Abstract
We describe a novel set of single particle based procedures for the structural analysis of electron microscope images of muscle thin filaments and other partially decorated actin based filaments. The thin filament comprises actin and the regulatory proteins tropomyosin and troponin in a 7:1:1 M ratio. Prior to our work, structure analysis from electron microscope images of the thin filament has largely involved either helical averaging defined by the underlying actin helix or the use of single particle analysis but using a starting model as a reference structure. Our single particle based approach yields an accurate structure for the complete thin filament by avoiding the loss of information from troponin and tropomyosin associated with helical averaging and also removing the potential reference bias associated with the use of a starting model. The approach is more widely applicable to sub-stoichiometric complexes of F-actin and actin-binding proteins.
Keywords: Actin, Thin filament, Electron microscopy, Image processing, Actin-binding proteins, Troponin
1. Introduction
1.1. Structure of the muscle thin filament
The contraction of vertebrate skeletal and cardiac muscle is regulated through the thin filament which is made up of the proteins actin, tropomyosin and troponin. Troponin and tropomyosin serve to control the interaction between myosin and actin so that contraction only takes place when the intracellular Ca2+ concentration is elevated. Understanding the molecular mechanism of this regulation requires a detailed knowledge of the structure of the thin filament and of the conformational changes associated with regulation.
The structure of the actin monomer (G-actin) has been determined by protein crystallography (Oda et al., 2009). In the thin filament G-actin monomers aggregate to form the F-actin filament, a two-stranded helical polymer of globular subunits. The high resolution structure of F-actin has been defined by modelling fibre diffraction pattern data with a resolution of 3.3 Å in the radial direction and 5.6 Å along the equator (Oda et al., 2009). This study revealed the conformational transition within the actin subunits associated with the transition from G to F-actin. Tropomyosin molecules are ∼400 Å in length and exist as a dimeric α-helical coiled-coil (Whitby and Phillips, 2000). In the thin filament tropomyosin molecules are joined end-to-end to form two helical strands which run along the filament following the long period helices of F-actin. Each tropomyosin molecule spans seven actin subunits and interacts with one troponin complex. Troponin contains the Ca2+ binding subunit, TnC, which acts as the Ca2+ switch, TnT which binds to tropomyosin and TnI which has an inhibitory role. Troponin is thought to consist of an extended tail formed by the N-terminal region of TnT and a globular core domain corresponding to the rest of TnT together with TnI and TnC (Flicker et al., 1982). Partial crystal structures exist for the globular core domain (Vinogradova et al., 2005; Takeda et al., 2003). Together the individual structures of F-actin, tropomyosin and the globular core domain of troponin provide almost a complete description of the thin filament provided that their relative configurations are known. Three-dimensional reconstruction of the thin filament from analysis of electron microscope images provides a framework in which these individual structures may be positioned.
1.2. Helical reconstruction
Historically, electron microscopy combined with image analysis has played a major role in the structural analysis of the thin filament and other actin filaments. Actin filaments were the first proteins to be subjected to Fourier based helical reconstruction methods applied to electron microscope images (Moore et al., 1970). In this work, helical reconstruction was used to reconstruct both F-actin and also the rigor complex (F-actin decorated fully with myosin heads) using images of negatively-stained particles. More recently, the helical reconstruction approach allowed the direct identification of tropomyosin movements within the thin filament associated with regulation (Lehman et al., 1995; Xu et al., 1999) which forms the basis of the steric blocking model of thin filament regulation originally proposed on the basis of fibre diffraction results (Huxley, 1972; Haselgrove and Huxley, 1973; Parry and Squire, 1973). Helical reconstruction leads to the averaging of each actin subunit and any associated protein. In general this approach is only likely to produce an accurate representation of associated proteins which are bound in a 1:1 molar ratio with actin. However, since the tropomyosin molecules extend over seven actin subunits, have a pseudo-repeating sequence and are likely to maintain a roughly equivalent coiled-coil conformation throughout, they are effectively recovered in these helical reconstructions despite the 1:7 molar ratio of tropomyosin and actin in the thin filament. The seven pseudo-repeats of the tropomyosin strands in the helical 3D reconstructions are essentially ‘averaged’ over each actin subunit so that any variation in the interaction between actin and different regions of tropomyosin are lost. This is also true of other discrete features of the tropomyosin strands such as the end to end interactions. Troponin complexes, which like tropomyosin are also present in a 1:7 molar ratio to actin monomers in the filament, are different from tropomyosin in that they are localised in pairs at discrete intervals of 385 Å along the thin filament and are thus not effectively recovered by helical reconstruction (Squire and Morris, 1998). Although helical reconstruction of electron microscope images of the thin filament has made a major contribution to the current understanding of the regulatory mechanism of tropomyosin, inevitably it has been much less informative on the mechanism by which troponin brings about this effect.
1.3. Single particle analysis
Helical reconstruction of helical filaments such as F-actin relies on the assumption that the globular subunits are positioned in exactly equivalent positions along a perfect helix leading to the presence of precisely defined rotational views of the filament at different axial levels. However, the angular deviation of the actin subunits from a truly helical arrangement and the intrinsic disorder of the actin filament have been well documented (Egelman et al., 1982; Bremer et al., 1991). In addition, actin has also been shown to possess structural polymorphism (Galkin et al., 2002). The application of single particle image processing techniques addresses these issues, potentially leading to significantly higher resolution. Here filamentous macromolecular complexes are divided into short segments or ‘particles’ which are then sorted into classes and averaged or directly aligned against a reference and used to reconstruct a 3D map.
