphorylation of serine residues 262 and 356. J Biol Chem. 1999;274:25481–25489. doi: 10.1074/jbc.274.36.25481. [DOI] [PubMed] [Google Scholar]
- Jin J, Meredith GE, Chen L, Zhou Y, Xu J, Shie FS, Lockhart P, Zhang J. Quantitative proteomic analysis of mitochondrial proteins: relevance to Lewy body formation and Parkinson’s disease. Mol Brain Res. 2005;134:119–138. doi: 10.1016/j.molbrainres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J. Identification of novel proteins associated with both alpha-synuclein and DJ–1. Mol Cell Proteomics. 2007;6:845–859. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- Johanson RA, Sarau HM, Foley JJ, Slemmon JR. Calmodulin Binding Peptide PEP–19 Modulates Activation of Calmodulin Kinase II in situ. J Neurosci. 2000;20:2860–2866. doi: 10.1523/JNEUROSCI.20-08-02860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Yu LR, Conrads TP, Kinoshita Y, Uo T, McBee JK, Veenstra TD, Morrison RS. The proteomics of neurodegeneration. Am J Pharmacogenomics. 2005;5:259–270. doi: 10.2165/00129785-200505040-00006. [DOI] [PubMed] [Google Scholar]
- Jorge I, Casas EM, Villar M, Ortega-Pérez I, López-Ferrer D, Martínez-Ruiz A, Carrera M, Marina A, Martínez P, Serrano H, Cañas B, Were F, Gallardo JM, Lamas S, Redondo JM, García-Dorado D, Vázquez J. High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: Application to biological studies. J Mass Spectrom. 2007;42:1391–1403. doi: 10.1002/jms.1314. [DOI] [PubMed] [Google Scholar]
- Kaur D, Yantiri F, Rajagopalan S, Kumar J, Mo JQ, Boonplueang R, Viswanath V, Jacobs R, Yang L, Beal MF, DiMonte D, Volitaskis I, Ellerby L, Cherny RA, Bush AI, Andersen JK. Genetic or pharmacological iron chelation prevents MPTP-induced neurotoxicity in vivo: A novel therapy for Parkinson’s disease. Neuron. 2003;37:899–909. doi: 10.1016/s0896-6273(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human. Biochem Biophys Res Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Burger C, Lundberg C, Johansen TE, Muzyczka N, Mandel RJ, Bjorklund A. Parkinson-like neurodegeneration induced by targeted overexpression of alpha-synuclein in the nigrostriatal system. J Neurosci. 2002;22:2780–2791. doi: 10.1523/JNEUROSCI.22-07-02780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klein C, Lohmann-Hedrich K. Impact of recent genetic findings in Parkinson’s disease. Curr Opin Neurol. 2007;20:453–464. doi: 10.1097/WCO.0b013e3281e6692b. [DOI] [PubMed] [Google Scholar]
- Klose J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 1975;26:231–243. doi: 10.1007/BF00281458. [DOI] [PubMed] [Google Scholar]
- Klucken J, Shin Y, Hyman BT, McLean PJ. A single amino acid substitution differentiates Hsp70-dependent effects on alpha-synuclein degradation and toxicity. Biochem Biophys Res Commun. 2004;325:367–373. doi: 10.1016/j.bbrc.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Kurosinski P, Guggisberg M, Götz J. Alzheimer’s and Parkinson’s disease - overlapping or synergistic pathologies? Trends Mol Med. 2002;8:3–5. doi: 10.1016/s1471-4914(01)02246-8. [DOI] [PubMed] [Google Scholar]
