Abstract
The underlying sensing mechanism of single–cell–based integrated microelectrode array (IMA) biosensors was investigated via experimental and modeling studies. IMA chips were microfabricated and single–cell–level manipulation was achieved through surface chemistry modification of IMA chips. Individual fibroblast cells (NIH3T3) were immobilized on either lysine–arginine–glycine–aspartic acid (KRGD) short peptide–modified or fibronectin extracellular–cell–adhesion–molecule–modified microelectrodes to record the impedance variations of cell–electrode heterostructure over a frequency range of 1 to 10 kHz. By fitting experimental data to an application–specific single–cell–level equivalent circuit model, important sensing parameters, including specific cell membrane capacity, cell membrane resistivity, and averaged cell–to–substrate separation, were determined. It was demonstrated that biofunctionalization of planar microelectrode surface by covalently linking short peptides or fibronectin molecules could achieve strong or tight cell adhesion (with an estimated averaged cell–to–substrate separation distance of 11–16 nm), which, in turn, improves the transduced electrical signal from IMA chips. Analyses on frequency–dependent characteristics of single– cell–covered microelectrode impedance and of IMA sensor circuitry response have revealed an addressable frequency band wherein electrical properties of single cells can be distinctively determined and monitored for cellular biosensing applications. The presented work addresses some major limitations in single–cell–based biosensing schemes, i.e. the manipulation of a single cell, the transduction of weak biological signals, and the implementation of a proper model for data analysis, and demonstrates the potential of IMA devices as single–cell biosensors.
Keywords: Single–cell, Microelectrode, Surface Modification, Sensor, Impedance, Equivalent Circuit Model
1. Introduction
Characterizing the frequency responses of the cell–electrode heterojunction impedance has been the fundamental mechanism of signal detection in electric cell substrate impedance sensing (ECIS) (Xiao et al., 2002) and electrochemical impedance spectroscopy (EIS) (Tlili et al., 2003) techniques. In addition, alterations to the impedance characteristics of cell–covered microelectrodes caused by exposing living cells, utilized as sensing bio–elements, to biologically active substances (Pancrazio et al., 1999; Tlili et al., 2003) render potential applications of single–cell–based biosensors in drug/bacteria detection and identification. In a traditional IMA biosensor device, a large cell population is often cultured over one large electrode because of the lack of control over cell isolation and patterning on the sensor substrate. Although averages of cell properties, such as proliferation, motility, and cell–cell separation, can be achieved over the cell population by traditional IMA biosensing schemes, it is impossible to examine individual cells and precisely monitor changes in cell membrane properties of individual cells. In order to obtain a more fundamental understanding of cell nature and cell metabolism, studies should also be emphasized on the single–cell level. However, there exist a number of challenges in single– cell–level studies including the time–consuming and low–yield manipulation of a single cell, the transduction of weak biological signals, and the implementation of proper modeling on a single– cell level to analyze the experimental data.
In a single–cell–based biosensor, it is very important that individual cells should be isolated and selectively immobilized onto individual cell–sized electrodes in a repeatable and controllable manner. Biological signals from impedance sensors are usually weak and occasionally unobservable even in multiple–cell level spectroscopy (Tlili et al., 2003). A significant factor that accounts for the observed weak signals is poor cell adhesion on the electrode (Lo and Ferrier, 1998; Tlili et al., 2003). Additionally, poor cell adhesion also results in larger paracellular currents, i.e., currents that flow between the bottom of the cell and the top surface of the substrate, and consequently, the measured impedance–related signal does not contain much information on the cell itself, especially the information about the cell membrane (Haung et al., 2004; Lo and Ferrier, 1998).
Efforts have been made to achieve tight cell–electrode adhesion by selectively and specifically modifying the surface chemistry of specially designed microelectrodes with dimensions comparable to those of single cells (Asphahani et al., 2008). This approach renders successful isolation and immobilization of individual cells on the microelectrodes as well as facilitating the precise control over cell adhesion, cell viability, and cell morphology in single– cell biosensors (Nam et al., 2004; Veiseh et al., 2004; Veiseh et al., 2007).
In this work, an array of single–cell–sized microelectrodes (30 µm in diameter) were fabricated and mouse fibroblast cells (NIH3T3) were immobilized onto microelectrodes through a lysine–arginine–glycine–aspartic acid (KRGD) short peptide–mediated or fibronectin extracellular–adhesion–molecule–mediated natural cell adhesion process (Asphahani et al., 2008). The impedance spectrum of single–NIH3T3–covered surface–chemistry–modified microelectrodes was recorded in the frequency range 1 to 10 kHz and numerically data–fitted with the equivalent circuit model (ECM) in order to extract important sensing parameters such as the specific cell membrane impedance and averaged cell–to–substrate separation distance. The effect of cell–to–substrate separation on sensor response was analyzed in order to optimize the transduction of cell–membrane–activity–related signal from the NIH3T3–based single–cell biosensor. The operating frequency band, within which the sensor achieves high sensitivity to cellular mechanisms/activities from a single NIH3T3 cell, was also determined from ECM simulations.
2. Material and Methods
2.1. Single–Cell Integrated Microelectrode Array Chip Fabrication
The single–cell integrated microelectrode array (IMA) chip used in the present study consists of an array of sensing/detecting circular–shaped planar Au microelectrodes (30 µm in diameter) and two large–dimension counter electrodes deposited on a glass substrate (Asphahani and Zhang, 2007). The sensing/detecting microelectrodes and counter electrodes are electrically linked to bonding/measurement pads located at the chip perimeter via Au interconnections. The interconnection metal is insulated from the tissue culture media with SiO2 overcoating. The planar microelectrodes are exposed to tissue culture media through small separate wells or via holes (see supplementary Fig. 1 for cross–sectional schematic of single–cell IMA chip).
