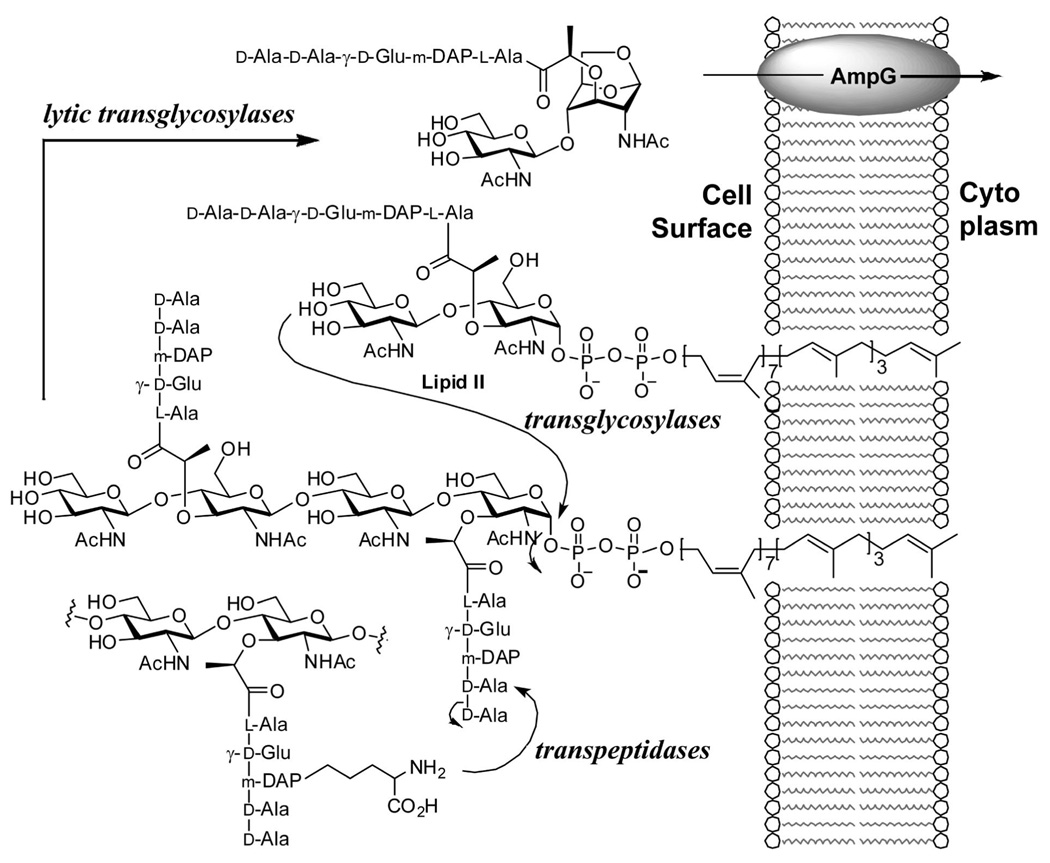
The cell wall is a critically important structural entity in bacteria.1–3 Because of its importance to bacterial survival, the cell wall itself and its biosynthetic enzymes are targets of antibiotics.4,5 The complex process of biosynthesis of the cell wall is initiated in the cytoplasm, leading to lipid II. Lipid II is translocated to the surface of the plasma membrane, where it is polymerized (by transglycosylases) and cross-linked (by transpeptidases) to give the cell wall structure that encases the entire organism, which incidentally exceeds the bacterial genome in mass (Figure 1).6 The backbone of the peptidoglycan, the major constituent of the cell wall, is a repeating unit of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). A special stem peptide, typically a pentapeptide, is appended to the NAM unit. The cross-linking is through the peptide portion of the structure (Figure 1).
Figure 1.
Functions of transglycosylases and transpeptidases in assembly of cell wall starting from lipid II, and its fragmentation by lytic transglycosylases. The minimal fragmented product is internalized by AmpG for recycling.
An intriguing observation on growth and biosynthesis of the cell wall is that on doubling of the bacterial population as much as 60% of the parental cell wall is recycled,7,8 entailing a degradative processing of the older cell wall, entry into the cell, and reuse of the components for the de novo biosynthesis. Much of this recycling process is not understood at the present state of knowledge. However, a key event is fragmentation of the parental cell wall by the family of lytic transglycosylases.9,10 In contrast to other known glycosidases, these enzymes are not hydrolytic. It has been proposed that they cleave the sugar backbone between the NAM and NAG saccharides by a process that goes through an oxocarbenium species, which in turn traps the 6-hydroxyl group of glucosamine to result in what is referred to as the 1,6-anhydromuramyl moiety (see conversion of compound 1 to 2 and 3, discussed below).11,12 This is a remarkable reaction, since the glucosamine rings in the peptidoglycan are shown to be in the chair conformation with all equatorial substituents, which become converted to an all axial arrangement upon reaction (2).
