Abstract
Until recently knowledge of genetic causes of glomerular disease was limited to certain rare or uncommon inherited diseases, and to a genes, either rare or with small effect, identified in candidate gene studies. These genetic factors accounted for only a very small fraction of kidney disease. However, the striking differences in frequency of many forms of kidney disease between African Americans and European Americans, which could not be completely explained by cultural or economic factors, pointed to a large unidentified genetic influence. Since FSGS and HIV-associated collapsing glomerulopathy (HVAN) have striking racial disparities, we performed an admixture mapping study to identify contributing genetic factors. Admixture mapping identified genetic variants in the non-muscle myosin gene MYH9 as having an extreme influence on both FSGS and HIVAN, with odds ratios from 4 to 8 and attributable fractions of 70–100%. Previously identified, rare inherited MYH9 disorders point to a mechanism by which MYH9 variation disrupts the actin-myosin filaments responsible for maintaining the structure of podocytes, the cells that provide one of three filtration barriers in the glomeruli. MYH9 variation has a smaller but still highly significant effect on non-diabetic kidney disease, and a weaker but significant effect on diabetic kidney disease; it is unclear whether underlying cryptic FSGS is responsible for the MYH9 association with these diseases. The strong predicted power of MYH9 variation for disease indicates a clear role for genetic testing for these variants in personalized medicine, for assessment of genetic risk, and potentially for diagnosis.
Introduction
In this bicentennial year of the birth of Charles Darwin, it is abundantly clear the findings from genetic and genomic studies are rapidly transforming medicine. This transformation is due largely to the success of the Human Genome and the HapMap Projects that has led to annotation of sequence variation and provided a road map to allelic and genomic organization structure of the human genome in world populations. The genetic basis for most monogenic, high penetrance diseases has been achieved—the remaining challenge is to understand the genetic and non-genetic correlates of common diseases that contribute most to human morbidity and mortality. Since 2005 several hundred genome wide associations for many common diseases (e.g. cancers and cardiovascular diseases) and traits (e.g., drug metabolism, height, obesity) have been reported. The major goal of these studies is the integration of personalized medicine into clinical practice and public health policies to mitigate and prevent human suffering. Personalized medicine has been defined as “The use of a person's clinical, genetic, genomic, and environmental information to select a medication, its dose, or to choose a therapy, or recommend preventive health measures” 1. Personalized medicine has arrived on the scene and will add new predictive power and clinical complexity to our diagnostic and therapeutic strategies. This review will summarize briefly the status of simple genetic causes of the podocytopathies and focus in greater depth on emerging data on the role of MYH9, encoding non-muscle myosin heavy chain IIA, in these and other kidney diseases.
Simple genetic inheritance
To date, 20 genes causing or associated with podocytopathies have been identified, as shown in Table 1; the genetics of the podocytopathies has been summarized in several recent reviews 2,3. Of these 20 genes, nearly all manifest Mendelian inheritance, including autosomal dominant, autosomal recessive, and X- linked inheritance, or maternal inheritance in the case of mitochondrial genes, and most disease-associated mutations change amino acid sequence. The underlying pathogenic mechanism and their effects varies among the disease genes with some showing complete penetrance with congenital onset and others incomplete penetrance with variable age of onset. They share in common apparent disruption of podocyte function. It is striking that several genes cause more than one form of podocytopathy, underscoring the likelihood of certain common mechanisms among diverse histologies but also pointing to how little we understand why similar mutations give rise to diverse histologies.
Table 1.
Genes causing glomerulopathies.
| Gene | Chromosome | Protein name | FSGS | Collapsing glomerulo- pathy |
Diffuse mesangial sclerosis |
Minimal change Nephropathy |
Kidney- limited or Syndromic |
Inheritance/ Penetrance |
|---|---|---|---|---|---|---|---|---|
| Slit diaphragm complex | ||||||||
| NPHS1 | 19q13.1 | Nephrin | + | + | Kidney | AR*/complete | ||
| NPHS2 | 1q25.2 | Podocin | + | + | + | Kidney | AR/ complete | |
| CD2AP | 6p12 | CD2 associated protein | + | Kidney | AR*/complete | |||
| Actinomyosin complex | ||||||||
| ACTN4 | 19q13 | Alpha-actinin 4 | + | Kidney | AD | |||
| MYH9 | 22q13.1 | Non-muscle myosin IIA heavy chain |
+ | + | Kidney | ND | ||
| Cell signaling | ||||||||
| TRPC6 | 11q21.22 | Transient receptor potential cation channel C6 |
+ | Kidney | AD | |||
| PLCE1 | 10q23 | Phospholipase epsilon 1 | + | Kidney | AR | |||
| Membrane repair and turnover and vesicle function | ||||||||
| DYSF | 2p13.2–13.1 | Dysferlin | + | Syndromic | AR | |||
| GLA | Xq22 | Alpha galactosidase | + | Syndromic | X linked | |||
| SCARB2 | 4q21.1 | Scavenger receptor class B, member 2 |
+ | Syndromic | AR | |||
| Transcription factors | ||||||||
| WT1 | 11p13 | Wilms tumor 1 | + | + | Both | Sporadic | ||
| PAX2 | 10q24 | Paired homeobox 2 | + | Syndromic | AR | |||
| LMX1B | 9q34 | LIM homeobox domain transcription factor 1 |
+ | Syndromic | AR | |||
| Extracellular matrix and receptors | ||||||||
| LAMB2 | 3p21 | Laminin Beta 2 | + | Syndromic | AR | |||
| ITGB4 | 17q25 | Integrin beta 4 | + | Syndromic | AR | |||
| COL4A3 | 2q36–37 | Collagen 4A3 | Both | AR | ||||
| COL4A4 | 2q36–37 | Collagen 4A4 | Both | AR | ||||
| COL4A5 | Xq22.3 | Collagen4A5 | Both | X-linked | ||||
| Mitochondrial function | ||||||||
| mtDNA tRNA leu |
mtDNA | Not applicable | + | Both | Maternal | |||
| COQ2 | 4q21.23 | Co-enzyme Q enzyme 2 | + | + | Syndromic? | AR | ||
Shown are 20 genes in which mutations (or a risk haplot ype, in the case of MYH9) are associated with one or more glomerulopathies as shown. AD, autosomal dominant. AR, autosomal recessive. ND, not determined. [note: bullets in table need to be deleted]
The gene products may be classified into seven functional clusters: slit diaphragm complex, actinomyosin and related proteins, cell signaling, membrane repair and turnover and vesicle function, transcription factors, extracellular matrix and receptors, and mitochondrial function. The most common of these genetic mutations are found in NPHS2, explaining ~20% of apparently-idiopathic FSGS in Europe but probably <5% of similar cases in the United States, even when analysis is limited to European descent individuals—likely due to ascertainment bias in patient selection European studies. Many of the European studies enrolled children or young adults while the USA studies tended to enroll mainly adults. The other 18 genes, excluding NPHS2 and MYH9, likely account for <1% of FSGS. Most of these genetic mutations are uncommon or rare and affect relatively few families and sporadic cases; however, the identity of the genes provide needed insights into mechanistic pathways of kidney function and pathology. Certainly, this list will continue to expand as more families are identified with familial glomerulopathies not attributable to the genes in Table 1.
