Abstract
We mapped two new recessive mutations causing circling behavior and deafness to the same region on chromosome 7 and showed they are allelic by complementation analysis. One was named ‘deaf circler’ (allele symbol dfcr) and the other ‘deaf circler 2 Jackson’ (allele symbol dfcr-2J). Both were shown to be mutations of the Ush1c gene, the mouse ortholog of the gene responsible for human Usher syndrome type IC and for the non-syndromic deafness disorder DFNB18. The Ush1c gene contains 28 exons, 20 that are constitutive and eight that are alternatively spliced. The dfcr mutation is a 12.8 kb intragenic deletion that eliminates three constitutive and five alternatively spliced exons. The dfcr-2J mutation is a 1 bp deletion in an alternatively spliced exon that creates a transcriptional frame shift, changing 38 amino acid codons before introducing a premature stop codon. Both mutations cause congenital deafness and severe balance deficits due to inner ear dysfunction. The stereocilia of cochlear hair cells are disorganized and splayed in mutant mice, with subsequent degeneration of the hair cells and spiral ganglion cells. Harmonin, the protein encoded by Ush1c, has been shown to bind, by means of its PDZ-domains, with the products of other Usher syndrome genes, including Myo7a, Cdh23 and Sans. The complexes formed by these protein interactions are thought to be essential for maintaining the integrity of hair cell stereocilia. The Ush1c mutant mice described here provide a means to directly investigate these interactions in vivo and to evaluate gene structure–function relationships that affect inner ear and eye phenotypes.
INTRODUCTION
As part of our research program at The Jackson Laboratory to identify genes causing hearing impairment in mice, we have been selecting and studying mutant mice that exhibit circling or head tossing behavior. This behavior is characteristic of vestibular dysfunction of the inner ear and often is associated with deafness. We mapped two new recessive mutations causing such circling behavior and deafness to the same region on chromosome (Chr) 7. The two independently occurring mutations were shown to be allelic by complementation analysis. The one first discovered was named ‘deaf circler’ (allele symbol dfcr), and the other was given the name ‘deaf circler 2 Jackson’ (allele symbol dfcr-2J), in accordance with nomenclature guidelines. In this paper we describe the phenotypes of these two new spontaneous mutations and show that both are mutations of the Ush1c gene, the mouse ortholog of the gene responsible for human Usher syndrome type IC and the non-syndromic deafness disorder DFNB18.
The Usher syndromes are a genetically heterogeneous group of recessive disorders that cause both deafness and blindness. They can be divided clinically into three subtypes based on the severity and progression of associated deafness, vestibular dysfunction, and retinitis pigmentosa (RP). Usher syndrome type I (USH1), the most severe form, is characterized by profound congenital sensory deafness, vestibular dysfunction, and progressive RP with onset by age 10. With the inclusion of the Ush1c mouse mutations described in this paper, mouse models now exist for all five of the known USH1 genes (Table 1). USH1C is a subtype of Usher syndrome type I that was localized to Chromosome 11 by linkage analysis of Acadian families in Louisiana (1,2). It was later shown to be caused by mutations in a gene designated USH1C, which encodes a PDZ-domain-containing protein named harmonin (3,4). Mutations of this same gene are also responsible for the non-syndromic recessive disorder DFNB18 (5–7).
Table 1.
Mouse models for human Usher syndromes. Some of these same genes are also responsible for non-syndromic deafness (marked by asterisks)
| Usher syndromes, type I | Locus | Gene (protein) | Human references | Mouse models (representative mutation) |
Mouse references |
|---|---|---|---|---|---|
| USH1B | 11q13 | MYO7A | (34–37) | Shaker 1 | (21) |
| DFNB2, DFNA11* | (myosin VIIa) | (Myo7ash1) | |||
| USH1C | 11p15 | USH1C | (3–6) | Deaf circler | This paper |
| DFNB18* | (harmonin) | (Ush1cdfcr) | |||
| USH1D | 10q21–q22 | CDH23 | (38,39) | Waltzer | (20,40) |
| DFNB12* | (cadherin 23) | (Cdh23v) | |||
| USH1F | 10q21–q22 | PCDH15 | (41,42) | Ames waltzer | (43) |
| (protocadherin 15) | (Pcdh15av) | ||||
| USH1G | 17q24–q25 | SANS | (11) | Jackson shaker | (23) |
| (sans) | (Sans js) |
The USH1C gene is composed of 28 exons, including 20 that are constitutive and 8 that are alternatively spliced in the mouse (4). These alternative transcripts encode multiple harmonin isoforms, including two type ‘a’ isoforms, which are the most abundant and widely expressed, and three longer type ‘b’ isoforms, which are more restricted in their tissue distribution. The human protein also exhibits multiple tissue-specific isoforms (8). The lack of RP in DFNB18 patients was attributed to the nature of the USH1C mutations, which occurred in the alternatively spliced exons that encode the longer harmonin b isoforms expressed in the inner ear but not the eye (6).
The two new Ush1c mutants described in this paper provide important mouse models for studying the pathogenesis and potential therapies for human USH1C. One of the mutations is a large deletion that includes both constitutive and alternatively spliced exons and alters both a and b isoforms; the other causes a frame-shift in one of the alternatively transcribed exons and alters only b isoforms. These qualitatively different mutations provide a means to evaluate the functional domains of the Ush1c gene. The mutations also will be valuable for identifying molecular pathways and gene interactions important in hearing and vision. The Ush1c gene product, harmonin, is thought to interact, by means of its PDZ-binding domain, with the products of three other Usher syndrome genes, Myo7a, Cdh23, and Sans (9–11). PDZ-domains are known to facilitate protein complex assembly at particular subcellular locations, especially at the plasma membrane (12,13). The Ush1c mutant mice described here can be used to directly investigate these molecular interactions in vivo, by producing and analyzing mice with compound mutations.
RESULTS
Origins and genetic mapping of dfcr and dfcr-2J
The dfcr mutation arose spontaneously at The Jackson Laboratory in a strain with a predominantly BALB/cByJ background (see Materials and Methods). The dfcr-2J mutation arose spontaneously in a stock derived from 129S4/SvJae ES cells and C57BL/6J mice that carries a targeted mutation of beta adducin, Add2 (14). The mutant strain has been crossed and maintained so that it no longer contains the Add2 targeted mutation.
All dfcr and dfcr-2J mutants exhibit head tossing and hyperactive circling behavior and are unable to swim. These phenotypes are inherited as recessive traits with full penetrance in homozygotes. Because of their similar phenotypes and map locations (see below), we tested the two mutations for allelism by complementation analysis. Two matings between pairs of homozygous mice (dfcr/dfcr × dfcr-2J/dfcr-2J) produced a total of nine progeny all with mutant phenotypes, confirming that the two mutations were allelic. Both mutant male and mutant female (dfcr/dfcr and dfcr-2J/dfcr-2J) mice have normal life spans and can breed.