The device fabrication process is as follows: Au microelectrodes and interconnections were patterned over 4” glass wafers with standard microfabrication techniques, which includes photolithography, physical vapor metal deposition (PVD), and a lift–off process. A thin titanium layer (~125Å) was evaporated prior to Au deposition (~5000 Å) for adhesion enhancement. The patterned Au microelectrodes were then buried underneath a layer of SiO2 (~1 µm in thickness) deposited via plasma enhanced chemical vapor deposition (PECVD). The surface of SiO2 was subsequently annealed at 250 °C for ~2 hrs in an oxygen environment. Next, openings or via holes were created over sensing microelectrodes and the large counter electrodes by reactive–ion etching (RIE). Finally, a second Au layer (~1000 Å) was deposited on top of the exposed electrodes in order to produce a fresh amorphous Au surface that was free of etching–induced surface impurities. This second Au coating was found to lower the cell–free baseline impedance of Au microelectrodes significantly as well as yielding high degree of cell coverage on the microelectrode after surface modifications. Thermal annealing has been avoided in producing impurity–free surface as it smoothens the surface of gold (Porath et al., 1996) and reduces the effective sensing surface of the planar electrode (Borkholder, 1998), which consequently results in much higher cell–free baseline impedance. Finally, individual chips were snapped off from the wafer through dicing, and cylindrical plastic wells, function as tissue culture media reservoirs during the sensor operation, were glue–mounted to enclose the microelectrodes of the chip.
2.2. Modification of Microelectrode Surface Chemistry
In order to immobilize individual cells on sensing/detecting Au microelectrodes, surfaces of IMA chips were modified or patterned. Surface topologies of Au electrodes and of SiO2 overcoating on IMA chips were used as bases to attain patterned surface chemistry. The patterning method (see supplementary materials) followed a previously reported method (Asphahani et al., 2008; Veiseh et al., 2004) with minor modifications.
2.3. Single–Cell Immobilization on Sensing/Detecting Microelectrodes
NIH3T3 cell line was cultured in 75 cm2 flasks (see supplementary materials). For cell adhesion, 1.0 mL of NIH3T3 cells at a concentration of 100 × 103 cells/mL was prepared and plated onto the KRGD– or fibronectin–patterned IMA chips. The cells were allowed to adhere to the substrates for 24 hrs under the standard culture conditions. Prior to the impedance measurements, NIH3T3–patterned IMA chips were inspected under a brightfield reflectance microscope (Nikon 80i Upright Microscope, Melville, NY) for cell attachment and coverage on the sensing electrodes. After electrical measurements were completed, the cells were fixed with Karnovsky’s fixative for 1 hr at room temperature for further optical imaging.
2.4. Experimental Setup and Measurement
The cell–electrode heterojunction impedance was characterized with voltage divider circuitry (Supplementary Fig. 2). The sensing microelectrode, on which a single NIH3T3 cell was immobilized, was electrically connected to a function generator (Agilent 33220A) through a current–limiting 1 MΩ resistor and the large counter electrode was grounded. All the contacts were made on Au bonding/measurement contact pads of the IMA chip using low parasitic capacitance tungsten probes. A 100 mV peak–to–peak sinusoidal voltage was applied from the function generator that injected a low–intensity ac probe current signal, clamped by the limiting resistor, into the interface between the sensing microelectrode and the testing cell. The resulting differential potential (comprising both magnitude and phase components) across the sensing and grounded counter electrodes of the IMA chip was lock–in detected with a phase sensitive digital lock–in amplifier (Stanford Research SR–810) for several frequencies (1 kHz, 2 kHz, 4 kHz, 6 kHz, 8 kHz, and 10 kHz), which provided an accurate measure of the NIH3T3’s membrane impedance and cell–electrode heterojunction impedance. The frequency band of interest (1–10 kHz) was determined based on the frequency–dependent dielectric properties of the biological membrane ensuring that the cell membrane impedance was measured.
The impedance characterization was performed with a four–step procedure as follows. First, the spreading resistance of the microelectrodes was measured in the culture medium prior to cell seeding in order to obtain the baseline data. For the second and third steps, the impedance measurements were carried out, respectively, after the surfaces of the microelectrodes were coated with SAM and after subsequent biofunctionalizion via covalently linking fibronectin proteins or KRGD peptides. The final measurement was conducted when the surface– chemistry–modified microelectrodes were covered with a single NIH3T3 cell after the cell seeding. The entire set of measurements (cell–free baseline, cell–free SAM–coated, cell–free surface–chemistry–modified, and single–NIH3T3–covered microelectrode) was recorded separately from a set of chips that consistently demonstrated approximately the same cell–free baseline impedance characteristics.