Gene discovery for kidney disease in African descent individuals
There are a number of diseases more common in African descent people compared to European descent individuals, as shown in Table 24–16. It has long been noted that kidney disease is both more frequent and progresses to end-stage kidney disease (ESKD) faster among populations with African ancestry. Family clustering of disparate etiologies of kidney disease (e.g., diabetes-attributed ESKD, hypertension-attributed ESKD, or HIVAN) has also been observed in African American families. This propensity to kidney disease is only partially explained by access to health care. For example, the predilection for African descent individuals to develop HIV-associated nephropathy (HIVAN) with collapsing glomerulopathy has been noted since the first recognition of the syndrome in 1984 17. Using data from the United States Renal Data Systems on ESKD attributed to HIV infection, it has been estimated that African descent individuals have a 60 fold increase compared to European descent individuals 18. Chronic and end stage kidney diseases associated with diabetes, hypertension, lupus, and obesity are also both more frequent and have a poorer prognosis in African descendents. Thus kidney disease represents a major USA and global health disparity not fully explained by non-genetic factors.
Table 2.
Diseases with disparate risks in European descent and African descent individuals.
| Disease class | Disease | Relative risk |
95% confidence interval |
P | Reference group |
|---|---|---|---|---|---|
| Cardiovascular | Hypertension | 1.4 | ND | ND | European descent |
| Congestive heart failure | 1.81* | 1.07, 3.07 | <0.05 | Other | |
| Endocrine | Diabetes (Males) | 1.04* | 0.92, 1.18 | ND | European descent males |
| Diabetes (Females) | 1.14* | 1.05, 1.24 | ND | European descent females |
|
| Diabetic nephropathy | 1.3* | 1.2, 1.4 | ND | European descent |
|
| Rheumatologic | Systemic lupus erythematosus | 2.65 | ND | ND | European descent females |
| Lupus nephritis | 3.13* | 1.21, 8.09 | 0.02 | Other | |
| Kidney | FSGS ESKD | 4.2 | ND | ND | European descent |
| HIV-associated nephropathy | 18 | ND | ND | European descent |
|
| HIV-attributed ESKD | 17.7 | 2.5, 127.0 | 0.004 | European descent |
|
| Hypertension-attributed ESKD | 2.42* | 1.52, 3.84 | ND | European descent males |
|
|
Pregnancy- related |
Preeclampsia | 1.2 | 0.8, 1.7 | NS | European descent females |
| Eclampsia | 1.6 | 0.9, 2.3 | NS | European descent females |
Shown are the relative risks for African descent individuals for a number of diseases associated with particular kidney diseases or associated conditions. ND, not determined.
denotes adjusted for relevant co-variates. References provided in text.
Genetic basis for non-familial glomerulopathies
The genetic basis for glomerulopathies in the general population and the increased risk of kidney disease in Africans has been elusive. United States-based population studies have excluded a major role of NPHS2 in HIV-collapsing glomerulosclerosis and mainly adult onset FSGS in African and European Americans19,20. Mouse models have been employed to identify genes associated with HIV-associated nephropathy. Mice transgenic for HIV genes develop a syndrome resembling HIV-associated glomerulopathy. Recently, Gharavi and colleagues have reported on the results of mouse strain inter-cross experiments, which identified a ~50 mb region on mouse chromosome 3A1-A3 that displays more severe glomerulopathy compared to the parental strain; the putative locus is termed HIVAN1 and comprises 126 genes, which will be interrogated as positional candidate genes 21.
While the relative risk for African descent individuals to develop HIVAN is dramatically high, there are a number of diseases that show racial and ethnic predilections14. Although at least a portion of the excess risk of these diseases may be attributed to socioeconomic status or access to health care, there is growing evidence that disparate rates for certain diseases (e.g., prostate cancer) among the world’s continental (racial) and ethnic populations may have a genetic basis22–24. Table 2 lists a number of kidney-related diseases with increased incidence among African descent individuals.
Genome wide associations studies (GWAS) are systemic genomic approaches to identify low to medium penetrant genetic polymorphisms associated with common traits and diseases25. Generally, DNA from thousands of (1000 to 10,000) cases and a similar number of controls or population-based studies is subjected to genotyping for 500 k to 1,000 k single nucleotide polymorphisms (SNPs). These large numbers are generally required for statistical power because the effect sizes for alleles associated with the majority of complex diseases are generally in the range of 1.1 to 1.526. Contemporary, commericially available gene arrays differ by SNP selection criteria, density of single nucleotide polymorphisms (SNPs) and copy number variation (CNV) for genome coverage, and inclusion of informative SNPs for African and Asians. For example the Illumina arrays incorporated haplotype information from the International HapMap Project to select SNPs tagging haplotypes to maximize coverage and to avoid highly correlated, redundant SNPs. Older SNP arrays based on European frequencies from the HapMap CEPH (European) samples did not capture well the variation in the more diverse African decent populations or in Asians, leading to a corresponding loss of power to detect associations in studies of non-Europeans 27. Recent SNP arrays have incorporated Yuruba HapMap data to increase power for genotype-phenotype association studies in African descent populations. It is important to note that the SNPs selected for arrays are usually proxies for the true causal alleles that they are tracking by linkage disequilibrium. The identity of the causal alleles is determined through additional fine or dense mapping in the region-of-interest to narrow the region of strongest association and by functional assays of implicated SNPs.
The large number of GWAS since 2005 have provided a number of surprises and lessons. Very few main-effect genes with odds ratios (OR)≥2 have been identified—notable exceptions are the complement factor risk alleles for age-related macular degeneration and the association of MYH9 with kidney disease which has OR in the 2–6 range for glomerulopathies28–34. Most high frequency (>5%) alleles tend to have small disease-causing effects in the OR range of 1.1–1.5), whereas most major-effect alleles tend to have low allele frequencies as might be expected for deleterious alleles32.
A potential confounder in many GWAS studies is disease heterogeneity—what appears to be a single gene or phenotype may actually have different etiologies or underlying mechanisms not resolved by clinical assessment methods. In very large studies, the power gained by large numbers may counteract the power lost through undetected or uncontrolled disease heterogeneity. However, different genes may predispose to the different etiologies of disease, resulting in a loss of power or the detection of one or more small effect genes associated with different underlying phenotypes. It is rare for polymorphic genes (population allele frequencies ≥1%) to have high penetrance—hence unaffected controls may also carry the risk alleles25,35. As a cause of low genetic penetrance, interactions among genetic, environmental factors, and the particular physiology of an individual likely all contribute to the manifestation of diseases such the glomerulopathies.
