Abstract
Penicillin-binding protein 5 (PBP 5) of Escherichia coli is a membrane-bound cell wall DD-carboxypeptidase, localized in the outer leaflet of the cytosolic membrane of this Gram-negative bacterium. Not only is it the most abundant PBP of E. coli, but it is as well a target for penicillins, and is the most studied of the PBP enzymes. PBP 5, as a representative peripheral membrane protein, is anchored to the cytoplasmic membrane by the 21 amino acids of its C-terminus. Although the importance of this terminus as a membrane anchor is well recognized, the structure of this anchor was previously unknown. Using natural isotope abundance NMR, the structure of the PBP 5 anchor peptide within a micelle was determined. The structure conforms to a helix-bend-helix-turn-helix motif and reveals that the anchor enters the membrane so as to form an amphiphilic structure within the interface of the hydrophilic/hydrophobic boundary regions near the lipid head groups. The bend and the turn within the motif allow the C-terminus to exit from the same side of the membrane that is penetrated. The PBP anchor sequences represent extraordinary diversity, encompassing both N-terminal and C-terminal anchoring domains. This study establishes a surface adherence mechanism for the PBP 5 C-terminus anchor peptide, as the structural basis for further study toward understanding the role of these domains in selecting membrane environments and in the assembly of the multi-enzyme hyperstructures of bacterial cell wall biosynthesis.
Introduction
Every cell has its boundary. For the unicellular bacterium, this boundary is a structurally complex cell wall integrating cytoskeletal proteins and a cytoskeletal polymer, the peptidoglycan, with membrane bilayer(s). Not surprisingly, the simultaneous fulfillment of the tasks of cytoprotection and facile communication with its environment occupy an appreciable portion of the bacterial genome: as many as 30% of the proteins of the Gram-negative bacterium Escherichia coli are membrane-bound.1–3 These membrane proteins may be divided among proteins that associate with the membrane, that embed into the membrane by a single membrane-spanning domain (bitopic proteins), and that embed into the membrane using multiple membrane-spanning domains (polytopic proteins). From the perspective of human control of infection, there is arguably no more important class of bacterial membrane proteins than the Penicillin-Binding Proteins (PBPs). These proteins catalyze the biosynthesis of the peptidoglycan polymer of the cell wall, and are the molecular targets of the β-lactam class of antibacterials. In the Gram-negative bacterium, the PBPs localize in the cytoplasmic membrane, where at least some assemble into multi-protein hyperstructures for cell wall biosynthesis.4,5
The genome of the E. coli bacterium encodes twelve PBP enzymes. This family of membrane proteins may be divided between the bitopic high-molecular-mass PBPs that catalyze the transglycosylase and transpeptidase activities of peptidoglycan biosynthesis, and the membrane-associated low-molecular-mass PBPs that engage in peptidoglycan maturation, separation, and recycling.6 The bitopic high-molecular-mass PBPs possess a single membrane-spanning α-helix domain, composed entirely of hydrophobic amino acids, and located at their N-terminus.7 In contrast, the membrane-associated low-molecular-mass PBPs use an amphipathic C-terminus as a membrane anchor, which incidentally is believed to be helical as well.8,9 The most abundant of the E. coli PBPs, found in approximately 800 copies per bacterium,10 is the low-molecular-mass PBP 5 carboxypeptidase. As with all of the low-molecular-mass PBPs of E. coli, the precise physiological role for this one PBP is uncertain.9 While deletion of PBP 5 is not lethal to the bacterium, its deletion correlates to diminished control over the morphology11 of the rod-shape of E. coli and sensitization of the bacterium to β-lactams.12 A key—and exceptionally poorly understood—aspect to the understanding of the specific tasks accomplished by the low-molecular-mass PBPs is the correlation of their temporal expression—PBP 5 is most abundant at early log phase growth—with their location in the cell membrane. The outstanding question in addressing these issues at the molecular level is the detailed understanding of how each PBP interacts with the bacterial membrane. It is now appreciated that protein-membrane interactions are mutually synergistic,13 and that one purpose of this synergism is membrane localization.14,15 Although the ability of membranes to induce a helix-like character in the C-terminus membrane anchor of PBP 5 has been long recognized,16–19 the structure of the PBP 5 membrane anchor when membrane-associated was not known previously. As the initial and critical step toward understanding the dynamics of the PBP 5-membrane interaction, we report herein the structural elucidation by NMR of the PBP 5 membrane anchor peptide, as the first solved example of the peripheral PBP anchors.
Materials and Methods
The cloning and purification of the anchor-free PBP 5 was reported earlier.20
Circular Dichroism Experiments
CD spectra of PBP 5 were recorded on a stopped-flow circular dichroism spectrometer (Aviv Instruments Inc-202 SF) with a 0.2 cm path length at 25 °C interfaced to an IBM 300GL personal computer. Samples of PBP 5 C-terminus peptide (53 µM, 750 µL), synthesized by Global Peptide, were prepared by dissolving the peptide in aqueous pH buffered micelle solution containing DPC (2.12 mM) or a lipid solution of POPE/POPG/CL 70:25:5 with a 20 mM phosphate buffer, pH values 5.5 or 7.5.
NMR Experiments
All NMR experiments were performed at 298.1 K using a 2 mM unlabeled peptide sample with a four-channel Bruker AVANCE II (800.13 MHz, 1H) spectrometer equipped with a 5-mm inverse triple-resonance (1H, 13C, 15N) cryoprobe. The standard experiments of DQF-COSY, TOCSY (31 and 61 ms), NOESY (80 and 200 ms), 13C HSQC, 15N HSQC and 13C HSQC-TOCSY were used to determine the structure. Relaxation rate constants R1 (R1 = 1/T1), and R2 (R2 = 1/T2) were determined from the cross-peak intensities of the corresponding 2D proton detected 15N HSQC like spectra.21,22 D2O exchange results were acquired from continuous alternating measurements of 1D 1H and 2D TOCSY spectra 15 minutes after addition of deuterated water. Peptide to micelle interactions were determined from 2D 1H NOESY spectra using non-deuterated DPC, purchased from Avanti Polar Lipids (Avanti Polar Lipids, Alabaster, AL). Details can be found in Supporting Information.
Structure and Dynamic Calculations
Dihedral angle restraints for Cyana were obtained from the chemical shifts using Talos.23 Peptide structure was determined using the program Cyana (LAS Systems Tokyo Japan).24 Peptide NMR Dynamics data were analyzed using Curvefit and Modelfree 4.2025,26 programs. Statistical structure information and hydrogen bonding were determined with the programs Cyana and Sybyl (Table S1,Supporting Information) using the 20 lowest energy structures from Cyana. The Ramachandran plot indicated that 94% of the residues were in the most favored or additionally allowed regions and 6% in the generously allowed regions. No residues were in the disallowed regions. Figure 5 and Figure S1 (Supporting Information) were made in ChemBioDraw. All other figures were produced using Molmol and PyMOL.
Figure 5.