3. Experimental Results
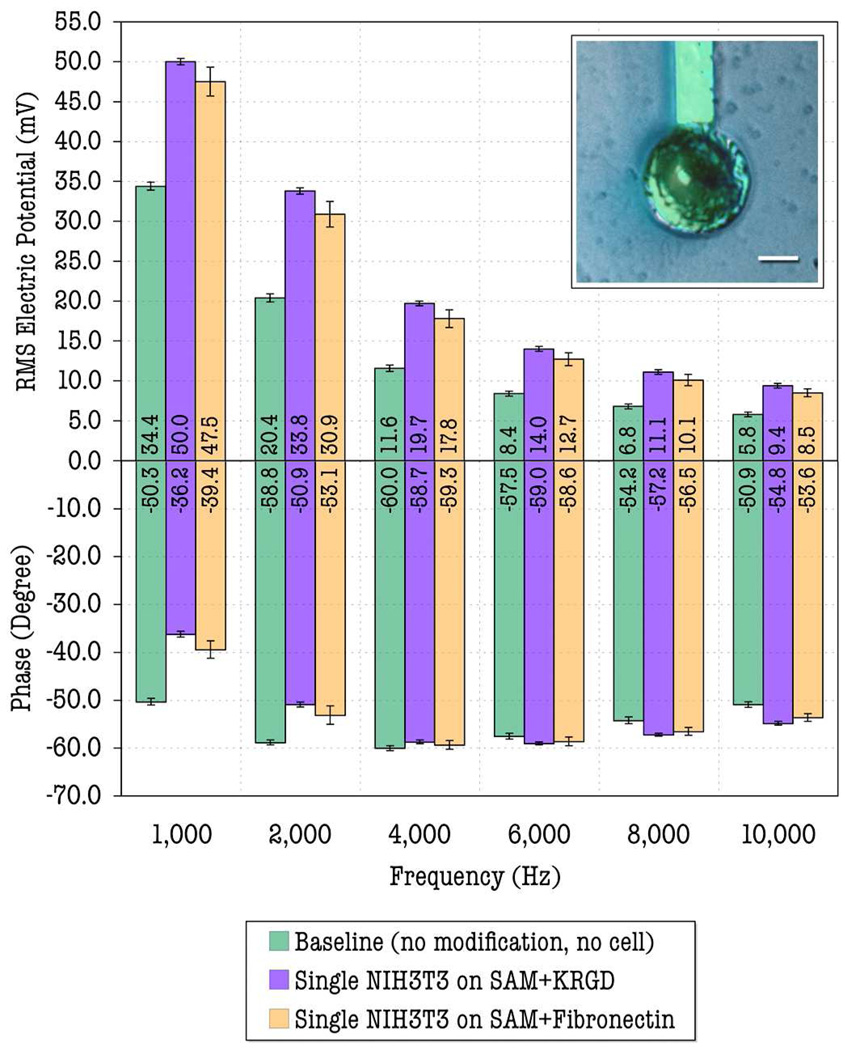
An optical photomicrograph of a single NIH3T3, immobilized on 30 µm diameter covalently–linked–KRGD–modified microelectrode, is shown in Fig. 1 (inset). The image shows the high degree of cell spreading and coverage achieved on the modified microelectrode. In addition, the picture also suggests that the PEG–modified SiO2 surface (surrounding area) facilitated cell–to–cell isolation by repelling a single NIH3T3 cell onto the modified microelectrode. The experimental measurements were carried out based on the protocols described in section 2.4. The recorded root–mean–square (RMS) voltages across the 30 µm– diameter sensing microelectrode and large counter–electrode (of both magnitude and phase), which reflect impedances of the sensing electrode for different conditions at six different frequencies (1 kHz, 2 kHz, 4 kHz, 6 kHz, 8 kHz, and 10 kHz), are plotted in Fig. 1. Each set of data is averaged over eight measurements taken from different electrodes. The reproducibility of the experimental data mainly depends on the degree of cell coverage and adhesion on the sensing microelectrode, which can be precisely controlled by our surface chemistry modification approaches (see supplementary materials) and elsewhere (Asphahani et al., 2008; Veiseh et al., 2004). As consistent and reproducible data are observed with higher degree of cell coverage, the impedance–related voltages were recorded from the electrodes on which the cell covers approximately > 95% of the electrode area. No significant difference in the cell coverage was observed before and after the recordings.
Fig. 1.
Measured RMS voltage signals across sensing/detecting microelectrodes and large counter electrodes for different conditions at 6 operating frequencies. Inset: a single NIH3T3 cell immobilized on the 30 µm–diameter covalently–linked–KRGD– modified microelectrode (The scale bar is 10 µm).
4. Data Fitting with the Equivalent Circuit Model
4.1. Modeling the cell–electrode heterojunctions with the area–contact–model–based distributive circuit network
It is well known that the cell membrane exhibits dielectric and insulating properties (capacitance and resistance) (Borkholder, 1998; Pancrazio et al., 1999). The impedance of the cell membrane can be modeled as a parallel combination of a capacitor that arises from the phospholipid bilayer and a variable resistor that represents the transmembrane ion transport proteins (Asphahani and Zhang, 2007). In addition to the cell membrane impedance, the cytoplasm is conductive and can be modeled as a variable resistor (Mossop et al., 2004; Pilwat and Zimmermann, 1985). It is also important to take into account of the planar electrode impedance in modeling of the cell–electrode impedance. When an electrode is immersed in conductive electrolyte solution such as tissue culture media, a reasonably approximated equivalent circuit model for the microelectrode–electrolyte interfacial impedance is a parallel combinatory circuit of a constant phase element (CPE) that represents the double–layer capacitance and a resistor that represents the charge transfer between the electrode and electrolyte solution (Franks et al., 2005; Lasseter et al., 2004; Panke et al., 2008).
The circular–shaped planar microelectrodes were fabricated in the present study and used in the model due to simple geometry and ease of analysis. It has been reported that there exists a very thin layer of tissue culture media between the bottom of the cell and the top surface of the electrode when a single cell covers over a microelectrode, (Braun and Fromherz, 2004; Haung et al., 2004; Iwanaga et al., 2001; Lo et al., 1995; Lo and Ferrier, 1998). When a probe current is injected through the microelectrode, only a portion of it will flow through the cell (transcellular current) while the rest of it will shunt through the cleft region, i.e., thin tissue culture media layer between the bottom of the cell and the substratum (supplementary Fig. 3). In the modeling effort, we use the first–order approximate of the radial shunt (or paracellular) currents flowing outwards from the center of the microelectrode due to the centrosymmetry (Lo and Ferrier, 1998). Constriction of shunt current path can be achieved through tight cell adhesion for the resistance of the shunt current path, i.e., the sealed resistance, is inversely proportional to the averaged cell–to–substrate separation distance (Asphahani et al., 2008; Lo and Ferrier, 1998).