Because of the importance of this recycling process, multiple genes exist for lytic transglycosylases in each bacterial genus. A quantitative analysis of these enzymic activities is absent from the literature, because of the lack of reagents and, in some cases, enzymes. There is also a paucity of information on the distinct roles of the individual enzymes in degradation of the cell wall. In this report, we disclose the cloning of the genes and purification of all six known lytic transglycosylases of Escherichia coli. We also document that all these recombinant enzymes are active in fragmentation of the sacculus (the whole cell wall), but only one, MltB, is able to process a short chain synthetic fragment of the bacterial cell wall. The quantitative analysis of this process reveals it to be a key event that leads to the component of the peptidoglycan that is transported into the cytoplasm for the recycling purpose in E. coli.
The genes for the six known lytic transglycosylases in E. coli were cloned without the sequences for the leader peptides to prevent them from being transported to the periplasmic space. The cysteine residue near the N-terminus that experiences lipidation responsible for membrane anchoring was left out to avoid membrane sequestration, and a sequence for C-terminal His-tag was added to each. There was good cytoplasmic expression of all six enzymes in water-soluble forms, which resulted in preparations of MltA, MltB (also known as Slt35), MltC, MltD, MltE (also known as EmtA), and Slt70 after a single Ni(II)-affinity chromatography purification (see Supporting Information). All six exhibited activity in fragmentation of the sacculus by a zymography assay. In this assay the isolated sacculus is embedded in a polyacrylamide gel under denaturing conditions. The proteins are run in the gel and are in turn renatured in situ. The degradative processing of the cell wall is indicated by the clearing of the gel at the location of the enzyme (Supporting Information).13
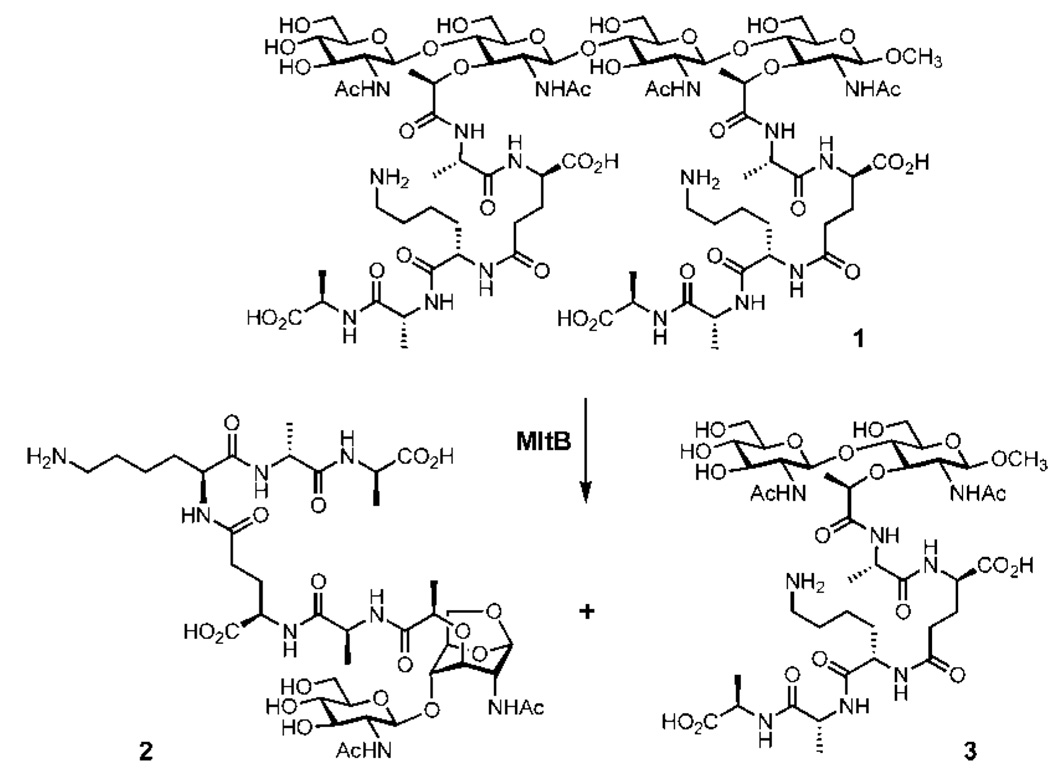
We subsequently investigated whether these purified enzymes would process a synthetic piece of the peptidoglycan (compound 1; lysine in the peptide moiety of this compound is seen in some bacteria, but others have diaminopimelate in its place). This synthetic fragment has a short chain backbone of only two repeats of NAG-NAM. This fragment was synthesized in 37 steps according to the method that we have published earlier.14 The screening (Supporting Information) with compound 1 was performed with reversed-phase HPLC monitoring. This analysis revealed that out of the six E. coli lytic transglycosylases only MltB was able to degrade the synthetic substrate. The two reaction products eluted on the chromatogram with retention times of 31.0 and 37.5 min, analyses of which by LC-MS (ESI) confirmed them as compounds 3 and 2, respectively. Authentic synthetic samples of 2 and 3 were prepared in our laboratory, which unequivocally confirmed the structure assignments for the two products of the MltB reaction (Figure 2). We hasten to add that conversion of 1 to 2 and 3 by MltB was clean (see Supporting Information), and no evidence exists for potential entrapment of a water molecule by the reactive intermediary oxocarbenium species.