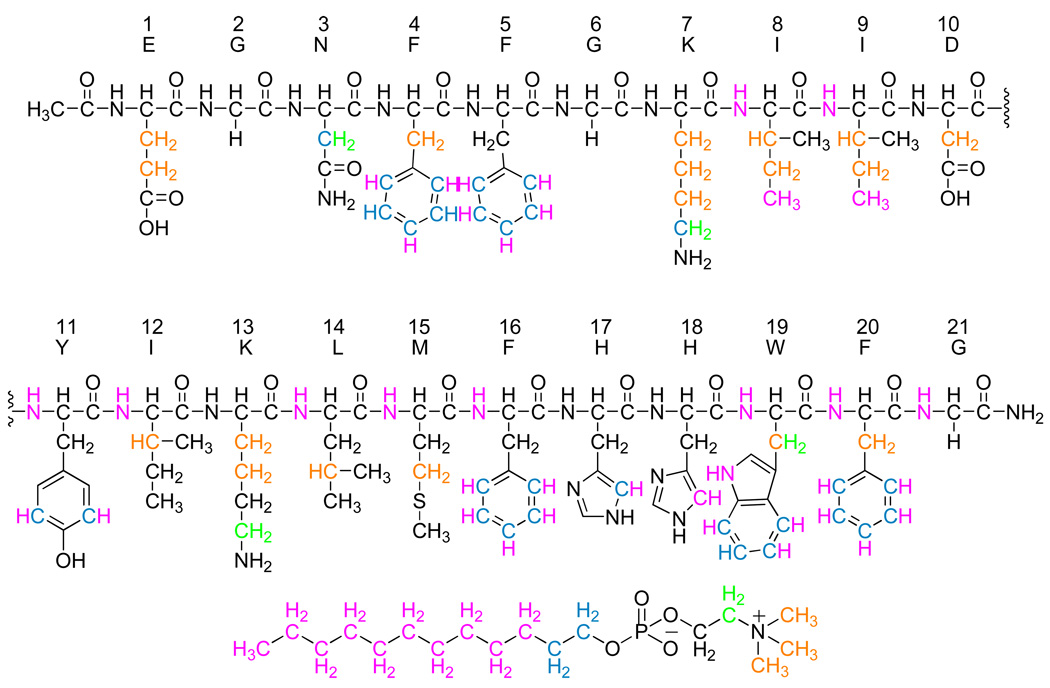
NOE interactions of PBP 5 anchor peptide protons to DPC micelle protons. The peptide atoms are colored according to the proton interactions with DPC. The atoms in black have no interaction with the micelle or the interactions overlap with the intra-peptide NOEs and can not be resolved. The peptide atoms where the carbon is one color and the hydrogen is another color indicates that those peptide protons are interacting with two colored regions of DPC and indicate with which regions the interactions occur. The structure of DPC is below the peptide sequence. The interacting regions are color coded according to resolved 1H NMR signals that interact with the peptide.
Cloning of the Full-length PBP 5
To clone the full-length PBP 5 (flPBP5), the dacA gene for PBP 5 was amplified by PCR from the chromosome of E. coli K12 using two custom-synthesized primers EcPBP5Nde: 5'-ATCATATGGATGACCTGAATATCAAAACTATG-3' and FullPBP5Hind: 5'-ATAAGCTTTTAACCAAACCAGTGATGGAACATTAATTTAATG-3' (NdeI and HindIII sites underlined) to remove its 29-residue-long N-terminal signal peptide. The resulting PCR products were digested respectively by the NdeI and HindIII, and ligated into the polylinker of pET24a(+) vector under the T7 promoter. The vectors were transformed into E. coli JM83. The selection of transformants was performed on LB agar supplemented with ampicillin (100 µg/mL). The nucleotide sequences of the dacA gene from several transformants were verified by sequencing of both DNA strands. The correct construct was transformed into E. coli BL21(DE3) cells for protein expression.
Purification of the Full-length PBP 5
Purification of the flPBP5 was done using a modification of a literature procedure.20 After cells were harvested by centrifugation, they were resuspended in 20 mM Tris pH 8.0 buffer with 0.75% Triton X-100, and were disrupted by sonication. Full-length PBP 5 was isolated from the supernatant by chromatography using an Ampicillin/CH-Sepharose 4B resin. The column fractions containing the flPBP5 were dialyzed at 4 °C against 25 mM NaHCO3, 0.15 M NaCl, 0.2% Triton X-100, pH 8.5 buffer. The protein solution was concentrated to approximately 6 mg/mL. The yield from a 500-mL cell culture was 10 to 15 mg. SDS-PAGE gave a purity of >95% for this flPBP5 enzyme. Concentrations of flPBP5 were measured spectrophotometrically using the BCA Protein Detection Kit (Pierce).
Coated Sensor Chips and SPR Measurements
Liposome was prepared from a 70:25:5 mixture of POPE/POPG/CL lipids, simulating the composition of an E. coli membrane,27 according to a literature procedure.28 The final concentration of total lipids was 0.5 mg/mL. All SPR experiments were performed at room temperature and at a flow rate of 50 µL/min in 10 mM HEPES pH 7.4 buffer containing 0.16 M KCl, using a Biacore 3000 (Biacore AB, Piscataway, NJ) instrument. The association was monitored for 100–120 s and dissociation for 4 min (flPBP5). The flPBP5 was not removed from the lipids using either high concentration salt or EDTA washing, and it also resisted extraction using 0.1 M NaOH. Thus, the immobilized liposome surface was regenerated for subsequent measurements using 20 µL of 8 M urea in 10 mM HEPES, pH 8.7 buffer. Data were acquired in two to four replicates at each concentration. The association rate constant ka and the dissociation rate constant kd were obtained from the experimental data using the BIAevaluation 3.0 software. The equilibrium dissociation constant (Kd) was calculated from the ratio of kd/ka.
Results
Structure of the Anchor Peptide Embedded in Micelles
The anchor peptide of full-length PBP 5 is 21 amino acids in length and is located at the C-terminus of the protein (Glu354 to Gly374). The polypeptide EGNFFGKIIDIKYLMFHHWFG (hereafter referred to as Glu1 to Gly21) corresponding to this terminus was synthesized, with an acetylated N-terminus, to >95% purity by standard solid-phase peptide synthesis. Circular dichroism (CD) spectra of this peptide were measured in dodecylphosphocholine (DPC) micelles at pH 5.5 (data not shown) and in the DPC micelles and in a 70:25:5 mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) and 1,1’,2,2’-tetraoleoyl cardiolipin (CL) as representative of the lipid composition of bacterial membranes, at pH 7.5 (Figure S2, Supporting Information) in order to compare the helicity of the peptide in the DPC micelles and POPE/POPG/CL lipid mixture. These spectra revealed, at a qualitative level, a helical peptide structure in these micelles at both pH values and that there is relatively no difference in peptide structure in the two different membrane mimics (Figure S2, Supporting Information). Since pH at 7.4 is more physiological, all subsequent NMR studies of the peptide in the DPC micelle were performed at this pH.
A brief discussion of the choice of the dodecylphosphocholine (DPC) for the NMR studies is warranted. Only two lipids, DPC and sodium dodecylsulfate (SDS), are commercially available in perdeuterated form. NMR analysis in SDS proved impossible as a result of the conductive properties of this lipid, a known limitation of SDS.29 Whereas DPC is a membrane mimetic, prior studies of bacterial proteins that interact with lipids validate it as a lipid surrogate for the bacterial membrane.30–32 Regardless, to confirm the suitability of DPC for the determination of the PBP 5 anchor peptide structure, membrane binding studies by surface plasmon resonance (SPR) were performed using the full-length PBP 5 (flPBP5). The synthetic anchor peptide was not suitable for these studies, as the instrument signal in response to the peptide was not sufficient. The experiments with the flPBP5 were performed in three different liposomes prepared from DPC, from a 70:25:5 mixture of POPE:POPG:CL, and from SDS. With all three of these liposomes, flPBP5 gave large rate constants for association (ka of 104 M−1s−1) and small rate constants for dissociation (kd ~10−3 s−1) and are comparable to what is found in literature.28,33 These rate constants (Table 1) translate to equilibrium dissociation constants in the nanomolar range, and demonstrate a high affinity of the anchor peptide for binding to these liposomes. Moreover, the rate constants are all similar (corresponding to energies within 1.1 kcal mol−1 of each other), indicating that anchoring occurs similarly in all three cases, irrespective of the nature of the lipid. These same experiments were repeated using a PBP 5 construct lacking its C-terminal anchor peptide in the POPE:POPG:CL mixture. Removal of the membrane anchor did not abolish the membrane interaction all together. The association constant (ka) of the anchor-free PBP 5 was 15 times smaller than that for flPBP5, and the equilibrium constant (Kd) of the anchor-free PBP 5 was 11 times smaller than that for flPBP5 (Table 1). The dissociation constants (kd) for anchor-free PBP 5 and flPBP5 were similar indicating that other parts of the protein, likely a hydrophobic patch at the membrane-proximal end of domain II, also interact with the lipids. Therefore, the anchor domain significantly influences the association rate constant. A reasonable conclusion from these determinations is that the nature of the lipid does not make a difference in interactions with the anchor and that the anchor itself comprises the major component of the membrane interaction. Edified with this knowledge, and the precedent for the use of DPC in bacterial systems,30–32 we conclude that determination of the structure of the peptide in DPC should be warranted.