- Krüger R, Kuhn W, Müller T, Woitalla D, Graeber M, Kösel S, Przuntek H, Epplen JT, Schöls L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Lee MY, Park SE, Chung KC, Oh YJ. Proteomic analysis reveals upregulation of calreticulin in murine dopaminergic neuronal cells after treatment with 6-hydroxidopamine. Neurosci Lett. 2003;352:17–20. doi: 10.1016/j.neulet.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Lee WD, Appel SH. Mutant genes responsible for Parkinson’s disease. Curr Opin Pharmacol. 2004;4:79–84. doi: 10.1016/j.coph.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Origins and effects of extracellular alpha-synuclein: Implications in Parkinson’s disease. J Mol Neurosci. 2008;34:17–22. doi: 10.1007/s12031-007-0012-9. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Limousin P, Krack P, Pollak P, Benazzouz A, Ardouin C, Hoffmann D, Benabid AL. Electrical stimulation of the subthalamic nucleus in advanced Parkinson’s disease. N Engl J Med. 1998;339:1105–1111. doi: 10.1056/NEJM199810153391603. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Lippa CF, Fujiwara H, Mann DM, Giasson B, Baba M, Schmidt ML, Nee LE, O’Connell B, Pollen DA, St George-Hyslop P, Ghetti B, Nochlin D, Bird TD, Cairns NJ, Lee VM, Iwatsubo T, Trojanowski JQ. Lewy bodies contain altered alpha-synuclein in brains of many familial Alzheimer’s disease patients with mutations in presenilin and amyloid precursor protein genes. Am J Pathol. 1998;153:1365–1370. doi: 10.1016/s0002-9440(10)65722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton MS, Pasa-Tolic L, Anderson GA. Global analysis of the deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci USA. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Miller E, van de Water B, Stevens JL. Endoplasmic reticulum stress proteins block oxidant-5nd 4ced Ca2+ increase and cell death. J Biol Chem. 2001;273:12858–12862. doi: 10.1074/jbc.273.21.12858. [DOI] [PubMed] [Google Scholar]
- Liu H, Zang Y, Wang J, Wang D, Zhou C, Cai Y, Qian X. Method for quantitative proteomics research by using metal element chelated tags coupled with mass spectrometry. Anal Chem. 2006;78:6614–6621. doi: 10.1021/ac060895j. [DOI] [PubMed] [Google Scholar]
- Lopes MF, Melov S. Applied proteomics: Mitochondrial proteins and effect on function. Circ Res. 2002;90:380–389. doi: 10.1161/hh0402.105757. [DOI] [PubMed] [Google Scholar]
- Manetto V, Perry G, Tabaton M, Mulvihill P, Fried VA, Smith HT, Gambetti P, Autilio-Gambetti L. Ubiquitin is associated with abnormal cytoplasmic filaments characteristic of neurodegenerative diseases. Proc Natl Acad Sci USA. 1988;85:4501–4505. doi: 10.1073/pnas.85.12.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning-Boğ AB, McCormack AL, Li J, Uversky VN, Fink AL, Di Monte DA. The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: paraquat and alpha-synuclein. J Biol Chem. 2002;277:1641–1644. doi: 10.1074/jbc.C100560200. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Vila M, Lincoln S, McCormack A, Picciano M, LaFrancois J, Yu X, Dickson D, Langston WJ, McGowan E, Farrer M, Hardy J, Duff K, Przedborski S, Di Monte DA. Lack of nigral pathology in transgenic mice expressing human alpha-synuclein driven by the tyrosine hydroxylase promoter. Neurobiol Dis. 2001;8:535–539. doi: 10.1006/nbdi.2001.0392. [DOI] [PubMed] [Google Scholar]
- Mercken M, Fischer I, Kosik KS, Nixon RA. Three distinct axonal transport rates for tau, tubulin and other microtubule-associated proteins: evidence for dynamic interactions of tau with microtubules in vivo. J Neurosci. 1995;15:8259–8267. doi: 10.1523/JNEUROSCI.15-12-08259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HE, Stuhler K. High-performance proteomics as a tool in biomarker discovery. Proteomics. 2007;7:18–26. doi: 10.1002/pmic.200700183. [DOI] [PubMed] [Google Scholar]
- Michell AW, Lewis SJG, Foltynie T, Barker RA. Biomarkers and Parkinson’s disease. Brain. 2004;127:1693–1705. doi: 10.1093/brain/awh198. [DOI] [PubMed] [Google Scholar]
- Miksys S, Tyndale RF. Nicotine induces brain CYP enzymes: Relevance to Parkinson’s disease. J Neural Transm Suppl. 2006;70:177–180. doi: 10.1007/978-3-211-45295-0_28. [DOI] [PubMed] [Google Scholar]
