SYNOPSIS
Influenza A viruses infect large numbers of warm-blooded animals, including wild birds, domestic birds, pigs, horses, and humans. Influenza viruses can switch hosts to form new lineages in novel hosts. The most significant of these events is the emergence of antigenically novel influenza A viruses in humans, leading to pandemics. Influenza pandemics have been reported for at least 500 years, with inter-pandemic intervals averaging approximately 40 years.
“We regret very much the fact that an influenza virologist is unable to live say 200 years, so that he himself would be able to see what has developed from his earlier assumptions.”
J. Mulder and J.F.P Hers, Influenza (1972)1
Ninety-one years after the “Spanish influenza” pandemic of 1918–1919, broad interest remains in understanding what happened, both to inform current pandemic planning and to advance basic science knowledge about how pandemic influenza viruses form and cause disease. Influenza viruses were not isolated until the 1930s. Characterization of the 1918 pandemic virus, which began in 1995 with the identification of 1918 autopsy and archeologic material, required an archaevirologic approach in which tiny fragments of the viral ribonucleic acid (RNA) were extracted from the preserved lung tissues of victims, amplified by reverse-transcription polymerase chain reaction (RT-PCR), and then sequenced. Obtaining the complete genome sequence and then reconstructing it in the laboratory took a decade.2–4 The recently declared swine-origin H1N1 pandemic5 and the ongoing H5N1 avian influenza epizootic—a pandemic-in-waiting according to some6—increases the importance of understanding pandemic viruses.
In addition to basic science understanding of the emergence and pathogenicity of the 1918 virus, we must also integrate such data within an historical framework of more than 500 years of influenza pandemicity. Major influenza epidemics have apparently occurred since at least the Middle Ages, if not since ancient times.7,8 In addition to periodic, seasonal, and regional epidemics, influenza pandemics have also occasionally appeared during this timeframe.3,9 When pandemics appear, 50% or more of an affected population can be infected in a single year, and the number of excess deaths can increase dramatically.10,11 Since 1500, there appear to have been 14 or more influenza pandemics; in the past 133 years of the “microbial era” (1876 to the present) there were undoubted pandemics in 1889, 1918, 1957, 1968, 1977, and 2009.8,12,13 In 1918, the worst pandemic in recorded history caused approximately 546,000 excess deaths in the United States (675,000 total deaths)3 and killed an estimated 50 million people or more worldwide.14
Unexpectedly, the first pandemic of the 21st century was caused by a novel H1N1 influenza virus, derived by reassortment of two preexisting swine influenza viruses.5,13 This virus was first detected as causing human illness in Mexico in March 2009, followed closely by cases in the United States. It has now spread globally, with millions of cases and at least 16,813 deaths documented by the World Health Organization (WHO) as of March 19, 2010.15
The continuing spread of H5N1 highly pathogenic avian influenza (HPAI) viruses into poultry populations on several continents, associated with a growing number of human “spill-over” infections, has also heightened interest in pandemics.6,16 These H5N1 HPAI viruses first caused a poultry epizootic in southern China in 1996, followed within a year by an epizootic in Hong Kong that produced 18 human spill-over cases and six deaths. Viral descendents continued to circulate thereafter in China, reappearing in epizootic form in, and spreading widely after, 2003. Geographical extension of H5N1 HPAI viruses was accompanied by the appearance and spread of genetically and antigenically different strains.17,18 Since 2003, dispersion of H5N1 viruses has led to epizootics in about 60 countries on three continents, and has caused 489 human cases and 289 deaths (as of March 19, 2010),19 millions of avian deaths, and infections and deaths in several other mammalian species.20
Despite uncertainties in the historical record of the pre-virology era, the study of previous pandemics may help to guide current and future pandemic planning and lead to a better understanding of the complex ecobiology that underlies the formation of pandemic strains of influenza A viruses. Although largely unappreciated by contemporary virologists and infectious disease experts, an enormous historical literature exists on influenza, spanning hundreds of years.8 As an interesting example, in 1820 the German physician Georg Friedrich Most published—more than 100 years prior to the first isolation of influenza viruses, and well before the modern microbiology era21—a book that asked key questions about influenza. On the title page, Influenza Europaea, oder die größeste Krankheits-Epidemie der neuern Zeit [European Influenza: Or the Greatest Disease Epidemic of the Modern Age], he asks the following:
What is influenza?
Where does influenza come from?
How did influenza behave in the past?
In what ways can we predict future occurrences […] and how will future outbreaks behave?
Through what means can its spread be halted?
These questions, posed almost 200 years ago, are still highly relevant today. Despite the tremendous progress made in virology, microbiology, immunology, pharmacology, epidemiology, vaccinology, and preventive medicine over the last century, they are still largely unanswerable. In this review, we use these questions as a framework to discuss what has been learned about the 1918 influenza pandemic through recent work, and what these and other studies may tell us about the nature of future influenza pandemics.
WHAT IS INFLUENZA?
In 1931, Rockefeller Institute investigator Richard Shope published the first of three landmark papers22–24 establishing the etiology of “swine influenza” or “hog flu”, the epizootic disease of pigs that had first been noted during the fall wave of the 1918 influenza pandemic.25,26 It is now believed that the pandemic virus appearing in 1918 was transmitted from humans to pigs early on, thereby splitting off into two lineages: one human, the other porcine.27,28 Both lineages persist today. The classical swine H1N1 influenza lineage has evolved continually since 1918. The human H1N1 lineage caused pandemic and endemic influenza from 1918 to 1956, then disappeared entirely around 1957 only to reappear in relatively low-level pandemic form in 1977.3 It has continued to circulate endemically in humans up to the present time (2009).