Table 1.
Rate constants for association and for dissociation, and the equilibrium dissociation constants for the flPBP 5 (first three entries) and anchor-free PBP 5 (fourth entry) and liposome in 10 mM HEPES PH 7.4a
| ka (M−1s−1) | kd(s−1) | Kd (M) | |
|---|---|---|---|
| POPE:POPG:CL | (1.1 ± 0.1) × 104 | (1.0 ± 0.2) × 10−3 | (9.6 ± 1.8) × 10−8 |
| DPC | (2.9 ± 0.2) × 104 | (6.2 ± 0.7) × 10−4 | (2.1 ± 0.5) × 10−8 |
| SDS | (4.0 ± 0.5) × 104 | (5.7 ± 0.8) × 10−4 | (1.4 ± 0.3) × 10−8 |
| POPE:POPG:CL | (6.4 ± 0.9) × 102 | (7.2 ± 1.8) ×10−4 | (1.1 ± 0.3) ×10−6 |
The sensograms for the data in this table are found in Figure S3 of the Supplemental Information.
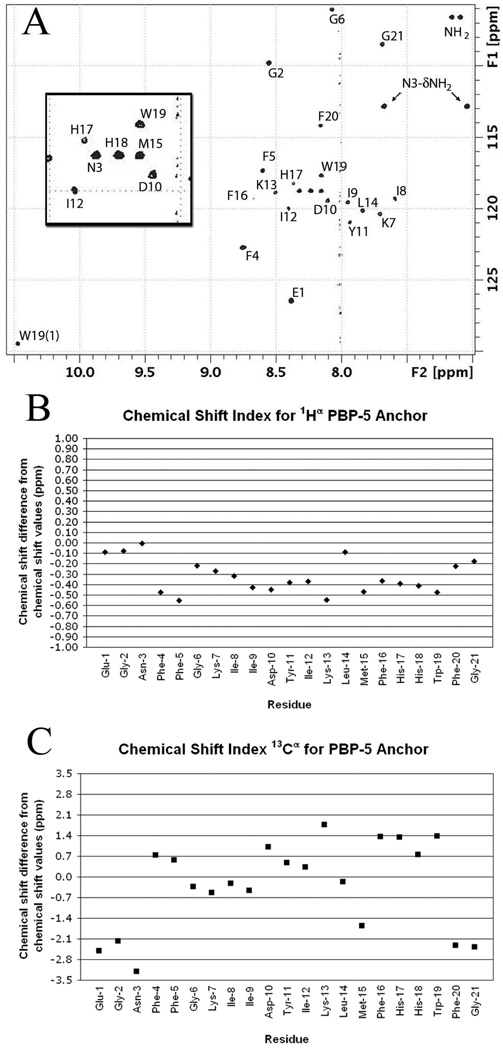
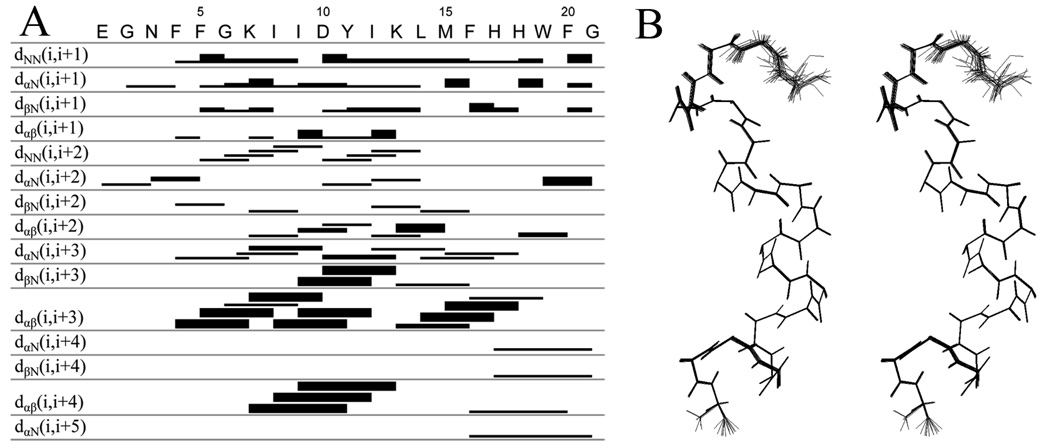
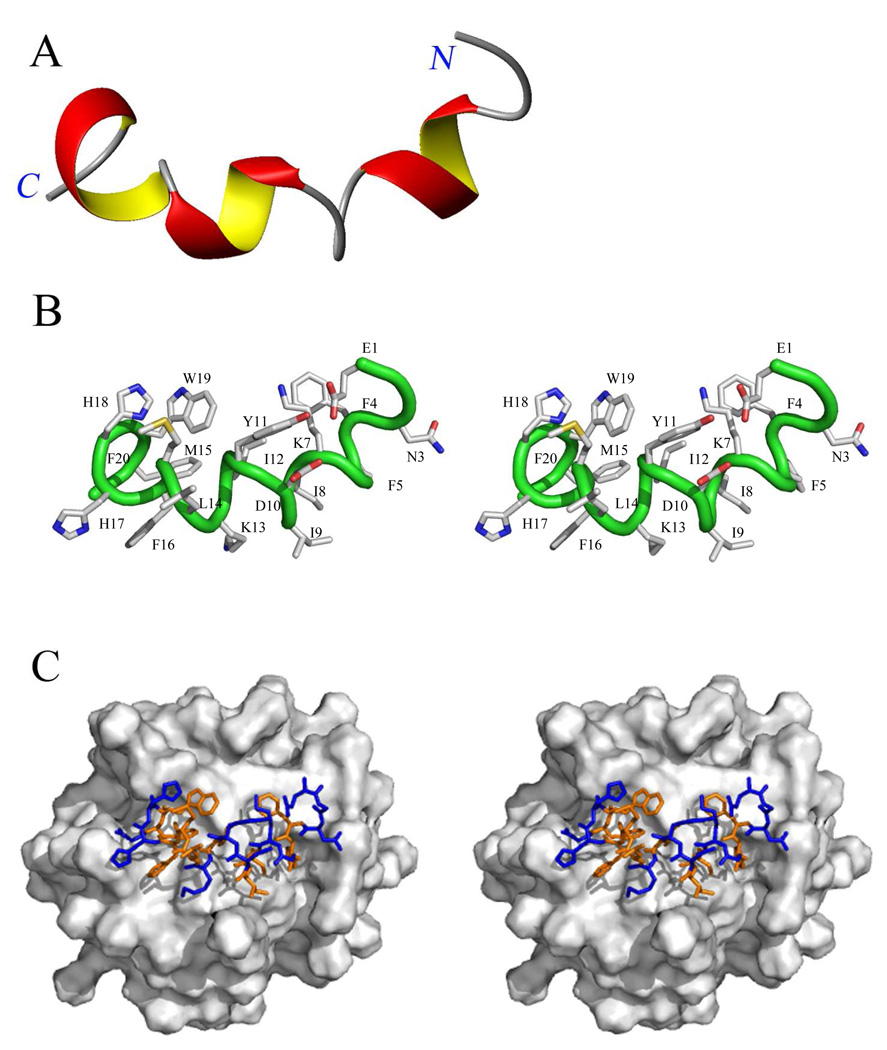
NMR spectra of the peptide were acquired in the presence of DPC-d38 micelles, at a relative stoichiometry of 55:1 lipid to peptide, in an aqueous 90:10 H2O/D2O BIS-TRIS/benzoic acid buffer pH 7.4 (see Supporting Information). The dispersion of the amide backbone 1H–15N chemical shifts in the 15N HSQC spectrum (Figure 1A) indicates a defined structure for the peptide in the micelle. The Hα and Cα chemical shift indices34 indicate a significant helical component for this structure spanning residues Phe4 to Trp19 (Figure 1B and 1C). The 1H, 13C and 15N chemical shifts for each residue (Table S2 and Table S3, Supporting Information) were assigned using DQF-COSY, TOCSY, NOESY, 13C HSQC, 13C HSQC–TOCSY and 15N HSQC spectra. The 1H NMR NOESY (mixing time 200 ms) spectrum provided many strong dαβ(i, i+3) and dαβ(i, i+4) cross-peaks. A total of 561 intra-residue NOEs and 120 interresidue NOEs were assigned. These NOEs (Figure 2A), together with dihedral angles obtained with the backbone 15N, Cα, Hα and Cβ chemical shift values from the program Talos, provided the constraints necessary for the determination of a structure using the Cyana program. The 20 lowest energy structures from this analysis are shown superimposed in Figure 2B. These structures confirm helical character within a helix-bend-helix-turn-helix motif (Figure 3A). The three α-helices in this motif are each single turns. The N-terminus is highly mobile until Phe4 of the peptide (Figure 2B and Figure 3A). The two sets of cross peaks for Glu1 and Gly2 observed in the TOCSY spectra indicate two conformational states for these residues. Residues Phe4–Lys7 form the first helix. A bend incurred between residues Ile8–Asp10 changes the direction of the backbone by approximately 33° relative to the first helix. The second helix stretch comprises residues Tyr11–Leu14. Subsequently, the backbone turns sharply at Met15, which reorients the backbone by approximately 70° relative to the second helical stretch. The third helix spans residues Phe16–Trp19, with the two C-terminus residues Phe20 and Gly21 bending back to cap the last helix. Thus, the backbone of the peptide forms an approximately 103° total inverted arch from residues Phe4–Trp19, conforming to an amphiphilic structure showing a 73:27 ratio of hydrophobic to hydrophilic surface area. The hydrophobic side chains of the peptide cluster mostly on one side and exhibit a poorly defined boundary between the hydrophobic and hydrophilic areas. Furthermore, the side chains of Lys13 and His17 protrude into the hydrophobic core of the peptide (Figure 3B).
Figure 1.
