Abstract
Most heritable traits, including many human diseases 1, are caused by multiple loci. Studies in both humans and model organisms, such as yeast, have failed to detect a large fraction of the loci that underlie such complex traits 2,3. A lack of statistical power to identify multiple loci with small effects is undoubtedly one of the primary reasons for this problem. We have developed a method in yeast that allows the use of dramatically larger sample sizes than previously possible and hence permits the detection of multiple loci with small effects. The method involves generating very large numbers of progeny from a cross between two strains and then phenotyping and genotyping pools of these offspring. We applied the method to 17 chemical resistance traits and mitochondrial function, and identified loci for each of these phenotypes. We show that the range of genetic complexity underlying these quantitative traits is highly variable, with some traits influenced by one major locus and others due to at least 20 loci. Our results provide an empirical demonstration of the genetic complexity of many traits and show that it is possible to identify many of the underlying factors using straightforward techniques. Our method should have broad applications in yeast and can be extended to other organisms.
Genome-wide association studies (GWAS) have recently detected many trait loci in humans 4. Despite the large number of loci that have been identified by GWAS, case studies, such as human height5, have shown that we remain unable to explain the genetic basis of complex traits in our population 2. Controlled crosses in model organisms can shed light on this problem by elucidating basic principles that govern the genetic basis of trait variation. However, akin to the problem in humans, mapping studies in model organisms typically detect only a fraction of the loci underlying heritable traits, implying that they lack statistical power 3.
Very large mapping populations are needed to comprehensively dissect the genetic basis of highly complex traits. In many cases, genotyping and phenotyping on a sufficient scale will not be feasible without the use of methods that examine pools of individuals. One such method, bulk segregant analysis (BSA), was first proposed nearly twenty years ago as an expeditious approach for mapping quantitative trait loci (QTLs) 6, and its modern implementations are commonly used to map major effect QTLs and Mendelian loci 7–11. However, BSA has yet to be effectively used to dissect a highly complex trait, even though simulations suggest it should be capable of detecting numerous small-effect loci with high resolution when > 105 cross progeny are used (Figures S1–S2). We have developed a powerful extension of BSA that can be used to comprehensively map complex traits in yeast. Extreme QTL mapping (X-QTL) has three key steps. The first is the generation of segregating populations of very large size. The second is selection-based phenotyping of these populations to recover large numbers of progeny with extreme trait values. This can be accomplished, for example, by selection for drug resistance or by cell-sorting. The final step is quantitative measurement of pooled allele frequencies across the genome, by either microarray-based genotyping or massively parallel sequencing.