Although several GW linkage and association scans have been performed for non-diabetic and diabetic kidney disease phenotypes, no major-effect genes have yet been identified that could account for the differential risk between African and European populations 32. This may have been the result of a combination of underlying disease heterogeneity, poor coverage of African alleles in the SNP arrays, or lack of power due to underrepresentation of African descent individuals with clearly defined kidney disease phenotypes.
Discovery of MYH9 as a major renal susceptibility locus by admixture mapping
A particular variant of a genome wide scans relies on an approach termed admixture mapping, which has been the subject of recent reviews 36–39. This mapping strategy leverages the differential occurrence of a disease between two ancestral populations for gene discovery. If the causal disease gene is more frequent in one ancestral population, the chromosomal segment bearing that disease allele should be elevated in frequency in the case group relative to the control group and to the rest of the genome. In a sense, admixture mapping resembles a combination of a family study and an association study. Both family and admixture studies rely on the identification of a parental/ancestral chromosomal segment, arising through meiotic recombination, that segregates with the risk allele40. The resolution of admixture mapping through linkage disequilibrium is intermediate (1–10 cM) between that of a family study (20–30 cM) and a traditional GWAS ≈0.1 cM 14. Unlike traditional genome wide studies employing 500K to a 1000k single nucleotide polymorphic (SNP) markers, this method requires only 1500 selected SNPS that are ancestry informative; that is the alleles differ in frequency by on average 30% between the ancestral populations 41. Statistical methods permit the inference of local ancestry at each marker; chromosomal segments showing significantly higher ancestry compared to either the control group or to the rest of the genome are considered a region-of-interest. It is worth noting that admixture mapping has several distinct advantages over GWAS: 1) since only 1500 or so ancestry informative markers are required the genotyping cost per DNA sample is correspondingly much less than 500k and 1000 k SNP chips; 2) perhaps less appreciated, the statistical cost is also much less; to achieve statistical significance after correction for multiple tests in GWAS studies, a genome wide statistical significance of p≤10−8 is the accepted threshold for securely identifying a locus.; 3) admixture mapping may be performed in the absence of an unaffected control group. At each locus, the extent of parental ancestry in the case group is compared to the genome wide average (with the rest of the genome serving as a control)42.
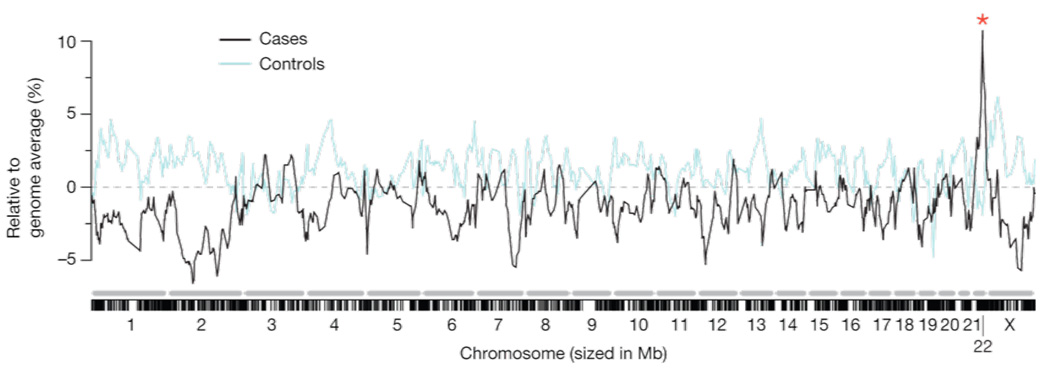
Due to the substantial differential in risk between African and European populations for certain forms of kidney diseases, genome wide admixture mapping scans were used by two independent groups to identify risk alleles associated with idiopathic FSGS, HIVAN, and ESKD among African Americans. Both groups identified a chromosomal segment of elevated African ancestry on chromosome 22, with biopsy-proven FSGS and HIV-associated collapsing nephropathy in Kopp et al. 31 and with non-diabetic ESKD Kao et al., 34. The Kopp et al. study found a highly significant peak of 93% African Ancestry compared to a genome wide average of only 83% African Ancestry in the case group (Figure 1). Further fine-mapping of MYH9 and neighboring genes securely identified MYH9 as a main effect disease gene for biopsy-proven FSGS and HIV-associated collapsing glomerulopathy (OR in the 4–7 range). The association of MYH9 risk alleles was confirmed in a group of European Americans (OR=7) and extended to non-diabetic end stage kidney disease (ESKD) attributed to hypertension (OR=2–3). The associations with MYH9 risk alleles in non-diabetic ESKD were replicated by the FIND (Family investigation of nephropathology in diabetes) consortium for non-diabetic end stage renal disease 34.
Figure 1. Admixture mapping for FSGS and HIVAN identifies a peak identifies a peak on chromosome 22.
A summary of the genome-wide admixture scan for 412 FSGS and HIVAN cases and controls showing 92% African ancestry on chromosome 22 compared to a genome-wide average of 81%. The genome-wide lod score and case control lod scores were 9.2 and 12.4, respectively. Lod scores >2 are considered significant. Figure reproduced from Kopp et a. Nature Genetics 2008 with permission.
The MYH9 risk alleles and the E1 extended haplotype had strikingly disparate frequencies between African Americans (>60%) and European Americans (<4%)31. The most frequent European-type haplotype (E2) (69%) was strongly protective against idiopathic FSGS, HIVAN, and hypertension-attributed ESKD (OR=0.24, 0.12, and 0.63, respectively)31. These results provide a genetic basis for the increased risk of major forms of chronic and end stage kidney disease in persons of Africa descent, a major USA and global health disparity.