Shope's studies were important in their own right, but also because they stimulated American and British research groups to take up the search for the cause of human influenza. In 1933, Alphonse Dochez and colleagues produced apparent influenza via human nasopharyngeal inoculation and succeeded in cultivating and serially passing a virus in primary chick embryo cultures, demonstrating that passage material still produced human disease.29 Several weeks later, a British group that had been collaborating with Dochez, led by Sir Christopher Andrewes, Wilson Smith, and Sir Patrick Laidlaw, reported isolation and serial propagation of human influenza virus in ferrets,30 introducing the great advantage of both a living culture medium and an animal model (the human virus was found to cause a catarrhal disease in ferrets after a two-day incubation period). The papers of these two influential groups, along with the ongoing work of Shope and colleagues,22–24 led to an explosion of research in the field of virology that has continued unabated until the present time.
We now know that influenza viruses (of the family Orthomyxoviridae) are enveloped negative-strand RNA viruses with segmented genomes.31 Of two genera, one includes influenza A and B viruses, and the other influenza C virus. The three virus types differ in host range and pathogenicity. A and B type viruses contain eight discrete gene segments, each coding for at least one protein. They are covered with projections of three proteins: hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2). Each influenza RNA segment is encapsidated by nucleoproteins to form ribonucleotide-nucleoprotein complexes.31
Types B and C influenza viruses are isolated almost exclusively from humans. Influenza A viruses, however, all circulate within or are derived from an avian reservoir, but can infect a wide variety of warm-blooded animals as well, including not only humans but also swine, horses, dogs, cats, and other mammals. Aquatic birds serve as the natural reservoir for all known subtypes of influenza A virus and probably are the ultimate source of human pandemic influenza strains.32 Influenza A viruses are subdivided by serologic or genetic characterization of the HA and NA surface glycoproteins that project from the virion. Sixteen HA (or “H”) and 9 NA (or “N”) subtypes are known,33 abbreviated H1-H16 and N1-N9. The subtype of an influenza A virus is given by listing together its HA and NA subtypes. The 1918 pandemic virus was an H1N1 strain of influenza A virus. Its descendants were replaced in 1957 by an H2N2 subtype pandemic strain. H2N2 viruses in turn circulated until 1968 when they were replaced by H3N2 pandemic viruses. As noted, in 1977, H1N1 strains from the pre-1957 period reappeared; since then, both influenza A subtypes H3N2 and H1N1 have co-circulated in humans.32
The influenza HA molecule initiates infection by binding to receptors on specific host cells. Antibodies against the HA protein may prevent re-infection with the same strain by blocking either attachment or cell fusion. Because their RNA polymerase complexes have no proofreading activity,31 high mutation rates (ranging from approximately 1×10−3 to 8×10−3 substitutions per site per year)34 led to accumulation of point mutations during replication. Mutations that change amino acids in the antigenic portions of surface glycoproteins may produce selective advantages for viral strains by allowing them to evade preexisting immunity (“antigenic drift”). The HA and NA can evade preexisting population immunity by either antigenic drift or antigenic shift, in which the virus acquires an HA of a new subtype by genetic reassortment with another influenza A virus.35
WHERE DOES INFLUENZA COME FROM?
Influenza A viruses bearing any one of the 16 known HA and nine NA subtypes exist in wild birds and provide a source of viral HA and NA subtypes antigenically novel to humans.32,36 Emergence into human circulation of an influenza strain with a novel subtype by antigenic shift caused both the 1957 and 1968 pandemics; in both cases, the previously circulating post-pandemic human virus imported an HA from an unidentified avian or avian-like virus.37 Although one of the absolute requirements for a pandemic seems to be that the HA must change, the extent to which the rest of the virus can or must change is not known. In 1957, three genes from the circulating H1N1 human influenza virus were replaced by avian-like genes: HA, NA, and a subunit of the polymerase complex (PB1). In 1968, only the HA and PB1 genes were replaced.37,38 The 1918 influenza pandemic virus has an avian-like genome and, unlike the 1957 and 1968 viruses, is hypothesized to have arisen by an entirely different mechanism: adaptation of all eight segments of a preexisting avian genome to humans.3,39–41 The pathways and mechanisms for this adaptation are unknown. The 2009 pandemic virus arose by yet another mechanism entirely.13
Although an antigenically novel HA subtype is a likely requirement for pandemic emergence, human infections with animal-adapted influenza viruses of novel HA subtype have usually not been transmitted efficiently from person to person, suggesting that human adaptation may be complex.42 For example, the 1976 U.S. military outbreak of a swine-adapted H1N1 influenza virus resulted in limited person-to-person transmission.43,44 In 2003, an HPAI H7N7 virus caused a poultry epizootic in the Netherlands and spread regionally. Before the epizootic was contained, at least 86 poultry workers and three contacts had been infected and developed conjunctivitis with or without an influenza-like illness.45 After reemergence in 2003, the ongoing H5N1 HPAI epizootic continues to produce spillover infections in humans, causing concern that human adaptation of this virus to humans could result in a pandemic.42 Such concerns hinge not only upon transmission between poultry and individual humans, but also upon potential development of sustained human-to-human transmission. Several case clusters of H5N1 infections have been reported.46 Although epidemiologic information has been limited, person-to-person transmission of H5N1 has been suggested in a few instances, usually involving family members. It is unknown whether this represents infection associated with particularly intimate or prolonged contact, or shared but unidentified host factors affecting either infection risk or virus transmissibility.
HOW DID INFLUENZA BEHAVE IN THE PAST?