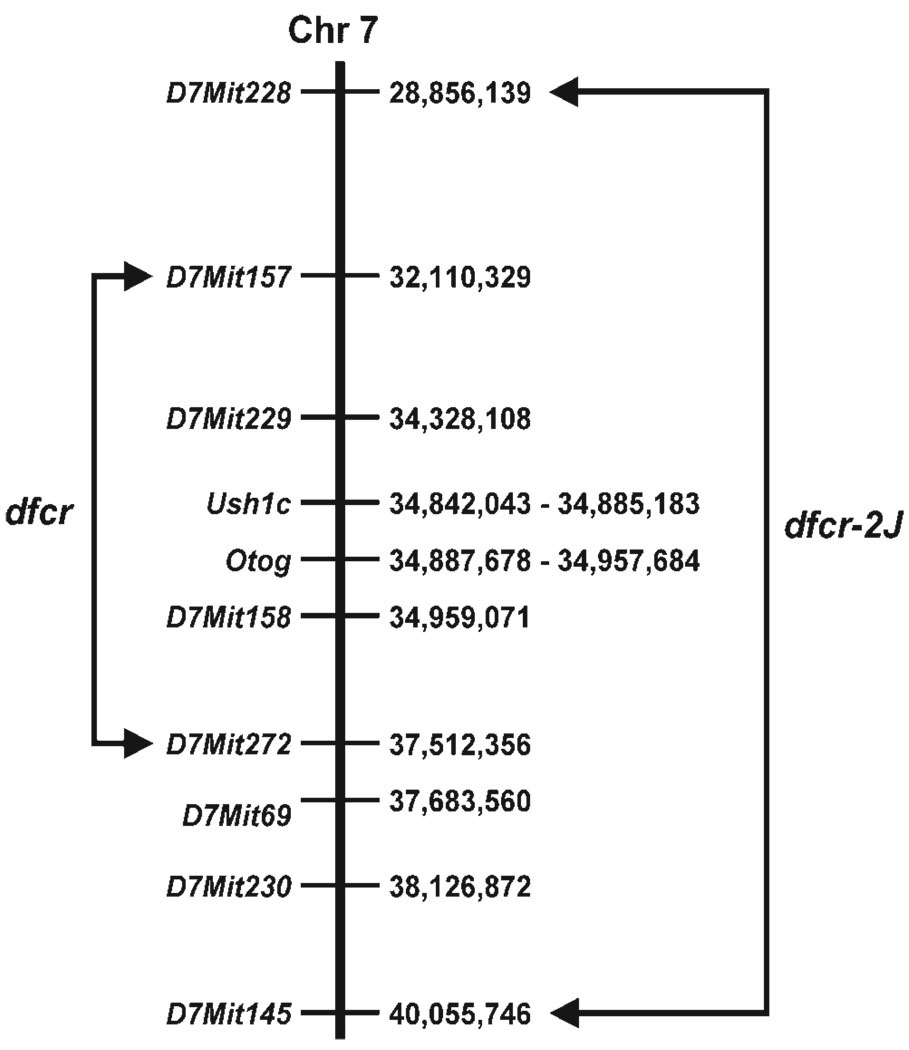
By linkage intercrosses with CAST/Ei, we mapped the dfcr and dfcr-2J mutations to the same location on Chr-7. Analysis of 54 F2 mice (108 meioses) positioned dfcr between D7Mit157 (3.7% recombination) and D7Mit272 (1.9% recombination), non-recombinant with D7Mit158. Analysis of 93 F2 mice (186 meioses) placed the dfcr 2J mutation between D7Mit228 (4.8% recombination) and D7Mit145 (5.4% recombination), non-recombinant with D7Mit229, D7Mit69 and D7Mit230.
Candidate genes
The map location of dfcr and dfcr-2J suggested they might be mutations of the otogelin gene (Otog), which is located within the genetically defined intervals containing these mutations (Fig. 1). Mice with targeted mutations of Otog (15) and mice with the spontaneous twister (twt) mutation of Otog (16) have vestibular and hearing impairment similar to dfcr and dfcr-2J mutants. To test for allelism, we set up a mating between a homozygous dfcr/dfcr female and a homozygous twister (Otogtwt/Otogtwt) male. All five offspring from this cross exhibited normal hearing and behavior; therefore, we concluded that dfcr is not a mutation of Otog.
Figure 1.
Genetically derived candidate gene intervals for dfcr and dfcr-2J. The Ensembl base pair locations (from the NCBI 30 Mouse Genome Assembly, May 12, 2003) along Chr 7 are shown to the right of each marker and candidate gene. Arrows connect recombinant flanking markers for each linkage cross, delimiting the candidate gene intervals for each mutation.
The Ush1c gene is also located in the dfcr and dfcr-2J candidate regions, less than 10 kb from Otog (Fig. 1). No mouse mutations of Ush1c have been reported, but human mutations of USH1C underlie both syndromic (Usher syndrome type IC) and non-syndromic (DFNB18) deafness disorders (3–6). Because of the map location and association with deafness in humans, we next considered Ush1c as a likely candidate for the dfcr and dfcr-2J mutations.
We designed gene-specific probes for Southern blots and PCR primers for genomic and cDNA sequence analyses according to the mouse Ush1c cDNA sequences in GenBank (accession numbers NM_023649 and AY103465) and the exon–intron structure and nomenclature described by Verpy et al. (4). Intronic DNA sequences were obtained from the public mouse genome sequence (Ensembl Gene ID ENSMUSG00000030838). The DNA sequences of all PCR primers used in this study are given in Table 2.
Table 2.
PCR primers used in this study. Primers designated ‘fex’ are located in intronic sequences immediately flanking an exon, those designated ‘ex’ are located within exons, and those designated ‘in’ are located within introns. An ‘F’ indicates a forward primer and an ‘R’ a reverse primer relative to Ush1c transcription
| Forward primer | Reverse primer | Product size (bp) | ||
|---|---|---|---|---|
| Mutation screening | ||||
| fex1-F | GTCCTGTCCTGGCAGCTAAC | fex1-R | GCCTCCACAGACCCTGTAAT | 221 |
| fex2-F | GATGTCTCCTCCCCAGGTCT | fex2-R | TGCCCACTCTGAATTCCTGT | 268 |
| fex3-F | CCACCCTCTGACATCTCCAA | fex4-R | CCCGCTCTGATGACTTACCT | 435 |
| fex5-F | CTGTGCCTGTTGCATCTCTC | fex7-R | TCCTTCTTCAAGGCCAGCTA | 547 |
| fex8-F | GGCCACTTGAAGGCATGTAG | fex8-R | TTGACCAGCTGTCACTGAGC | 225 |
| fex9-F | CAGAAGGCTACAGGCTGGTC | fex9-R | GACGCAGCATCTGTCAGTGT | 247 |
| fex10-F | CTGTGGGATCACTGGCCTAT | fex10-R | CTGGAAGACGTGTGTGGAGA | 250 |
| fex10-F | CACACACTGACCCGTTTTTCT | fex12-R | TCAGTTGTGTCTGGTGTGGAA | 594 |
| fex13-F | CAGAACCACACCCACAGTTG | fex13-R | GGTGGATTTGAGCCAATGAA | 358 |
| fex14-F | AGGTTCTGCCATCAATCACA | fex14-R | ATCCCGGACAGGTCATACAT | 197 |
| fex15-F | CACACCAGAGCAAGGAGACC | fex15-R | ATGGTCCAGCTCTTTCAAGC | 322 |
| fex16-F | CCATCCCAACCACAGCTACT | fex16-R | CCCTCTGAGCTGGAGAGTTC | 221 |
| fex17-F | AGGCCACACATGCTCTTACC | fex17-R | AGGGAACCAAGACGACAGAG | 178 |
| fex18-F | CTATCTGTGTGGCTGCCTGA | fex18-R | TCTGCTCTCCTGGGACATCT | 212 |
| fex19-F | GAGTCAGGCACAGGTCACAA | fex19-R | TTGTAAGGCCAGGGTCTGAG | 225 |
| fex20-F | GTCAGAACACCAGGCAGGTT | fex20-R | GGACATCCCTTTCCCTGAAT | 220 |
| fex21-F | GCAAGTCCTGTCCACTCCAT | fex21-R | TGCTGTCCACACAATTCACC | 454 |
| fexA-F | ACATACCTTGGGAACGCAAC | fexA-R | CACATCAAACTTGGCAGCAC | 173 |
| fexB-F | CACAATGCTCCATCTGCTCA | fexB-R | ATCTGGCCCTTGGAGGTATG | 232 |
| fexC-F | TACCCATGTGACTCGGTGTG | fexC-R | GTGACTGTCAGGAGGGTCCA | 226 |
| fexD-F | GGATCCAGCAGCAAGAGAAA | exD-2R | CGGGTAGGAGTGAGGTCTTG | 413 |
| exD-2F | CACACCACTGACCTGGATGA | fexD-R | CTTCCTTTGCAAGCACAACA | 437 |
| fexE-F | ATGACGCCACATGGGTATCT | fexE-R | TCACTTCCCTTCCAGAGGTG | 248 |
| fexF-F | CCGAACATCAGCCTATCTGG | fexF-R | CCAGACACACGGGTCTCATT | 202 |
| fexG-F | CCTCTTGTCTCCTGCTCCAC | fexG-R | CCAGTCCAAAGAACCTCCAG | 227 |
| dfcr genotyping | ||||
| In11-F | GAGCCGTAAGTCCAGAATGC | ex12-R | TGCTCCTGGAGGATCTTGTT | |
| inD-R | TCATCAGAGGCTGCTGACAC | |||
| dfcr-2J genotyping | ||||
| mutC-F | TACCCATGTGACTCGGTGTG | mutC-R | AAGGTCTTCCCGCTCAGTCTGTG | |
| Transcription analysis | ||||
| ex8-F | CGGACAACCAAGGAGAAGAA | ex11-R | GGCTGCTCTTCAGGACATTC | |
| ex10-F | ACTTCACCAACCTGGACCAC | exG-R | GCTGCAGGAAAAACCCGTGTCT | |
| ex21-R | TGTGGGTCAGCTTCCAGGGAGACTTT |
Molecular characterization of the dfcr and dfcr-2J mutations
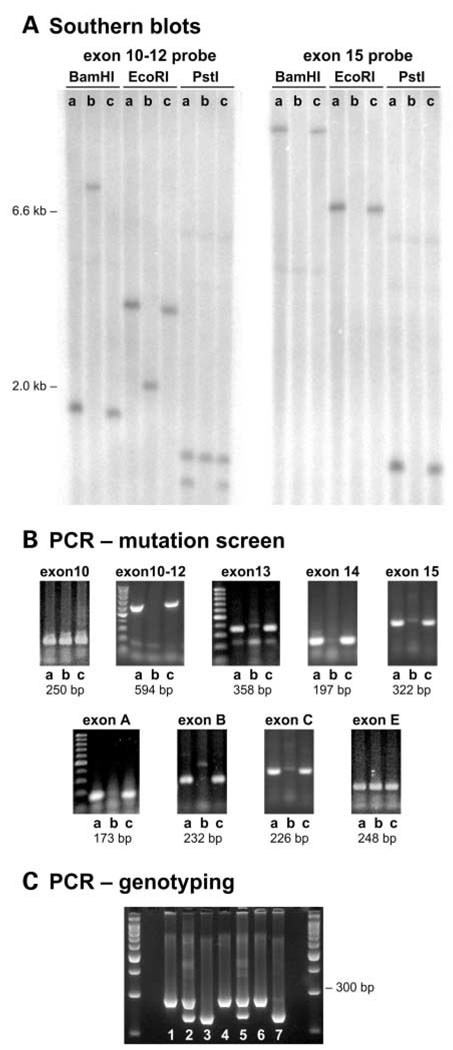
Genomic DNA from dfcr and dfcr-2J mutant mice and their controls was examined for gross alterations in the Ush1c gene by Southern blot analysis. DNA from dfcr-2J mutant mice gave the same Ush1c restriction fragment lengths as wild-type control mice for all restriction enzymes and probes used. DNA from dfcr mutant mice, however, showed altered sizes or missing fragments when hybridized with probes containing Ush1c exons 12, 13, 14, 15, A, B, C or D (Fig. 2A). PCR primers flanking these same exons failed to amplify products from genomic DNA of dfcr/dfcr mice, but amplified the expected-sized products from DNA of wild-type control and dfcr-2J/dfcr-2J mice (Fig. 2B). These results indicate that the dfcr mutation is a large deletion of Ush1c including exons 12-D.