- Miller N, Noble E, Jones D, Burn D. Hard to swallow: dysphagia in Parkinson’s disease. Age Ageing. 2006;35:614–618. doi: 10.1093/ageing/afl105. [DOI] [PubMed] [Google Scholar]
- Mitsumoto A, Nakagawa Y. DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radic Res. 2001;35:885–893. doi: 10.1080/10715760100301381. [DOI] [PubMed] [Google Scholar]
- Mitsumoto A, Nakagawa Y, Takeuchi A, Okawa K, Iwamatsu A, Takanezawa Y. Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radic Res. 2001;35:301–310. doi: 10.1080/10715760100300831. [DOI] [PubMed] [Google Scholar]
- Montgomery EB, Jr, Koller WC, LaMantia TJ, Newman MC, Swanson-Hyland AW, Kaszniak AW, Lyons K. Early detection of probable idiopathic Parkinson’s disease I. Development of a diagnostic test battery. Mov Disord. 2000a;15:467–473. [PubMed] [Google Scholar]
- Montgomery EB, Jr, Lyons K, Koller WC. Early detection of probable idiopathic Parkinson’s disease II. A prospective application of a diagnostic test battery. Mov Disord. 2000b;15:474–478. doi: 10.1002/1531-8257(200005)15:3<474::AID-MDS1008>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Moseley MA. Current trends in differential expression proteomics: Isotopically coded tags. Trends Biotechnol. 2001;19:S10–S16. doi: 10.1016/S0167-7799(01)01793-0. [DOI] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Müller M. SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- Murakami T, Shoji M, Imai Y, Inoue H, Kawarabayashi T, Matsubara E, Harigaya Y, Sasaki A, Takahashi R, Abe K. Pael-R is accumulated in Lewy bodies of Parkinson’s disease. Ann Neurol. 2004;55:439–442. doi: 10.1002/ana.20064. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Bossy-Wetzel E, Burns K, Fadel MP, Lozyk M, Goping IS, Opas M, Bleackley RC, Green DR, Michalak M. Changes in endoplasmic reticulum luminal environment affect cell sensitivity to apoptosis. J Cell Biol. 2000;150:731–740. doi: 10.1083/jcb.150.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Muller V, Gorner K, Kretzschmar HA, Haas C, Kahle PJ. Pathological properties of the Parkinson’s disease associated protein DJ-1 in alpha-synucleinopathies and tauopathies: relevance for multiple system atrophy and Pick’s disease. Acta Neuropathol (Berl) 2004;107:489–496. doi: 10.1007/s00401-004-0834-2. [DOI] [PubMed] [Google Scholar]
- Nissbaum RL, Ellis C. Alzheimer’s disease and Parkinson’s disease. N Engl J Med. 2003;348:1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- Nunez MT, Osorio A, Tapia V, Vergara A, Mura CV. Iron-induced oxidative stress up-regulates calreticulin levels in intestinal epithelial (Caco-2) cells. J Cell Biochem. 2001;82:660–665. doi: 10.1002/jcb.1194. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Olanow CW, Tatton WG. Etiology and pathogenesis of Parkinson’s disease. Annu Rev Neurosci. 1999;22:123–144. doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Andersen JR, Nielsen PA, Nielsen ML, Figeys D, Mann M, Wisniewski JR. HysTag–a novel proteomic quantification tool applied to differential display analysis of membrane proteins from distinct areas of mouse brain. Mol Cell Proteomics. 2004;3:82–92. doi: 10.1074/mcp.M300103-MCP200. [DOI] [PubMed] [Google Scholar]
- Onn SE, Mann M. Mass spectrometry-based proteomics turn quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Opiteck GJ, Lewis KC, Jorgenson JW, Anderegg RJ. Comprehensive on-line LC/LC/MS of proteins. Anal Chem. 1997;69:1518–1524. doi: 10.1021/ac961155l. [DOI] [PubMed] [Google Scholar]