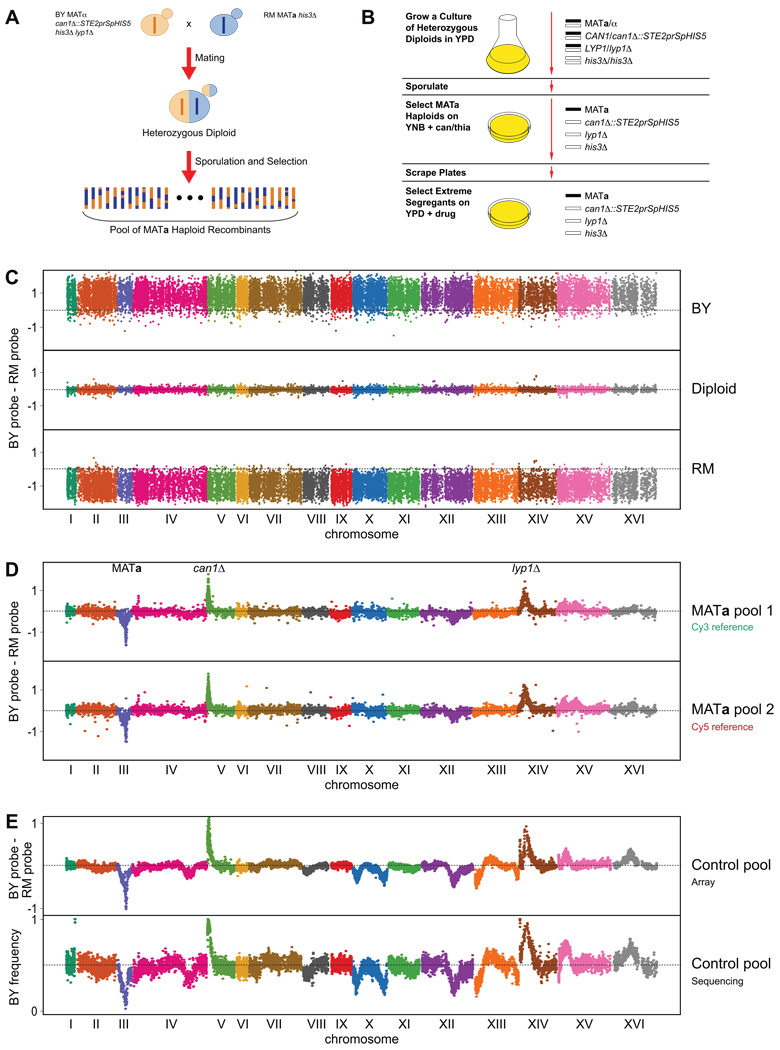
To generate the pools of segregants that form the starting point for X-QTL, we implemented the Synthetic Genetic Array (SGA) marker scheme 12,13, which enables the recovery of MATa haploids from a cross of appropriately marked parental strains (Figure 1A,B). We used BY4716 (hereafter BY), a lab strain, and RM11-1a (hereafter RM), a wine strain, as the progenitors of the pools. We crossed these strains to form a diploid, sporulated the diploid, and selected for ~107 unique BYxRM MATa haploid segregants. We designed an allele-specific genotyping microarray with isothermal probes 14 that assays ~18,000 SNPs between BY and RM. We tested the array by hybridizing the haploid and diploid progenitor strains, as well as multiple MATa pools, and found that we could discriminate the parental strains and reproducibly identify deviations in allele frequencies associated with the SGA markers and other loci in the segregating pools (Figure 1C,D,E). Comparable results were obtained by sequencing pools to ~180X coverage with the Illumina Genome Analyzer (Figure 1E).
Figure 1. X-QTL design and quantitative allele frequency measurement in DNA pools.
The crossing design used for X-QTL is shown in (A), while the selection scheme used to generate segregating pools is shown in (B). Genotyping of parental strains (C), two segregating pools (D), and an unselected control pool grown on rich medium (E). Dashed lines at zero indicate no difference between the BY and RM allele-specific probes. Enrichment of the BY allele is indicated by deviations above 0 and enrichment of the RM allele is indicated by deviations below 0. For the segregating pools, both the control loci involved in MATa selection and the dye used for reference labelling are denoted. In (D), we use a dye-swap experiment to show that the dye used for labelling does not cause any bias in allele frequency measurement. (D) and (E) differ in that (D) shows a MATa pool prior to plating on rich medium and (E) shows a MATa pool after two days of growth on rich medium. In (E), the same pool was hybridized to the genotyping microarray and was sequenced to ~180X coverage with the Illumina Genome Analyzer. The results in (C) and (D) are plots of raw data with no sites removed, while in (E) raw data was plotted with sites more than 1.5 standard deviations away from the local average of the 10 nearest data points removed for clarity.
We first used X-QTL to map the genetic basis of sensitivity to 4-nitroquinoline (4-NQO), a DNA damaging agent. We previously showed that sensitivity to 4-NQO is a complex trait in the BYxRM cross 15. BYxRM segregants show varying degrees of sensitivity, and the parental strains are both intermediate relative to their progeny, suggesting contributions of multiple alleles from each parent. Conventional QTL mapping with 123 genotyped segregants detected a single significant locus on chromosome 12, and subsequent experiments identified an amino acid substitution in the DNA repair gene RAD5 as the underlying causative polymorphism. A backcrossing strategy identified a smaller contributing effect of a polymorphism in the gene MKT1. The BY allele of RAD5 and the RM allele of MKT1 conferred 4-NQO resistance, but these loci did not fully explain the observed 4-NQO responses of the segregants, implying that additional loci must exist.