Two forms of single particle analysis for filaments have emerged. In the first approach filament segments are aligned to reference 3D maps and helical symmetry is imposed on the 3D maps calculated from the segments. The IHRSR procedure (Egelman, 2000) uses this approach whilst determining and refining helical parameters as part of the analysis. Helical single particle analysis has achieved resolutions of 14 Å for F-actin decorated with myosin-S1 (Holmes et al., 2003) and 12 Å for F-actin, using the IHRSR approach (Galkin et al., 2008). However, as for Fourier–Bessel helical analysis, proteins which are not bound to actin in a 1:1 molar ratio, such as troponin in the thin filament, will not be recovered correctly.
The second approach to single particle analysis of filament segments avoids the imposition of helical symmetry and is thus appropriate for recovery of components which either are not helically arranged or which adopt a helical distribution which does not lend itself to helical averaging. This approach is typified by the analysis of the vertebrate skeletal thin filament (Paul et al., 2004, 2009) and vertebrate skeletal (Al-Khayat et al., 2006) and cardiac (Al-Khayat et al., 2008) thick filaments. For vertebrate thick filaments the only symmetry imposed is the underlying threefold rotational symmetry. For the thin filament the only imposed symmetry is the screw symmetry which relates the two strands of the filament (Paul et al., 2009) (see below). For analysis of the thin filament we have adopted this second ‘model free’ approach. Since troponin represents ∼16% of the mass of the thin filament, we find it important to adopt a segmentation strategy where troponin density is placed at the centre of the boxed region and hence at the centre of the reconstruction. This is achieved by visual identification in the raw images and by monitoring of the axial density profile. Our preferred strategy is to follow an ab-initio approach when studying such systems. Model-based single particle image processing methods have been applied not only to the thin filament (Pirani et al., 2005, 2006) but also to study other proteins bound to actin like the C-terminal domain of talin (Gingras et al., 2008). Whilst it is informative to incorporate the structural information available in an analysis of this type, by avoiding the use of a starting model we deliberately avoid any potential for biasing the final reconstruction towards the initial model. We have shown in previous work that the assignment of Euler angles to projection images of actin with additional protein density such as troponin is accurately achieved by using only the backbone structure of actin itself as a reference (Paul et al., 2004).
1.4. Imposing screw symmetry
Despite the incompatibility of the actin helical symmetry and the periodic location of troponin in the thin filament, the regular repeating structure of the actin backbone can still be exploited. Actin can be described as a two-stranded helix where both strands are identical: the tropomyosin–troponin complex conforms to this internal symmetry. We have developed procedures in a cylindrical co-ordinate system to identify the rotation and translation which relates one strand to its neighbour and to average the two strands together, thereby improving the signal to noise ratio (SNR) of our reconstructions. This approach has allowed the identification of troponin and tropomyosin in 3D maps of goldfish muscle thin filaments leading to a reinterpretation of the polarity of the troponin complex (Paul et al., 2009).
1.5. Analysis of F-actin in complex with actin-binding proteins
The regulation of actin filament function is a fundamental biological process. Actin and actin-binding proteins play a vital role in many cellular processes. The analysis of F-actin structure in complex with actin-binding proteins is thus highly relevant and has been the subject of numerous studies by electron microscopy and image analysis (Holmes et al., 2003; Galkin et al., 2008; Janssen et al., 2006; Hanein et al., 1998; Hodgkinson et al., 1997). The majority of such studies have involved helical analysis which is entirely appropriate for cases where there is 1:1 stoichiometry and equivalent binding. However, other cases such as the Arp2/3 complex (Rouiller et al., 2008) and myosin in weak-binding states do not readily allow 1:1 binding and so require alternative strategies for structural analysis. We suggest that the ab-initio non-helical single particle analysis approach we have developed for the thin filament has strong potential for the analysis of this type of system.
Here after defining in Section 2 how the thin filaments were prepared, how the electron microscopy was carried out, and which programs were used for image processing, in the main part of the paper we present a detailed description of the procedures used in our reference-free single particle analysis of partially decorated actin filaments.
2. Materials and methods
Thin filament preparation and viewing. Thin filaments were isolated from two sources; goldfish (Carassius auratus) body muscle and mouse cardiac muscle. Muscles were prepared in relaxing solution as described in Kensler and Stewart (1989). Electron micrographs were recorded of negatively-stained filaments applied to a thin carbon film support over the holes of holey carbon grids. The goldfish images were collected on a JEOL 1200EX at a nominal magnification of 20,000× and the mouse cardiac images recorded on an FEI T 12 at a nominal magnification of 42,000× under low dose conditions. The images were digitised on a Nikon Coolscan 8000ED with a step size of 6.35 μm. A 3D reconstruction of the goldfish data has been published in Paul et al. (2009).