- Paciello O, Wojcik S, Engel WK, McFerrin J, Askanas V. Parkin and its association with alpha-synuclein and AbetaPP in inclusion-body myositis and AbetaPP—overexpressing cultured human muscle fibers. Acta Myol. 2006;25:13–22. [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Pan S, Rush J, Peskind ER, Galasko D, Chung K, Quinn J, Jankovic J, Leverenz JB, Zabetian C, Pan C, Wang Y, Oh JH, Gao J, Zhang J, Montine T, Zhang J. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7:720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Pannese E, Procacci P, Ledda M. Ultrastructural localization of actin in then cell biology of rat spinal ganglion neurons. Anat Embryol. 1996;194:527–531. doi: 10.1007/BF00187466. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Parker WD, Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- Parker WD, Parks JK, Swerdlow RH. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Res. 2008;1189:215–218. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali C, Fialka I, Huber LA. Preparative two-dimensional gel electrophoresis of membrane proteins. Electrophoresis. 1997;18:2573–2581. doi: 10.1002/elps.1150181413. [DOI] [PubMed] [Google Scholar]
- Paweletz CP, Trock B, Pennanen M, Tsangaris T, Magnant C, Liotta LA, Petricoin EF., III Proteomic patterns of nipple aspirate fluids obtained by SELDI-TOF: potential for new biomarkers to aid in the diagnosis of breast cancer. Dis Markers. 2001;17:301–307. doi: 10.1155/2001/674959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M, Corti O, Jacquier S, Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J Neurochem. 2005;95:1259–1276. doi: 10.1111/j.1471-4159.2005.03442.x. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Praprotnik D, Smith MA, Richey PL, Vinters HV, Perry G. Filament heterogeneity within the dystrophic neurites of senile plaques suggests blockage of fast axonal transport in Alzheimer’s disease. Acta Neuropathol. 1996;91:226–235. doi: 10.1007/s004010050420. [DOI] [PubMed] [Google Scholar]
- Racette BA, Tabbal SD, Jennings D, Good L, Perlmutter JS, Evanoff B. Prevalence of parkinsonism and relationship to exposure in a large sample of Alabama welders. Neurology. 2005;64:230–235. doi: 10.1212/01.WNL.0000149511.19487.44. [DOI] [PubMed] [Google Scholar]
- Rego AC, Oliveira CR. Mitochondrial dysfunction and reactive oxygen species in excitotoxicity and apoptosis: implications for the pathogenesis of neurodegenerative diseases. Neurochem Res. 2003;28:1563–1574. doi: 10.1023/a:1025682611389. [DOI] [PubMed] [Google Scholar]
- Reinders J, Lewandrowski U, Moebius J, Wagner Y, Sickmann A. Challenges in mass spectrometry-based proteomics. Proteomics. 2004;4:3686–3703. doi: 10.1002/pmic.200400869. [DOI] [PubMed] [Google Scholar]
- Richard S, Brion J-P, Couck AM, Flament-Durand J. Accumulation of smooth endoplasmic reticulum in Alzheimer’s disease: new morphological evidence of axoplasmic flow disturbances. J Submicrosc Cytol Pathol. 1989;21:461–467. [PubMed] [Google Scholar]
- Riederer P, Wuketich S. Time course of nigrostriatal degeneration in parkinson’s disease: a detailed study of influential factors in human brain amine analysis. J Neural Transm. 1976;38:277–301. doi: 10.1007/BF01249445. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Quantitative proteomics: a review of different methodologies. Eur J Mass Spectrom. 2004;10:335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- Romeo M, Espina V, Lowenthal M, Espina BH, Petricoin EF, Liotta LA. CSF proteome: a protein repository for potential biomarker identification. Expert Rev Proteomics. 2005;2:57–70. doi: 10.1586/14789450.2.1.57. [DOI] [PubMed] [Google Scholar]