Figure 2.
The dfcr mutation of Ush1c. Genomic DNA samples from BALB/cByJ +/+control (lane a), dfcr/dfcr (lane b), and dfcr-2J/dfcr-2J (lane c) mice were compared. (A) Southern blot analysis. Restriction fragments from dfcr/dfcr mice are differently sized than those of controls and dfcr-2J/dfcr-2J mice when cDNA corresponding to exons 10–12 of Ush1c is used as a probe and absent when an exon 15 probe is used. (B) RT–PCR analysis. PCR primers flanking Ush1c exons 10–12, 13, 14, 15, A, B, and C amplified products with +/+ and dfcr-2J/dfcr-2J DNA but not with dfcr/dfcr DNA. Primers flanking exons 10 and E amplified products in all three DNA samples, indicating that the dfcr deletion lies between these two exons. (C) Genotyping mice with the dfcr mutation. Two primers flanking the deletion (in11-F and inD-R, Table 2), amplify a 282 bp product only in mutant DNA. The in11-F primer and a primer within the deletion (ex12-R, Table 2), amplify a 234 bp product only in wildtype DNA. The three primers were used together in a single PCR reaction to amplify genomic DNA from +/+ (lanes 3 and 7), +/dfcr (lanes 2 and 5), and dfcr/dfcr (lanes 1, 4, and 6) mice. A 3% agarose gel was used to resolve the products shown here; the two outer lanes contain 100 bp size ladders.
We designed PCR primers (in11-F and inD-R; Table 2) on either side of the deleted region and sequenced the resulting product from dfcr/dfcr genomic DNA. The DNA sequence exactly matched the mouse Ush1c gene sequence (Ensembl Gene ID ENSMUSG00000030838) from the forward primer position in intron 11 through the first 25 bp of exon 12; it then skipped to a position 110 bp past the 3′ end of exon D and continued to the position of the reverse primer in intron D. This result indicates that 12 770 bp are deleted in the mutant DNA, including 118 bp from exon 12 and exons 13, 14, 15, A, B, C and D. No other genomic rearrangements were detected that are associated with this deletion.
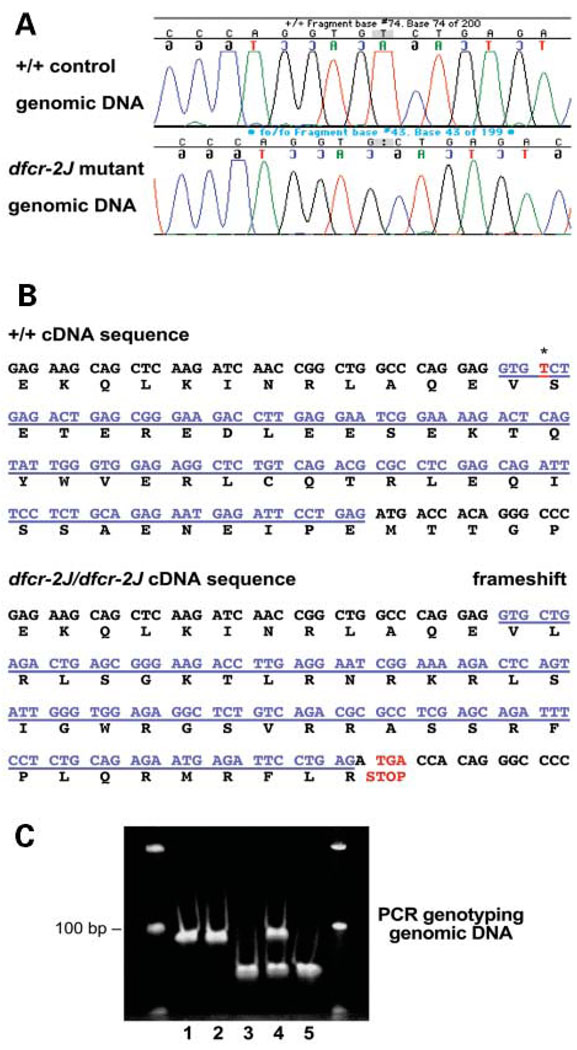
The dfcr-2J mutation was not detected by Southern blot analysis, so the DNA sequence of all 28 Ush1c exons and their splice junctions was compared between mutant mice and +/+ controls. PCR primers flanking each exon (Table 2) were designed to amplify genomic DNA for sequence analysis. The only sequence difference detected was a 1 bp deletion at position 4 of exon C in mutant DNA (Fig. 3A). The deletion creates a transcriptional frame shift that changes the codons for 38 amino acids before introducing a stop codon at the beginning of exon D (Fig. 3B). DNA from two additional dfcr-2J/dfcr-2J mutant mice was sequenced to confirm this 1 bp deletion, which was not observed in DNA from any of the inbred strains we examined (C57BL/6J, 129S1/SvImJ or BALB/cByJ), nor in any of the Genbank database entries for mouse, rat, or human genomic DNA, mRNAs, or ESTs.
Figure 3.
The dfcr-2J mutation of Ush1c. (A) DNA sequence chromatogram of C57BL/6J +/+ control compared with homozygous dfcr-2J mutant, illustrating the deletion of a single bp (T/A) in mutant DNA. (B) The 1 bp deletion is at position 4 (indicated by an asterisk) of exon C (underlined in blue), which causes a frameshift and results in a transcription stop codon (shown in red) at the beginning of exon D. (C) Genotyping mice with the dfcr-2J mutation. Primers mutC-F and mutC-R, flanking the dfcr-2J mutation in exon C (Table 2), were used to amplify genomic DNA from +/+ (lanes 1 and 2), +/dfcr-2J (lane 4), and dfcr-2J/dfcr-2J (lanes 3 and 5) mice, progeny from matings between +/dfcr-2J parents. The reverse primer, mutC-R, ends immediately adjacent to the point of the 1 bp deletion in exon C. Two nucleotides in this primer (underlined in Table 2) were changed (to GT from CA) to create an ApaL1 restriction site (GTGCAC) in mutant but not wildtype DNA. After incubation with the restriction endonuclease ApaL1, the wild-type 93 bp product is unaffected, whereas the mutant product is cleaved into 70 and 22 bp fragments. A 6% acrylamide gel was used to resolve the DNA fragments shown here; the two outer lanes contain 100 bp size ladders.