Application of modern criteria to identify disease outbreaks as influenza pandemics8 suggests that there may have been at least 14 pandemics over the past 500 years (1509 to 2009), or approximately one pandemic every 36 years. These pandemics may not have occurred randomly,8 and some (but not all) have been followed by periods of high respiratory disease activity associated with large outbreaks and high mortality over a number of years. It may thus be helpful to think not only about pandemics as events that occur at specific points in time, but also to consider the occurrence of pandemic eras. For example, the 90 years since 1918 can be said to comprise a pandemic era because all of the influenza A viruses circulating since that time, up to the present, are direct descendants of the 1918 virus, and because seasonal influenza activity has been detected continuously during that period.3,8,13 Yet, clearly in that interval there have been five pandemics, including 1918.8
The Spanish influenza (1918–1919)
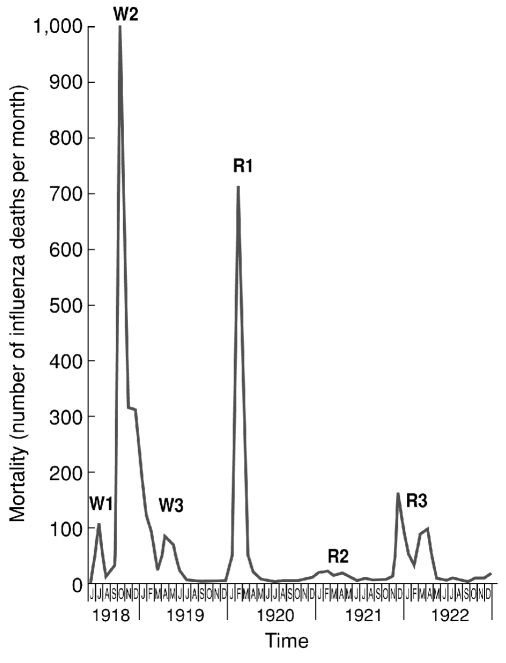
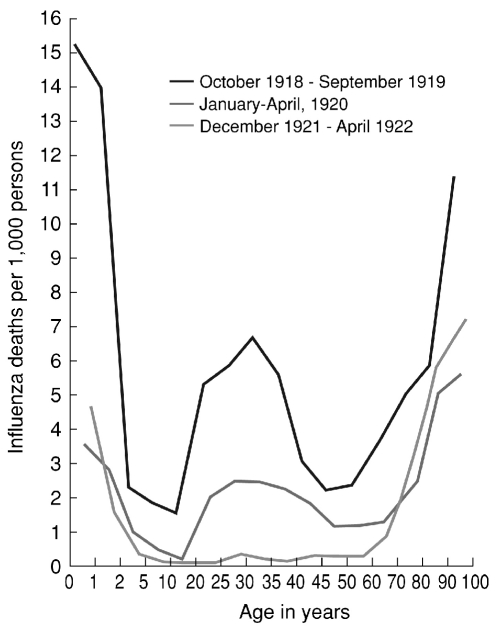
The Spanish influenza pandemic, which stands as the single most fatal event in human history, killed an estimated 50 million people or more globally.14 As noted, the causative agent was an avian-descended H1N1 virus and a direct progenitor of all of the influenza A viruses circulating in humans today.2,3 The high mortality associated with the 1918 virus appears to have been largely a result of bacterial pneumonia, but the co-pathogenic mechanisms responsible for such fatal outcomes remain unknown.47 Epidemiological features of the pandemic were also unprecedented, including its appearance in up to three waves within the first year (Figure 1)48 and a “W-shaped,” age-specific mortality curve featuring an unexplained peak in healthy young adults (Figure 2).3,12 Evidence of a lower-than-expected mortality elevation in people older than age 65 is consistent with a protective effect that ended around the early 1850s, apparently corresponding to the circulation of pandemic viruses appearing in the 1830s and/or 1840s.8
Figure 1.
Three waves of the 1918–1919 influenza pandemic, Breslau, June 1918–December 1922
Note: Three pandemic waves were observed in many locales in 1918–1919, as in these data from Breslau, Silesia (now Wroclaw, Poland), documenting monthly influenza mortality from June 1918 through December 1922. The figure is reproduced from data of Lubinski,a upon which we have superimposed indications of the three 1918–1919 “waves” (W1, W2, and W3) and the first three annual winter post-pandemic recurrences, of 1919–1920 (R1), 1920–1921 (R2), and 1921–1922 (R3).
Source: Lubinski H. Statistische Betrachtungen zur Grippepandemie in Breslau 1918-22. Zentralblatt fur Bakteriologie, Parasitenkunde und Infektionskrankheiten 1923–1924;91:372-83.
Figure 2.
Age-specific influenza mortality, Breslau, July 1918–April 1922
Note: The top line combines influenza mortality in Wave 2 (W2) and Wave 3 (W3) of 1918–1919. The middle line reflects influenza mortality in the first winter recurrence of January–April 1920 (R1). The bottom line reflects influenza mortality in the R3 winter recurrence of December 1921–April 1922. The young adult mortality peak, documented worldwide, is evident in the W2, W3, and R1 curves of 1919–1921, but completely disappeared by 1922.
Source: Lubinski H. Statistische Betrachtungen zur Grippepandemie in Breslau 1918-22. Zentralblatt fur Bakteriologie, Parasitenkunde und Infektionskrankheiten 1923–1924;91:372-83.
The place of origin of the 1918 virus is unknown despite the moniker “Spanish” applied to it in 1918,3,4,49 but there is little evidence of directionality of spread other than chaotic multi-directionality during the second of the three major waves. By about 1920, the virus had begun to settle down into a pattern of seasonal endemic recurrences and remained so as it “drifted” for nearly 40 years. When the next pandemic appeared in 1957, the H1N1 virus disappeared from circulation, although it returned in 1977 to cause a (low-grade) pandemic that disproportionately affected people younger than 20 years of age. The virus continues to co-circulate globally today, along with H3N2 influenza A viruses descended from the 1968 pandemic.
Viral sequence data suggest that the entire 1918 virus was novel to humans in, or shortly before, 1918, and thus unlikely to have been produced from previously circulating human influenza strains that acquired one or more new gene segments by reassortment, as was the case in 1957 and 1968.37,38 On the contrary, data suggest that the 1918 virus was an avian-like influenza virus derived in toto from an unknown source3,40,41 that was ultimately avian. Whether the adaptation pathway between birds and humans was direct, involved adaptation in an intermediate mammalian or other animal host, or involved gradual adaptation during pre-pandemic human circulation, is unknown.