(A) The 1H-15N HSQC spectrum of DPC micelle-bound PBP 5 anchor for the 1H δ 10.5–7.0 ppm and 15N δ 105–130 regions. The cross-peaks are labeled by the amino acid residue. The inset is an expansion of the 1H δ 8.5–8.0 ppm and 15N δ 117–121 ppm region. (B) Chemical shift index34 for the α-protons of the PBP 5 anchor. Values between –0.10 and 0.10 are considered zero. Negative values indicate helical structure and positive values indicate beta structure. (C) Chemical shift index for 13C α-carbons of the PBP 5 anchor. Values between –0.7 and 0.7 are considered zero. Negative values indicate beta sheet structure and positive values indicate helical structure.
Figure 2.
NOE-connectivity table and stereo view of the peptide backbone. (A) Summary of NOE connectivities from NOESY spectrum (mixing time 200 ms) of PBP 5 anchor in DPC micelle (1:55) at pH 7.4 and 25.0 °C. The thickness of the band (strong, medium and weak) corresponds to the intensity of the NOE interaction between residues. The first four rows indicate interactions between adjacent amino acids. The remaining rows show interactions between the residues at each end of the bar. (B) Stereo view of the 20 overlaid backbone structures of the PBP 5 anchor from the Cyana calculation. The N-terminus of the peptide is on the top. The first three N-terminal residues display several conformations, while two distinct conformations are seen for the Gly21 C-terminus residue of the peptide.
Figure 3.
Structure of the E. coli PBP 5 anchor peptide. (A) The ribbon structure of the PBP 5 anchor displaying its secondary structure. The surface domain of the protein connects to the N-terminus (right side) of the peptide. The α-helical portions of the peptide are colored red and yellow and the non-α-helical portions are displayed as wires in gray. (B) Stereo view of the peptide (backbone as a wire in green) with side chains displayed as capped sticks (C in gray; O in red; N in blue; S in yellow). Hydrogens are removed and the residues are labeled for clarity. (C) Stereo view of the computational model of the peptide interaction with the DPC micelle (Connolly surface in gray) from Sybyl. The top view of the peptide on the surface of the micelle is depicted. The peptide is colored according to hydrophobic (orange) and hydrophilic (blue) residues, with the N-terminus to the right and C-terminus to the left (the same perspective as in panel A).
Dynamics of the Anchor Peptide Ingrained in Micelles
To assess the peptide backbone dynamics within the micelle, we measured the 15N spin-lattice relaxation time (T1) and spin-spin relaxation time (T2) as well as the steady-state [1H–15N] NOE values of the backbone nitrogen atoms. These data revealed greater R1 (= 1/T1) values for the Gly1–Ile8 and His17–Gly21 residues relative to the intervening residues. On the other hand, the R2 (= 1/T2) values were smaller for Gly1–Phe4 and Phe20– Gly21 (Figure S3, Supporting Information), compared to the rest of the residues. These observations indicate greater backbone motion in the terminal portions of the peptide. These results complement the smaller number of 1H NOE interactions within these two regions relative to the rest of the peptide (Figure 2A). The significant motion at the N-terminus of the anchor seen for this peptide may not occur in the full-length protein, as this terminus tethers the surface domains of PBP 5 to the membrane anchor peptide.