To map the genetic basis of sensitivity to 4-NQO using X-QTL, we first plated segregating pools across a range of drug doses in order to find a highly selective 4-NQO concentration. We then conducted 4-NQO selections at this concentration, while in parallel growing control populations on rich medium without the drug. 4-NQO-resistant and control pools were harvested, and the extracted DNA was hybridized to genotyping microarrays. To identify loci that confer resistance to 4-NQO, we scanned the genome for locations at which allele frequencies in selected pools were significantly different than in the control pools (Supplementary Methods). Using this approach, we identified 14 loci in the 4-NQO selection at a false discovery rate (FDR) of 0.05. Similar deviations in allele frequency in the selected pools were observed when the genotyping step was carried out by either arrays or short-read sequencing (X-QTL-seq; Figure S3).
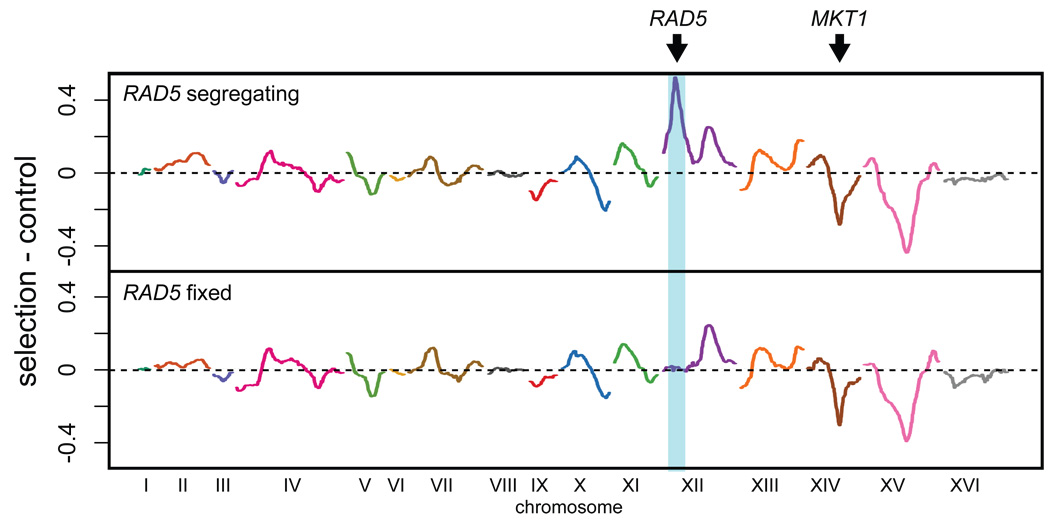
We examined whether the loci identified by X-QTL for 4-NQO resistance correspond to real biological effects. Using X-QTL, we observed peaks at RAD5 and MKT1, with both loci selected in the expected direction (Figure 2). We confirmed that the peak overlapping RAD5 was actually due to this gene by repeating the BYxRM cross with an RM parent strain that had the BY version of RAD5. When 4-NQO resistance was mapped in the selected pool with RAD5 fixed, the resulting segregating pool exhibited increased resistance to 4-NQO, and no RAD5 peak was observed by X-QTL (Figure 2). Next, we isolated 96 individual progeny from the same cross used to generate the segregating pools, phenotyped them for 4-NQO sensitivity, and genotyped them at the 14 loci identified by X-QTL. Nine of the loci showed significant effects in this independent data set (p < 0.05), five of which were highly significant (p < 0.001). The loci jointly explained 59% of the phenotypic variance in 4-NQO sensitivity in an additive model (Figure S4). Because we measured the heritability of this trait to be 0.84, the loci explained 70% of the genetic variance, indicating that we have explained most of the genetic basis of this trait with the loci detected by X-QTL.
Figure 2. X-QTL detection of loci for 4-NQO resistance.