The 1918 pandemic was the most lethal influenza pandemic on record. Most communities experienced morbidity of 25% to 40%, but the majority of cases in open populations (97% to 99%) were self-limited. Age-specific morbidity patterns were similar to pandemics before and since, with children younger than 15 years of age experiencing the highest rates of symptomatic infection.49 Clinically, the 1918 pandemic presented generally the same symptoms and course as influenza of other years and, pathologically, the disease was similar to other pandemics in that severe complications were confined largely to the respiratory tract.47,50,51
However, the 1918 pandemic differed from other pandemics in several important clinical and epidemiologic aspects. Although the clinical course was usually self-limited, a substantially higher percentage of cases developed severe pneumonic complications. As a result, the case mortality rate in the United States averaged 2.5%, several times higher than the current rate. Moreover, mortality during the 1918 pandemic was concentrated in an unusually young age group.3 People younger than age 65 accounted for more than 99% of excess influenza-related deaths in 1918. In contrast, in the 1957 and 1968 pandemics, people younger than 65 years of age accounted for only 36% and 48% of excess deaths due to influenza, respectively.11 The age group affected most severely by the 1918 pandemic was that between 20 and 40 years, accounting for almost half of influenza deaths during the pandemic. Despite the progress made to sequence,2,40 reconstruct,52 and evaluate the 1918 influenza virus in experimental animals,53 the reasons for these unexpected patterns remain obscure.
The Asian influenza (1957–1958)
The pandemic virus that emerged in 1957–1958 was a lineal descendant of the 1918 H1N1 pandemic virus that had somehow acquired three novel gene segments. The gene segments encoding the two surface proteins, HA and NA, were replaced by avian-like H2 and N2 subtypes, respectively. The gene segment encoding the PB1 polymerase was also replaced with an avian-like gene segment.37,38 Even though this pandemic occurred in the era of influenza virology, it is not known in what host the reassortment event(s) occurred. It is also not known how long it took from the initial reassortment event(s) for the virus to evolve into the efficiently transmissible, human-adapted influenza A virus that caused the pandemic.
The 1957–1958 pandemic followed the by-now typical pattern of appearance in Southeast Asia and subsequent global spread, although its movement and mortality rate were not as impressive as those of the two previous pandemics, in 1889 and 1918.8 Emergence of the H2N2 Asian influenza virus was first detected in April 1957, when it was reported that the strain responsible for epidemic outbreaks throughout Southeast Asia was antigenically distinct from the prevailing H1N1 strain.
Contemporary observers noted its easily traceable geographic spread, a characteristic that was shared with the pandemic of 1889,8 but was not readily apparent in 1918 or during interpandemic influenza epidemics.54 As the first pandemic to occur in the era of modern virology, the 1957 pandemic was studied scientifically with the latest virological and bacteriological methods. Its pathology and clinical appearance were similar or identical to those caused by the 1918 virus, although some of the unusual epidemiologic features of the 1918 pandemic were not seen in 1957. As was true for the 1918 pandemic, after about two years the virus became seasonally endemic and sporadic, disappearing entirely within 11 years. To date (2009) it has not returned.
The Hong Kong influenza (1968–1969)
Like the pandemic that preceded it, the 1968–1969 H3N2 pandemic (“Hong Kong flu”) was caused by a virus that had been “updated” from the previously circulating virus by reassortment of avian genes, in this case the HA and PB1, to create yet another new generation of 1918 viral descendants. Spreading again from Southeast Asia, the 1968 pandemic was so modest in its mortality impact that in some locales fewer influenza deaths occurred than in certain non-pandemic years.8 As had been the case in 1957, the virus quickly became endemic and sporadic in its appearance, and it has now (in 2009) circulated globally for 41 years.
The 1968 H3N2 pandemic virus replaced the previous H2N2 subtype virus. A molecular analysis of the H3N2 virus demonstrated that the H2 HA had been replaced by reassortment with an avian-like H3 HA and that the PB1 polymerase gene segment had also been replaced, again by reassortment with an avian-like PB1.37,38 The other six gene segments, including the NA gene segment, were retained from the 1957 H2N2 virus. It has been suggested that the relative mildness of the 1968 pandemic in comparison with previous pandemics was the result of the retention of the previously circulating NA,55 to which most of the population was at least partially immune. Antibodies to NA, while not preventing infection, have been shown to limit virus replication and the duration and severity of illness.56
The 2009 swine-origin influenza
The 2009 H1N1 pandemic virus was derived by reassortment between two preexisting swine influenza viruses—a North American swine H1N2 “triple reassortant” lineage virus and a Eurasian H1N1 swine lineage virus—although whether the novel virus first emerged in humans or swine is not currently known.5,28 Like the 1957 and 1968 pandemic viruses described previously, the 2009 pandemic virus is also a descendant of the 1918 pandemic virus,13 through the “classical” swine H1N1 lineage that circulated enzootically in swine in North America since 1918. That the 1957, 1968, 1977, and 2009 pandemic viruses all share genetic ancestry with the 1918 pandemic virus suggests that we have been living in a specific pandemic era since the 1918 pandemic.13
The novel H1N1 virus was first detected in a widespread outbreak in Mexico in March–April 2009,57,58 but may have been circulating in people as early as late 2008.59,60 While severe pneumonias have been described, especially associated with the initial Mexican outbreak,58,61 most cases in the United States and in other countries have been self-limited, and appear clinically similar to seasonal influenza.62 While the WHO has reported millions of cases and at least 16,813 documented deaths (as of March 19, 2010), the number of deaths is undoubtedly higher. In fact, a recent estimate of H1N1 cases in the United States placed the figure at more than 41 million cases with at least 8,330 deaths.63 The full impact of this pandemic cannot yet be predicted.
It will be important to follow the pandemic as it enters the 2010 Southern Hemisphere winter influenza season, as well as to prepare for further recurrences in the fall-winter of 2010–2011 in the Northern Hemisphere.
IN WHAT WAYS CAN WE PREDICT FUTURE OCCURRENCES […] AND HOW WILL FUTURE OUTBREAKS BEHAVE?