MYH9 encodes non-muscle myosin heavy chain IIA
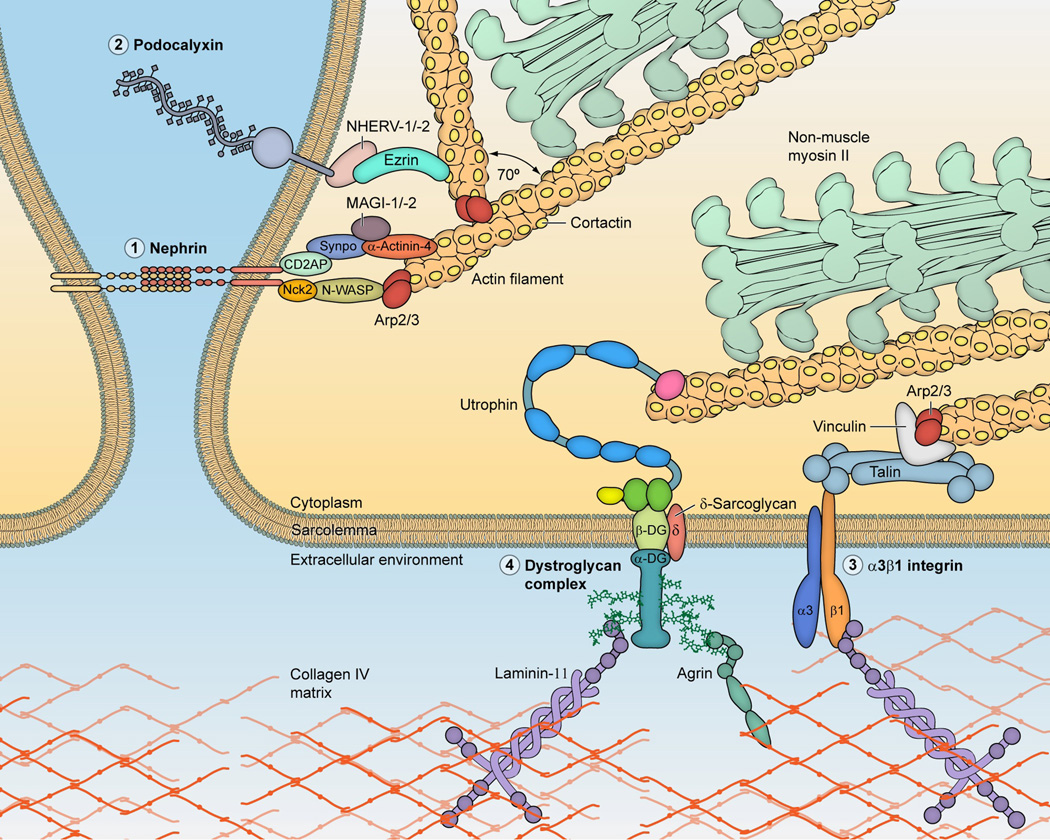
Located on the short arm of C22q12, the 110kb MYH9 gene comprises 40 exons encoding a protein of 1960 amino acid residues, the heavy chain of nonmuscle myosin heavy chain IIA. Myosin IIA is a widely-distributed cellular motor that is essential for cytoskeletal rearrangement, cell motility, division, and cell-cell adhesion. In the podocyte, the actinomyosin complex is linked to several important protein complexes, including the slit diaphram and podocalyxin on the apical domain and alpha3/beta1 integrin and dystroglycan complex on the basal domain (Figure 2). Dysfunction of Exons 1–19 specify the myosin globular head containing an actin binding site, the ATP binding site, and a short neck. Exons 20–40 encode the coiled coil tail domain.
Figure 2. Actin-myosin and molecular partners in the podocyte.
The actinomyosin complex is composed of an actin filament and a multimeric myosin assembly, composed of non-muscle myosin IIA or IIB molecules. The actinomyosin functional cycle is as follows: the myosin head binds ATP and this enables release from the actin filament, ATP is hydrolyzed to ADP and Pi, the myosin head domain binds actin when a site becomes available, the myosin head generates a power stroke that pushes the actin filament and simultaneously ADP and Pi are released, and myosin detaches from actin. This actinomyosin complex maintains cell tension and cytoarchitecture, and facilitates contraction (perhaps in response to increased hydrostatic glomerular capillary pressure) and migration (if podocyte foot processes are indeed mobile under physiologic or pathologic conditions). The figure shows links between the actinomyosin complex and four molecular complexes linking the actinomyosin complex to the extra-cellular environment: the slit diaphragm and podocalyxin (on the apical domain) and dystroglycan complex and a3b1 integrin (on the basal domain). The figure is not exhaustive in the molecular complexes nor in the inter-molecular interactions depicted. Molecular structures are depicted in varying levels of details and differing scales, and those interacting with nephrin are shown schematically. Further details on molecular interactions are available in an excellent review by Faul et al, Trends in Cell Biology 2007. Figure expanded from a previous figure, with permission (Kopp, Clin J Am Soc Nephrol 2009).
Autosomal dominant mutations in MYH9 cause four over-lapping syndromes, perhaps best described as MYH9-related platelet disorder spectrum: May-Hegglin anomaly, Flechtner syndrome, Epstein syndrome, and Sebastian syndrome. These MYH9-related syndromes show complete penetrance for congenital macrothrombocytopenia and variable penetrance for hearing impairment, renal impairment, and pre-senile cataracts with adult onset. Although the pathological mutations span exons 1 through 40, most, but not all, of those showing the full spectrum of syndromic manifestations are found in exon 30 and kidney involvement is noted for missense mutations in exons 16, 25, and 30.
MYH9 associations with kidney disease
Following the Kopp et al and Kao et al. reports of MYH9 associations with idiopathic FSGS, HIVAN, hypertensioin-attributed ESKD, and non-diabetic ESKD, additional papers have since replicated and extended the associations of MYH9 to hypertension-attributed ESKD, microalbuminuria in subjects with hypertension 43, and diabetes-attributed ESKD 44. The effect is easiest to detect in African descent populations, where the high frequencies and strong effects of the risk alleles provide excellent statistical power, even with small sample sizes. An association with MYH9 can be seen in European Americans with FSGS and although the strength of the association is very strong (OR=7), very few people of European-descent would be expected to be homozygous (<0.2%), given the low allele frequencies for MYH9 risk alleles in the European population. Interestingly, in the original study, three of the four self-identified European American individuals with two copies of the MYH9 risk haplotype had substantial African ancestry 31.