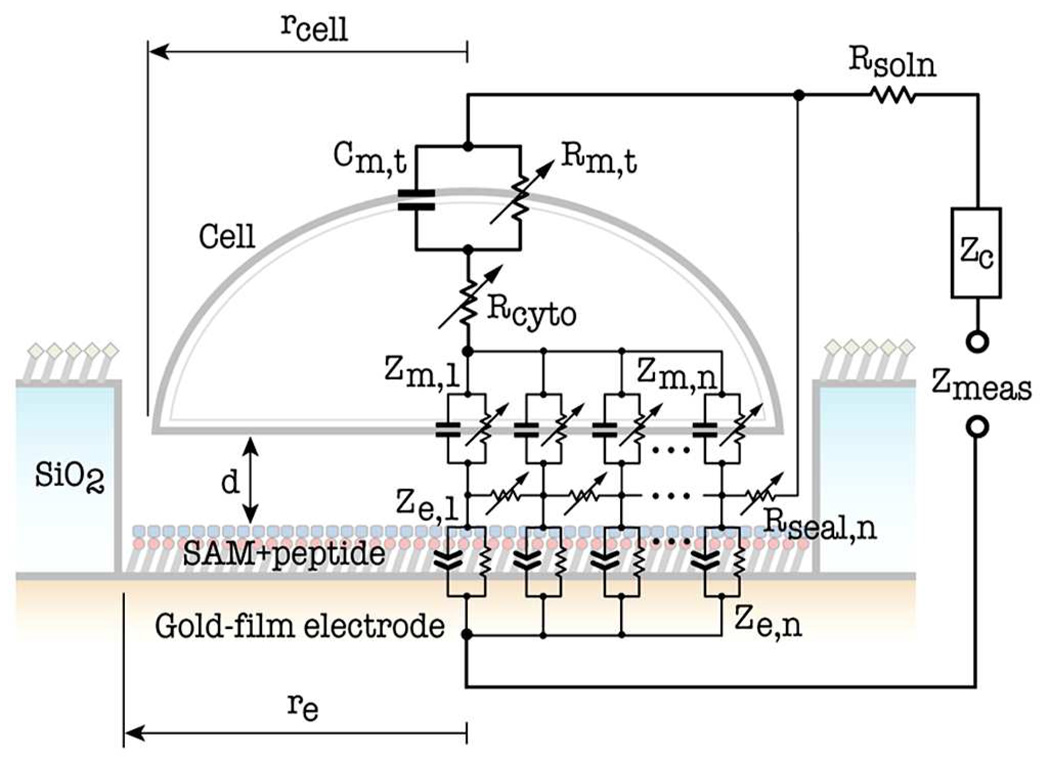
An area–contact model, described by weighted circuit elements in a distributed circuit network, is employed in the development of the equivalent circuit for a single–cell–covered microelectrode, which takes into account of shunt/paracellular currents, transcellular currents, and the probe current from the microelectrode as shown in Fig. 2 (Borkholder, 1998; Gordon et al., 1989; Weis et al., 1996; Weis and Fromherz, 1997). The cell–electrode heterojunction in this model is partitioned into a series of peripheral domains or segments with microelectrode and cell radius increment (Δre and Δrcell) and denoted with the subscript number n. Each domain or segment is represented by weighted equivalent–circuit components (Zn’s, Cn’s and Rn’s). The counter–electrode impedance (Zc) is negligible due to its very large surface area as compared to the sensing microelectrode (Borkholder, 1998; Lo et al., 1995; Lo and Ferrier, 1998; Xiao et al., 2002). The following assumptions were made in the derivation of formulas for weighted equivalent–circuit elements (see supplementary Table 1): (1) the cell is centered over and completely covers the surface–chemistry–modified microelectrode, and therefore, the immobilized cell’s radius (rcell) is equal to the planar microelectrode’s radius (re); (2) the current spreads through the cytoplasm of the immobilized cell.
Fig. 2.
Equivalent circuit of single–cell–covered microelectrode based on the area–contact model.
4.2. Data Fitting
At the given frequency, sinusoidal voltage input, and current clamping resistance, the RMS voltage across the sensing microelectrode and counter–electrode, which is also a function of the physical and electrical parameters (Supplementary Table 1 and 2), were derived from the area– contact–model–based equivalent circuit network shown in Fig. 2. These model parameters were then extracted by iteratively least–square–fitting the function to the measured impedance–related RMS output voltages shown in Fig. 1 (Bodmer et al., 2005). A multiple–step–fitting approach was employed in our multi–parameter extraction process in order to minimize the errors (Gordon et al., 1989; Lo et al., 1995). For computing efficiency, the number of weighted segments (n) used in the area–contact model was 5. The estimated model parameters and their respective error values (Supplementary Table 2) give the best fit of the model to the experimental measured data since they were estimated based on both the reproducibility of experimental measured data and the stability of the model parameters (Bodmer et al., 2005; Lo and Ferrier, 1998). The cell membrane resistivity (ρm), cell membrane capacity (cm), and cell–to–substrate separation distance (d), are of particular importance among all the equivalent–circuit parameters as the accurate determination of their values is vital to the sensing function and mechanism of the single–cell IMA biosensors under study (the estimated ρm and cm of a single NIH3T3 immobilized on KRGD–modified microelectrode is 0.0312 ± 0.0016 kΩ–cm2 and 97.9 ± 4.9 µF/cm2 respectively and those of a single NIH3T3 immobilized on fibronection–modified microelectrode is 0.0286 ± 0.0014 kΩ–cm2 and 83.7 ± 4.2 µF/cm2 respectively). In other words, as the cell membrane resistivity and capacity contain information related to cell membrane activities and dynamics, alterations in the cell membrane due to the external or internal stimuli will be accurately represented by the changes in the values of these two model parameters.