A 1931 editorial in the Journal of the American Medical Association64 stated, “. . . it does not seem possible, with our present knowledge, to make any prediction as to whether or not an epidemic might be expected in the near future.” Unfortunately, despite more than 70 years of intensive study of influenza virus biology since this editorial was written, and 189 years after the question was posed by Georg Friedrich Most,21 we are still unable to predict future pandemics, as evidenced by the completely unexpected emergence of the 2009 swine-origin H1N1 virus.8,13,42
H5N1 avian influenza and the continuing risk of a future pandemic
The continuing spread of H5N1 HPAI viruses in poultry and wild bird populations on several continents since 2003 has led to repeated human spill-over infections and broad interest in pandemic prediction.6,16 Although overshadowed by H5N1, during the past decade at least eight other major poultry epizootics have occurred, caused either by emergence of novel H5 or H7 subtype HPAI viruses unrelated to Asian H5N1 viruses, or in one case by an H9N2 low pathogenic avian influenza (LPAI) virus. Some of these epizootics have featured human infections and, rarely, human deaths.65
In the past decade, several H7 subtype HPAI viruses have independently emerged in poultry in which there have also been human zoonotic infections, perdominantly involving viral conjuctivitis.45,66 Since the mid-1990s, strains of H9N2 LPAI viruses have become enzootic in domestic poultry populations on several continents,65,67 leading to a small number of human infections. As with H5N1,16 different genetic lineages of H9N2 have been established. Some H9N2 viruses have even acquired enhanced specificity for the human form of the HA receptor.68
HPAI H5N1 epizootics are unique, however, in causing infections and deaths in a large number of wild bird species, occasional infections in wild and domestic mammals, more frequently severe and fatal spill-over infections in humans, and in rare instances, possible “dead-end” human-to-human transmission.46 Do these unique features of epizootic H5N1 viruses predict an impending pandemic? There is little consensus among experts. Despite significant research, fundamental questions about how influenza A viruses switch hosts from wild avian species to domestic poultry and mammals, and subsequently to human hosts, remain unanswered. Also incompletely understood are the viral genetic changes that underlie human adaptation; even less well understood are those genetic changes that would allow human-to-human transmissibility and the viral, host, or environmental cofactors that may contribute to human pathogenesis.3 Given its potential for high human morbidity and mortality, the likelihood that the H5N1 virus could become adapted to efficient human-to-human transmission is a critical unknown in pandemic preparedness planning. In this regard, even though historical observations support the inevitability of future pandemics, data accumulated over the past decade may not strongly point to emergence of an H5N1 influenza pandemic. Examination of current and historical information leads us to the following reflections.
Evidence suggests that H5N1 viruses are evolving rapidly; however, the direction of this evolution, which is driven by incompletely understood selection pressures, is unclear. While current strains of Southeast Asian H5N1 HPAI viruses are descendants of the 1996 Chinese epizootic virus, significant genetic and antigenic evolution has since occurred, involving drift in the H5 HA, mutations in other genes, and reassortment with other avian influenza viruses.17 It is not yet clear which of these many changes are associated with lethality in wild birds, or with pathogenicity and transmissibility in poultry or other species. At the same time, adaptation of H5N1 HPAI strains associated with asymptomatic, endemic infection of domestic ducks is probably contributing to continuing spill-over into poultry, leading to the maintenance of an enzootic pool of viruses to which humans will be continually exposed.69 Nevertheless, there are limited data relating to whether any H5N1 influenza strain is evolving in the direction of human adaptation.
Only H5 and H7 viruses are known to acquire the requisite polybasic insertional mutation at the HA cleavage site that makes them highly pathogenic to poultry. The last five human pandemic viruses, which contained HA genes of H1 (1918, 1977, 2009), H2 (1957), and H3 (1968) subtypes, were thus by definition not HPAI viruses. Neither is there evidence that a human pandemic or even an epidemic has been caused by any of the many other HPAI viruses. Furthermore, while HPAI outbreaks have been described in poultry for more than 130 years, none of the last five pandemics is known to have been temporally associated with an epizootic in poultry or wild birds, leaving no historical data to support the possibility that poultry are capable of serving as intermediate hosts in the development of a pandemic.
Biological barriers to the fitness of viruses with particular gene segment combinations are still poorly understood; however, virulence/pathogenicity, host adaptation, and host-to-host transmissibility are likely to be independent properties that are associated with different, and possibly competing, mutational changes. The role of virulence and pathogenicity in evolutionary virus/host relationships is, therefore, unclear; pandemic viruses of comparatively low (e.g., 1968), intermediate (e.g., 1957), and high (e.g., 1918) pathogenicity have all adapted to humans and exhibited efficient pandemic transmissibility. The impact of the current H1N1 pandemic virus is currently unknown.
To cause a pandemic, an avian virus would have to adapt at least to human HA receptors and separately acquire human transmissibility properties. This appears to be a difficult challenge that is rarely met by influenza A viruses. Despite the likelihood that humans and other mammals have been exposed to countless avian viruses over many centuries, the last three pandemics have resulted from reassortment of preexisting human-adapted or swine-adapted viruses with imported genes derived from avian influenza viruses, not from de novo adaptation of avian viruses to humans.13 When genes from a 1997 H5N1 virus were experimentally reassorted in various combinations with those from a human H3N2 virus, some reassortant combinations resulted in viral replication in ferrets, but none was efficiently transmitted between animals,70 prompting critical questions about whether H5N1 viruses may be limited in their potential to adapt to, and be transmitted between, humans.
The mutational changes associated with the binding of H5N1 viruses to receptors in different hosts are proving to be complex.71 Adaptation of the viral HA receptor-binding site from a form optimized for binding the “avian” receptor to a form binding efficiently to the “human” receptor seems to require some loss of specificity for a2,3-linked (putative “avian-like”) sialic acids in favor of increased specificity for a2,6-linked (putative “human-like”) sialic acids. Experiments suggest that only two mutations in the receptor-binding site converted the H1, H2, and H3 HAs of the past three influenza pandemic viruses from avian receptor-binding patterns to human receptor-binding patterns. Several mutations have been reported to enhance the binding of H5 to the human form of the receptor; however, none has been reported to induce a complete switch in specificity.