Image processing. Image processing software packages were used as follows: Imagic (van Heel et al., 1996) the MRC suite of programs (Crowther et al., 1996); Spider (Frank et al., 1996) and ImageJ (http://rsb.info.nih.gov/nih-imageJ). The straightening of filament images was achieved using an in-house straightening program and the straighten plugin for ImageJ (Kocsis et al., 1991). Helical reconstruction was carried out using adapted programs from the MRC Image package.
Single particle analysis and imposing screw symmetry. The image processing package Imagic provided the framework for the single particle analysis (van Heel et al., 2000). Adaptations to the standard procedures, specific to filamentous particles, were implemented including the modified back-projection algorithm (Paul et al., 2004).
Rendering in PyMol. 3D models, electron density maps and PDB structures were rendered using PyMol (www.pymol.org). A hybrid method (using Imagic and PyMol) has been established that allows EM density maps to be displayed in PyMol at a theoretical volume corresponding to the appropriate molecular weight.
3. Procedures for reference-free single particle analysis
3.1. Image processing protocol
We describe our methodology fully in the context of the muscle thin filament. The image processing involved in the 3D structure analysis was divided into three main areas: (i) steps leading to particle selection, (ii) generation of the initial 3D map and (iii) the refinement cycle. The generation of the initial map used the single particle techniques of classification, Euler angle assignment & 3D reconstruction. The refinement cycle was where the screw symmetry of the underlying actin was applied to the initial 3D reconstruction and this map was then used as a reference structure to which the alignment of the data was refined. The image processing steps are described in detail below.
3.2. Steps leading to particle selection
3.2.1. Initial selection of filaments
The selection of a filament for processing depends on the identification of regions with visible extra density, in our case, due to troponin. It is not always possible to directly locate regions of extra density along the entire length of the filament since in some orientations the superposition of actin on troponin sometimes makes the complex hard to identify. In these cases it becomes necessary to interpolate positions for troponin. Once a filament has been selected the region was boxed, resulting in a filament length of ∼2000–3000 Å. Straightening and characterisation of the filaments was carried out on these long sections. This length of filament provides 73–110 actin monomers, resulting in a strong signal for helical averaging. Hence such filament regions can also be used to provide the layer-lines required for helical reconstruction which was used as an initial reference for angular assignment.
3.2.2. Rotational alignment and straightening
Prior to straightening it was necessary to rotationally align the long filaments. A direct alignment of each filament against a white streak of density, roughly the same width as the filament, running vertically down the centre of a similar sized box was carried out. This in-plane rotational alignment produces roughly vertical filaments. Despite avoiding obviously bent filaments in the initial selection, computational unbending of the long filament lengths was necessary at this point. Filaments were straightened using in-house software and the straightening plugin for ImageJ (Kocsis et al., 1991). The reasons for straightening were threefold. Firstly, the initial translational alignment that straightening achieves results in well-centred particles and consequently initial class averages with a high level of detail. Secondly, straightening avoids any potential misclassification due to filament curvature. Finally, straightening was an essential first step in the helical reconstruction of the data (see below). A set of nodes was placed along the axis of the filament and a non-uniform cubic spline was fitted to these points. An example of a filament before and after straightening can be seen in Fig. 1(A and B). Straightening or unbending algorithms serve to correct the filament geometry by reversing an assumed two-dimensional elastic deformation. For protein filaments made up of discrete subunits such as F-actin, this is likely to be an approximate, rather than an exact description of the physical deformation associated with curvature. Hence, we limited our analysis to filaments with only small amounts of curvature. The effectiveness of the procedure was assessed by inspection of the layer-lines in the power spectrum of the filament. These became sharper and more symmetrical after straightening. Incorrect positioning of nodes (i.e. away from the filament axis) led to splitting of layer-lines In principle, straightening or unbending is not necessary if the filaments are already sufficiently straight and might lead degradation in the quality of the projection images due to misplacement of nodes. In practice, however, virtually all the filament segments we have processed exhibit a small degree of curvature and the straightening procedure led to improvements in the power spectrum.
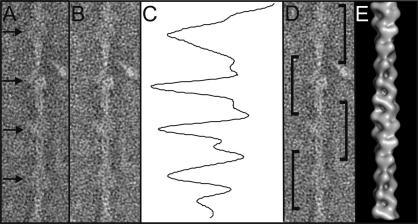
Fig. 1.
Pre-treatment of filaments. (A) Electron microscope image of a region of negatively-stained cardiac muscle thin filament exhibiting slight curvature and readily identifiable troponin densities. (B) Thin filament region shown in A after computational straightening. (C) Axial density profile of Fourier-filtered version of B in which spatial frequencies ≤200 and ≥500 Å were suppressed, allowing the identification and location of regular peaks arising from troponin. (D) Filament segments centred on the troponin density derived from B. (E) Helical reconstruction from the data used for angular assignment.
3.2.3. Helical reconstruction
A helically-averaged reference structure can then be created from the long, straight filament images using the well-established Fourier–Bessel 3D reconstruction techniques (DeRosier and Moore, 1970) and the MRC suite of programs for helical reconstruction (Crowther et al., 1996). The reasons for this approach are also threefold. Firstly, some sort of starting model is necessary to correctly assign initial viewing angles to our class averages (Paul et al., 2004). For other protein filaments with core components less readily analysed by helical methods, other forms of starting model such as tomographic reconstructions or atomic models could be used for this purpose. Secondly, by making a helical reconstruction from the same data, a direct comparison could be drawn between the single particle and helical methods. Finally, a helical analysis allows the quantitative determination of the filament polarity (Fig. 1E). The polarity defined in this way could be compared to that assigned in subsequent steps and used as means of quality control.