5. Analysis and Discussions
5.1. Signal Enhancement via Cleft Thickness Control over Surface–Modified Sensing Microelectrodes
It is well known that the accuracy of cell–electrode impedance characterization will be enhanced effectively by reducing the “cleft” thickness at the interface. When cells are immobilized on planar metal electrodes, cell adhesion is mediated by protein molecules that protrude from the cell membrane (integrins, glycocalix) and those that are deposited on the substrate (extracellular matrix proteins). These proteins keep the lipid core of the membrane at a certain distance from the substrate, stabilizing a cleft between the cell and the chip, which is filled with electrolyte solution (Fromherz, 2003). The resulting shunt resistance that arises from the cleft may seriously degrade the actual impedance characterization of the cell membrane, and, in some cases, even dominate the measured data.
The estimated averaged cell–to–substrate separation distance for NIH3T3 cell on the surfaces of both covalently–linked–KRGD–modified and covalently–linked–fibronectin–modified microelectrodes is in the range of 11–16 nm, which is obtained via numerically data–fitting the ECM model to the experimentally recorded data in the present study (Fig. 1) and suggests very tight adhesion between the single NIH3T3 cell and the surface–chemistry–modified Au microelectrode (Braun and Fromherz, 1998; Braun and Fromherz, 2004). In comparison, the averaged cell–to–substrate separation distance for fibroblast cells and rat neural cells on the surfaces of substrates coated with physically–adsorbed fibronectin or laminin was estimated to be approximately 50–100 nm according to previous studies (Braun and Fromherz, 1998; Iwanaga et al., 2001; Garcia et al., 1997). Our result, therefore, indicates that covalently linking biocompatible peptides or cell–adhesion molecules on the substrate surface improves the cell adhesion significantly. This observation is also consistent with the previous report that covalently–linked KRGD peptides on the Au surfaces mediates a higher degree of cell spreading than physically–adsorbed KRGD peptides (Asphahani et al., 2008).
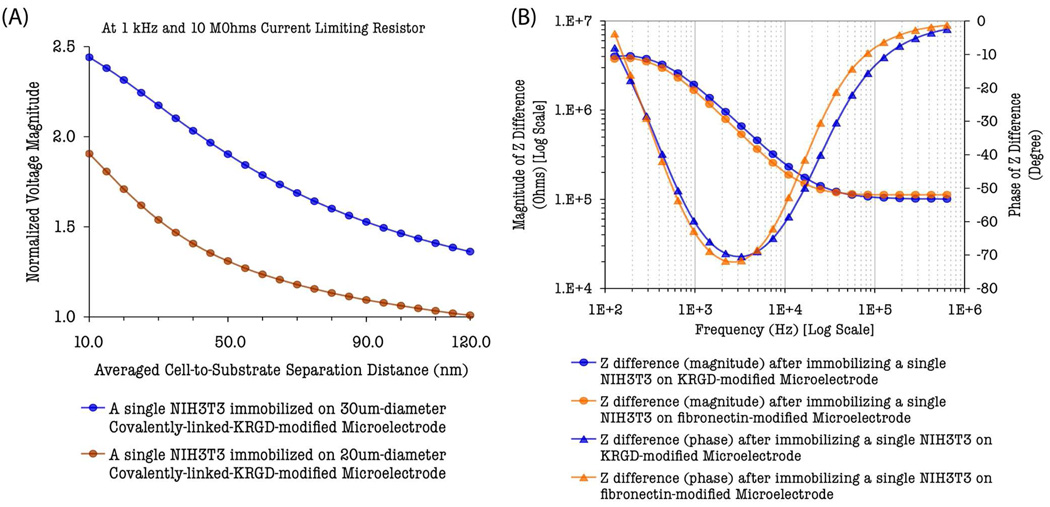
Stronger cell adhesion also improves the transduction of signal from cell–based biosensor circuitry. The shunt resistance increases with the electrode radius or cleft disc radius and is inversely proportional to the averaged cell–to–substrate separation distance according to the area–contact model. Therefore, stronger cell adhesion on the surface of the electrode results in smaller averaged cell–to–substrate separation distance that, in turn, yields larger values of shunt resistances. In order to understand and analyze the effect of cell adhesion on the single–cell– based sensor circuit response, the normalized RMS voltage magnitudes across the sensing microelectrode (20 µm or 30 µm in diameter) and the counter electrode (i.e., ν = /Vcell-covered / Vbaseline/), which represent the strength of the single–cell biosensor response, were calculated and plotted [Fig. 3(A)] for different averaged cell–to–substrate separation distances (d), ranging from 10 nm to 120 nm, under the probing condition of 1 kHz frequency, 10 MΩ current clamping resistance, and an input impedance (Zin) of 20 GΩ ‖ 25 pF for the lock–in detection (estimated ECM model parameters from supplementary Table 2 were used in the simulation). The computed plot shows that when the cell–to–substrate separation distance is reduced from 120 nm to 10 nm, the single–cell biosensor response, represented by the normalized RMS voltage magnitude, is enhanced by the factors of approximately 1.8 and 2.4 for 20 µm and 30 µm electrodes, respectively. The fact that the calculated normalized RMS voltage magnitudes for the same type of cell is smaller in the 20 µm–diameter electrode compared to that of the 30 µm– diameter electrode indicates the strong cell adhesion, and hence the minimized cell–to–substrate separation distance is absolutely essential for smaller electrodes suitable for single–cell–based sensing schemes, which utilize small–sized biological cells. A plausible explanation for this phenomenon is that cell–to–substrate separation must be dominantly smaller in order to obtain a large shunt resistance due to shortening of paracellular or shunt current paths in smaller electrodes covered with a small–sized cell. Otherwise, the injected ac current will shunt through the paracellular paths and the signal becomes hardly distinguishable from that of cell–free baseline.