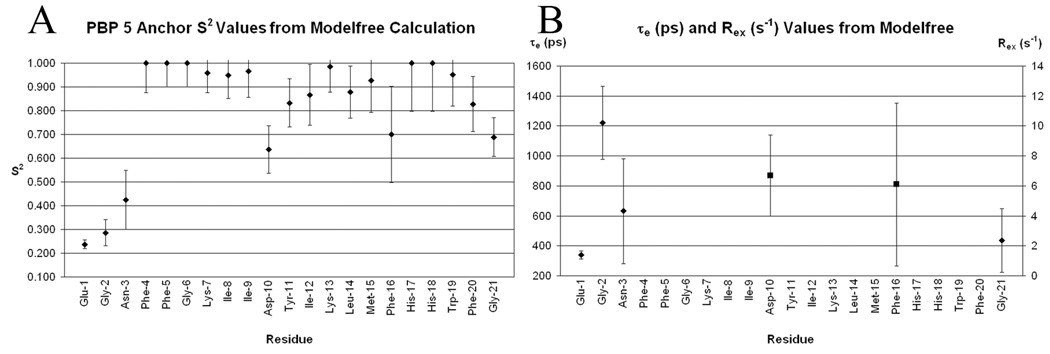
In order to provide deeper insight on the peptide backbone dynamics of the peptide in the DPC micelles, the measured relaxation parameters were analyzed in terms of extended model-free parameters using the program Modelfree.25,26 The program fits the experimental R1, R2, and NOE values to five dynamic models that differ in the number of motional parameters.25,35 The variable parameters for the five models are: model 1: the square of the generalized order parameter S2; model 2: S2 and the correlation time τe characterizing fast internal motions; model 3: S2 and the chemical exchange term Rex characterizing conformational exchange processes occurring on the µs-ms time scale; model 4: S2, Rex, and the correlation time τe for fast internal motions; and model 5: S2, the square of the order parameter for internal motions on the fast time scale Sf2, and the correlation time τe characterizing slow internal motions. The values of S2 and Sf2 span from zero, corresponding to isotropic internal motion, to one, corresponding to entirely restricted internal motion. The peptide experimental relaxation data were analyzed assuming anisotropic axially symmetric overall reorientation of the peptide micelle complex. The initial values of the correlation time characterizing overall reorientation τm = 6.54 ns and the ratio of the corresponding diffusion tensor components 2Dzz/(Dxx + Dyy) = 1.63 were determined from the Stokes-Einstein formula for the viscosity of water calculated for an anisotropic micelle and from published data for DPC micelles,36 respectively. To establish a proper motional model for each residue, a grid search for the optimal values of these parameters was first carried out with model 1 selected for all residues for which the relaxation parameters were measured, followed by 300 Monte Carlo simulations using the Modelfree program. The optimized values (τm = 8.60 ns and 2Dzz/(Dxx + Dyy) = 1.44) were used in the model selection procedure, which furnished models for 17 residues. No suitable model could be obtained for four residues (Phe4, Gly6, His17 and His18). Dynamics of the majority of residues (Phe5, Lys7–Ile9, Tyr11–Met15, Trp19 and Phe20) could be characterized by model 1, while residues Glu1–Asn3 and Gly21 required model 2. The dynamics of residues 10 and 16 were best characterized by model 3. The S2 values (Figure 4A) for N-terminal residues Glu1–Asn3 and Gly21 indicate that the amplitudes of the backbone motions for these residues are large and the correlation times of the motions are in the range of 300 to 1200 ps (Figure 4B). Also, residues Asp10 and Phe16 exhibit large amplitude fast motions (Figure 4A). However, they also undergo conformational exchange with the Rex values of 6.68 and 6.11 Hz, respectively (Figure 4B). Interestingly, both residues are found where the peptide backbone substantially changes its direction. The S2 values for all of the remaining residues are >0.8, indicating that the amplitudes of fast internal backbone motions are small and therefore the interior of the peptide backbone is well-ordered.
Figure 4.
Plots of PBP 5 residue backbone N-H dynamic values. Error bars indicate standard deviations. (A) Plot of S2 from the Modelfree calculation for each residue. The lower the S2 value the greater amplitude of motion for the N–H vector. (B) Plot of effective correlation time (τe, ♦) and conformational exchange (Rex, ■) from the Modelfree calculation for each residue that is described by the parameter. Data are only presented if the motional model for the residues is contained in these terms. The parameter τe describes fast motions on the ps time scale and Rex describes conformational exchange on the ms time scale.
The incorporation of the peptide within the DPC micelle environment was also addressed by evaluating the kinetics of solvent deuterium-exchange of the peptide amide hydrogens. This experiment gave t½ values for NH proton exchange. The t½ values for fourteen of the backbone amide hydrogens were faster than the first 15 minutes after the addition of D2O to the PBP 5 peptide. The seven exceptions were the HN resonances of Ile9 (t½ = 38 min), Asp10 (t½ = 15 min), Lys13 (t½ = 204 min), Leu14 (t½ = 15 min), Met15 (t½ = 15 min), Phe16 (t½ = 204 min) and Ile12 (t½ = 63.5 h). Solvent exposed residues generally exchange with rate orders of 103 s−1.37 The exchange rate is often as much as 105 times slower when the proton is hydrogen bonded, while it is about 102 times slower if it is blocked.38 Examples of slow exchange have also been attributed to immersion in an environment where the hydrogen is protected from the solvent.39–41 In the Cyana structure, hydrogen bonding in the PBP 5 anchor peptide can account for the slow exchange of residues Ile9, Lys13, Met15 and Phe16. It may not account, however, for the slow exchange of residues Asp10, Leu14 and Ile12 (Table S4). The slow exchange in these residues may indicate internalization of their amide hydrogens in the hydrophobic medium of the micelle, and thus the poor contact with water. Although we could not confirm the result for Asp10, the results are confirmed for Leu14 and Ile12 by using the peptide to micelle NOE interactions (Figure 5), which indicate that the amide hydrogens are interacting with the hydrophobic portion of the micelle.
To further investigate the interactions of the peptide to the micelle, we measured 2D 1H NOESY spectra using non-deuterated DPC micelles. The data indicate that the peptide side chains interact mostly with the hydrophilic head group and the upper hydrophobic portions of the lipids (Figure 5, Table S5 and Figure S1, Supporting Information). None of the side chains of the peptide show an interaction with just the polymethylenes of DPC.
Discussion
E. coli expresses twelve PBPs, each of varied molecular mass and each with different membrane affinities. Its high-molecular-mass PBPs use an N-terminal transmembrane anchor for tight membrane adherence,7 while its low-molecular-mass carboxypeptidase-type PBPs (including PBP 4, PBP 5, PBP 6 and PBP 6b) use an amphipathic C-terminus for the purpose of moderate to strong membrane adherence.6,9 Each of these four carboxypeptidase-type PBPs has very similar nucleic acid coding sequences, and in vitro each has either carboxypeptidase or endopeptidase activity with respect to the d-Ala-d-Ala terminus of the peptide stem of the bacterial peptidoglycan. Although there are extensive (and powerful) circumstantial arguments that each low-molecular-mass PBP has a specific physiological role, the precise identification of these role(s) remains an unsolved problem.9 For example, while the low-molecular-mass PBPs are not critical for growth of planktonic E. coli in the laboratory,42 the evolutionary conservation of their genes implies that specific roles do exist, and that these roles are important. Of the low-molecular-mass PBPs, PBP 5 is the most abundant, is the best studied structurally and mechanistically, and it is the only low-molecular-mass PBP correlated to a specific phenotypic response. PBP 5 has carboxypeptidase activity in vitro20,43 and in vivo,44 and this catalytic activity contributes in the maintenance of the rod morphology of E. coli.45,46 The molecular basis for this accomplishment is not known. The PBP 5 protein structure shows three domains: a catalytic domain at the N-terminal (Domain I, strongly conserved among all PBPs), a central β-sheet-rich domain (Domain II) having much lower sequence homology, and a carboxy-terminus domain conferring membrane association. The observation that replacement of Domain II of PBP 5 by Domain II of PBP 6 preserves the ability of PBP 5 to maintain bacterial shape,47 strongly argues that the role of Domain II is primarily structural (that of elevating the catalytic domain into the periplasmic space, and into contact with the cell wall). In contrast to the probable structural role of Domain II, the essentiality of the C-terminus for membrane association and a direct correlation of PBP 5 membrane-association in order to accomplish its role in the maintenance of the E. coli shape, are well proven.17,47,48 Without its C-terminal membrane anchor, PBP 5 loses its ability to contribute to the preservation of cell shape, and indeed is lethal to the cell at even modest levels of expression.46 Hence, the role of the C-terminus is not merely that of enabling membrane association, but also includes control of the catalytic activity of Domain I. How may both of these roles be interpreted in terms of the C-terminus structure?