Results for 4-NQO resistance with RAD5 segregating (top panel) and fixed (bottom panel) are shown. The difference between the average of the selections and the average of the controls generated on the same day is plotted, with enrichment of the BY allele indicated by deviations above 0 and enrichment of the RM allele indicated by deviations below 0. Arrows point to MKT1 and RAD5. The RAD5 fixed population was generated by using a RM parent strain in which the RAD5RM allele was replaced with a RAD5BY::NatMX allele. This strain was constructed by crossing strain EAY1467 15 to the RM parent strain used for X-QTL.
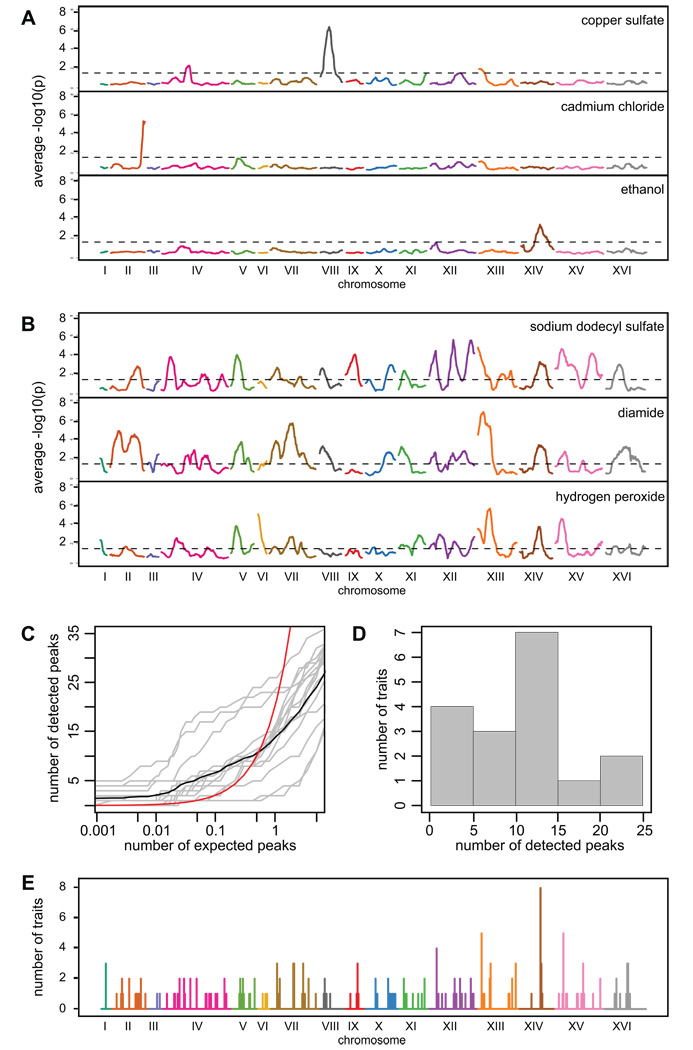
We next applied X-QTL to resistance to 16 diverse chemical agents (Table S1), including a detergent and a number of antifungal compounds, using the same methodology that was employed for 4-NQO. At a global FDR of 0.05, we mapped 177 total loci for these 16 traits. We detected between 1 and 24 peaks in pools selected on these agents. Including 4-NQO, we detect an average of 11 peaks per trait, suggesting high genetic complexity for many traits. The 17 traits show striking differences in their genetic architectures (Figure 3 and Figure S5). At the simpler end of the range, resistance to cadmium chloride, copper sulfate, and ethanol is controlled by one major locus for each trait (Figure 3A). At the other extreme, we identified more than 20 loci in the diamide, hydrogen peroxide, and sodium dodecyl sulfate selections (Figure 3B). Other traits show intermediate levels of complexity (Figure 3C,D).
Figure 3. Genetic architecture of chemical resistance traits.