3.2.4. Particle selection – segmenting filaments
After straightening, the long filament sections were inspected to locate the troponin complex on the filament prior to segmentation. Characterising the filaments in this way was an important step to ensure the creation of segments that were accurately centred on the troponin complex. The locations of troponin complexes were initially estimated by visual inspection of the filament sections (Fig. 1B). Further assessment was made using Fourier-filtered images of the long filaments in which spatial frequencies ≤200 and ≥500 Å are suppressed. Axial density profiles from these filtered images (Fig. 1C) showed peaks originating from the troponin density which could be seen to clearly correspond to the visible troponin ‘bumps’ in the unfiltered image. This technique has been used previously to identify crossover regions in F-actin where a modulation was detected in data filtered to mask out a similar range of spatial frequencies (Bremer et al., 1991). The spacing between the peaks was also taken into consideration and should agree reasonably well with the known 385 Å periodicity.
Filaments with strong densities attributable to troponin were then segmented into particles (Fig. 1D) making use of a digital ruler overlaid on the image. The result of picking particles in this way was that the origin of reconstruction is placed at or very close to the density peak due to troponin. The optimum length of filament for this type of single particle analysis was previously found to be ∼840 Å (Paul et al., 2004).
3.3. Generation of the initial 3D map
3.3.1. Classification
Large numbers of individual particle images (2000–3000) were created by segmenting the long sections of filaments. A classification step was then applied to these particles to obtain images with a higher SNR and as a form of data compression. Classification was carried out using multivariate statistical analysis (MSA) (van Heel and Frank, 1981) and hierarchical ascendance classification (HAC) (Borland and van Heel, 1990). Classes of similar particles were identified and averaged together creating a set of 2D class averages. In the analysis of conventional protein complexes by MSA, a circular mask that surrounds the particle is normally used. We trialled a number of different MSA masks encompassing various lengths of particle and found the most effective to be a circular one which limits the classification to the central region around the troponin complex. By classifying this central region we were focusing on the troponin complex itself rather than on the underlying actin. Classifying the data in this way provides an internal consistency check. Class averages containing high levels of detail for the entire length of the filament (i.e. all the actin subunits are well defined) are evidence of a successful classification. Typically we used masks with a radius of ∼90 Å. Large numbers of classes were created (typically ∼5–7 particles per class in the initial rounds) thereby reducing potential over-averaging which might obscure any heterogeneity.
3.3.2. Initial angle assignment
A reference model was used to assign the initial angles to the class averages (Fig. 2). Both angular reconstitution and projection matching procedures (van Heel, 1984) were used for assignment of angles and where a consistent value was assigned to the azimuthal rotation angle (Euler angle gamma) this was used. It is possible to assign Euler angles to a near tomographic data set of filament projections using angular reconstitution and a reference model providing there is a degree of out of plane tilt in the reference projections. Model studies have shown that for an ab-initio angle assignment ±20° is the minimum amount of out of plane tilt required to reconstruct a model to 25 Å (19). We have shown that this procedure for angular assignment works with model data and applied it to our analysis of native thin filament data using a helical reconstruction as the reference model The references generated for projection matching were calculated as projection images finely sampled (1°) around the filament axis. Due to the filamentous nature of the particles the predicted tilt about the short axis is small and in the first round of angular assignment using projection matching such tilt was neglected. In later rounds of processing tilts about the short axis were also considered up to ±15°. The results from angular assignment were also checked for consistency with the polarity established by helical reconstruction of the long filament sections. Class averages with inconsistent assignment were not used for reconstruction.
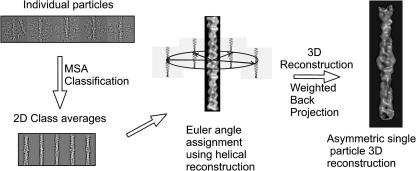
Fig. 2.
The initial 3D reconstruction. Classification of the individual particles was carried out using MSA. Classes of similar particles were determined and summed together creating a set of 2D class averages. The relative angles of these class angles were then determined with reference to projections of a helical reconstruction about the filament axis. Finally, a weighted back-projection algorithm is applied to reconstruct the 3D density map.
3.3.3. 3D reconstruction
To calculate the first 3D reconstruction the modified exact filter back-projection algorithm was used. This filter allows the width of the filament or the extent to which it fills the reconstructed volume to be specified (Paul et al., 2004). This parameter permits the anisotropic dimensions of the filament to be taken into account for the correct weighting. The initial 3D map can then be used as a starting point for the refinement cycle.
3.4. The refinement cycle
3.4.1. Multi-reference alignment
The screw symmetry averaged 3D reconstruction (method described later) could then be used to generate references for a multi-reference alignment. By aligning the particles to the calculated 3D map it was possible to reduce noise due to rotational and translational misalignment. The map was re-projected out in a specified set of directions and the reference images generated. In the early rounds of refinement we neglected tilt about the short axis. By creating references that correspond to rotations about the filament axis in the first round we were able to reduce the number of degrees of freedom that we were considering. In subsequent rounds of alignment we progressively relaxed this restriction and allowed up to ±15° tilt about the short axis. The multi-reference alignment was performed with the Spider routine APSH with an inner radial ring of ∼14 Å and an outer radial ring of ∼324 Å.