Figure 2.
Degradation of synthetic peptidoglycan 1 by MltB.
Nonlinear regression of the data at pH 7.0 for either the consumption of substrate 1 or formation of the products 2 and 3 furnished the kinetic parameters Km of 1.8 ± 0.5 mM and kcat of 7.7 ± 1.0 s−1. The millimolar Km value is not atypical for many important metabolic enzymes. For instance, of the 21 enzymes for glycolysis and the tricarboxylic acid cycle, 10 have millimolar Km values for their respective substrates. The effective concentration of the peptidoglycan near the surface of the plasma membrane (an entropic contribution) is expected to be high; hence saturation of the membrane-anchored MltB by the peptidoglycan should not be a problem. This level of activity is sufficient to allow turnover of the peptidoglycan as many as 14 000 times within a single bacterial doubling time of 30 min for each MltB molecule.
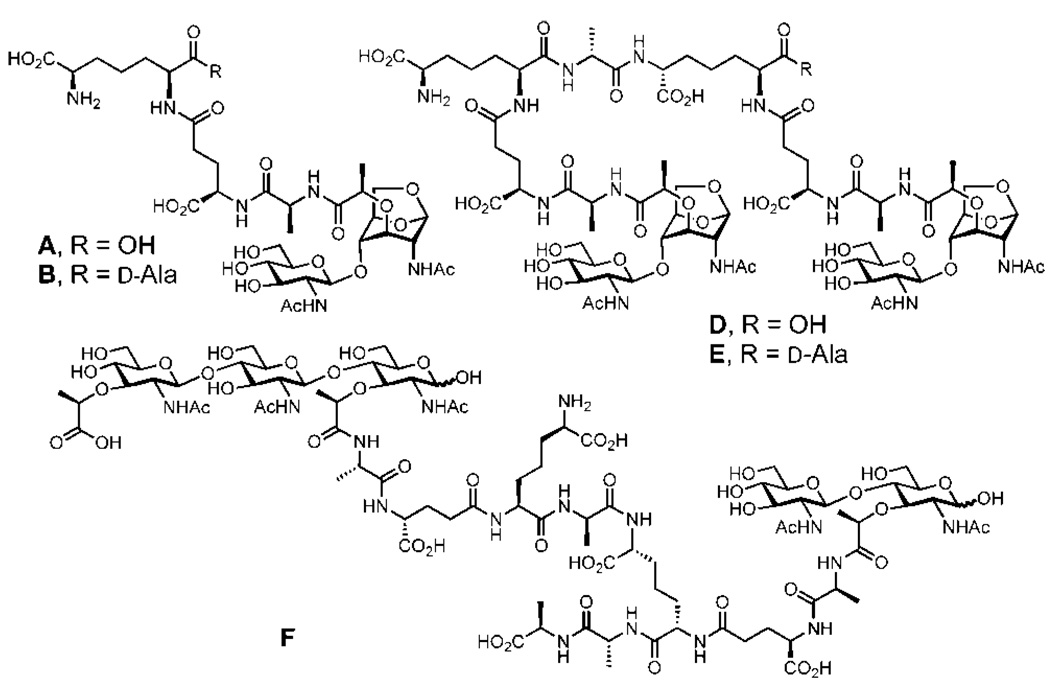
When MltB was incubated with the purified E. coli sacculus, the enzyme fragmented it, as would be expected from the aforementioned zymography assay. LC-MS (ESI) analysis of the resulting fragments is shown in Figure 3 (see Supporting Information for the HPLC elution profile). Six major fragments were discernible, of which four (fragments A, B, D, and E) possessed the 1,6-anhydromuramyl moiety, the products of the MltB reaction, whereas one (fragment F) did not. Fragment C could not be characterized. The structures of these fragments indicate that the cell wall is extensively modified in the peptide portions, as there are multiple enzymes that process the peptide stems in E. coli. The two major products are A and B, which differ from one another by one amino acid. Fragments D and E are products of MltB processing of two cross-linked strands of peptidoglycan. It is important to note that fragment F is not the result of the catalytic function of MltB, but it would appear to be “left over” from the exhaustive processing of the sacculus by this enzyme. We had noted from the turnover of compound 1 by MltB that no entrapment of water by the oxocarbenium ion took place. Hence, the observation of “reducing sugars” in fragment F is indicative of processing of peptidoglycan by a hydrolytic enzyme in the course of maturation of the cell wall. Compound 2 and its congeners from the sacculus, fragments A and B, are known to be transported into the cytoplasm for recycling by AmpG (Figure 1), an integral membrane protein.15 Diffusion of a portion of these products of the MltB reaction into the host tissue causes the inflammation associated with bacterial infections.16–18 Hence, the formation of these key chemical species is responsible for both the economy of the biosynthetic processes of the cell wall and the pathological manifestations associated with infections. The steps mediated by MltB in E. coli are likely to be processive, from one or the other end of the peptidoglycan. We hasten to add that it is likely that compound 1 might be the minimal substrate for MltB, requiring occupation of the seat of the reaction by one NAG-NAM repeat on each side of the scissile bond. We conclude this based on the observation that the β-glycosidic compound 3 (methoxy at C1) is not a substrate for the enzyme. This was seen both by the lack of processing of 3 in the product mixture from the reaction of turnover of 1 by MltB and by the addition of the authentic compound 3 to the enzyme in a separate attempt at the reaction.