Effects of the Ush1c mutations on transcription and translation
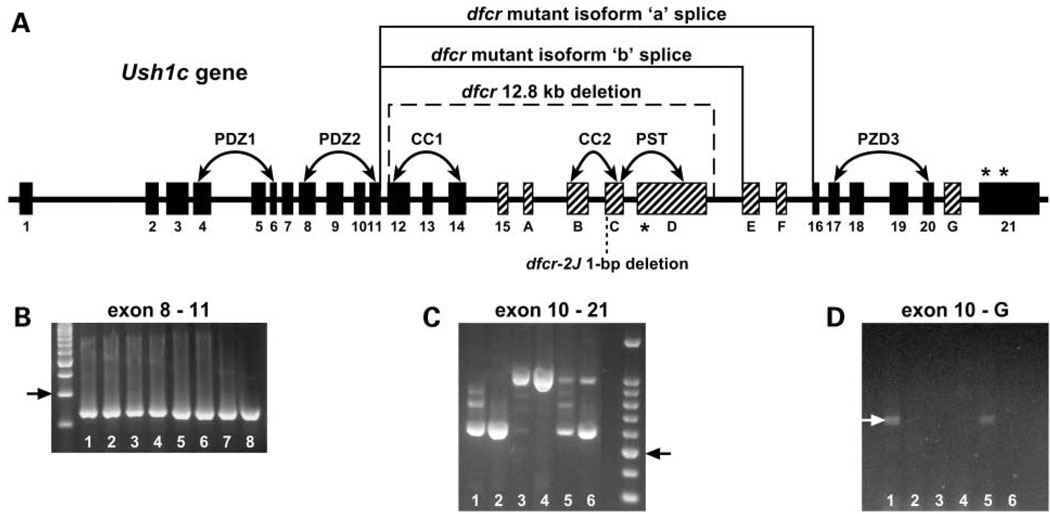
The mouse Ush1c gene consists of 28 exons spanning more than 43 kb of genomic DNA. Eight of the 28 exons are alternatively spliced: exons 15, A, B, C, D, E, F, and G (4). These alternative transcripts encode multiple harmonin isoforms, including two type a isoforms, which are the most abundant and widespread forms and usually include exon 15. Three type b isoforms are more restricted in their tissue distribution and include exons A, B, C, D, E, F and sometimes G or G′. Transcripts containing exons A–F and exon G/G′ appear to be expressed only in the inner ear. The exon–intron structure of the Ush1c gene and the positions of the dfcr and dfcr-2J mutations are diagramed in Figure 4A.
Figure 4.
Transcription of dfcr and dfcr-2J mutations. (A) Ush1c exon–intron structure. Exons are represented as rectangles and introns as lines. Exons (greatly exaggerated in size relative to introns) are designated according to Verpy et al. (4), with the 20 constitutive exons shown in black and the eight alternatively spliced exons in gray. A dashed line indicates the extent of the dfcr deletion and a dotted line marks the position of the dfcr-2J mutation. The dfcr deletion causes an inframe splicing of exon 11 with exon 16 in transcripts encoding the ‘a’ isoform of harmonin and an in-frame splicing of exon 11 with exon E in transcripts encoding the ‘b’ isoform. An asterisk in exon D marks the stop codon created by the dfcr-2J mutation, and asterisks in exon 21 denote the normal stop codons, unchanged by the dfcr mutation. Connected arrows indicate genomic regions encoding the PDZ, CC (coiled-coil), and PST (proline, serine, threonine-rich) domains of the harmonin protein. (B) Ush1c transcripts are detected in both dfcr and dfcr-2J mutant mice. The primers ex8-F (located within exon 8) and ex11-R (located within exon 11) amplified the expected 250 bp product in cDNA derived from inner ear (lanes 1–4) and kidney (lanes 5–8) of +/dfcr mice (lanes1 and 5), dfcr/dfcr mice (lanes 2 and 6), +/dfcr-2J mice (lanes 3 and 7), and dfcr-2J/dfcr-2J mice (lanes 4 and 8). Other tissues examined but not shown here (eye, brain, intestine) gave similar results. The arrow to the left of the figure indicates the position of the 300 bp DNA size marker. (C) Exons12, 13, 14 and 15 (396 nt) are deleted from isoform ‘a’ transcripts in dfcr/dfcr mice. The PCR products amplified with primers ex10-F (located within exon 10) and ex21-R (located within exon 21) from cDNAs derived from inner ear (lanes 1, 3 and 5) and intestine (lanes 2, 4 and 6) were smaller in dfcr/dfcr mice (574 bp; lanes 1 and 2) than those from +/+ mice (970 bp; lanes 3 and 4); both sizes were amplified in heterozygotes (lanes 5 and 6). Sequence analysis of the mutant 574 bp PCR product revealed an in-frame splicing of exon 11 with exon 16. The arrow to the right of the figure indicates the position of the 500 bp DNA size marker. (D) Exons 12, 13, 14, A, B, C and D (1137 nt) are deleted from isoform ‘b’ transcripts in dfcr/dfcr mice. A faint ~700 bp PCR product was amplified from inner ear cDNA with primers ex10-F and exG-R (located within exon G) of dfcr/dfcr (lane 1) and +/dfcr (lane 5) mice, but not from +/+ mice (lane 3) because of the large distance between exons 10 and G (>1800 nt) in wildtype cDNA. No product was amplified from intestine-derived cDNA of any genotype (lanes 2, 4 and 6). Sequence analysis of the mutant product from inner ear (indicated by white arrow) revealed an in-frame splicing of exon 11 with exon E.
To evaluate how the dfcr and dfcr-2J mutant genes are transcribed, RNA was extracted from mutant and control mice and converted to cDNA for PCR analysis. Ush1c transcripts were detected in dfcr/dfcr and dfcr-2J/dfcrf-2J mice in all tissues examined (eye, inner ear, brain, kidney and intestine); results for inner ear and kidney are shown in Figure 4B. The RT–PCR product sizes from dfcr-2J/dfcrf-2J mice were the same as those from control mice for all Ush1c regions examined. Transcript size is not be affected by the dfcr-2J mutation, but the translated protein would be altered as described above (Fig. 3B).
For dfcr/dfcr mutants, primers corresponding to DNA sequences located 5′ of the deletion (Fig. 4B) gave the same sized products as controls, whereas primers that spanned the deleted region amplified smaller products (Fig. 4C). Sequence analysis of the mutant 574 bp PCR product (Fig. 4C, lanes 1 and 2) that was amplified with primers specific to exons 10 and 21 revealed an in-frame splicing of exon 11 with exon 16. This mutant transcript, present in all tissues examined, lacks exons 12, 13, 14 and 15 (396 nt), which encode 132 of the 548 amino acids of harmonin isoform a1 (4). Sequence analysis of a faint ~700 bp PCR product amplified from inner ear cDNA of dfcr/dfcr mice with primers specific to exon 10 and exon G (Fig. 4D) revealed an in-frame splicing of exon 11 with exon E. This mutant inner ear-specific transcript lacks exons 12, 13, 14, A, B, C and D (1137 nt), which encode 379 of the 852–910 amino acids of harmonin isoform b (4). The dfcr mutant splice forms do not change the normal reading frame of the Ush1c transcripts and, therefore, do not introduce any new amino acid or stop codons.
Inner ear pathology
The vestibular dysfunction of mutant mice suggested that they might also be hearing impaired. We assessed hearing in mutant and control mice by auditory-evoked brainstem response (ABR) thresholds to broad-band clicks and to 8, 16 and 32 kHz pure-tone stimuli (Table 3), according to previously established criteria (17). ABR tests of eight homozygous dfcr mutants (one tested at 21 days, four at 32 days, and three at 57 days of age) indicated they were completely deaf (no detectable ABR with 100 dB SPL stimuli), whereas seven heterozygotes (one tested at 21 days, two at 32 days, two at 88 days and two at 176 days of age) had ABR thresholds in the normal-hearing range. Two mice homozygous for the dfcr-2J mutation were tested by ABR analysis at 98 days of age and found to be completely deaf, whereas four age-matched heterozygotes had normal hearing.
Table 3.