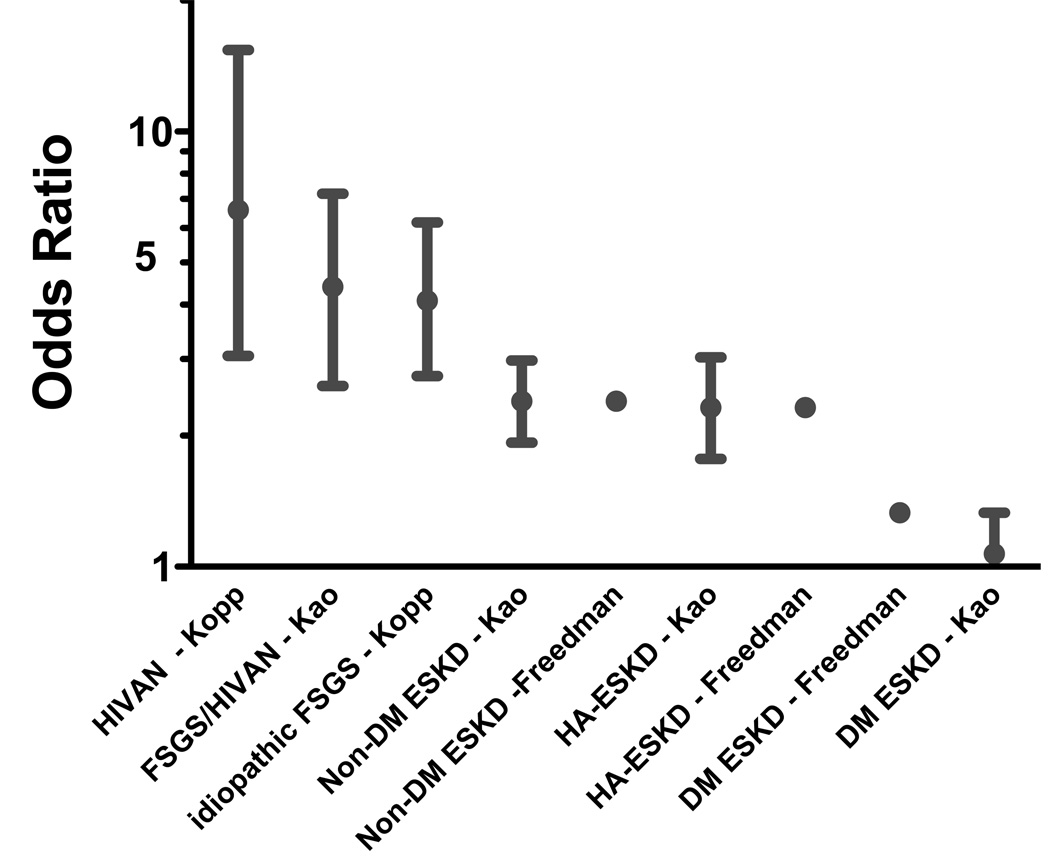
It is notable that the strength of the association of MYH9 risk alleles is very similar between different independent studies (Figure 3). The association with rs4821481 (typed across all studies) has an identical odds ratio for non-diabetic end stage renal disease (2.4) and for a subgroup of non-diabetic ESKD attributed to hypertension (2.32) in Freedman et al., and Kao et al. Similar consistency was observed for FSGS between the Kopp et al. and Kao et al. (OR=4.09 and 4.3, respectively). The role of MYH9 in ESKD attributable to diabetes is not resolved. Using a similar large number of cases and controls, the Kao et al. study found no association while Freedman et al., observed a 30%–40% increase in the odds of developing ESKD risk for diabetes in MYH9 risk allele homozygotes 34,45. These results suggest that at least a portion of diabetic ESKD may be due to other underlying glomerulopathies (particularly idiopathic FSGS and hypertension-attributed kidney disease) and may account for the higher incidence of ESKD among diabetics of African descent.
Figure 3.
A comparison of odds ratios for HIVAN, idiopathic FSGS, non-diabetic ESKD, hypertension-attributed ESKD attributable, and diabetes-attributed ESKD in four published studies31,34,43,61.
The majority of the most highly associated SNP risk alleles are located in the 15.5 kb extended (E) haplotype block spanning introns 12–23 and are tracking each other through linkage disequilibrium (Figure 4). The risk extended haplotype (named E1) is tagged by just two SNPs (rs4821481 plus rs3752462) and is more informative than any single SNP. The E1 haplotype is the most frequent in African descent individuals. There are also associated SNP alleles elsewhere in the gene that may have independent effects. Follow-up fine and dense mapping of the region, together with resequencing, have narrowed the region of strongest association, but purely statistical methods indicate only correlation and do not prove causality between an allele and phenotype (Nelson et al., submitted). Most, but not all of the associated alleles with this haplotype block best fit a recessive genetic model, but this may change with further knowledge of the causal MYH9 allele or variation. It is noteworthy that although the majority (73%) of 58 HIV infected persons with collapsing glomerulosclerosis carried two copies of the E 1 haplotype, each of the remaining individuals (27%) carried one risk allele, suggesting that MYH9 risk alleles may be additive. Under the null hypothesis, one would expect 36% of the group to be homozygous for the risk haplotype. While this pattern best fits a recessive model, the MYH9 E1 haplotype is also highly significant for additive and dominant genetic models. The true genetic model that best fits MYH9 will be determined only after the causal allele or variation has been identified. It is also possible that MYH9 alleles may have epistatic interactions with other genes.
Figure 4. MYH9 linkage disequilibrium and associations.
Location and strength of association of 17 MYH9 SNPs, shown with map of exons of MYH9 and HapMap plot of MYH9 linkage disequilibrium (LD) in Yoruba from Nigeria. Strength of association for SNPs (colored wedges) is keyed by colors as shown. Black triangles on HapMap plot indicate HapMap-defined LD blocks. Haplotype E1 (for “extended haplotype 1”) extends through three HapMap LD blocks. F
Resequencing of MYH9 has excluded an exonic codon-changing mutation associated with FSGS and HIVAN (Nelson et al. submitted); all of the associated SNPs are within introns. Intronic SNPs may affect expression levels or alternative splicing in a tissue- or cell-specific manner. We speculate that the MYH9 genetic variation leading to glomerulopathies is either not expressed or is compensated for in other tissues. Perturbations in protein structure due to mutations or insertion/deletions throughout the MYH9 gene cause giant platelet syndromes, characterized by congenital macrothrombocytopenia, sensorineural deafness, and glomerulonephritis 46. Platelet abnormalities have not been reported or observed for MYH9 risk alleles (Kopp et al, unpublished data), nor are there reports in the literature of platelet abnormalities being a common clinical finding in FSGS or HIVAN syndromes. We therefore speculate that MYH9 intron variants alter MYH9 expression levels or alternative splicing in a podocyte-cell specific manner. A recent genome-wide screen for polymorphisms that influence genetic control of transcription provided convincing evidence that protein complexity is achieved in part by tissue-specific regulation of alternative splicing leading to different protein isoforms in various tissues 47.
Continental Distribution of MYH9 risk alleles
The MYH9 risk alleles have strikingly different allele frequencies among world populations (Figure 5). The international HapMAP Project data indicate that the MYH9 risk allele rs4821481, a tag SNP for the extended E haplotype is quite frequent in Nigerian Yoruba (76%), less frequent in Europeans (6%), and absent in Japanese and Chinese Asians. Thus MYH9 variation provides a genetic basis for the kidney disease risk disparities observed for the African-descent population.
Figure 5.