Figure 3.
Products of sacculus fragmentation by the action of MltB and the structure assignment by ESI-MS.
Supplementary Material
Acknowledgment
This work was supported by the National Institutes of Health.
Footnotes
Supporting Information Available: Experimental procedures of cloning, purification, zymography, kinetics, mass spectrometry, and degradation of sacculus. This material is available free of charge via the Internet at http://pubs.acs.org. the structure assignment by ESI-MS.
References
- 1.Coyette J, van der Ende A. FEMS Microbiol. Rev. 2008;32:147–148. doi: 10.1111/j.1574-6976.2008.00108.x. [DOI] [PubMed] [Google Scholar]
- 2.Suvorov M, Fisher JF, Mobashery S. Bacterial Cell Wall: Morphology and Biochemistry. In: Goldman E, Green LH, editors. Practical Handbook of Microbiology. 2nd ed. CRC Press; 2008. pp. 153–183. [Google Scholar]
- 3.Höltje J-V. Cell Walls, Bacterial. In: Schaechter M, editor. Desk Encyclopedia of Microbiology. San Diego, London: Elsevier Academic Press; 2004. pp. 239–250. [Google Scholar]
- 4.Van Bambeke F, Mingeot-Leclercq MP, Struelens MJ, Tulkens PM. Trends Pharmacol. Sci. 2008;29:124–134. doi: 10.1016/j.tips.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Silver LL. Biochem. Pharmacol. 2006;71:996–1005. doi: 10.1016/j.bcp.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Vollmer W, Höltje J-V. J. Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JT, Uehara T. Microbiol. Mol. Biol. Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Pedro MA, Donachie WD, Höltje J-V, Schwarz H. J. Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vollmer W, Joris B, Charlier P, Foster S. FEMS Microbiol. Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 10.Scheurwater E, Reid CW, Clarke AJ. Int. J. Biochem. Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Powell AJ, Liu ZJ, Nicholas RA, Davies C. J. Mol. Biol. 2006;359:122–136. doi: 10.1016/j.jmb.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 12.van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, Dijkstra BW. Structure. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer JM, Strating H, Weadge JT, Clarke AJ. J. Bacteriol. 2006;188:902–908. doi: 10.1128/JB.188.3.902-908.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hesek D, Lee M, Morio K-I, Mobashery S. J. Org. Chem. 2004;69:2137–2146. doi: 10.1021/jo035583k. [DOI] [PubMed] [Google Scholar]
- 15.Cheng QM, Park JT. J. Bacteriol. 2002;184:6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 17.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Nat. Rev. Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 18.Kohler PL, Cloud KA, Hackett KT, Beck ET, Dillard JP. Microbiology. 2005;151:3081–3088. doi: 10.1099/mic.0.28125-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.