Examples of genetically simple traits (A), examples of genetically complex traits (B), relationship between the number of expected and detected peaks (C), the number of loci detected per trait (D), and a map of compound-specific and pleiotropic loci across the genome (E). In (A) and (B), the −log10(p) values are shown for t-tests comparing selected samples to control samples. The sliding averages within 50 kilobase windows for these tests are plotted. In (C), the global relationship between expected and detected peaks is plotted as a black line and the trait-specific relationships are plotted as grey lines. The red line plots the relationship between expected and detected peaks at an FDR of 0.05. The expected counts were generated from permutations of the chemical resistance dataset. The histogram in (D) was made using loci significant at a global FDR of 0.05. In (E), detected loci were grouped within 20 kb windows across the genome.
We compared the 191 peaks detected across the 17 traits. The genome was divided into 20 kb bins and all loci within a bin were grouped together. Using this procedure, we found 123 distinct loci (Figure 3E). Of these, 82 loci (~67%) were trait-specific. For instance, a peak was detected at RAD5 on Chromosome XII only in our analysis of resistance to 4-NQO. Similarly, the major locus for copper sulfate resistance, which was previously mapped in a screen for QTLs involved in resistance to small molecules in the BYxRM cross 16, coincides with the location of the CUP1 genes on Chromosome VIII and was detected only in the copper sulfate selection. Of the 41 remaining loci, 40 were detected for two to five traits and one locus, which overlaps MKT1, was detected for eight different compounds. An amino acid polymorphism in MKT1 is known to be involved in a large number of trait differences between BY and other strains, including 4-NQO resistance 15, sensitivity to dipropyldopamine and phenylephrine 17, high temperature growth 18, sporulation efficiency 19, gene expression 20, and growth of petite colonies 21. Our results suggest that in addition to these previously studied phenotypes, MKT1 also has a broadly pleiotropic effect on drug resistance under the conditions of our study. Furthermore, our results suggest that X-QTL detects loci at a fine resolution, as the locations of the peaks corresponding to MKT1 and RAD5 were estimated to be within two kilobases of these genes themselves (Table S2). The loci we have detected across 17 compounds thus provide a foundation for comprehensively studying the molecular mechanisms that shape phenotypic variation in response to chemical agents among yeast strains.
Selections for resistance to chemical agents permit only one extreme tail of the phenotype distribution to be sampled. Additional insights can be gained from selections where both high and low extreme segregants can be recovered. Fluorescence-activated cell sorting (FACS) provides a straightforward approach to such two-tailed selections, as large numbers of individuals exhibiting high and low values for a stain or reporter can easily be recovered. To pilot this approach, we used the dye Mitotracker Red, which stains cells depending on the mitochondrial proton gradient and mitochondrial volume. We harvested a MATa pool, stained it with Mitotracker Red, and then sorted out extreme cells by FACS. We sorted a population of ~5 × 106 cells and selected 3 × 104 cells from each tail. These selected cells were then grown up on agar plates with rich medium to generate enough cells from which to extract DNA. DNA pools from both tails, as well as from a subsample of the whole population, were hybridized to the genotyping microarray.