Global distribution of MHY9 E1–5 extended haplotypes. Frequencies of risk (E1), protective (E2), and neutral MYH9 haplotypes (E3-E5) in the Human Genome Diversity Panel 1050 individuals in 51 populations, and grouped by the continental origins: African, European, Middle Eastern, South-Central Asian, East Asian, Oceanian, and American. African samples carry the largest proportion of risk haplotype, while most of the Europeans and the peoples in the Middle East in this study feature protective haplotype (E2). A large proportion of the South-Central Asian populations also have the protective haplotype, while the risk haplotype is at very low levels. East Asian and Oceanian populations are represented mainly by the neutral haplotype (E3), some protective haplotype, but with the risk haplotype virtually absent
The risk alleles and the E1 haplotype associated with kidney disease show high-level population stratification. The kidney disease risk SNPs located within the E haplotype block, as well as the E1 haplotype itself, are the most common in sub-Saharan Africa (allele frequencies ~60%) but much less frequent in Europe (<4%) and virtually absent in Asians. Rs4821481, a E1 Haplotype tagging SNP, has a frequency of 76% in the Nigerian Yoruba. Figure 4 shows the relative frequencies of MYH9 risk alleles based on data available from the International HapMap Phase II Project in major continental and geographic populations 48. It is striking that with distance away from African, the E1 haplotype is lost and replaced by the protective E2 European-type haplotype. Another measure of population differentiation or genetic distance is the fixation index (FST); it measures the proportion of the genetic diversity that can be attributed to the allele frequency differences among populations 49. The MYH9 SNPs with the highest associations with FSGS and HIV-associated glomerulopathy show have some of the highest FST scores (based on Happlotter, a program for exploring the HapMap database 50) between African and non-African samples in HapMap populations 51, a putative sign of selection 52.
Is MYH9 a selected gene?
What is the role of natural selection in producing the particular distribution of risk and non-risk alleles that characterize the world’s populations? Natural selection takes three forms. First, in directional selection, an allele with greater fitness steadily increases in frequency until it becomes fixed and dominates the population. Second, on the opposite side of the coin, in stabilizing selection, an allele with lower fitness steadily decreases in frequency until it vanishes from the population. Third, balancing selection occurs when individuals with extreme trait values, on both sides of any particular dichotomy, have an advantage over individuals with intermediate trait values53. Steven Pinker has analogized this concept as follows:
“A better way of thinking about genetic diversity is that if everyone were a tinker, it would pay to have tailor genes, and the tailor genes would start to make an inroad, but then as society filled up with tailor genes, the advantage would shift back to the tinkers. A result would be in equilibrium with a certain proportion of tinkers and a certain proportion of tailors. Biologists call this process balancing selection: two designs for an organism are equally fit, but in different physical or social environments, including the environments that consist of other members of the species.”54
Another form of balancing selection involves heterozygote advantage, which maintains a variant at a higher population frequency of an otherwise deleterious allele because it would confer survival advantage in a heterozygote. In a classic example, the B-globin sickle cell allele S, found in 25% of west Africans, reduces susceptibility to cerebral malaria 55. Heterozygotes have a survival advantage over the individuals homozygous for the normal A allele who are at increased risk for cerebral malaria, while individuals homozygous for the sickle cell S allele will develop sickle cell anemia.
Aside the population differentiation described in the previous section, is there other evidence that deleterious MYH9 alleles have been maintained in the general population by the process of natural selection? At this time, a specific agent, such as particular pathogen or an environmental factor, can be held responsible for selection in MYH9 is unknown. However, there are some arguments about this issue that warrant further investigation.
First, the MYH9 E1 haplotype (and many of the SNPs comprising the haplotype) has a frequency of approximately 60% in Yoruba, with the expected frequency of heterozygosity for the E1 haplotype of 48%. A possible explanation of this observation is the action of balancing selection. In a survey of natural selection on genes, Bustamante et al. 56 reported that 6 of 9 nonmuscle myosin heavy chain genes, including MYH9, show lower than expected amino acid divergence between chimps and humans, yet maintain high levels of amino acid polymorphisms in human populations. This means that these actin-binding genes may harbor an excess of mildly deleterious variation, possibly maintained by negative or balancing selection 56. However, signatures of balancing selection based on comparisons of DNA coding region polymorphisms within populations to divergence between humans and chimpanzees are less likely to detect recent or ongoing selection episodes. In a more recent report, the non-muscle myosin genes were not among the 60 genes showing the strongest evidence of balancing selection 57.
Second, the expansion of non-risk alleles in European descent populations might indicate either relaxation of selective pressure or a founder effect. On the other hand, the fact that the frequency of non-risk allele alleles has risen so significantly in European descent populations, and have become fixed in Asian descent populations (reaching 100% frequency) may be pointing to an evolutionary adaptation for surviving in a new environment or founder effect.
Third, the gene may be involved in a complex regional interplay within its genomic neighborhood 51. MYH9 exhibits at least three recombination hotspots; SNPs on either side of these hotspots are in genetic equilibrium, which can be interpreted as evidence against the case for the recent selection; however, it does not exclude old selection that may have occurred between the time of human-primate divergence and dispersal of humans out of African 100,000 years ago. In fact, several of the most strongly associated alleles are derived and are not shared with our primate cousin, the chimpanzee50.
Proposed models for MYH9 disease mechanisms
As noted above, MYH9 variation is associated with podocyte injury, both podocyte loss (FSGS) and podocyte proliferation (HIV-associated collapsing glomerulopathy). While it might seem puzzling that a single genetic variant should be associated with these two contrasting forms of podocyte injury, there is one precedent for this, with regard to COQ2 mutation.
At present, two unifying models can be proposed to explain common presentations for MYH9-associated kidney disease. Both models incorporate the concept of a podocyte made “fragile” by the expression of MYH9 risk alleles. This fragility manifests as reduced tolerance for local stress of various kinds. These models are not mutually exclusive. In the glomerulosclerosis-first model, a second factor (another genetic mutation, a virus, an environmental chemical, obesity, or other factor) interacts with MYH9 risk alleles expressed in fragile podocyte to induce FSGS, either in childhood or in adulthood. Alternatively, in the hypertension-first model, essential systemic hypertension (or less commonly secondary hypertension due to non-renal causes) or intra-glomerular hypertension (associated with glomerulomegaly and hyperfiltration) triggers injury to the fragile podocyte, perhaps acting via increased glomerular capillary pressures (transmitted via a failure of renal auto-regulation) or increased GBM stretch due to these increased pressures or other mechanisms.
In both models, podocytes undergo cell death (apoptosis or necrosis) or detach from the GBM and are lost into the urinary space, segmental or global scarring ensues (perhaps by mis-directed glomerular filtration as outlined by Kriz and colleagues 58). In both models, glomerulosclerosis and later associated interstitial fibrosis may progress insidiously, leading to CKD and hypertension, and may therefore present in as unexplained CKD or hypertension with subnephrotic proteinuria. If and when a kidney biopsy is done, it may show FSGS or focal global glomerulosclerosis (perhaps primarily of the solidified rather than ischemic type) and if hypertension is present these changes may be mistakenly attributed to hypertensive nephrosclerosis (particularly among African descent individuals).