ABR thresholds of mutant and control mice. The number, gender (f = female, m = male), and ages of mice tested are given for each genotype. ABR threshold values below 55 (click, 8 kHz) 35 (16 kHz) or 60 (32 kHz) are considered within the normal range for mice. The maximum amplitude presented for each test stimulus was 100 dB
| Genotype | Number | Age (days) | Average ABR thresholds (dB SPL) | |||
|---|---|---|---|---|---|---|
| Click | 8 kHz | 16 kHz | 16 kHz | |||
| dfcr/+ | 7 (4 f, 3 m) | 21–176 | 37 | 46 | 21 | 50 |
| dfcr/dfcr | 8 (4 f, 4 m) | 21–82 | 100 | 100 | 100 | 100 |
| dfcr-2J/+ | 4 (3 f, 1 m) | 98 | 40 | 50 | 20 | 50 |
| dfcr-2J/dfcr-2J | 2 (2 f) | 98 | 100 | 100 | 100 | 100 |
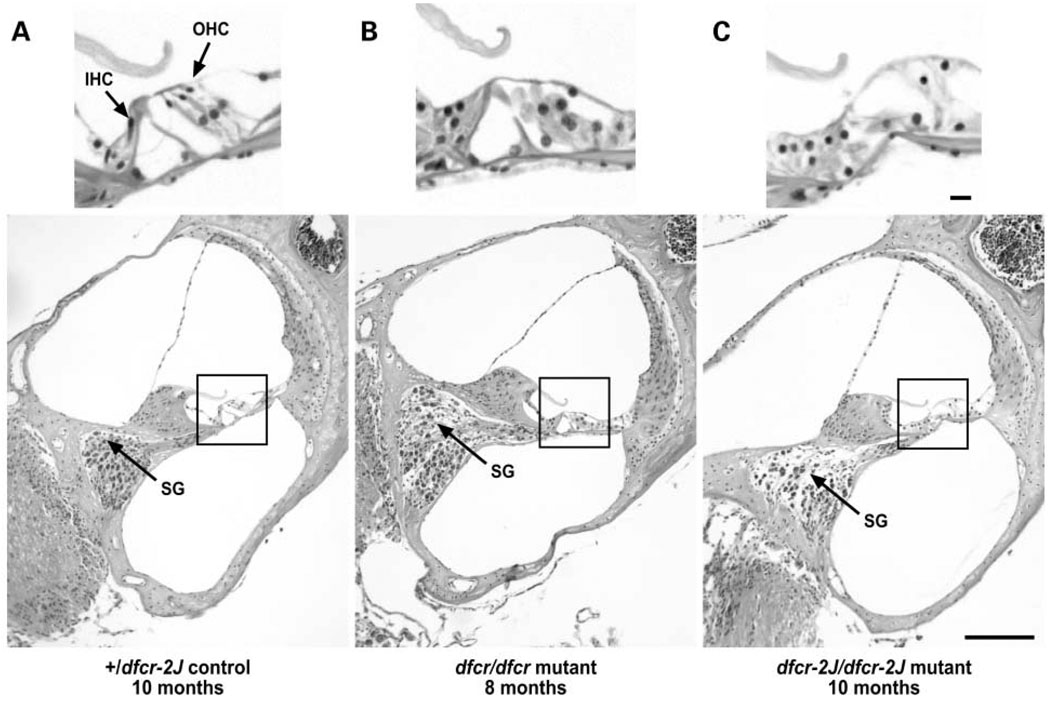
The deafness and balance impairment of mutant mice indicated an inner ear dysfunction. We therefore examined whole mounts and cross sections of inner ears from mutant and control mice by light microscopy and organ of Corti surface preparations by scanning electron microscopy (SEM). Careful examinations of whole mounts of inner ears from adult mice revealed no discernable morphological abnormalities. Examinations of cross sections through the cochleae of both dfcr/dfcr and dfcr-2J/dfcr-2J mutant mice revealed a progressive loss of hair cells and a secondary loss of spiral ganglion cells. At 3 months of age, mutant cochleae already exhibited noticeable hair cell degeneration, especially in the basal and middle turns. By 8 months of age there was severe degeneration of hair cells, and spiral ganglion cell loss was apparent (Fig. 5). Degeneration appeared specific to the hair cells and not to the supporting cells of the organ of Corti. All other cochlear structures, including Reissner’s membrane and stria vascularis, appeared normal in mutant mice.
Figure 5.
Hair cell and spiral ganglion cell loss in cochleae of mutant mice. Cross sections through the basal turn of cochleae of a 10-month-old +/dfcr-2J control (A), an 8-month-old dfcr/dfcr mutant (B), and a 10-month-old dfcr-2J/dfcr-2J mutant (C). The upper panels show magnified images of the organ of Corti corresponding to the boxed areas in the lower panels. IHC, inner hair cell; OHC, outer hair cell; SG, spiral ganglia. Scale bars, 10 µm for upper panels and 100 µm for lower panels.
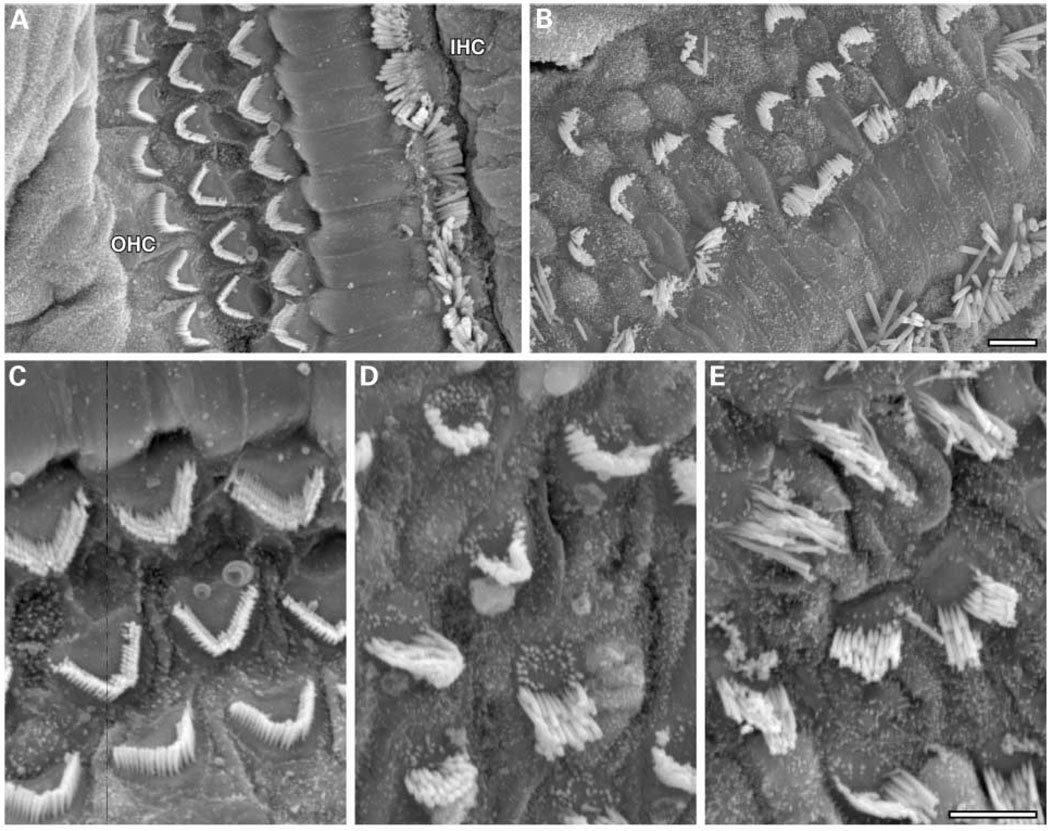
The Ush1c mutant mice are congenitally deaf (at least by 21 days, the earliest age tested by ABR), but hair cell degeneration was not apparent in cochlear cross sections until much later, indicating that hair cell dysfunction preceded degeneration. To examine earlier indicators of hair cell dysmorphology that might be associated with dysfunction, we examined surface preparations of the organ of Corti by SEM, with particular attention to the stereocilia (Fig. 6). At 3 weeks of age, outer hair cell stereocilia of both dfcr/dfcr and dfcr-2J/dfcr-2J mutant mice appeared disorganized and splayed compared with the well-organized pattern and rigid structure typical of normal stereocilia. Stereocilia of inner hair cells of mutant mice also showed a disorganized appearance, but less so than that of outer hair cells.
Figure 6.
Stereocilia disorganization in cochlear hair cells of mutant mice. Scanning electron micrographs of hair cell stereocilia in cochleae from 3-week old mice. The three rows of outer hair cells (OHC) and row of inner hair cells (IHC) in a cochlea from a +/dfcr heterozygous mouse (A) show a normal, highly organized pattern of stereocilia, as compared with stereocilia of a dfcr/dfcr mutant mouse (B). Scale bar, 5 µm. Hair cell loss in the mutant cochlea underlies the disrupted appearance of the hair cell pattern (B). Higher magnification of OHCs better illustrates the intricate pattern and rigid morphology of stereocilia of a normal mouse (C) compared with the disorganized and splayed stereocilia of dfcr/dfcr (D) and dfcr-2J/dfcr-2J (E) mutant mice. Scale bar, 5 µm.