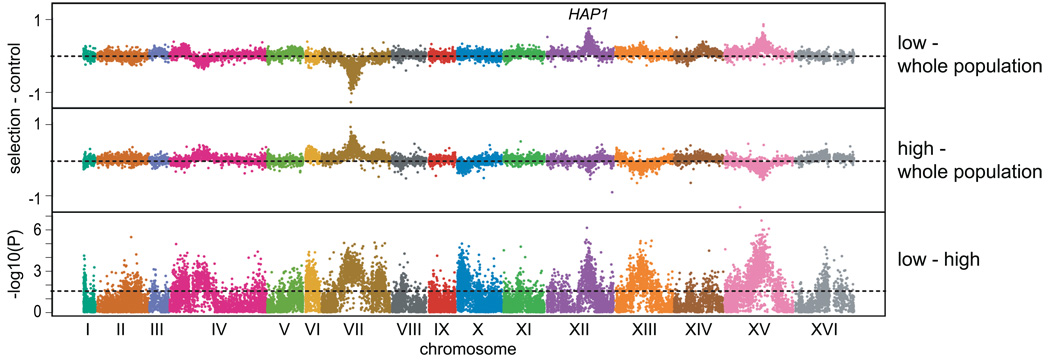
Comparison of the high and low extremes found multiple major peaks at an FDR of 0.05 (Figure 4). These peaks exhibited similar heights but opposite directions in the two tails. The location of one of the peaks provided a strong candidate for the causal gene. The peak on chromosome XII spans HAP1, a zinc finger transcription factor involved in response to oxygen. HAP1 was previously shown to be a hotspot for trans regulation of gene expression differences in the BYxRM cross 22,23. BY has a partially functional allele of HAP1 due to a Ty transposon insertion in the HAP1 coding region 24, whereas RM has a fully functional HAP1 allele. Consistent with HAP1’s function, segregants carrying the RM allele of HAP1 show increased oxidative capacity based on X-QTL mapping. Comparison of BY with a partially functional HAP1 to BY with a fully functional HAP1 shows that HAP1 plays a causal role in variation in Mitotracker Red staining (Figure S6).
Figure 4. X-QTL mapping of mitochondrial activity by cell sorting.
Segregants were stained with the dye Mitotracker Red. The comparisons of high and low pools to the entire population are shown, in addition to −log10(p) values for the difference between these groups. The dashed lines in the high or low - control plots indicate zero difference in a comparison, whereas the dashed line in the final plot indicates the probe-level threshold for an FDR of 0.05.
X-QTL represents a powerful method for rapidly and cost-effectively mapping the multiple QTLs underlying a trait difference between two yeast strains. We have used X-QTL to empirically demonstrate that many traits have a highly complex genetic basis. These results are consistent with previous studies in yeast, such as those focused on transcript levels 3, protein abundance 25, and sensitivity to chemical agents 16, in which genetic complexity was inferred from trait distributions and a lack of mapped loci, rather than from direct detection of multiple loci as we have accomplished here. Our results agree with those from the comprehensive genetic dissection of a small number of traits in other model organisms, such as bristle number in Drosophila 26 and flowering time in maize 27, which have shown that dozens of loci can underlie a difference between two individuals. Importantly, whereas these studies required substantial labor, time, and resources, X-QTL is a quick and easy approach to achieve a comparable level of genetic dissection. The levels of complexity observed here (e.g. 14 loci explaining 70% of the genetic variance for 4-NQO resistance) are still dramatically lower than those seen in for some human traits in GWAS (e.g. 40 loci explaining 5% of the variance for height 2,5). One obvious explanation is the difference in experimental designs (line crosses vs. population association studies), but differences in genetic architectures among species and traits may also contribute. The comprehensive genetic dissection of complex traits by X-QTL makes it possible to empirically answer many of the basic questions about the genetic architecture of complex traits, including the number of loci underlying a trait and the distribution of their allele frequencies in a population. High-resolution mapping of these loci also enables identification of the underlying genes and sequence variants, as well as investigation of allelic effect sizes and genetic interactions. We anticipate that general insights from such studies will be applicable to understanding the genetics of complex traits in other organisms, including humans, and that variants of X-QTL can be developed for other species.
Methods Summary
Microarray hybridizations
DNA was extracted from segregating pools using Qiagen Genomic-tip 100/G columns. DNA was labelled using array comparative genomic hybridization reagents from Invitrogen and Cy3- or Cy5-labelled dUTP from Enzo. Hybridization, scanning, and feature extraction were done using Agilent equipment and software. Normalization of arrays was done using the rank invariant method within the Agilent software.
Statistical analysis
For a given SNP, the difference in log2 ratios of the intensities of the BY and RM allele-specific probes on a single array was computed, and this metric was used in downstream analyses. In cases where a SNP was represented by two probe sets, the probe sets were used as separate data points. For the drug selections, selection and control experiments were compared using t-tests with equal variances. A regression-based peak-finding approach was then used, which scans the genome for locations where the slope in –log10(P) values changes signs. Significance levels were determined by permutation (Figure 3C). For the Mitotracker Red study, the high- and low-staining pools were compared using t-tests with equal variances. QVALUE 28 was then used to determine an FDR based on the observed p-values.