An explanation for the first of these roles—that of membrane association—has been discussed extensively, from the perspective that helical wheel representations of this terminus (and as well, those of others with PBP C-termini anchors) correlate well to a contiguous amphipathic helix.18,46,47,49–54 The assumption of an amphipathic helix at the C-terminus is further supported by CD and IR spectroscopies.54,55 This NMR study of the PBP 5 C-terminus confirms the presence of amphipathic structure, but decisively excludes the contiguous single amphipathic helix, previously assumed for this terminus, within the membrane-mimetic environment of the DPC micelle used for the NMR study. Rather, we find that the PBP 5 anchor peptide forms a helix-bend-helix-turn-helix motif, with a mobile and solvent-exposed N-terminus consistent with the attachment of surface Domain II of the full-length protein at this site. The anchor peptide forms a hydrophobic surface to interact with the hydrophobic portion of the membrane, notwithstanding the presence of two hydrophilic residues, Lys13 and His17. The amide of Lys13 is oriented parallel to the micelle surface and is into the hydrophobic region of the micelle. The lysine side chain is oriented 90° to the side of the peptide but also parallels the micelle surface. The peptide-micelle NOE connectivities confirm an interaction of the lysine side chain near the phosphate of the lipid head group, which likely stabilizes the immersion of this residue in the hydrophobic region of the micelle. Although the energetic cost of insertion of a Lys or a His into a non-polar membrane at neutral pH is well known,56 the insertion of the hydrophilic side chains into the hydrophobic region of the membrane is not uncommon7,57,58 and has been proposed to prevent excessive adherence of the protein to the hydrophobic surface. Prior studies indicated strong adherence of PBP 5 to the membrane,46,59 and our results (Table 1) confirm this finding. The anchoring ability is not affected by 4 M NaCl solution, only small quantities of the protein (~18%) are removed from the membrane at pH >9 or in combination at pH >9 with 4 M urea (~35%),46 and only a small portion (~18%) of the protein is released by extraction with a solution of a chaotropic molecule, 2 M sodium thiocyanate, at pH 8.0.59 Phoenix et al. concluded from these results that hydrophobic interactions are a major component involved in anchoring59 with additional but minor electrostatic components involving the cationic region of the anchor.52,60
The short helical sequence …[GN]FFGK… that follows the mobile EGN N-terminus has the potential of special significance. Within the C-terminal anchor peptides of PBP 5 homologs (including PBP 6 and DacD of E. coli) are several strongly conserved amino acids.47 These amino acids include Gly2 (using the numbering of the excised helix as shown in Figure 2A, and corresponding to G385 of full-length PBP 5, including the N-terminal signal portion), Phe5, Lys7 (or Arg7 of the other homologs), followed by a hydrophobic amino acid (I, V, L or A) at position 8, and Asp10. The remaining sequences of PBP 5 homolog C-termini following Asp10 are highly variable. The …FFGK… sequence of the first short helix includes Phe5 and Lys7 and the hydrophobic Ile at position 8 and Asp10 are involved in the bend of the peptide. While Ile8 and Asp10 are both conserved residues, at this moment we cannot ascertain the basis for this sequence conservation. Both PBP 5 and PBP 6 show a GXXXG motif at their N--termini (…EGNFFGK… for PBP 5, …EGGFFGR… for PBP 6) of their anchor peptides. Although the GXXXG motif is used for pairwise recognition of transmembrane α-helices,61 there is strong circumstantial evidence that such recognition does not occur with full-length PBP 5. Addition of the PBP 5 anchor peptide to the C-terminus of the otherwise soluble and periplasmic β-lactamase, results in retention of this enzyme to the membrane and gives a fully viable (and β-lactam-resistant) phenotype.62,63 If any sequence (or structural motif) within the PBP 5 anchor peptide was intended to participate in essential protein-protein (such as helix-helix) recognition, a different outcome for membrane-bound β-lactamase phenotype is expected. Likewise, the retention of shape-contributing catalytic activity upon point mutations of the aspartate and lysine in the conserved …FX[K/R]XXD… motif, and for some (but not all) phenylalanine point mutants in this same motif,47 strongly suggest that it is the anchor structure as a whole (as distinct from its sequence), that is important to the membrane adherence it imposes upon PBP 5.
The substantial sequence variability at the C-terminus among the low-molecular-mass PBPs (corresponding to the last ten amino acids of the PBP 5 anchor peptide) cannot be assessed. If the present assumption that each of the low-molecular-mass PBPs has specific roles with respect to the cell wall is correct—there are powerful arguments that this is the case—then the sequence variability might relate to these different roles.9 For the specific example of the PBP 5 anchor peptide, two critical questions are presented. First, taken as a whole, what is the orientation of the peptide within the membrane? Second, how may this orientation be understood to enable control (or communication with) the spatially distant and catalytic Domain I? Our NMR study presents an answer to the first question, and is the basis for the future experiments to address the second.
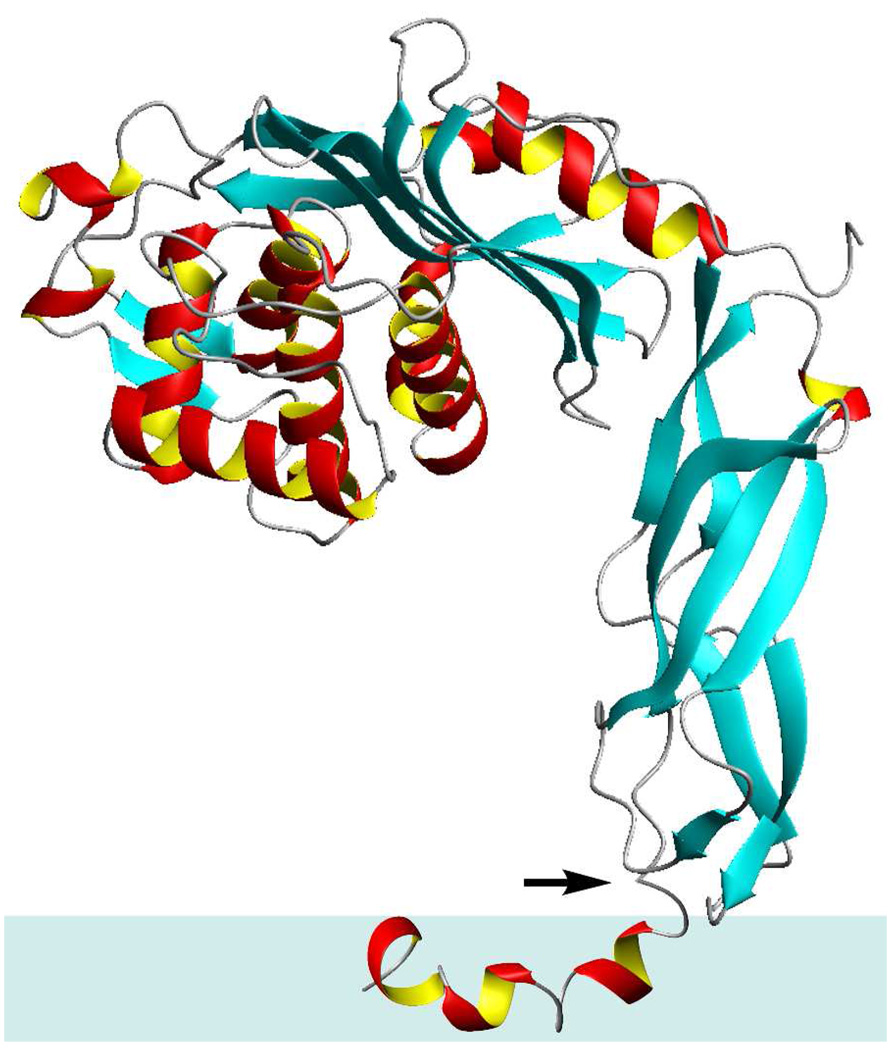
An amphipathic peptide may insert perpendicular to, or parallel to, the surface of the membrane. In the former case, the hydrophilic amino acids Lys7, Asp10, Lys13, His17 and His18 of the PBP 5 anchor peptide would be membrane-buried, in an electrostatically unfavored environment, in the absence of aggregation of the peptide so as to mask the peptide hydrophilic surfaces within the membrane (Figure 3B). Our NOE measurements between the peptide and the micelle entirely exclude the self-aggregation possibility. In such a case, NOE correlations between the peptide and the hydrogens internal to the DPC micelle would be found solely on several residue side chains and of the peptide. No such correlations are seen (Figure 5, Table S5 and Figure S1, Supporting Information). The alternative structure for the anchor peptide is parallel insertion into the membrane, allowing most of the hydrophobic surface to interact with the interior of the lipid (Figure 3C). This second model is consistent with our other NMR experiments. The kinetics of amide NH exchange with D2O sharply stratify the individual amides. Those amide bonds with outright exposure to the aqueous milieu exchanged immediately, while the amide bonds that are sheltered by the micelle exchanged much more slowly. The exchange occurred according to the orientation of the backbone relative to the surface of the micelle. For example, if the peptide were to penetrate vertically into the hydrophobic region of the micelle, the exchange would have occurred from one end of the peptide towards the center, with the hydrogen-bonded and buried residues experiencing protection from exchange. This is simply not seen. The exchange time decreases and increases in an alternating manner as the backbone structure rotates towards the surface and then away, indicating the proximity of the backbone residues to the surface of the micelle. Collectively, our results indicate that the anchor peptide—as a monomer—lies horizontally on top of the membrane, with the hydrophobic core buried and the two ends exposed to the aqueous milieu due to the central bend in the structure. Hence, the anchor peptide behaves as a “raft” (Figure 6) that enables the anchor (and thus the full-length PBP 5 as well) to sequester in (or perhaps even to glide along) the membrane surface, as has been proposed for another PBP.6,64 The amphiphilic membrane interaction by the PBP 5 C-terminal anchor is in contrast to the recent crystal structure of E. coli PBP 1b in which the uncleaved N-terminal signal peptide spans the cytoplasmic membrane and acts as the anchor for the protein.7
Figure 6.