3.4.2. Procedures for assigning projection angles
Both projection matching and angular reconstitution were used to assign angles in the refinement rounds of processing and the results compared. However, after the initial round of angle assignment the angles determined using both methods were strikingly similar. It was helpful to use both methods in the early stages of the analysis to avoid the known limitations of each method. Assigning angles using projection matching against a helically-averaged structure results in 3D maps with a tendency to give smeared out contributions from troponin. On the other hand, angular reconstitution may have difficulties in assigning angles at particular points on the Euler sphere. In refinement rounds of angle assignment a larger range of Euler angles was considered when using projection matching. A tilt about the short axis corresponding to ±15° (in β) was also incorporated into the reference images resulting in a reference set of 7200 images. The number of class averages generated increased (and hence the number of raw images in each class decreased) in each subsequent round of refinement reflecting the increased SNR in the 3D map. Towards the end of the refinement, assignment of angles to unaveraged images has been observed to give accurate results judged on the agreement between angular reconstitution and projection matching.
The refinement procedure involving reprojection of the 3D map, multi-reference alignment, angular assignment and 3D reconstruction was cyclical (Fig. 3). The workflow described above was iterated until no significant changes could be observed in the screw symmetry averaged 3D reconstruction from one round to the next.
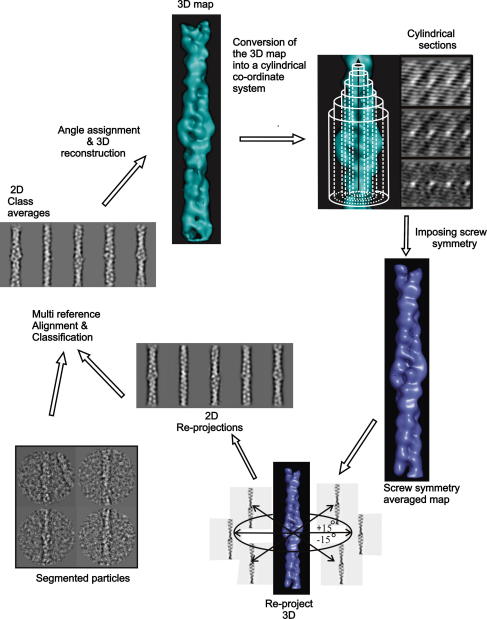
Fig. 3.
The refinement cycle. The initial 3D was converted into a cylindrical co-ordinate system to perform the two strand averaging that results in a screw symmetry averaged map. This averaged map was then used to calculate reference projections with up to +/−15° tilt about the short axis. These projections were used as references in the multi-reference alignment of the “raw” particles. The particles were the subject to classification and projections from the averaged structure were used to assign Euler angles to the class averages. Finally a new single particle 3D reconstruction was calculated. The cycle was repeated until no further improvements in the 3D reconstruction was observed.
3.4.3. Imposing screw symmetry
3D reconstructions, calculated using the adapted single particle methods described above, have no symmetry applied. It was possible to increase the degree of averaging and raise the SNR by calculating an average of the two strands. Actin’s helical symmetry is commonly used and profited from in helical 3D reconstructions. The roughly 13/6 helix effectively allows each subunit in the filament to be averaged, thereby allowing 13-fold averaging. We cannot impose this form of averaging without losing information on the troponin density (Squire and Morris, 1998), but the helical arrangement of the troponin complexes can be exploited to improve the SNR of our analysis. Within the functional unit each strand is made up of 7 actin subunits, 1 tropomyosin molecule and 1 troponin complex. If the two strands are brought into alignment by translation and rotation to match troponin’s helical path they can then be averaged. These steps can be conveniently carried out in a cylindrical polar co-ordinate system.
3.4.4. Preparation for averaging; conversion into cylindrical co-ordinates
Fig. 4 shows how a thin filament was described in a cylindrical co-ordinate system. Sets of concentric cylinders at increasing radii could be defined at regular intervals throughout the volume (Fig. 4A). These cylinders were then flattened out to form cylindrical sections. In each cylindrical section the horizontal axis represents an angle of rotation. The complete structure was described by one revolution or 360°, but it was also useful to display two revolutions as in Fig. 4B and C in order to avoid wraparound problems. The vertical axis is a distance measurement along the z-axis of the filament. The conversion into cylindrical co-ordinates was achieved using the locally developed program carttocyl. A typical 3D map described in a Cartesian co-ordinate system consists of X–Y slices in Z. The program converts these slices into a set of cylindrical sections regularly spaced along the radius. Finer radial sampling could be achieved by magnifying the map prior to conversion; a factor of two was applied in this example. The number of angular segments (angular sampling step), the number of cylindrical sections and the location of the axis were required for conversion. To ensure that the exact position of the axis was determined, a grid of points close to the theoretical axis position was trialled and visually inspected. If the axis was not located correctly an uneven density distribution was seen in the surfaces. The cylindrical surface shown in Fig. 4Bi is at a radius of 42 Å. At this radius the contributions to the density are predominantly from actin and tropomyosin. Cylindrical sections at radii less than 21 Å (data not shown) show little information: close to the filament axis the low frequency components of the structure dominate. Fig. 4Bi clearly shows the right-handed long-pitch helix of F-actin which runs diagonally from bottom left to top right. The shallow diagonal stripes inclined in the opposite direction correspond to the short pitch left-handed helical path of F-actin. The contribution from the actin diminishes at higher radius and the troponin density becomes the strongest feature as can be seen at in Fig. 4Bii, corresponding to a radius of 56 Å. At this radius the troponin density appears elongated in a diagonal direction approximately parallel to the F-actin long-pitch helix. At higher radius this elongation is reduced. The troponin density is axially staggered on adjacent actin strands (Fig. 4Bii). Slight differences between the strands prior to averaging are apparent.