While it is possible that additional unknown mutations could result and cause such a switch, there is no evidence that this has happened after at least 13 years of exposure of thousands of humans to H5N1, and no evidence that this has happened after human exposure to other HPAI or LPAI viruses of the H5 subtype over many decades. Changes in HA receptor binding during host adaptation must therefore be extremely complex, and must differ from subtype to subtype. The H5 viruses and other subtypes may well face unappreciated biological barriers in achieving efficient binding to human receptors.
The next pandemic
No one predicted the emergence of the 2009 H1N1 swine-origin pandemic virus; with current knowledge, we doubt that anyone will be able to accurately predict any future pandemic either, including when or where it will occur, what subtype it will be, and what morbidity/mortality impact it will have. While concern over the emergence of an H5N1 pandemic is clearly warranted, if for no other reason than its current high case fatality rate, many other possibilities for future pandemic emergence must also be anticipated and planned for.
The majority of the world's population (those younger than age 41) has no protective immunity to the H2 subtype-bearing influenza viruses that circulated between 1957 and 1968. Isolates of H2N2 viruses from that era are still maintained in countless laboratory freezers, while circulating human-adapted H3N2 viruses presumably remain susceptible to importation of avian H2 by reassortment; this suggests obvious potential origins of future pandemics. Current H9N2 viruses, some with the ability to bind to human receptors, and already capable of causing human disease, are another potential source of a future pandemic.
Since 1977, H1N1 and H3N2 viruses have co-circulated globally to produce seasonal epidemics that cause an average of 36,000 deaths annually in the U.S.72 Moreover, recent data have made it clear that evolution of circulating human influenza viruses occurs not just by gradual antigenic drift but also by intra-clade reassortment resulting in the importation of new HAs of the same subtype to which there is a lesser degree of population immunity, and which creates, at the same time, novel constellations of viral gene segments.73–75 It is unclear whether the 2009 pandemic H1N1 virus will replace the seasonal H1N1 and H3N2 lineages, or co-circulate with them. It is also unclear whether continued co-circulation and accelerated evolution of different post-pandemic viruses, coupled with the growing use of influenza vaccines against them, will increase or decrease future pandemic risk or influence the HA or NA subtype of the next pandemic virus. The co-circulation of post-pandemic H1 and H3 viruses for three consecutive decades seems to be unprecedented over the past 125 to 160 years. If only H1, H2, or H3 viruses have pandemic potential, the question arises whether such co-circulation limits, in the future, the next pandemic to only H2 viruses. At present there are no data to answer such a question; however, over the past several decades the dogma regarding pandemics has been so radically overturned that it is now important to rethink and restudy all aspects of this issue.
The past decade has demonstrated how difficult it is to contain HPAI outbreaks, given high-intensity poultry production and the movement of poultry between countries. The H5N1 viruses are likely to remain enzootic in domestic bird populations in many countries indefinitely. This poses numerous agricultural and economic problems. While it might provide an opportunity for H5N1 viruses to acquire efficient human-to-human transmission (if such a change is in fact possible), it might, on the other hand, provide a better opportunity for viruses to adapt to poultry and wild birds, the chief spill-over hosts. The use of antiviral drugs in agricultural settings has made many H5N1 viruses resistant to adamantanes, while there has also been evidence for H5N1 resistance to neuraminidase inhibitors.76 The evolution of H5N1 into antigenically distinct clades, probably driven in part by the use of poultry vaccines, greatly complicates the situation and makes it more difficult to predict where H5N1 evolution is going, what to expect next, and how to plan for it.6
Understanding and predicting pandemic emergence is a difficult challenge that we are far from being able to meet in 2010. As our understanding of influenza viruses has increased dramatically in recent decades, we have moved ever further from certainty about the determinants of, and possibilities for, pandemic emergence. Planning efforts must consider a range of possibilities that cannot yet be prioritized in terms of their likelihood, and must also address unpredictable ranges of pandemic morbidity and mortality impacts. Until such time as “universal” influenza vaccines77 or better drug treatments become available,78 there is a need for strong basic public health approaches to pandemic control.
THROUGH WHAT MEANS CAN ITS SPREAD BE HALTED?
Pandemic planning envisions that if a virus with pandemic potential emerges, initial human-to-human transmission can be spotted quickly and contained by non-pharmaceutical interventions79 and by rapid community administration of antiviral agents and vaccines.80,81 This did not happen with the 2009 pandemic virus, which had spread internationally before it was recognized.60 Most national stockpiles have appropriately favored NA inhibitors (mainly orally administered oseltamivir) over ion-channel blockers (oral adamantanes) for pandemic preparedness given the well-recognized rapidity of emergence of resistance to the latter when used in treatment. Currently, transmissible oseltamivir resistance in human A/H1N1 strains82 makes this strategy problematic on many levels, including concern about efficacy in a pandemic and about pandemic reassortants containing resistance genes.83 In fact, small numbers of oseltamivir-resistant pandemic H1N1 virus isolates have already been observed.84,85 Whether such resistance will become more common in pandemic H1N1 viruses is currently unknown. A further complicating factor is the increasing recognition that secondary bacterial pneumonias have caused most deaths in past pandemics.47 Circulation of clinically aggressive community-acquired methicillin-resistant Staphylococcus aureus is an additional factor to be considered in planning for pandemic response. Taken together, these several developments suggest a need to continually examine and periodically reconfirm or update pandemic response strategies.78
CONCLUSIONS
How might we prevent and manage a future influenza pandemic? The most obvious requirement is a rapid and expansive influenza surveillance and response network86 with open global communication and data sharing.87 Such surveillance activity also needs to include humans, domestic animals, and wild birds. Second, we must develop further, effective intervention strategies to reduce transmission and disease. This must include implementation of effective non-pharmaceutical interventions.79 The development of vaccines against H5N1 strains, and ultimately against all subtypes, is also a clear priority.88 New vaccine methodologies are in reach, but international agreements on production, intellectual property, distribution, and administration must be pursued aggressively. Antiviral drug stockpiles are limited. We must also begin to think about strategies to reduce the probability of pandemics. This will require a multitude of basic scientific information, including the probability and mechanism of reassortment; a measure of the exposure rates of influenza viruses at the human/animal interface; and, most critically, an understanding of how avian viruses evolve to develop sustained transmission networks in humans. It is therefore essential to conduct global surveillance of genetic diversity in avian influenza viruses,33 sequencing complete genomes from not only avian but also mammalian strains to explore the polygenic nature of host adaptation.28 Although a unified political effort is essential to avert or mitigate a major influenza pandemic, it must proceed in parallel with advances in basic science.