Conclusions and future prospects
Rapidly accumulating evidence indicates that MYH9 genetic variation contributes to the excessive burden of kidney disease among African descent individuals. Many questions remain, at all levels of clinicopathologic study. There following is a partial list of these questions that we and others are actively investigating:
Causality and functional consequence of MYH9 risk alleles
What are the causal allele within the MYH9 predisposing to podocyte injury?
What is the function of these alleles—do they regulate alternative splicing, transcription expression levels, or mRNA stability by altering microRNA expression or binding?
Are MYH9 isoforms or expression levels regulated in a cell- or tissue-specific fashion?
Cellular function
Does the MYH9 risk allele correlate with gain in function (increased non-muscle myosin activity IIA) or loss of function?
Does the MYH9 risk allele correlate with altered interaction with the many molecular partners of the podocyte actinomyosin complex?
Clinical Applications
Do MYH9 risk alleles tend to be associated with particular FSGS histologic variants?
Do MYH9 risk alleles predict resistance to glucocorticoids or other immunosuppressive therapy?
Does MYH9 glomerular disease more typically present with the sudden onset of heavy proteinuria or the slow development of proteinuria over a period of many years, from microalbuminuria to subnephrotic proteinuria and finally to nephrotic proteinuria?
Do MYH9 alleles predict progression rates to ESKD?
What is the impact of angiotensin pathway antagonism in proteinuria and progressive loss of glomerular filtration rate in MYH9 associated FSGS and hypertension-attributed kidney disease?
Do MYH9 risk alleles contribute to the increased risk for other kidney diseases more common among African descent individuals, including lupus nephritis, sickle cell nephropathy, or pre-eclampsia?
Does the renal allograft donor risk allele influence allograft survival in the recipient or the appearance of albuminuria or the extent of kidney hypertrophy or FSGS in the donor?
Epidemiology of MYH9-associated kidney disease
What fraction of MYH9 risk allele homozygotes and heterozygotes will develop clinical kidney disease during their lifetimes?
What is the role of MYH9 testing in asymptomatic individuals? Would such individuals benefit from annual measurement of urine albumin? If microalbuminuria were detected how shoould it be treated?
What is the role of MYH9 variation in sub-Saharan Africa where over 22 million people are infected with HIV-1? What is the role of MYH9 variation in the reported high incidence of chronic kidney disease in African countries with rates ranging from 6 in South Africa to 20–48% in Uganda (reviewed in 59)?
Summary
The discovery of MYH9 as a major renal susceptibility gene has enhanced our understanding of the epidemiology of a major USA and global health disparity for people of African descent. It has also underscored the importance of non-muscle myosin heavy chain IIA in previously under-considered biologic pathways leading to podocytopathies associated with several etiologies of glomerusclerosis. The strength of MYH9 disease associations and the frequency of the risk alleles will no doubt lead to a push for genetic screening for individualized risk assessment. The National Institutes of Health and Centers for Disease Control and Centers for Disease Control and Prevention Workshop recently formulated recommendations for evaluating the clinical validity and utility of a genetic screening so that evidence-based guidelines can be developed60. Prospective studies are needed to assess the benefits, effectiveness, and clinical utility of MYH9 genetic risk assessment for kidney donors and recipients, for patients with known renal disease risk factors, and for targeted public health policies to reduce the burden of disease at the community and national levels.
Acknowledgements
This Research was supported in part by the Intramural Research Programs of the NIH, National Cancer Institute, Center for Cancer Research and of the National Institute of Diabetes and Digestive and Kidney Disease. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The authors wish to acknowledge Ethan Taylor preparing figure 2, Dr. Laura Barisoni for helpful discussions and Dr. James Balow for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest. Three authors (CAW, GN, JBK) and the NIH have applied for a patent on SNPs associated with the diseases described here.
References
- 1.DukeInstituteofGenomeSciencesandPolicy. [Accessed 20 Sep 2009];Website Glossary. http://www.genomestohealth.org/glossary.php.
- 2.Barisoni L, Schnaper HW, Kopp JB. Advances in the biology and genetics of the podocytopathies: implications for diagnosis and therapy. Arch Pathol Lab Med. 2009;133(2):201–216. doi: 10.1043/1543-2165-133.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woroniecki RP, Kopp JB. Genetics of focal segmental glomerulosclerosis. Pediatr Nephrol. 2007;22(5):638–644. doi: 10.1007/s00467-007-0445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastian HM, et al. Systemic lupus erythematosus in three ethnic groups. XII. Risk factors fro lupus nephritis after diagnosis. lupus. 2002;11:152–160. doi: 10.1191/0961203302lu158oa. [DOI] [PubMed] [Google Scholar]
- 5.Burt VL, Whelton P, Roccella EJ, Brown C, Cutler JA, Higgins M, Horan MJ, Labarthe D. Prevalence of Hypertension in the US Adult Population : Results From the Third National Health and Nutrition Examination Survey, 1988–1991. Hypertension. 1995;25(3):305–313. doi: 10.1161/01.hyp.25.3.305. [DOI] [PubMed] [Google Scholar]
- 6.Davey smith G, Neaton JD, Wentworth D, Stamler R, Stamler J. Mortality differences between black and white men in the USA: contribution of income and other risk factors among men screened for the MRFIT. Lancet. 1998;351:934–939. doi: 10.1016/s0140-6736(00)80010-0. [DOI] [PubMed] [Google Scholar]
- 7.Gupta V, et al. Racial differences in thoracic aorta atherosclerosis among ischemic stroke patients. Stroke. 2003;34:408–412. doi: 10.1161/01.str.0000050643.32175.89. [DOI] [PubMed] [Google Scholar]
- 8.Harper MA, et al. Racial disparty in pregnancy-related mortality following a live birth outcome. Ann. Epidemiol. 2004;14:274–279. doi: 10.1016/S1047-2797(03)00128-5. [DOI] [PubMed] [Google Scholar]
- 9.Kitiyakara C, Eggers P, Kopp JB. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. American Journal of Kidney Diseases. 2004;44(5):815–825. [PubMed] [Google Scholar]
- 10.Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Stamler J. End-stage renal disease in African-American and white men. 16-year MRFIT findings. JAMA. 1997;277(16):1293–1298. [PubMed] [Google Scholar]
- 11.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans1. Kidney Int. 2003;63(S83):S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 12.Lucas GM, Lau B, Atta MG, Fine DM, Keruly J, Moore RD. Chronic Kidney Disease Incidence, and Progression to End,ÄêStage Renal Disease, in HIV,ÄêInfected Individuals: A Tale of Two Races. The Journal of Infectious Diseases. 2008;197(11):1548–1557. doi: 10.1086/587994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signorello LB, et al. Comparing Diabetes Prevalence Between African Americans and Whites of Similar Socioeconomic Status. American Journal of Public Health. 2007;97(12):2260–2267. doi: 10.2105/AJPH.2006.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. 2005;6(8):623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 15.Young BE, Maynard C, Boyko EJ. Racial differences in diabetic nephropathy, cardiovascular disease, and mortality in a national population of veterans. Diabetes Care. 2003;26(8):2393–2399. doi: 10.2337/diacare.26.8.2392. [DOI] [PubMed] [Google Scholar]