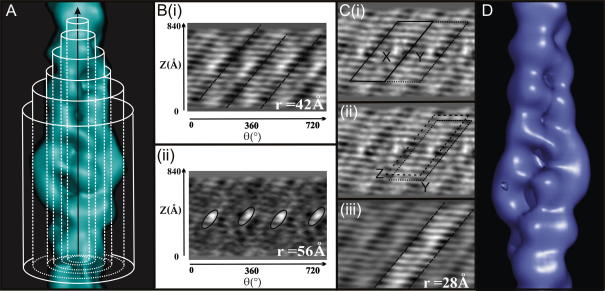
Fig. 4.
Imposing screw symmetry. (A) The 3D density map (panel A) was converted into layers of concentric cylindrical surfaces. Panel B displays the cylindrical surfaces flattened into planes corresponding to r = 42 (Bi), & 56 Å (Bii) of the map shown in A. The radius and the distance along the filament axis are shown as the two axes r and z, respectively. The entire length of the reconstructed volume (840 Å) and two revolutions of the filament are shown in each cylindrical surface (0–720°). A single strand cannot be outlined in a single revolution of the filament. The distinct long-pitch right handed tracks of the actin strands are highlighted (Bi). The cylindrical surfaces show how the two strands differed prior to averaging and how at higher radii the contribution from the troponin density became more evident (ellipses). (C) A radial projection (average of the cylindrical surfaces from a radius of 21 to 86 Å) of the 3D map was calculated and used in the alignment of the two actin strands. The ‘short’ strand X used in the alignment procedure is indicated in Ci. The initial 180° shift of strand X to position Y is shown in Ci and the final alignment of the strands from position Y to position Z is shown in Cii. The relative shift needed to translate one strand of actin directly on top of the second strand was calculated using the radial projection. The translation was imposed on individually selected strands and each strand was added to a copy of the cylindrical surface from which it originated. The sum of the two strands at radii of 28 Å is shown in Ciii. The region where the two strands are added together appears brighter. The sum of the two strands was computationally cut from every cylindrical surface and four copies were used to create composite surfaces again applying the calculated translation. By converting the composite cylindrical surfaces back into Cartesian co-ordinates the screw symmetry averaged map was generated (D).
3.4.5. Defining and aligning strands
The distinct actin density seen in the cylindrical surfaces corresponding to the long-pitch F-actin helix was used to isolate one strand. The natural shape of the strand in cylindrical co-ordinates is a parallelogram. The white stripes of protein density run from bottom left to top right in the cylindrical surfaces, separated by black stripes corresponding to low density regions. A radial projection, the sum of the cylindrical surfaces between a radius of 21 and 86 Å, (Fig. 4Ci and ii) was used to define and align the strands: the projection was used in preference to an individual cylindrical surface due to the higher SNR. A single strand (labelled X in Fig. 4Ci) can be extracted by using a parallelogram shaped window. The short side of the parallelogram selected corresponds to a 180° rotation along the horizontal or angular axis. No prior information about the pitch was used to select the strand. A minimum of two revolutions of the filament are needed to define a strand of actin. The strand was used as a test object in the alignment and ‘cut’ from the radial projection. To ensure edge effects do not affect the subsequent alignments the parallelogram window ‘cut out’ was shorter (520 Å along the filament axis) than the whole filament. The radial projection displaying four revolutions of the entire filament was used as a reference image (only two revolutions are shown in Fig. 4C) in the subsequent alignment. The parallelogram X was shifted by 180° (i.e. strand X was shifted to position Y) and then aligned to the reference image. The Imagic program ALI-DIR was used to align the shifted strand, in position Y, with respect to the radial projection to an aligned position Z. By restricting the maximum shift, the strand is not permitted to revert to its original position. The calculated shift gives the small additional axial rise and rotation that aligns the two strands exactly.
3.4.6. Averaging the two strands
The shift calculated by aligning the ‘short’ strands was applied to each cylindrical surface. The parallelogram windows were then extended to encompass the entire length of the filament. Fig. 4Ciii shows a cylindrical surface at a radius of 28 Å in which the aligned strand has been highlighted. The average of the two parallelogram strands was computationally cut out of each cylindrical surface. The calculated axial and azimuthal shifts were applied to a copy of this average. Four of the averaged parallelograms were used to construct one complete revolution of the filament. The set of composite cylindrical surfaces formed the screw symmetry averaged 3D: these surfaces were then converted back into Cartesian co-ordinates. The final screw symmetry averaged 3D is shown in Fig. 4D: both strands of the thin filament are now identical and the SNR of the reconstruction is improved.