Fig. 3.
Simulated plots: (A) Normalized RMS voltage magnitudes, measured across the sensing microelectrode and the counter electrode, as a function of averaged cell–to– substrate separation distance. (B) Frequency–dependent impedance difference, referenced to the cell–free baseline, after a single NIH3T3 cell was immobilized on the microelectrode (30 µm–diameter) via different surface–chemistry–mediated cell adhesion processes.
5.2. Frequency–dependent Characteristics
To investigate the frequency–dependent characteristics of the IMA–based single–cell sensing scheme, the spectrum of impedance difference between the single–NIH3T3–covered electrode and the unmodified cell–free baseline electrode was calculated using the area–contact–model–based circuit network over the frequency range between 100 Hz to 1 MHz. Fig. 3(B) plots the spectrum of the computed impedance difference (ΔZ = Zcell-covered– Zcell-free baseline), which is associated with the covalently–linked–KRGD– or –fibronectin–mediated cell adhesion of single NIH3T3 cells on 30µm–diameter microelectrodes and provides a mean of measure of the cell– electrode interfacial impedance as well as the cell membrane impedance. The frequency– dependent characteristic of the magnitudes of the impedance difference (|ΔZ|) approximates to a linear function in log scale up to the frequency of 50 kHz. This characteristic is governed by the resistive and capacitive properties of the cell membrane, the binding–specific cell adhesion molecules (KRGD or fibronectin) and their regulated cell adhesion, and the intracellular cytoplasm. The cell membrane becomes more capacitive and leaky at higher frequencies (≥ 50 kHz) and, thus, the capacitive elements of NIH3T3 are effectively short–circuited at higher frequencies (Gordon et al., 1989). The impedance change due to the present of a single NIH3T3 on the microelectrode, therefore, becomes frequency independent at high frequencies (≥ 50 kHz) as it is dominated by the resistance of cytoplasm (Rcyto). This resistive nature of the single– NIH3T3–covered microelectrode impedance at high frequencies is also revealed by the reduced phase angles of the impedance difference in the spectral diagram [Fig. 3(B)].
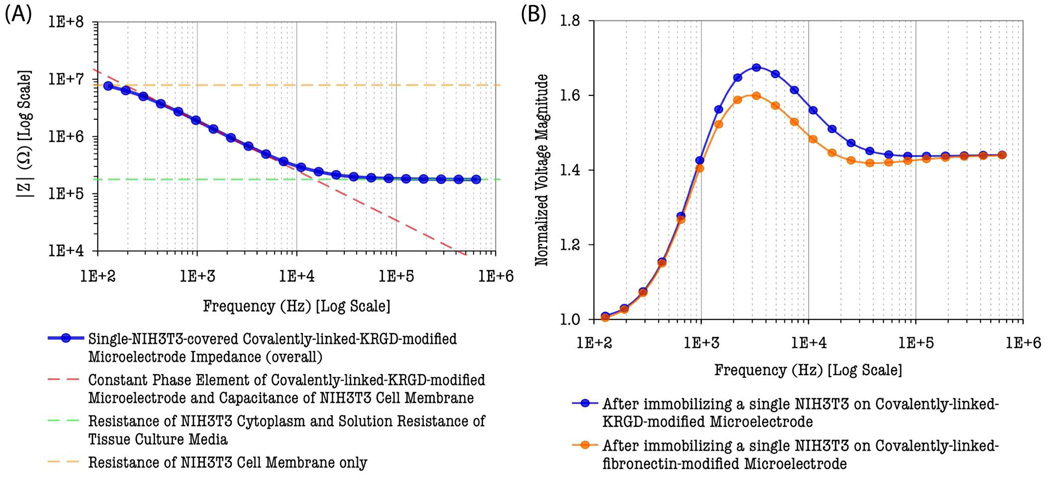
An important conclusion drawn from the spectrum analysis is that there exists a suitable frequency band within which the electrical properties of multiple cellular components become dominant in measured signals. The computed spectrum of overall impedance magnitude of single–NIH3T3–covered microelectrode in Fig. 4(A) is found to trace along the asymptotic approximation line of the NIH3T3’s membrane capacitance and the microelectrode constant phase element (CPE) within the frequency band of 100 Hz to 50 kHz. In addition, the phase shifts of the impedance due to the presence of a morphologically–controlled single NIH3T3 on the microelectrode, represented by the phase angles of the impedance difference in Fig. 3(B), are very prominent within the same frequency band. These characteristics suggest the constriction of shunt currents due to tight cell adhesion and confirm that the measured signal in this frequency range is mostly dominated by NIH3T3’s cell membrane impedance as well as the surface–modified microelectrode’s CPE. The most important aspect of these characteristics for single–cell–based sensing schemes is that real–time changes and variations in NIH3T3’s cell membrane properties (such as membrane permittivity and conductivity) due to external or internal stimuli can be transduced and revealed in this frequency band.