SEM analysis of the 3-week-old mouse cochleae revealed a degree of hair cell loss that was not apparent by light microscopic examination of cross sections at this age. The expansion of supporting cells into the regions previously occupied by degenerated hair cells disrupts the positions of the remaining hair cells and results in the disorganized pattern of hair cells seen in Figure 6. This scarring process seals the reticular lamina, thus maintaining the barrier between endolymph and perilymph (18).
The circling and head tossing behaviors of the dfcr and dfcr-2J mutant mice are probably caused by similar hair cell defects in the vestibular end organs of the inner ear. These structures were not examined in this study but will be the focus of future investigation.
Eye phenotype
We examined the eyes of Ush1c mutant mice because mutations of the homologous gene in humans cause progressive retinitis pigmentosa. The eyes of dfcr/dfcr and dfcr-2J/dfcr-2J mutant mice appear clinically normal on gross examination, having a normal fundus with thin retinal vessels at 1 and 8 months of age, the same as littermate controls. ERGs of eyes from homozygous dfcr and dfcr-2J mutants appeared normal compared with those of heterozygous littermate controls at 9 months of age.
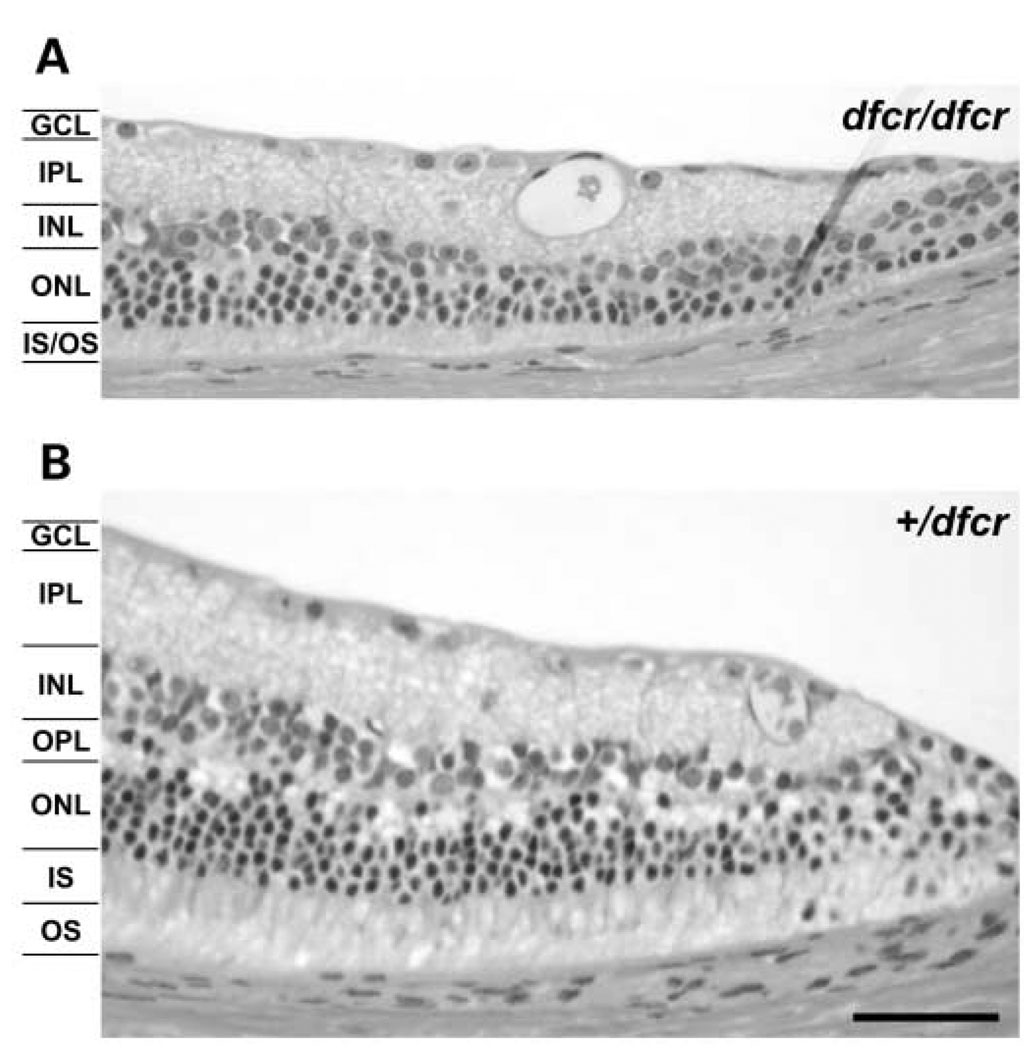
Histological examination of eyes from both dfcr and dfcr-2J mutant homozygotes showed no gross abnormalities; however, a slight peripheral retinal degeneration was noticed in the three homozygous dfcr mutants that were examined at 9 months of age, but not in three littermate controls (Fig. 7). The dfcr-2J homozygous mutants had normal retinas at that age.
Figure 7.
Retinal histology of eyes from dfcr mutant mice compared with controls. Histologic sections of retinas from eyes of three BALB/cBy-dfcr/dfcr mutant mice showed evidence of a slight peripheral retinal degeneration at 9 months of age (A), compared with the normal retinal histology seen in eyes from three litter-mate BALB/cBy-+/dfcr controls (B). The overall thickness of cells at the periphery of the retina is reduced in mutant eyes compared with controls. Each layer is indicated as follows: ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), inner segments (IS) and outer segments (OS). At about 200 µm from the outer edge of the retina, the thickness of the INL-OS layers in mutant eyes was 40–80% that of normal eyes, and mutant eyes appeared to be missing the OPL. Scale bar, 50 µm.
DISCUSSION
The dfcr mutation is a large deletion that includes both constitutive and alternatively spliced exons, whereas the dfcr-2J mutation causes a frame-shift in a single alternatively transcribed exon. These qualitatively different mutations provide a means to evaluate the functional domains of the Ush1c gene and the harmonin isoforms it encodes. Alternative splicing of the Ush1c gene results in multiple harmonin isoforms (4). The short isoform a transcripts are the most abundant and are present in most tissues. The inner ear is one of the few tissues that express the longer isoform b transcripts, particularly those that include exon G. Both isoform a and isoform b transcripts are affected by the dfcr deletion; however, the dfcr-2J mutation affects only isoform b transcripts. The deafness and vestibular dysfunction that is common to both mutations, therefore, must be the result of altered isoform b transcripts, emphasizing the importance of this harmonin isoform to normal inner ear function. Isoform b contains a PST (proline, serine, threonine-rich) domain that is not present in isoform a. This PST region may be critical for the F-actin binding property of harmonin b and thus may help to stabilize the actin fibers in hair cell stereocilia (9).
The dfcr mutation, though more severe than dfcr-2J, may not be completely null. Altered harmonin isoforms retaining partial function might be translated from dfcr mutant transcripts. The reading frames are not changed in the shortened dfcr transcripts of either isoform a or isoform b, and transcription proceeds to the normal stop codons (Fig. 4A). The exons deleted by the dfcr mutation in isoform a transcripts encode the first coiled coil (CC1) domain of harmonin, and the exons deleted in the mutant isoform b transcripts encode the two coiled-coil domains (CC1 and CC2) and the PST domain of harmonin. None of the three PDZ-encoding domains are deleted in dfcr mutant transcripts, but if translated the spatial relationships of these domains would be altered in mutant proteins, with possible functional significance.
The presence of RP in USH1C and its absence in DFNB18 has been attributed to the nature of the underlying USH1C mutations (6). All of the USH1C mutations that cause type I Usher syndrome (3,4,19) have occurred in constitutive exons that are expressed in both eye and cochlea of mice (4), whereas mutations in alternatively transcribed exons that are not expressed in the eye have been associated with nonsyndromic deafness (6). If the same relationship held for mutant Ush1c phenotypes in the mouse, we would expect the dfcr mutation to cause both deafness and retinal degeneration (because it affects all transcripts) and the dfcr-2J mutation to cause only deafness (because it affects isoform b transcripts found in the inner ear but not the eye). We found no gross retinal abnormalities or ERG evidence for retinal dysfunction in dfcr mutants, which indicates that mutational disruption of constitutive Ush1c exons is not sufficient to cause this pathology in the mouse. Mouse mutations in other Usher syndrome-related genes (Myo7a, Cdh23, Pcdh15 and Sans), even those presumed to be null, also lack retinal phenotypes (20–23), suggesting a species difference in manifestation. However, upon more refined analyses, some physiological anomalies recently have been detected in the retinas of mice with Myo7a mutations (24,25). Subtle retinal abnormalities may also occur in the other mouse models of Usher syndromes, including dfcr mutant mice. Indeed, we found some histological evidence of a slight peripheral degeneration in 9-month-old dfcr mutants (Fig. 7). It is not clear if this peripheral degeneration is caused by the dfcr mutation or by a natural aging process including light toxicity, since the dfcr mutation is carried on a BALB/cBy albino background. However, because the degeneration was not seen in BALB/cBy littermate controls, the association of this retinal pathology with the dfcr mutation warrants further investigation.