4. Discussion
4.1. A novel approach
We have described an ab-initio single particle approach developed for the analysis of the muscle thin filaments which is also likely to be suited to the analysis of actin filaments partially decorated with actin-binding proteins. The approach limits helical averaging to the true asymmetric unit and avoids the use of externally derived starting models thereby avoiding potential model bias in our final 3D reconstruction. Our image processing protocol includes careful selection of the individual particles via extensive pre-processing steps, the assignment of angles using an initial model generated from the data, and a refinement cycle that maximizes the potential for averaging over the asymmetric/functional unit of the system. We do not impose the helical parameters of actin at any point in our analysis as it is not appropriate unless there is a 1:1 binding stoichiometry of the binding protein to actin.
4.2. Cylindrical averaging
Averaging of actin subunits has historically been a successful method for structural analysis of EM images of muscle thin filaments and other actin based filaments. However, by imposing the helical symmetry of the underlying actin filament any density that is not lying on the same helical path as actin will be smeared out. Contributions from actin-binding proteins such as troponin in our case will be reduced if not lost completely. By identifying the true asymmetric, and in this case functional, unit of the filament and using this as our repeating structure we were able to reinforce any density resulting from the binding protein as well as the SNR of the resulting map.
4.3. Wider application of methodology
We have applied our image processing strategy successfully to the analysis of the muscle thin filament (Paul et al., 2009). However, we believe that this methodology has the possibility for a substantially wider range of applications. Other actin based systems can be studied in a similar manner and to this end we propose this methodology as a general protocol for the analysis of F-actin sparsely decorated with actin-binding proteins. Examples of actin-binding proteins found in muscle that could be suitable for this type of analysis when complexed with actin include caldesmon, calponin and myosin heads in different nucleotide states. Other interesting cytoskeletal proteins such as the Talin and the Arp2/3 complexes could also be studied using this methodology (Gingras et al., 2008; Rouiller et al., 2008). Defining the asymmetric unit of a filament for averaging may also have a role in the analysis of microtubules where there would be the possibility of identifying protofilaments more readily in a cylindrical co-ordinate system and analysing sub-stoichiometric complexes with microtubule binding proteins.
4.4. Biological significance of our results
The application of our novel strand averaging technique directly established the axial stagger of the troponin complexes for the first time in both the native system (Paul et al., 2009) and in reconstituted thin filament data (Paul et al., 2009). The stagger, 27.5 Å, was the same for both systems. Due to the different assembly mechanisms of the two systems this led us to conclude that the stagger between the two troponin complexes and ultimately the assembly of the thin filament can occur correctly without any other sarcomeric proteins playing a role. Also, as part of our analysis of the goldfish data (Paul et al., 2009), we interpret the polarity of the troponin tail as reversed from the model of the thin filament which was previously widely accepted. If we had used the earlier starting model as a reference structure for alignment it would have been significantly harder if not impossible to recover such a difference and successfully refine the resulting 3D map. In addition to recovering density for troponin our single particle approach allows the definition of the tropomyosin strands and individual actin subunits of the regulatory unit of the thin filament with no internal averaging. Hence, the actin subunits directly associated with the globular core of troponin can be compared with other actin subunits. We are investigating these and related phenomena in our current higher resolution structural analysis of reconstituted thin filaments.
Acknowledgments
We would like to thank Dr R. Kensler for the preparation of the native cardiac thin filaments (Fig. 1) and the native goldfish thin filament data. DP was supported by a BHF project grant (#23480) to EPM and JMS and JMS acknowledges support from the European MYORES network on muscle development.