Fig. 4.
(A) Overall impedance magnitude spectrum of single–NIH3T3–covered covalently– linked–KRGD–modified gold microelectrode (30 µm–diameter). Dashed lines are asymptotic approximation lines representing dominant electrical properties in the measured data. (B) Characteristic voltage response of IMA sensor after single cell immobilization.
The uncovered frequency band also provides maximum sensitivity in the single–cell–based IMA sensing scheme and can be utilized to optimize the operation of the sensor circuit in real–time analysis so that the time needed for impedance recording and processing can be reduced. The optimal frequency range for the maximum sensor response and sensitivity in the present study was also investigated by simulating the frequency–dependent sensor response characteristics of the IMA sensing circuitry. The normalized RMS voltage magnitudes, ν = /Vnon–baseime/ Vbaseiine/, were calculated (with 1 MΩ current limiting resistor) over the frequency range between 100 Hz to 1 MHz and plotted for NIH3T3 cells immobilized on KRGD– or fibronectin–modified microelectrodes, respectively, as shown in Fig. 4(B). A small impedance difference between the two cell–electrode configurations, resolved by the difference in normalized RMS voltage magnitude plots, is observed due to two different surface chemistries. This difference is unambiguously resolved over the frequency range of 1 kHz to 100 kHz. This suggests the frequency–dependent impedance–specific sensitivity of the single–cell–based IMA impedance sensor circuitry. The reduced impedance–specific resolution and sensitivity at lower (< 1 kHz) frequencies, where resistive properties dominate, also suggests that modifying the surface chemistry with KRGD peptides or fibronectin proteins alters significantly on the double– layered capacitance of the electrode and cell membrane capacitance of NIH3T3 (see supplementary Table 2). The sensor response remains constant or frequency independent at higher frequencies (>100 kHz) as shown in Fig. 4(B) for the impedances are mainly contributed by the resistance of the cytoplasm at higher frequencies. No difference in sensor responses between two different types of modified electrodes is observed at higher frequencies above 100 kHz. This could also suggest that the resistance of NIH3T3 cytoplasm remains unaltered in substituting from fibronectin–mediated to KRGD–mediated cell adhesion.
The plots in Fig. 4(B) also suggest that the probing frequency must be within 1–100 kHz if the real–time variations in NIH3T3 cell membrane due to external or internal stimuli are to be monitored. It is also expected that utilizing different types of cells as a sensing bio–element and different surface chemistries could shift the frequency band and optimum operating/probing frequencies. Therefore, a proper equivalent circuit model is necessary and prior experimental measurement and analysis must be carried out to obtain the insight knowledge of the contribution of each property of a single cell to the overall impedance, as well as determining optimum sensor circuit operating/probing frequencies for single–cell–based biosensing applications (e.g., real–time constant–frequency analysis of the single–cell response to a given dose of a pharmaceutical agent).
5.3. Present and Future Perspectives of the Application Potential of Single–Cell–Based IMA Sensors
The methodology we have demonstrated lays the foundation for many sensing and screening applications in which responses from the cell membranes are being focused and monitored. Multiple–Cell–based impedance sensors, recently, exhibit the capability of monitoring toxic substances and, thus, the commercialization of these sensors is becoming feasible (Curtis et al., 2009). However, the response of a single cell to a given harmful toxin or a pharmacological drug can be more significant than that of multiple cells. Additionally, in a single–cell–based impedance sensor, cell response to toxin or drug alone, without cell–to–cell interaction, will be accurately monitored and, as a result, it could also detect lower doses of toxic substances or drugs. This will be the primary advantage of single–cell–based sensors over the traditional multiple–cell–based sensors. We anticipate that the methodology presented in this work and our single–cell–based IMA chips could be employed in the high–throughput screening of: substances that alter the ionic channel conductance and the integrity of the cell membrane, ion–channel blocker drugs that block the passage of specific ions across the cell membrane, and anti–cancer drugs that trigger apoptosis.
6. Conclusion
A systematic analysis on the response characteristic of the single–cell integrated microelectrode array (IMA) chips was performed in the presented study. It is demonstrated that surface engineering on sensing microelectrodes optimizes single–cell level manipulations such as cell isolation and cell selectivity, cell morphological control, cell spreading, and cell adhesion, which, in turn, improves the sensor’s transduced signals related to cell membrane properties and mechanisms. The estimated value of the averaged cell–to–substrate separation distance from the experimental data indicates that covalently linking cell–adhesion molecules to the substrate increases the overall cell adhesion strength. The implementation of a proper equivalent circuit model in the single–cell level provides insightful information on electrical properties cellular components and cell–substrate interactions at the single–cell level and reveals optimal sensor operating/probing frequencies. These eliminate the complexity of single–cell–based biosensing as well as uncovering its great potential.