Orientation of the anchor peptide in the membrane and its relationship to the surface domains of PBP 5. The arrow indicates the connection point between the anchor and the surface domains. The relative disposition of the surface and the membrane domains is likely, although it cannot be surmised from the work presented in this report.
There are several differences between these anchors. The secondary structure of the PBP 1b anchor is relatively linear, while the PBP 5 anchor structure is sharply curved over most of is backbone. In addition, the PBP 1b anchor is comprised of 94% hydrophobic residues compared to 71% for the PBP 5 anchor. The greater hydrophobic surface area for PBP 1b also increases the interaction for the hydrophobic region of the membrane compared to the PBP 5 anchor. The PBP 1b anchor contains only one hydrophilic residue, Lys71, internalized in the hydrophobic region of the membrane, while PBP 5 has two such residues, Lys13 and His17, inserted into the hydrophobic region. However, in the structures of both anchors, the side chains of the lysine residues lie parallel with the surface of the membrane.
This conclusion for the PBP 5 anchor defines the functional roles of the amphipathic helix-bend-helix-turn-helix C-terminus of PBP 5 as primarily membrane adherence. This role is consistent with our current (although limited) understanding of the functional role of the enzyme itself, which is believed to be the control of the population of acyl-D-Ala-D-Ala termini of the peptide stems of the nascent peptidoglycan, and thus control of the global extent of cross-linking within the peptidoglycan.65,66 There remains, however, a significant mechanistic dilemma. While defining the functional roles of the amphipathic C-terminus of PBP 5 in terms of membrane insertion and membrane mobility is attractive for the simplicity of the definition, this definition omits a probable third functional role for this terminus. While the catalytic mechanism of PBP 5 is fully consistent with an involvement in active peptidoglycan biosynthesis, we do not know where PBP 5 localizes within the bacterium, nor precisely how PBP 5 contributes to cell wall elongation and division. Given that control of all PBP localization within the bacterium is a highly probable biological imperative, is it possible to conceptualize a role for the amphipathic C-terminus of PBP 5 in this critical aspect? We are now well aware that profound structural heterogeneity occurs within bacterial membranes,67 and that the properties of these membranes—such as curvature68,69 and lipid composition70–74—directly affect membrane protein localization. The PBP 5 anchor peptide is responsive to its lipid environment,52 and to the catalytic integrity of its companion PBPs.5 Its replacement by a transmembrane α-helix results in loss of the ability of PBP 5 to contribute to control of the rod shape of the E. coli bacterium.47 Hence, the role of the anchor is not merely sequestration to the membrane surface. Our determination of the unprecedented helix-bend-helix-turn-helix amphipathic structure of the PBP 5 C-terminus is the necessary first step for further experimental design to validate the expectation of a direct relationship between the C-terminal structures of the PBPs, and lipid microenvironment positioning of the PBPs for catalysis.
Our elucidation of the structure of the E. coli PBP 5 anchor is the first of the PBP C-terminus anchors, and to our knowledge of any such anchor domain for a peripheral membrane protein. Although each PBP anchor might represent a separate structural inquiry, consistent with each PBP having separate roles in the fabrication of the cell wall, this study gives the first structure-based insight into the membrane interaction and backbone dynamics of a PBP anchor. With the increased recognition of the multi-enzyme assemblies involved in bacterial growth and division, the structure of the PBP 5 C-terminal anchor peptide is the basis for further exploration of the structural basis for communication between the bacterial membrane environment and the control of PBP catalysis.
Supplementary Material
Acknowledgements
We thank F. J. Castellino, M. Prorok, Z. Sheng and M. Figuera-Losada for the use of the CD instrument. We acknowledge P. Güntert for help with Cyana. We thank A. Palmer for corrections to the Modelfree programs. This work was supported by a grant from the National Institutes of Health.
Footnotes
Supporting Information Available: Tables of NMR and refinement statistics, 1H NMR chemical shifts, 15N and 13C NMR chemical shifts, hydrogen bond distances and t½ Exchange times and NOE interactions between the PBP 5 anchor peptide and DPC micelle protons. Figures of the DPC structure with protons color coded according to 1H NMR NOE interactions with the peptide, CD spectrum of PBP 5 in different lipids, 15N plots of R1 and R2, SPR plots, and Supporting Materials and Methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wallin E, Von Heijne G. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiner JH, Li L. Biochim. Biophys. Acta. 2008;1778:1698–1713. doi: 10.1016/j.bbamem.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Bernsel A, Daley DO. Trends Microbiol. 2009;17:444–449. doi: 10.1016/j.tim.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Norris V, den Blaauwen T, Doi RH, Harshey RM, Janniere L, Jiménez-Sánchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Misevic G, Ripoll CMS, Jr, Skarstad K, Thellier M. Annu. Rev.Microbiol. 2007;61:309–329. doi: 10.1146/annurev.micro.61.081606.103348. [DOI] [PubMed] [Google Scholar]
- 5.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 6.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. FEMS Microbiol. Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 7.Sung M-T, Lai Y-T, Huang C-Y, Chou L-Y, Shih H-W, Cheng W-C, Wong C-H, Ma C. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishida H, Unzai S, Roper DI, Lloyd A, Park S-Y, Tame JRH. Biochemistry. 2006;45:783–792. doi: 10.1021/bi051533t. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh AS, Chowdhury C, Nelson DE. Trends Microbiol. 2008;16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty TJ, Kennedy K, Kessler RE, Pucci MJ. J. Bacteriol. 1996;178:6110–6115. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh AS, Young KD. J.Bacteriol. 2003;185:2178–2186. doi: 10.1128/JB.185.7.2178-2186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar SK, Chowdhury C, Ghosh AS. Inter. J. Antimicrob. Agents. 2010;35:244–249. doi: 10.1016/j.ijantimicag.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Phillips R, Ursell T, Wiggins P, Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro L, McAdams HH, Losick R. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drin G, Antonny B. FEBS Lett. 2010 doi: 10.1016/j.febslet.2009.10.022. published on the Web. [DOI] [PubMed] [Google Scholar]
- 16.Phoenix DA. Biochem. Soc. Trans. 1993;21:225S. doi: 10.1042/bst021225s. [DOI] [PubMed] [Google Scholar]