But whatever strategies are adopted, it is clear that additional anti-influenza therapeutics are urgently needed.78,83 So far, vaccines and antivirals have targeted three influenza envelope proteins: HA, NA, and the M2 ion channel protein. We need new classes of antivirals that interfere with other necessary viral processes (e.g., polymerase complex activity, interferon antagonist activity, and viral assembly), and further exploration of passive immunotherapy approaches to treat severe influenza cases.89,90 The desired outcomes of existing and future therapies (reduced severity, mortality, viral shedding, and transmission) should be considered with respect to both seasonal and pandemic influenza.
The unpredictable nature of influenza presents a challenge for both research and pandemic preparedness planning.42 Our ability to anticipate pandemic events is poor and our anti-pandemic armamentarium weak. In an ever-shifting landscape of influenza evolution, we must be far-sighted and forceful in optimizing pandemic response capacity.
Footnotes
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Mulder J, Hers JFP. Influenza. Groningen (Netherlands): Wolters-Noordhoff Publishing; 1972. [Google Scholar]
- 2.Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir Ther. 2007;12(4 Pt B):581–91. [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Reid AH, Fanning TG. Capturing a killer flu virus. Sci Am. 2005;292:48–57. [PubMed] [Google Scholar]
- 5.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–7. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch A. Handbook of geographical and historical pathology. London: New Sydenham Society; 1883. [Google Scholar]
- 8.Taubenberger JK, Morens DM. Pandemic influenza—including a risk assessment of H5N1. Rev Sci Tech. 2009;28:187–202. doi: 10.20506/rst.28.1.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–21. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 10.Beveridge W. Influenza: the last great plague, an unfinished story. New York: Prodist; 1977. [Google Scholar]
- 11.Simonsen L. The global impact of influenza on morbidity and mortality. Vaccine. 1999;17(Suppl 1):S3–10. doi: 10.1016/s0264-410x(99)00099-7. [DOI] [PubMed] [Google Scholar]
- 12.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–28. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 13.Morens DM, Taubenberger JK, Fauci AS. The persistent legacy of the 1918 influenza virus. N Engl J Med. 2009;361:225–9. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–15. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. Pandemic (H1N1) 2009—update 92. 2009. [cited 2010 Mar 22]. Available from: URL: http://www.who.int/csr/don/2010_03_19/en/index.html.
- 16.Guan Y, Poon LL, Cheung CY, Ellis TM, Lim W, Lipatov AS, et al. H5N1 influenza: a protean pandemic threat. Proc Natl Acad Sci USA. 2004;101:8156–61. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Smith GJ, Li KS, Wang J, Fan XH, Rayner JM, et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc Natl Acad Sci USA. 2006;103:2845–50. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster RG, Peiris M, Chen H, Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12:3–8. doi: 10.3201/eid1201.051024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. 2010. [cited 2010 Mar 21]. Available from: URL: http://www.who.int/csr/disease/avian_influenza/country/cases_table_2010_03_16/en/index.html.
- 20.Webster RG, Hulse-Post DJ, Sturm-Ramirez KM, Guan Y, Peiris M, Smith G, et al. Changing epidemiology and ecology of highly pathogenic avian H5N1 influenza viruses. Avian Dis. 2007;51(1) Suppl:269–72. doi: 10.1637/7641-050206R.1. [DOI] [PubMed] [Google Scholar]
- 21.Most GF. Influenza Europaea, oder die Größeste Krankheits-Epidemie der neuern Zeit Hamburg: Perthes und Besser. :1820. [Google Scholar]
- 22.Shope RE. Swine influenza. I. Experimental transmission and pathology. J Exp Med. 1931;54:349–59. doi: 10.1084/jem.54.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis PA, Shope RE. Swine influenza: II. A hemophilic bacillus from the respiratory tract of infected swine. J Exp Med. 1931;54:361–71. doi: 10.1084/jem.54.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shope RE. Swine influenza. III. Filtration experiments and etiology. J Exp Med. 1931;54:373–85. doi: 10.1084/jem.54.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun J. Influenza including its infection among pigs. Natl Med J. 1919;5:34–44. [Google Scholar]
- 26.Koen J. A practical method for field diagnosis of swine diseases. Am J Vet Med. 1919;14:468–70. [Google Scholar]
- 27.Taubenberger JK, Reid AH, Janczewski TA, Fanning TG. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:1829–39. doi: 10.1098/rstb.2001.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunham EJ, Dugan VG, Kaser EK, Perkins SE, Brown IH, Holmes EC, et al. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J Virol. 2009;83:5485–94. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dochez A, Mills K, Kneeland Y. Studies of the etiology of influenza. Proc Soc Exp Biol Med. 1934–1935;30:1017–22. [Google Scholar]
- 30.Smith W, Andrewes C, Laidlaw P. A virus obtained from influenza patients. Lancet. 1933;2:66–8. [Google Scholar]
- 31.Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. 5th ed. Philadelphia: Lippincott: Williams & Wilkins; 2007. pp. 1647–90. [Google Scholar]
- 32.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, Holmes EC. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol. 2006;23:2336–41. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- 35.Wright PF, Neumann G, Kawaoka Y. Fields virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. Orthomyxoviruses. In: Knipe DM, Howley PM, editors; pp. 1691–740. [Google Scholar]