- Ross GW, Abbott RD, Petrovitch H, Morens DM, Grandinetti A, Tung KH, Tanner CM, Masaki KH, Blanchette PL, Curb JD, Popper JS, White LR. Association of coffee and caffeine intake with risk of Parkinson’s disease. JAMA. 2000;283:2674–2679. doi: 10.1001/jama.283.20.2674. [DOI] [PubMed] [Google Scholar]
- Schapira AHV, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial dysfunction in neurodegenerative disorders. Biochem Biophys Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Disease modification in Parkinson’s disease. Lancet Neurol. 2004;3:362–368. doi: 10.1016/S1474-4422(04)00769-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- Schulenborg T, Schmidt O, van Hall A, Meyer HE, Hamacher M, Marcus K. Proteomics in neurodegeneration—disease driven approaches. J Neural Transm. 2006;113:1055–1073. doi: 10.1007/s00702-006-0512-8. [DOI] [PubMed] [Google Scholar]
- Seniuk NA, Tatton WG, Greenwood CE. Dose-dependent destruction of the coeruleus-cortical and nigral-striatal projections by MPTP. Brain Res. 1990;527:7–20. doi: 10.1016/0006-8993(90)91055-l. [DOI] [PubMed] [Google Scholar]
- Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Internat. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shahani N, Brandt R. Functions and malfunctions of the tau-proteins. Cell Mol Life Sci. 2002;59:1668–1680. doi: 10.1007/PL00012495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Cookson M. Mitochondria and dopamine: new insights into recessive parkinsonism. Neuron. 2004;43:301–304. doi: 10.1016/j.neuron.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheta EA, Appel SH, Goldknopf IL. 2-D gel blood serum biomarkers reveal differential clinical proteomics of the neurodegenerative diseases. Expert Rev Proteomics. 2006;3:45–62. doi: 10.1586/14789450.3.1.45. [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Watts RL, Juncos JL, Torroni A, Wallace DC. Mitochondrial oxidative phosporylation defects in Parkinson’s disease. Ann Neurol. 1991;30:332–339. doi: 10.1002/ana.410300304. [DOI] [PubMed] [Google Scholar]
- Simpson RJ. Purifying proteins for proteomics: a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2003. [Google Scholar]
- Sjögren M, Davidsson P, Tullberg L, Minthon L, Wallin A, Wikkelso C, Granérus AK, Vanderstichele H, Vanmechelen E, Blennow K. Both total and phosphorylated tau are increased in Alzheimer’s disease. J Neurol Neurosurg Psych. 2001;70:624–630. doi: 10.1136/jnnp.70.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold K, Svensson M, Nilsson A, Zhang X, Nydahl K, Caprioli RM, Svenningsson P, Andren PE. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res. 2006;5:262–269. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM-Y, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Spiro RG, Zhu Q, Bhoyroo V, Soling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J Biol Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- Stühler K, Joppich C, Stephan C, Jung K, Müller M, Schmidt O, van Hall A, Hamacher M, Urfen W, Meyer HE, Marcus K. Pilot study of the Human Proteome Organisation Brain Proteome Project: applying different 2-DE techniques to monitor proteomic changes during murine brain development. Proteomics. 2006;6:4899–4913. doi: 10.1002/pmic.200600089. [DOI] [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, Butterfield DA. Proteomics analysis of the Alzheimer’s disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- Svensson M, Skold K, Nilsson A, Falth M, Svenningsson P, Andren PE. Neuropeptidomics: expanding proteomics downwards. Biochem Soc Trans. 2007;35:588–593. doi: 10.1042/BST0350588. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Parks JK, Miller SW, Tuttle JB, Trimmer PA, Sheehan JP, Bennett JP., Jr Origin and functional consequences of the complex I defect in Parkinson’s disease. Ann Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The deep-brain stimulation for Parkinson study group. Deep-brain stimulation of the subthalamic nucleus or the pars interna of the globus pallidus in Parkinson’s disease. N Eng J Med. 2001;345:956–963. doi: 10.1056/NEJMoa000827. [DOI] [PubMed] [Google Scholar]