- 17.Jackson ME, Pratt JM. Mol. Microbiol. 1987;1:23–28. doi: 10.1111/j.1365-2958.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 18.Phoenix DA. Biochem. Soc. Trans. 1990;18:948–949. doi: 10.1042/bst0180948a. [DOI] [PubMed] [Google Scholar]
- 19.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Shi Q, Meroueh SO, Valkulenko SB, Mobashery S. Biochemistry. 2007;46:10113–10121. doi: 10.1021/bi700777x. [DOI] [PubMed] [Google Scholar]
- 21.Kay LE, Torchia DA, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 22.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu G, Delaglio F, Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Guntert P, Mumenthaler C, Wuthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 25.Mandel AM, Akke M, Palmer AG., III J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 26.Palmer AG, III, Rance M, Wright PE. J. Am. Chem. Soc. 1991;113:4371–4380. [Google Scholar]
- 27.Dowhan W. Annu. Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 28.Stahelin RV, Cho W. Biochemistry. 2001;40:4672–4678. doi: 10.1021/bi0020325. [DOI] [PubMed] [Google Scholar]
- 29.Kelly AE, Ou HD, Withers R, Dotsch V. J. Am. Chem. Soc. 2002;124:12013–12019. doi: 10.1021/ja026121b. [DOI] [PubMed] [Google Scholar]
- 30.Evanics F, Hwang PM, Cheng Y, Kay LE, Prosser RS. J. Am. Chem.Soc. 2006;128:8256–8264. doi: 10.1021/ja0610075. [DOI] [PubMed] [Google Scholar]
- 31.Liang B, Tamm LK. Proc. Natl. Acad. Sci.U.S.A. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourbigot S, Dodd E, Horwood C, Cumby N, Fardy L, Welch WH, Ramjan Z, Sharma S, Waring AJ, Yeaman MR, Booth V. Biopolymers. 2009;91:1–13. doi: 10.1002/bip.21071. [DOI] [PubMed] [Google Scholar]
- 33.Blatner NR, Stahelin RV, Diraviyam K, Hawkins PT, Hong W, Murray D, Cho W. J. Biol.Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]
- 34.Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 35.Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM. Biochemistry. 1990;29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- 36.Lipfert CJ, Columbus L, Chu VB, Lesley SA, Doniach S. J. Phys.Chem. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 37.Fernfindez I, Ubach J, Andreu D, Pons M. Colloid Surface A. 1996;115:39–45. [Google Scholar]
- 38.Englander SW, Kallenbach NR. Q. Rev.Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 39.Zalkin A, Forrester JD, Templeton DH. J. Am. Chem.Soc. 1966;88:1810–1814. doi: 10.1021/ja00960a040. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan RP, Levy RM, Englander SW. Proc. Natl. Acad. Sci.U.S.A. 1983;80:5569–5572. doi: 10.1073/pnas.80.18.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mason SA, Bentley GA, McIntyre GJ. Deuterium Exchange in Lysozyme at 1.4Å Resolution. In: Schoenborn BP, editor. Neutrons in Biology: Neutron Scattering Analysis for Biological Structures. Vol. 27. New York: Plenum; 1983. pp. 323–334. [Google Scholar]
- 42.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. J. Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stefanova ME, Davies C, Nicholas RA, Gutheil WG. Biochim. Biophys.Acta. 2002;1597:292–300. doi: 10.1016/s0167-4838(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 44.Priyadarshini R, Popham DL, Young KD. J. Bacteriol. 2006;188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson DE, Young KD. J. Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson DE, Young KD. J. Bacteriol. 2001;183:3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson DE, Ghosh AS, Paulson AL, Young KD. J. Bacteriol. 2002;184:3630–3639. doi: 10.1128/JB.184.13.3630-3639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratt JM, Jackson ME, Holland IB. EMBO J. 1986;5:2399–2405. doi: 10.1002/j.1460-2075.1986.tb04510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris F, Chatfield L, Phoenix DA. Biochem. Soc.Trans. 1995;23:32S. doi: 10.1042/bst023032s. [DOI] [PubMed] [Google Scholar]
- 50.Phoenix DA. Biochem. Soc.Trans. 1995;23:31S. doi: 10.1042/bst023031s. [DOI] [PubMed] [Google Scholar]
- 51.Harris F, Phoenix DA. Biochimie. 1997;79:171–174. doi: 10.1016/s0300-9084(97)83502-x. [DOI] [PubMed] [Google Scholar]
- 52.Harris F, Demel R, de Kruijff B, Phoenix DA. Biochim. Biophys. Acta., Biomembranes. 1998;1415:10–22. doi: 10.1016/s0005-2736(98)00173-4. [DOI] [PubMed] [Google Scholar]
- 53.Phoenix DA, Harris F. Biologicheskie Membrany. 2001;18:38–41. [Google Scholar]
- 54.Brandenburg K, Harris F, Phoenix DA, Seydel U. Biochem. Biophys. Res.Commun. 2002;290:427–430. doi: 10.1006/bbrc.2001.6198. [DOI] [PubMed] [Google Scholar]
- 55.Siligardi G, Harris F, Phoenix DA. Biochim. Biophys. Acta., Biomembranes. 1997;1329:278–284. doi: 10.1016/s0005-2736(97)00117-x. [DOI] [PubMed] [Google Scholar]
- 56.Bond PJ, Wee CL, Sansom MSP. Biochemistry. 2008;47:11321–11331. doi: 10.1021/bi800642m. [DOI] [PubMed] [Google Scholar]
- 57.Fukushima D, Yokoyama S, Kezdy FJ, Kaiser ET. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2732–2736. doi: 10.1073/pnas.78.5.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ladokhin AS, White SH. Biochemistry. 2004;43:5782–5791. doi: 10.1021/bi0361259. [DOI] [PubMed] [Google Scholar]
- 59.Phoenix D, Pratt JM. Eur. J. Biochem. 1990:365–369. doi: 10.1111/j.1432-1033.1990.tb15584.x. [DOI] [PubMed] [Google Scholar]
- 60.Harris F, Chatfield LK, Phoenix DA. FEMS Microbiol. Lett. 1995;129:215–220. doi: 10.1111/j.1574-6968.1995.tb07582.x. [DOI] [PubMed] [Google Scholar]
- 61.Russ WP, Engelman DM. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 62.Phoenix DA, Pratt JM. FEBS Lett. 1993;322:215–218. doi: 10.1016/0014-5793(93)81572-h. [DOI] [PubMed] [Google Scholar]
- 63.Suvorov M, Vakulenko SB, Mobashery S. Antimicrob. Agents Chemother. 2007;51:2937–2942. doi: 10.1128/AAC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morlot C, Pernot L, Le Gouellec A, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. J. Biol. Chem. 2005;280:15984–15991. doi: 10.1074/jbc.M408446200. [DOI] [PubMed] [Google Scholar]
- 65.Varma A, de Pedro MA, Young KD. J. Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kraus W, Holtje J-V. J. Bacteriol. 1987;169:3099–3103. doi: 10.1128/jb.169.7.3099-3103.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto K, Kusaka J, Nishibori A, Hara H. Mol. Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 68.Mukhopadhyay R, Huang KC, Wingreen NS. Biophys. J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. J. Bacteriol. 2005;187:2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mileykovskaya E. Mol. Microbiol. 2007;64:1419–1422. doi: 10.1111/j.1365-2958.2007.05766.x. [DOI] [PubMed] [Google Scholar]
- 72.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. Mol. Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 73.Romantsov T, Stalker L, Culham DE, Wood JM. J. Biol. Chem. 2008;283:12314–12323. doi: 10.1074/jbc.M709871200. [DOI] [PubMed] [Google Scholar]
- 74.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. J. Biol. Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.