- 36.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–80. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 37.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 38.Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–8. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid AH, Taubenberger JK, Fanning TG. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Natl Rev Microbiol. 2004;2:909–14. doi: 10.1038/nrmicro1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–93. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 41.Rabadan R, Levine AJ, Robins H. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J Virol. 2006;80:11887–91. doi: 10.1128/JVI.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taubenberger JK, Morens DM, Fauci AS. The next influenza pandemic: can it be predicted? JAMA. 2007;297:2025–7. doi: 10.1001/jama.297.18.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaydos JC, Top FH, Jr., Hodder RA, Russell PK. Swine influenza A outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis. 2006;12:23–8. doi: 10.3201/eid1201.050965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sencer DJ, Millar JD. Reflections on the 1976 swine flu vaccination program. Emerg Infect Dis. 2006;12:29–33. doi: 10.3201/eid1201.051007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci USA. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352:333–40. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 47.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–70. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morens DM, Taubenberger JK. Understanding influenza backward. JAMA. 2009;302:679–80. doi: 10.1001/jama.2009.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan EO. Chicago: American Medical Association; 1927. Epidemic influenza: a survey. [Google Scholar]
- 50.Kuiken T, Taubenberger JK. The pathology of human influenza revisited. Vaccine. 2008;26(Suppl 4):D59–66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Ann Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 53.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–81. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmuir AD, Serfling RE, Shermann IL, Dauer CC, Epidemiology Branch CDC . Washington: Department of Health, Education, and Welfare, Public Health Service, Communicable Disease Center (US); 1960. The epidemiology of Asian influenza 1957–1960: a descriptive brochure. [Google Scholar]
- 55.Kilbourne ED. Perspectives on pandemics: a research agenda. J Infect Dis. 1997;176(Suppl 1):S29–31. doi: 10.1086/514171. [DOI] [PubMed] [Google Scholar]
- 56.Kilbourne ED, Pokorny BA, Johansson B, Brett I, Milev Y, Matthews JT. Protection of mice with recombinant influenza virus neuraminidase. J Infect Dis. 2004;189:459–61. doi: 10.1086/381123. [DOI] [PubMed] [Google Scholar]
- 57.Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March–April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:1–3. (Dispatch) [PubMed] [Google Scholar]
- 58.Perez-Padilla R, de la Rosa-Zamboni D, Ponce de Leon S, Hernandez M, Quinones-Falconi F, Bautista E, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361:680–9. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 59.Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–61. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–5. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 61.Chowell G, Bertozzi SM, Colchero MA, Lopez-Gatell H, Alpuche-Aranda C, Hernandez M, et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N Engl J Med. 2009;361:674–9. doi: 10.1056/NEJMoa0904023. [DOI] [PubMed] [Google Scholar]
- 62.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 63.Centers for Disease Control and Prevention (US) CDC estimates of 2009 H1N1 influenza cases, hospitalizations and deaths in the United States, April 2009–January 16, 2010. [cited 2010 Mar 21]. Available from: http://www.cdc.gov/h1n1flu/estimates/April_January_16.htm.
- 64.Occurence of epidemic influenza in cycles. JAMA. 1931;96:711. Editor. [Google Scholar]
- 65.Alexander DJ. Avian influenza viruses and human health. Dev Biol (Basel) 2006;124:77–84. [PubMed] [Google Scholar]
- 66.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10:2196–9. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol. 2004;78:8609–14. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–62. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 69.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–79. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci USA. 2006;103:12121–6. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taubenberger JK. Influenza hemagglutinin attachment to target cells: “birds do it, we do it . . .”. Future Virol. 2006;1:415–8. doi: 10.2217/17460794.1.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 73.Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008;4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–9. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, Nguyen KH, et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature. 2005;437:1108. doi: 10.1038/4371108a. [DOI] [PubMed] [Google Scholar]
- 77.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg Infect Dis. 2006;12:569–74. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Memoli MJ, Morens DM, Taubenberger JK. Pandemic and seasonal influenza: therapeutic challenges. Drug Discov Today. 2008;13:590–5. doi: 10.1016/j.drudis.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Markel H, Lipman HB, Navarro JA, Sloan A, Michalsen JR, Stern AM, et al. Nonpharmaceutical interventions implemented by US cities during the 1918–1919 influenza pandemic. JAMA. 2007;298:644–54. doi: 10.1001/jama.298.6.644. [DOI] [PubMed] [Google Scholar]
- 80.Halloran ME, Ferguson NM, Eubank S, Longini IM, Jr, Cummings DA, Lewis B, et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci USA. 2008;105:4639–44. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monto AS. The risk of seasonal and pandemic influenza: prospects for control. Clin Infect Dis. 2009;48(Suppl 1):S20–5. doi: 10.1086/591853. [DOI] [PubMed] [Google Scholar]
- 82.Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–41. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 83.Layne SP, Monto AS, Taubenberger JK. Pandemic influenza: an inconvenient mutation. Science. 2009;323:1560–1. doi: 10.1126/science.323.5921.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oseltamivir-resistant pandemic (H1N1) 2009 influenza virus October 2009. Weekly Epidemiol Rec. 2009;84:453–9. [PubMed] [Google Scholar]
- 85.Pandemic (H1N1) 2009 briefing note 1. Viruses resistant to oseltamivir (Tamiflu) identified. Weekly Epidemiol Rec. 2009;84:299–399. [PubMed] [Google Scholar]
- 86.Holmes EC, Taubenberger JK, Grenfell BT. Heading off an influenza pandemic. Science. 2005;309:989. doi: 10.1126/science.1117128. [DOI] [PubMed] [Google Scholar]
- 87.Barry JM. Pandemics: avoiding the mistakes of 1918. Nature. 2009;459:324–5. doi: 10.1038/459324a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Subbarao K, Murphy BR, Fauci AS. Development of effective vaccines against pandemic influenza. Immunity. 2006;24:5–9. doi: 10.1016/j.immuni.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 89.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]