- Tinazzi M, Del Vesco C, Fincati E, Ottaviani S, Smania N, Moretto G, Fiaschi A, Martino D, Defazio G. Pain and motor complications in Parkinson’s disease. J Neurol Neurosurg Psych. 2006;77:822–825. doi: 10.1136/jnnp.2005.079053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribl F, Marcus K, Bringmann G, Meyer HE, Gerlach M, Riederer P. Proteomics of the human brain: sub-proteomes might hold the key to handle brain complexity. J Neural Transm. 2006a;113:1041–1054. doi: 10.1007/s00702-006-0513-7. [DOI] [PubMed] [Google Scholar]
- Tribl F, Marcus K, Meyer HE, Bringmann G, Gerlach M, Riederer P. Subcellular proteomics reveals neuromelanin granules to be a lysosome-related organelle. J Neural Transm. 2006b;113:741–749. doi: 10.1007/s00702-006-0452-3. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Lee VM. Aggregation of neurofilament and alpha-synuclein proteins in Lewy bodies: implications for the pathogenesis of Parkinson disease and Lewy body dementia. Arch Neurol. 1998;55:151–152. doi: 10.1001/archneur.55.2.151. [DOI] [PubMed] [Google Scholar]
- Truong DD, Bhidayasiri R, Wolters E. Management of non-motor symptoms in advanced Parkinson disease. J Neurol Sci. 2007;266:216–228. doi: 10.1016/j.jns.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Tseng H-M, Su PC, Liu H-M, Liou H-H, Yen R-F. Bilateral subthalamotomy for advanced Parkinson disease. Surg Neurol. 2007;68:S43–S50. doi: 10.1016/j.surneu.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Utal AK, Stopka AL, Roy M, Coleman PD. PEP-19 immunohistochemistry defines the basal ganglia and associated structures in the adult human brain and is dramatically reduced in Huntington’s disease. Neuroscience. 1998;86:1055–1063. doi: 10.1016/s0306-4522(98)00130-4. [DOI] [PubMed] [Google Scholar]
- Utton MA, Connell J, Asuni AA, Van Slegtenhorst M, Hutton M, De Silva R, Lees AJ, Miller CC, Anderton BH. The slow axonal transport of the microtubule-associated protein tau and the transport rates of different isoforms and mutants in cultured neurons. J Neurosci. 2002;22:6394–6400. doi: 10.1523/JNEUROSCI.22-15-06394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren FGG, Bergeron JJM, Vandesande F, Arckens L, Quirion R. Proteomic approaches in brain research and neuropharmacology. Eur J Pharmacol. 2004;500:385–398. doi: 10.1016/j.ejphar.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Vila-Carriles WH, Zhou ZH, Bubien JK, Fuller CM, Benos DJ. Participation of the chaperone Hsc70 in the trafficking and functional expression of ASIC2 in glioma cells. J Biol Chem. 2007;282:34381–34391. doi: 10.1074/jbc.M705354200. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O, Schober A, Krieglstein K. Genes, proteins, and neurotoxins involved in Parkinson’s disease. Prog Neurobiol. 2004;73:151–177. doi: 10.1016/j.pneurobio.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Waragai M, Nakai M, Wei J, Fujita M, Mizuno H, Ho G, Masliah E, Akatsu H, Yokochi F, Hashimoto M. Plasma levels of DJ-1 as a possible marker for progression of sporadic Parkinson’s disease. Neurosci Lett. 2007;425:18–22. doi: 10.1016/j.neulet.