Supplementary Material
Acknowledgements
The authors acknowledge the funding support from the National Institutes of Health (NIH/GMS) (Grant No. R01GM075095), and technical assistance from Dr. Fan Zhang (Ph.D.) for PECVD. Thanks are also due to process engineers at Penn State Nanofabrication Facility for tool training and technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arndt S, Seebach J, Psathaki K, Galla H, Wegener J. Biosens. Bioelectron. 2000;19:583–594. doi: 10.1016/s0956-5663(03)00269-0. [DOI] [PubMed] [Google Scholar]
- Asphahani F, Zhang M. Analyst. 2007;132:835–841. doi: 10.1039/b704513a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asphahani F, Thein M, Veiseh O, Edmondson D, Kosai R, Veiseh M, Xu J, Zhang M. Biosens. Bioelectron. 2008;23(8):1307–1313. doi: 10.1016/j.bios.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer JE, English A, Brady M, Blackwell K, Haxhinasto K, Fotedar S, Borgman K, Bai E, Moy AB. Am. J. Physiol. Cell Physiol. 2005;289:C735–C747. doi: 10.1152/ajpcell.00103.2005. [DOI] [PubMed] [Google Scholar]
- Borkholder DA. Ph.D. Dissertation. Stanford, CA: Stanford University; 1998. [Google Scholar]
- Braun D, Fromherz P. Phys. Rev. Lett. 1998;81(23):5241–5244. [Google Scholar]
- Braun D, P. Fromherz P. Biophys. J. 2004;87:1351–1359. doi: 10.1529/biophysj.104.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TM, Widder MW, Brennan LM, Schwager SJ, van der Schalie WH, Fey J, Salazar N. Lab Chip. 2009;9:2176–2183. doi: 10.1039/b901314h. [DOI] [PubMed] [Google Scholar]
- Franks W, Schenker I, Schmutz P, Hierlemann A. IEEE Trans. Biomed. Eng. 2005;52(7):1295–1302. doi: 10.1109/TBME.2005.847523. [DOI] [PubMed] [Google Scholar]
- Fromherz P. Physica E, Low-Dimensional Systems & Nanostructures. 2003;16:24–34. [Google Scholar]
- Garcia AJ, Ducheyne P, Boettiger D. Tissue Eng. 1997;3(2):197–206. [Google Scholar]
- Giaever I, Keese CR. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LGM, Kottra G, Frömter E. Methods Enzymol. 1989;171:642–663. doi: 10.1016/s0076-6879(89)71036-3. [DOI] [PubMed] [Google Scholar]
- Greve DW, Huang X, Nguyen D, Domach MM. Proc. IEEE. 2003;2:1358–1363. [Google Scholar]
- Huang X, Nguyen D, Greve DW, Domach MM. IEEE Sens. J. 2004;4(5):576–583. [Google Scholar]
- Iwanaga Y, Braun D, Fromherz P. Eur. Biophys. J. 2001;30:17–26. doi: 10.1007/s002490000119. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Iwaki K, Hashimoto K, Mochizuki S, Ogasawara Y, Sato M, Tsujioka K, Kajiya F. Proc. Natl. Acad. Sci. U.S.A. 2002;99(24):15638–15643. doi: 10.1073/pnas.242590799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasseter TL, Cai W, Hamers RJ. Analyst. 2004;129:3–8. doi: 10.1039/b307591e. [DOI] [PubMed] [Google Scholar]
- Lo C, Keese CR, Giaever I. Biophys. J. 1995;69:2800–2807. doi: 10.1016/S0006-3495(95)80153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C, Ferrier J. Phys. Rev. E. 1998;57(6):6982–6987. [Google Scholar]
- Mossop BJ, Barr RC, Zaharoff DA, Yuan F. IEEE Trans. Nanobioscience. 2004;3(3):225–231. doi: 10.1109/tnb.2004.833703. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chang JC, Wheeler BC, Brewer GJ. IEEE Trans. Biomed. Eng. 2004;51(1):158–165. doi: 10.1109/TBME.2003.820336. [DOI] [PubMed] [Google Scholar]
- Pancrazio JJ, Whelean JP, Brokholder DA, Ma W, Stenger DA. Ann. Biomed. Eng. 1999;27:697–711. doi: 10.1114/1.225. [DOI] [PubMed] [Google Scholar]
- Panke O, Balkenhohl T, Kafka J, Schafer D, Lisdat F. Adv. Biochem. Eng. Biotechnol. 2008;109:195–237. doi: 10.1007/10_2007_081. [DOI] [PubMed] [Google Scholar]
- Pilwat G, Zimmermann U. Biochim. Biophys. Acta. 1985;820:305–314. doi: 10.1016/0005-2736(85)90125-7. [DOI] [PubMed] [Google Scholar]
- Porath D, Millo O, Gersten JI. J. Vac. Sci. Technol. B. 1996;14(1):30–37. [Google Scholar]
- Tlili C, Reybier K, Géloën A, Ponsonnet L, Martelet C, Ouada HB, Lagarde M, Jaffrezic-Renault N. Anal. Chem. 2003;75:3340–3344. doi: 10.1021/ac0340861. [DOI] [PubMed] [Google Scholar]
- Urdapilleta E, Bellotti M, Bonetto FJ. Phys. Rev. E. 2006;74:041908. doi: 10.1103/PhysRevE.74.041908. [DOI] [PubMed] [Google Scholar]
- Veiseh M, Wickes BT, Castner DG, M. Zhang M. Biomaterials. 2004;25(16):3315–3324. doi: 10.1016/j.biomaterials.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Veiseh M, Veiseh O, Martin MC, Asphahani F, Zhang M. Langmuir. 2007;23(8):4472–4479. doi: 10.1021/la062849k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener J, Keese CR, Giaever I. Exp. Cell Res. 2000;259:158–166. doi: 10.1006/excr.2000.4919. [DOI] [PubMed] [Google Scholar]
- Weis R, Muller B, Fromherz P. Phys. Rev. Lett. 1996;76(2):327–330. doi: 10.1103/PhysRevLett.76.327. [DOI] [PubMed] [Google Scholar]
- Weis R, Fromherz P. Phys. Rev. E. 1997;55(1):877–889. [Google Scholar]
- Xiao C, Lachance B, Sunahara G, Luong JHT. Anal. Chem. 2002;74:1333–1339. doi: 10.1021/ac011104a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.