In the inner ear of the mouse, the Ush1c gene is expressed only in sensory hair cells (4), consistent with the cochlear abnormalities we observed for dfcr and dfcr-2J mutants (Fig. 5). The abnormal stereocilia morphology observed in the Ush1c mutant mice (Fig. 6) is similar to that reported in mice with mutations of Myo7a (26), Cdh23 (20), Pcdh15 (22), and Sans (23), which are models for other forms of human USH1 (Table 1). The products of these genes may form a functional network that maintains stereocilia cohesion (9–11). Disruption of this network would result in disorganization of the hair bundle and the splayed stereocilia seen in mutant mice. The PDZ-domains of harmonin are thought to be of central importance to the formation and maintenance of this transmembrane complex. In particular, the harmonin isoform b, and not a, was shown to interact with the actin cytoskeleton and to bind with cadherin 23 and myosin VIIa (9). Harmonin isoform a (which is expressed in most tissues, including the eye) is altered by the dfcr mutation but not by the dfcr-2J mutation. Harmonin isoform b (which is expressed in the inner ear but not the eye) is disrupted by both mutations. The mouse Ush1c mouse mutants, thus, will provide a direct means to examine the roles of the different harmonin isoforms in these interactive relationships.
MATERIALS AND METHODS
Mice
All mice were from production or research facilities of The Jackson Laboratory, and all procedures were approved by the institutional Animal Care and Use Committee. Both the dfcr and the dfcr-2J mutations arose spontaneously at The Jackson Laboratory. The dfcr mutation occurred in the CBySmn.CB17-Prkdcscid//J strain. It was then backcrossed twice to BALB/cByJ to remove the scid mutation from the strain prior to maintaining by sibling matings. The dfcr-2J mutation arose in a mixed 129;B6 colony segregating a targeted mutation of the beta adducin 2 gene (Add2) and since has been crossed and maintained so that it no longer contains the Add2 mutant allele.
Preparation of genomic DNA, RNA and cDNA
Genomic DNA for genotyping mice was rapidly prepared from tail tips by the hot sodium hydroxide and Tris (Hot SHOT) procedure (27). High-quality genomic DNA for mutation screening was prepared from mouse spleens by standard phenol/chloroform extraction and ethanol precipitation methods. Total RNA was purified from adult mouse eye inner ear, brain, kidney and intestines with TRIzol reagent, according to manufacturer’s protocol (Invitrogen Corp., Carlsbad, CA, USA). DNA and RNA concentrations were estimated by spectrophotometric measurements of absorbance. Mouse cDNA was prepared from total RNA extracted with the SuperScript First Strand cDNA Synthesis System for RT–PCR (Invitrogen Corp., Carlsbad, CA, USA).
Genetic mapping
PCR primer pairs (MapPairs) for MIT microsatellite markers distributed throughout the mouse genome (28) were purchased from Research Genetics (Huntsville, AL, USA) or from Integrated DNA Technologies (Coralville, IA, USA). PCR reactions were composed of 50 ng genomic DNA in 10 µl containing 50mm KCL, 10mm Tris–HCL (pH 9.0 at 25°C), and 0.01% Triton-X-100, 2.25mM MgCl2, 100 nm of each primer (forward and reverse), 100 µm of each of four deoxyribonucleoside triphosphates, and 1.0 units of Taq DNA polymerase (Amplitaq from Applied Biosystems no. N808-0145). Amplification consisted of one cycle of denaturation at 97°C for 30 s followed by 40 cycles, each consisting of 94°C for 30 s, denaturation, 55°C for 30 s of annealing, and 72°C for 30 + 1 s per cycle of extension. After the 40 cycles, the final product was extended for 10 min at 72°C. A pooled DNA strategy (29) was used to efficiently localize the mutations to Chr 7, and then individual DNAs were typed to refine the map position. Gene order, determined by minimizing the number of obligate crossover events, and recombination frequency estimates were calculated with the aid of the Map Manager computer program (30).
Mutation screening
PCR primers used to amplify portions of the mouse Ush1c gene for sequence comparisons are given in Table 2. PCR conditions were the same as described above for genetic mapping except that reaction volumes were 25 µl rather than 10 µl. PCR-amplified products from cDNA and genomic DNA templates were purified with the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA, USA). DNA was sequenced using an Applied Biosystems 373A DNA Sequencer and an optimized DyeDeoxy Terminator Cycle Sequencing method. The same primers used for PCR amplification were also used for cycle sequencing.
For Southern blot analysis, DNA probes for specific regions of the Ush1c gene were generated by PCR amplification of genomic DNA from BALB/cByJ mice with the primers listed in Table 2. The PCR fragments were separated on agarose gels, extracted and purified with the QIAquick Gel Extraction Kit (Qiagen, Inc., Valencia, CA, USA). Preparation of Southern blots, probe labeling, and hybridization methods followed previously described procedures (31).
Assessment of hearing by ABR
Briefly, mice are anesthetized and body temperature is maintained at 37–38°C by placing them on an isothermal pad in a sound-attenuating chamber. Sub-dermal needles are used as electrodes, inserted at the vertex, and ventrolaterally to each ear. Stimulus presentation, ABR acquisition, equipment control and data management are coordinated using the computerized Intelligent Hearing Systems (IHS; Miami, FL, USA). A pair of high frequency transducers is coupled with the IHS system to generate specific acoustic stimuli. Clicks, and 8, 16 and 32 kHz tone-bursts are respectively channeled through plastic tubes into the animal’s ear canals. The amplified brainstem responses are averaged by a computer and displayed on the computer screen. Auditory thresholds are obtained for each stimulus by reducing the sound pressure level (SPL) at 10 dB steps and finally at 5 dB steps up and down to identify the lowest level at which an ABR pattern can be recognized.
Histological analyses of inner ears
Anesthetized mice were perfused through the left ventricle of the heart with phosphate-buffered saline (PBS) followed by Bouin’s fixative. For microscopic analysis of cross-sections, inner ears from mutant and control mice were dissected, perfused with Bouin’s fixative, immersed in same for 24–48 h, decalcified with Cal-EX solution for 6 h, and embedded in paraffin. Sections (7 µm) were cut, mounted on glass slides and counterstained in hematoxylin/eosin (H&E).
For SEM analysis, eight inner ears from mutant mice and six ears from controls were dissected and fixed in 2.5% glutaraldehyde, 0.25% tannic acid, in 0.1m phosphate buffer (pH 7.2) for 5 h at 4°C. The temporal bone surrounding the cochlea and the tectorial membrane were removed to expose the organ of Corti. An osmium tetroxide–thiocarbohydrazide (OTOTO) procedure was used to stain prior to dehydrating and critical-point drying. Specimens were sputter coated with gold and examined at 15 kV under a Hitachi 3000N scanning electron microscope.
Clinical evaluation of eyes and electroretinography
All mice had pupils dilated with 1% atropine ophthalmic drops and were evaluated by indirect ophthalmoscopy with a 78 diopter lens. Signs of retinal degeneration, such as vessel attenuation, alterations in the RPE, and presence or absence of retinal dots were noted (32).
Methods for ERG were as previously described (33). After at least 2 h of dark-adaptation, mice were anesthetized with an intraperitoneal injection of normal saline solution containing ketamine (15 mg/g) and xylazine (7 mg/g body wt). Electroretinograms (ERGs) were recorded from the corneal surface of one eye after pupil dilation (1% atropine sulfate) using a gold loop electrode referenced to a gold wire in the mouth. A needle electrode placed in the tail served as ground. A drop of methylcellulose (2.5%) was placed on the corneal surface to ensure electrical contract and to maintain corneal integrity. Body temperature was maintained at a constant temperature of 38°C using a heated water pad. All stimuli were presented in a Ganzfeld dome (LKC Technologies, Gaithersburg, MD, USA) whose interior surface was painted with a highly reflective white matte paint (no. 6080; Eastman Kodak, Rochester, NY, USA). Stimuli were generated with a Grass Photic Stimulator (model PS33 Plus; Grass Instruments, Worcester, MA, USA) affixed to the outside of the dome at 90µ to the viewing porthole. Rod-dominated responses were recorded to short-wavelength (λmax = 470 nm; Wratten 47A filter) flashes of light over a 4.0 log unit range of intensities (0.3 log unit steps) up to the maximum allowable by the photic stimulator. Cone-dominated responses were obtained with white flashes (0.3 steps) on the rod-saturating background after 10 min of exposure to the background light to allow complete light adaptation. Responses were amplified (Grass CP511 AC amplifier, ×10 000; 3 dB down at 2 and 10 000 Hz) and digitized using an I/O board (model PCI-1200; National Instruments, Austin, TX, USA) in a personal computer. Signal processing was performed with custom software (LabWindows/CVI; National Instruments). Signals were sampled every 0.8 ms over a response window of 200 ms. For each stimulus condition, responses were computer-averaged with up to 50 records averaged for the weakest signals. A signal rejection window could be adjusted online to eliminate electrical artifacts.