Full Methods and any associated references are described in the Supplement to the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
The authors are grateful to members of the Kruglyak lab for comments on this manuscript, David Botstein, Justin Gerke, Greg Lang, and Ethan Perlstein for input regarding experiments, Joshua Bloom, Dipen Sangurdekar, and John Storey for advice regarding analyses, and Eric Alani for sharing the RM RAD5BY::NatMX (EAY1467) strain. This work was supported by NIH grant R37 MH59520, a James S. McDonnell Centennial Fellowship, and the Howard Hughes Medical Institute (L.K.), a NIH postdoctoral fellowship F32 HG51762 (I.M.E.), and NIH grant P50 GM071508 to the Center for Quantitative Biology at the Lewis-Sigler Institute of Princeton University.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Author Contributions
Experiments were designed by I.M.E., A.A.C., and L.K. Strains were constructed by I.M.E., Y.J., S.M., and A.A.C. Microarrays were designed by I.M.E. and D.G. Experiments were performed by I.M.E., N.T., Y.J., J.K., S.M., and A.A.C. Simulation scripts were written by I.M.E. and J.A.S. Analyses were conducted by I.M.E. The manuscript was written by I.M.E. and L.K., and incorporates comments by all other authors.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009 doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci U S A. 2005;102(5):1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40(5):489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 6.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolyn DJ, et al. Light-response quantitative trait loci identified with composite interval and eXtreme array mapping in Arabidopsis thaliana. Genetics. 2004;167(2):907–917. doi: 10.1534/genetics.103.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brauer MJ, Christianson CM, Pai DA, Dunham MJ. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics. 2006;173(3):1813–1816. doi: 10.1534/genetics.106.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segre AV, Murray AW, Leu JY. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol. 2006;4(8):e256. doi: 10.1371/journal.pbio.0040256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai CQ, et al. Speed-mapping quantitative trait loci using microarrays. Nat Methods. 2007;4(10):839–841. doi: 10.1038/nmeth1084. [DOI] [PubMed] [Google Scholar]
- 11.Schneeberger K, et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6(8):550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 12.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 13.Tong AH, Boone C. Methods in Microbiology. Vol. 36. Elsevier Ltd.; 2007. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae in Yeast Gene Analysis - Second Edition; pp. 369–707. [Google Scholar]
- 14.Gresham D, et al. Optimized detection of sequence variation in heterozygous genomes using DNA microarrays with isothermal-melting probes. PNAS. doi: 10.1073/pnas.0913883107. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demogines A, Smith E, Kruglyak L, Alani E. Identification and dissection of a complex DNA repair sensitivity phenotype in Baker's yeast. PLoS Genet. 2008;4(7):e1000123. doi: 10.1371/journal.pgen.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat Genet. 2007;39(4):496–502. doi: 10.1038/ng1991. [DOI] [PubMed] [Google Scholar]
- 17.Kim HS, Fay JC. A Combined Cross Analysis Reveals Genes With Drug-specific and Background-dependent Effects on Drug-sensitivity in Saccharomyces cerevisiae. Genetics. 2009 doi: 10.1534/genetics.109.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinmetz LM, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416(6878):326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 19.Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37(12):1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 20.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6(4):e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics. 2009;183(1):365–383. doi: 10.1534/genetics.109.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296(5568):752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 23.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35(1):57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 24.Gaisne M, Becam AM, Verdiere J, Herbert CJ. A 'natural' mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1) Curr Genet. 1999;36(4):195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- 25.Foss EJ, et al. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39(11):1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- 26.Mackay TF, Lyman RF. Drosophila bristles and the nature of quantitative genetic variation. Philos Trans R Soc Lond B Biol Sci. 2005;360(1459):1513–1527. doi: 10.1098/rstb.2005.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325(5941):714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 28.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.