- 16.Tucker MJ, Berg CJ, Callaghan WM, Hsia J. The Black-White disparity in pregnancy-related mortality from 5 conditions: differences in prevalence and case-fatality rates. Am J Public Health. 2007;97(2):247–251. doi: 10.2105/AJPH.2005.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao TK, Filippone EJ, Nicastri AD, Landesman SH, Frank E, Chen CK, Friedman EA. Associated focal and segmental glomerulosclerosis in the acquired immunodeficiency syndrome. N Engl J Med. 1984;310(11):669–673. doi: 10.1056/NEJM198403153101101. [DOI] [PubMed] [Google Scholar]
- 18.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans. Kidney Int Suppl. 2003;(83):S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 19.Tonna SJ, Needham A, Polu K, Uscinski A, Appel GB, Falk RJ, Katz A, Al-Waheeb S, Kaplan BS, Jerums G, et al. NPHS2 variation in focal and segmental glomerulosclerosis. BMC Nephrol. 2008;9:13. doi: 10.1186/1471-2369-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenzie LM, Hendrickson SL, Briggs WA, Dart RA, Korbet SM, Mokrzycki MH, Kimmel PL, Ahuja TS, Berns JS, Simon EE, et al. NPHS2 variation in sporadic focal segmental glomerulosclerosis. J Am Soc Nephrol. 2007;18(11):2987–2995. doi: 10.1681/ASN.2007030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan KT, Papeta N, Martino J, Zheng Z, Frankel RZ, Klotman PE, D'Agati VD, Lifton RP, Gharavi AG. Accelerated development of collapsing glomerulopathy in mice congenic for the HIVAN1 locus. Kidney Int. 2009;75(4):366–372. doi: 10.1038/ki.2008.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bock CH, Schwartz AG, Ruterbusch JJ, Levin AM, Neslund-Dudas C, Land SJ, Wenzlaff AS, Reich D, McKeigue P, Chen W, et al. Results from a prostate cancer admixture mapping study in African-American men. Hum Genet. 2009 doi: 10.1007/s00439-009-0712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 25.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 26.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diergaarde B, Bowen DJ, Ludman EJ, Culver JO, Press N, Burke W. Genetic information: Special or not? Responses from focus groups with members of a health maintenance organization. Am J Med Genet A. 2007;143(6):564–569. doi: 10.1002/ajmg.a.31621. [DOI] [PubMed] [Google Scholar]
- 28.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 30.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 31.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. A Catalog of Published Genome-Wide Association Studies. 2009 [Google Scholar]
- 33.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40(10):1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends Ecol Evol. 2008;23(12):686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Cooper RS, Tayo B, Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet. 2008;17(R2):R151–R155. doi: 10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Divers J, Moossavi S, Langefeld CD, Freedman BI. Genetic admixture: a tool to identify diabetic nephropathy genes in African Americans. Ethn Dis. 2008;18(3):384–388. [PubMed] [Google Scholar]
- 39.Zhu X, Tang H, Risch N. Admixture mapping and the role of population structure for localizing disease genes. Adv Genet. 2008;60:547–569. doi: 10.1016/S0065-2660(07)00419-1. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty R, Kamboh MI, Ferrell RE. 'Unique' alleles in admixed populations: a strategy for determining 'hereditary' population differences of disease frequencies. Ethn Dis. 1991;1(3):245–256. [PubMed] [Google Scholar]
- 41.Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O'Brien SJ. Markers for mapping by admixture linkage disequilibrium in african american and hispanic populations. Am J Hum Genet. 2001;69(5):1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74(5):965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freedman BI, Kopp JB, Winkler CA, Nelson GW, Rao DC, Eckfeldt JH, Leppert MF, Hicks PJ, Divers J, Langefeld CD, et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: the HyperGEN study. Am J Nephrol. 2009;29(6):626–632. doi: 10.1159/000194791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freedman BI, Hicks PJ, Bostrom MA, Comeau ME, Divers J, Bleyer AJ, Kopp JB, Winkler CA, Nelson GW, Langefeld CD, et al. Non-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRD. Nephrol Dial Transplant. 2009 doi: 10.1093/ndt/gfp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freedman BI, Bowden DW, Rich SS, Xu J, Wagenknecht LE, Ziegler J, Hicks PJ, Langefeld CD. Genome-wide linkage scans for renal function and albuminuria in Type 2 diabetes mellitus: the Diabetes Heart Study. Diabet Med. 2008;25(3):268–276. doi: 10.1111/j.1464-5491.2007.02361.x. [DOI] [PubMed] [Google Scholar]
- 46.Althaus K, Greinacher A. MYH9-related platelet disorders. Semin Thromb Hemost. 2009;35(2):189–203. doi: 10.1055/s-0029-1220327. [DOI] [PubMed] [Google Scholar]
- 47.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6(12):e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10(9):639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oleksyk TK, Nelson GW, Jamba M, Winkler CA, Smith MW. Interrogating genomic region containing major effect risk gene for renal disease for signatures of recent selection in human populations. Philadelphia, PA: 2008. p. 2523. [Google Scholar]
- 52.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. 2. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinker S. [Accessed 21 Sept 2009];My Genome, My Self. New York Times. 2009 http://www.nytimes.com/2009/01/11/magazine/11Genome-t.html?pagewanted=6&_r=3.
- 55.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci U S A. 1978;75(11):5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437(7062):1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 57.Andres AM, Hubisz MJ, Indap A, Torgerson DG, Degenhardt JD, Boyko AR, Gutenkunst RN, White TJ, Green ED, Bustamante CD, et al. Targets of balancing selection in the human genome. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kriz W. The pathogenesis of 'classic' focal segmental glomerulosclerosis-lessons from rat models. Nephrol Dial Transplant. 2003;18 Suppl 6:vi39–vi44. doi: 10.1093/ndt/gfg1064. [DOI] [PubMed] [Google Scholar]
- 59.Fabian J, Naicker S. HIV and kidney disease in sub-Saharan Africa. Nat Rev Nephrol. 2009;5(10):591–598. doi: 10.1038/nrneph.2009.141. [DOI] [PubMed] [Google Scholar]
- 60.Khoury MJ, McBride CM, Schully SD, Ioannidis JP, Feero WG, Janssens AC, Gwinn M, Simons-Morton DG, Bernhardt JM, Cargill M, et al. The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009;11(8):559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freedman BI, Hicks PJ, Bostrom MA, Cunningham ME, Liu Y, Divers J, Kopp JB, Winkler CA, Nelson GW, Langefeld CD, et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009;75(7):736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]