Histological and electron microscopy of eyes
A total of 10 eyes were studied histologically. The eyes for light microscopy were immediately removed and immersed in cold fixative (1% paraformaldehyde, 2% glutaraldehyde, and 0.1m cacodylate buffer). Eyes were left in fixative for 24 h, after which time they were transferred to cold 0.1m cacodylate buffer solution for an additional 24 h. Samples were embedded in methacrylate historesin, and sections were cut and stained with hematoxylin and eosin (H&E) according to previously published methods (33).
ACKNOWLEDGEMENTS
We thank Verity Letts and Richard Libby for critical review of this manuscript, Heping Yu for technical expertise in ABR measurements, Peter Finger for EM imaging assistance, Sandra Gray for husbandry skills and A. Sawyer for initially identifying the dfcr mutant mouse. This work was supported by National Institutes of Health (NIH) grants DC62108, DC04301, DC04376, EY07758, RR01183, DK07449 and HL64885. TJL institutional shared services are supported by NIH grant CA34196.
REFERENCES
- 1.Keats BJ, Nouri N, Pelias MZ, Deininger PL, Litt M. Tightly linked flanking microsatellite markers for the Usher syndrome type I locus on the short arm of chromosome 11. Am. J. Hum. Genet. 1994;54:681–686. [PMC free article] [PubMed] [Google Scholar]
- 2.Smith RJ, Lee EC, Kimberling WJ, Daiger SP, Pelias MZ, Keats BJ, Jay M, Bird A, Reardon W, Guest M, et al. Localization of two genes for Usher syndrome type I to chromosome 11. Genomics. 1992;14:995–1002. doi: 10.1016/s0888-7543(05)80122-3. [DOI] [PubMed] [Google Scholar]
- 3.Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat. Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- 4.Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat. Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, Menon PS, Deshmukh D, Griffith AJ, Friedman TB, et al. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum. Genet. 2002;110:527–531. doi: 10.1007/s00439-002-0732-4. [DOI] [PubMed] [Google Scholar]
- 6.Ouyang XM, Xia XJ, Verpy E, Du LL, Pandya A, Petit C, Balkany T, Nance WE, Liu XZ. Mutations in the alternatively spliced exons of USH1C cause non-syndromic recessive deafness. Hum. Genet. 2002;111:26–30. doi: 10.1007/s00439-002-0736-0. [DOI] [PubMed] [Google Scholar]
- 7.Jain PK, Lalwani AK, Li XC, Singleton TL, Smith TN, Chen A, Deshmukh D, Verma IC, Smith RJ, Wilcox ER. A gene for recessive nonsyndromic sensorineural deafness (DFNB18) maps to the chromosomal region 11p14-p15.1 containing the Usher syndrome type 1C gene. Genomics. 1998;50:290–292. doi: 10.1006/geno.1998.5320. [DOI] [PubMed] [Google Scholar]
- 8.Scanlan MJ, Williamson B, Jungbluth A, Stockert E, Arden KC, Viars CS, Gure AO, Gordan JD, Chen YT, Old LJ. Isoforms of the human PDZ-73 protein exhibit differential tissue expression. Biochim. Biophys. Acta. 1999;1445:39–52. doi: 10.1016/s0167-4781(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 9.Boeda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners J, et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siemens J, Kazmierczak P, Reynolds A, Sticker M, Littlewood-Evans A, Muller U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl Acad. Sci. USA. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum. Mol. Genet. 2003;12:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 12.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilligan DM, Lozovatsky L, Gwynn B, Brugnara C, Mohandas N, Peters LL. Targeted disruption of the beta adducin gene (Add2) causes red blood cell spherocytosis in mice. Proc. Natl Acad. Sci. USA. 1999;96:10717–10722. doi: 10.1073/pnas.96.19.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmler MC, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou JC, Petit C, Panthier JJ. Targeted disruption of otog results in deafness and severe imbalance. Nat. Genet. 2000;24:139–143. doi: 10.1038/72793. [DOI] [PubMed] [Google Scholar]
- 16.Simmler MC, Zwaenepoel I, Verpy E, Guillaud L, Elbaz C, Petit C, Panthier JJ. Twister mutant mice are defective for otogelin, a component specific to inner ear acellular membranes. Mamm. Genome. 2000;11:960–966. doi: 10.1007/s003350010197. [DOI] [PubMed] [Google Scholar]
- 17.Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear. Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raphael Y. Cochlear pathology, sensory cell death and regeneration. Br. Med. Bull. 2002;63:25–38. doi: 10.1093/bmb/63.1.25. [DOI] [PubMed] [Google Scholar]
- 19.Zwaenepoel I, Verpy E, Blanchard S, Meins M, Apfelstedt-Sylla E, Gal A, Petit C. Identification of three novel mutations in the USH1C gene and detection of thirty-one polymorphisms used for haplotype analysis. Hum. Mutat. 2001;17:34–41. doi: 10.1002/1098-1004(2001)17:1<34::AID-HUMU4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 20.Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, Steel KP, Noben-Trauth K. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 21.Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- 22.Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 23.Kikkawa Y, Shitara H, Wakana S, Kohara Y, Takada T, Okamoto M, Taya C, Kamiya K, Yoshikawa Y, Tokano H, et al. Mutations in a new scaffold protein Sans cause deafness in Jackson shaker mice. Hum. Mol. Genet. 2003;12:453–461. doi: 10.1093/hmg/ddg042. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl Acad. Sci. USA. 2003;100:6481–6486. doi: 10.1073/pnas.1130432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libby RT, Steel KP. Electroretinographic anomalies in mice with mutations in Myo7a, the gene involved in human Usher syndrome type 1B. Invest. Ophthal. Visual Sci. 2001;42:770–778. [PubMed] [Google Scholar]
- 26.Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 27.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich WF, Miller JC, Steen RG, Merchant M, Damron D, Nahf R, Gross A, Joyce DC, Wessel M, Dredge RD, et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat. Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 29.Taylor BA, Navin A, Phillips SJ. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 30.Manly KF, Cudmore RH, Jr, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- 31.Johnson KR, Cook SA, Davisson MT. Chromosomal localization of the murine gene and two related sequences encoding high-mobility-group I and Y proteins. Genomics. 1992;12:503–509. doi: 10.1016/0888-7543(92)90441-t. [DOI] [PubMed] [Google Scholar]
- 32.Hawes NL, Smith RS, Chang B, Davisson M, Heckenlively JR, John SW. Mouse fundus photography and angiography: a catalogue of normal and mutant phenotypes. Mol. Vis. 1999;5:22. [PubMed] [Google Scholar]
- 33.Hawes NL, Chang B, Hageman GS, Nusinowitz S, Nishina PM, Schneider BS, Smith RS, Roderick TH, Davisson MT, Heckenlively JR. Retinal degeneration 6 (rd6): a new mouse model for human retinitis punctata albescens. Invest. Ophthal. Visual Sci. 2000;41:3149–3157. [PubMed] [Google Scholar]
- 34.Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat. Genet. 1997;16:191–193. doi: 10.1038/ng0697-191. [DOI] [PubMed] [Google Scholar]
- 35.Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat. Genet. 1997;16:188–190. doi: 10.1038/ng0697-188. [DOI] [PubMed] [Google Scholar]
- 36.Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat. Genet. 1997;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- 37.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 38.Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del CSCM, Vila MC, Molina OP, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 39.Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson SM, Householder DB, Coppola V, Tessarollo L, Fritzsch B, Lee EC, Goss D, Carlson GA, Copeland NG, Jenkins NA. Mutations in Cdh23 cause nonsyndromic hearing loss in waltzer mice. Genomics. 2001;74:228–233. doi: 10.1006/geno.2001.6554. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene pcdh15 cause usher syndrome type 1f. Am. J. Hum. Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1f. Hum. Mol. Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 43.Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse ames waltzer hearing-loss mutant is caused by mutation of pcdh15, a novel protocadherin gene. Nat. Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]