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Yates JR. Analysis of the microbial proteome. Curr Opin Microbiol. 2000;3:292–297. doi: 10.1016/s1369-5274(00)00092-8. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- Wiederkehr F. Analysis of cerebrospinal fluid proteins by electrophoresis. J Chromatogr. 1991;569:281–296. doi: 10.1016/0378-4347(91)80234-4. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Ochstrasser DF, Williams KL. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19–50. doi: 10.1080/02648725.1996.10647923. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Wolters EC, Francot C, Bergmans P, Winogrodzka A, Booij J, Berendse HW, Stoof HC. Preclinical (premotor) Parkinson’s disease. J Neurol. 2000;247:II103–II109. [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, Yates JR. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- Wood-Allum CA, Barber SC, Kirby J, Heath P, Holden H, Mead R, Higginbottom A, Allen S, Beaujeux T, Alexson SE, Ince PG, Shaw PJ. Impairment of mitochondrial anti-oxidant defence in SOD1-related motor neuron injury and amelioration by ebselen. Brain. 2006;128:1686–1706. doi: 10.1093/brain/awl118. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr Opin Mol Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- Wu TL. Two-dimensional difference gel electrophoresis. Methods Mol Biol. 2006;328:71–95. doi: 10.1385/1-59745-026-X:71. [DOI] [PubMed] [Google Scholar]
- Xun Z, Sowell RA, Kaufman TC, Clemmer DE. Lifetime proteomic profiling of an A30P α-synuclein drosophila model of Parkinson’s disease. J Proteome Res. 2007;6:3729–3738. doi: 10.1021/pr0700504. [DOI] [PubMed] [Google Scholar]
- Yao H, Sem DS. Cofactor fingerprinting with STD NMR to characterize proteins of unknown function: identification of a rare cCMP cofactor preference. FEBS Lett. 2005;579:661–666. doi: 10.1016/j.febslet.2004.11.110. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman E, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson’s disease. Proc Natl Acad Sci USA. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang L, Toy DP, Hancock WS, Sgroi DC, Karger BL. J Prot Res. 2004;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- Zetterberg H, Ruetschi U, Portelius E, Brinkmalm G, Andreasson U, Blennow K, Brinkmalm A. Clinical proteomics in neurodegenerative disorders. Acta Neurol Scand. 2008 doi: 10.1111/j.1600-0404.2007.00985.x. (in press) [DOI] [PubMed] [Google Scholar]
- Zigmond MJ, Burke RE. Pathophysiology of Parkinson’s disease. In: Davis KL, Coyle J, Charney D, Nemeroff C, editors. Fifth Generation of Progress. Lippincott Williams & Wilkins; Philadelphia: 2002. pp. 1781–1794. [Google Scholar]
- Zhang B, Higuchi M, Yoshiyama Y, Ishihara T, Forman MS, Martinez D, Joyce S, Trojanowski JQ, Lee VM. Retarded axonal transport of R406 W mutant tau in transgenic mice with a neurodegenerative tauopathy. J Neurosci. 2004;24:4657–4667. doi: 10.1523/JNEUROSCI.0797-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J. Analysis of α-synuclein associated proteins by quantitative proteomics. J Biol Chem. 2004;279:39155–39164. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T α-synuclein toxicity. J Biol Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu M, Wilson MA, Petsko GA, Fink LA. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J Mol Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Zhu M, Qin ZJ, Hu D, Munishkina LA, Fink AL. Alpha-synuclein can function as an antioxidant preventing oxidation of unsaturated lipid in vesicles. Biochemistry. 2006;45:8135–8142. doi: 10.1021/bi052584t. [DOI] [PubMed] [Google Scholar]