References
- Al-Khayat, H.A., Morris, E.P., Kensler, R.W., Squire, J.M., 2006. 3D structure of relaxed fish muscle myosin filaments by single particle analysis. J. Struct. Biol. [DOI] [PubMed]
- Al-Khayat H.A., Morris E.P., Kensler R.W., Squire J.M. Myosin filament 3D structure in mammalian cardiac muscle. J. Struct. Biol. 2008;163:117–126. doi: 10.1016/j.jsb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland L., van Heel M. Classification of image data in conjugate representation spaces. J. Opt. Soc. Am. 1990:601–610. [Google Scholar]
- Bremer A., Millonig R.C., Sutterlin R., Engel A., Pollard T.D., Aebi U. The structural basis for the intrinsic disorder of the actin filament: the “lateral slipping” model. J. Cell. Biol. 1991;115:689–703. doi: 10.1083/jcb.115.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R.A., Henderson R., Smith J.M. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- DeRosier D.J., Moore P.B. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 1970:335–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- Egelman E.H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman E.H., Francis N., DeRosier D.J. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Flicker P.F., Phillips G.N., Jr., Cohen C. Troponin and its interactions with tropomyosin. An electron microscope study. J. Mol. Biol. 1982;162:495–501. doi: 10.1016/0022-2836(82)90540-x. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Galkin V.E., VanLoock M.S., Orlova A., Egelman E.H. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Curr. Biol. 2002;12:570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- Galkin V.E., Orlova A., Cherepanova O., Lebart M.C., Egelman E.H. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.R., Bate N., Goult B.T., Hazelwood L., Canestrelli I., Grossmann J.G., Liu H., Putz N.S., Roberts G.C., Volkmann N., Hanein D., Barsukov I.L., Critchley D.R. The structure of the C-terminal actin-binding domain of talin. Embo J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Volkmann N., Goldsmith S., Michon A.M., Lehman W., Craig R., DeRosier D., Almo S., Matsudaira P. An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat. Struct. Biol. 1998;5:787–792. doi: 10.1038/1828. [DOI] [PubMed] [Google Scholar]
- Haselgrove J.C., Huxley H.E. X-ray evidence for radial cross-bridge movement and for the sliding filament model in actively contracting skeletal muscle. J. Mol. Biol. 1973;77:549–568. doi: 10.1016/0022-2836(73)90222-2. [DOI] [PubMed] [Google Scholar]
- Hodgkinson J.L., el-Mezgueldi M., Craig R., Vibert P., Marston S.B., Lehman W. 3-D image reconstruction of reconstituted smooth muscle thin filaments containing calponin: visualization of interactions between F-actin and calponin. J. Mol. Biol. 1997;273:150–159. doi: 10.1006/jmbi.1997.1307. [DOI] [PubMed] [Google Scholar]
- Holmes K.C., Angert I., Kull F.J., Jahn W., Schroder R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- Huxley H.E. Structural changes in actin- and myosin-containing filaments during contraction. Cold Spring Harbor Symp. Quant. Biol. 1972:361–376. [Google Scholar]
- Janssen M.E., Kim E., Liu H., Fujimoto L.M., Bobkov A., Volkmann N., Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Kensler R.W., Stewart M. An ultrastructural study of crossbridge arrangement in the fish skeletal muscle thick filament. J. Cell Sci. 1989;94(Pt 3):391–401. doi: 10.1242/jcs.94.3.391. [DOI] [PubMed] [Google Scholar]
- Kocsis E., Trus B.L., Steer C.J., Bisher M.E., Steven A.C. Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J. Struct. Biol. 1991;107:6–14. doi: 10.1016/1047-8477(91)90025-r. [DOI] [PubMed] [Google Scholar]
- Lehman W., Vibert P., Uman P., Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J. Mol. Biol. 1995;251:191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- Moore P.B., Huxley H.E., DeRosier D.J. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J. Mol. Biol. 1970;50:279–295. doi: 10.1016/0022-2836(70)90192-0. [DOI] [PubMed] [Google Scholar]
- Oda T., Iwasa M., Aihara T., Maeda Y., Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Parry D.A., Squire J.M. Structural role of tropomyosin in muscle regulation: analysis of the X-ray diffraction patterns from relaxed and contracting muscles. J. Mol. Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- Paul D., Patwardhan A., Squire J.M., Morris E.P. Single particle analysis of filamentous and highly elongated macromolecular assemblies. J. Struct. Biol. 2004;148:236–250. doi: 10.1016/j.jsb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Paul D.M., Morris E.P., Kensler R.W., Squire J.M. Structure and orientation of troponin in the thin filament. J. Biol. Chem. 2009;284:15007–15015. doi: 10.1074/jbc.M808615200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D.M., Lehman W., Pirani A., Craig R., Tobacman L.S., Squire J.M., Morris E.P. Reference free single particle analysis of reconstituted thin filaments. Biophys. J. 2009;96:376a. [Google Scholar]
- Pirani A., Xu C., Hatch V., Craig R., Tobacman L.S., Lehman W. Single particle analysis of relaxed and activated muscle thin filaments. J. Mol. Biol. 2005;346:761–772. doi: 10.1016/j.jmb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Pirani A., Vinogradova M.V., Curmi P.M., King W.A., Fletterick R.J., Craig R., Tobacman L.S., Xu C., Hatch V., Lehman W. An atomic model of the thin filament in the relaxed and ca(2+)-activated States. J. Mol. Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Rouiller I., Xu X.P., Amann K.J., Egile C., Nickell S., Nicastro D., Li R., Pollard T.D., Volkmann N., Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire J.M., Morris E.P. A new look at thin filament regulation in vertebrate skeletal muscle. Faseb J. 1998;12:761–771. doi: 10.1096/fasebj.12.10.761. [DOI] [PubMed] [Google Scholar]
- Takeda S., Yamashita A., Maeda K., Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- van Heel M. Three-dimensional reconstructions from projections with unknown angular relationship. Proc. 8th Eur. Cong. EM, Budapest. 1984;2:1347–1348. [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz G., Orlova E.V., Schmidt R., Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- van Heel M., Gowen B., Matadeen R., Orlova E.V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Vinogradova M.V., Stone D.B., Malanina G.G., Karatzaferi C., Cooke R., Mendelson R.A., Fletterick R.J. Ca(2+)-regulated structural changes in troponin. Proc. Natl. Acad. Sci. USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F.G., Phillips G.N., Jr. Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins. 2000;38:49–59. [PubMed] [Google Scholar]
- Xu C., Craig R., Tobacman L., Horowitz R., Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys. J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]