Summary
An understanding of sleep requires the identification of distinct cellular circuits that mediate the action of specific sleep:wake-regulating molecules, but such analysis has been very limited. We identify here a circuit that underlies the wake-promoting effects of octopamine in Drosophila. Using MARCM, we identified the ASM cells in the medial protocerebrum as the wake-promoting octopaminergic cells. We then blocked octopamine signaling in random areas of the fly brain and mapped the post-synaptic effect to insulin-secreting neurons of the pars intercerebralis (PI). These PI neurons show altered potassium channel function as well as an increase in cAMP in response to octopamine, and genetic manipulation of their electrical excitability alters sleep:wake behavior. Effects of octopamine on sleep:wake are mediated by the cAMP-dependent isoform of the OAMB receptor. These studies define the cellular and molecular basis of octopamine action and suggest that the PI is a sleep:wake-regulating neuroendocrine structure like the mammalian hypothalamus.
Introduction
Although sleep occupies a large part of daily life for many organisms, it remains a poorly understood phenomenon. Analysis of sleep regulation would be facilitated by a focus on specific cellular loci, but knowledge of such loci is limited (Kryger et al., 2005). Molecules known to affect sleep are expressed widely and function in many different processes (Kryger et al., 2005; Saper et al., 2005). Delineating the cellular basis of their effect on sleep is complicated in mammals because of the complexity of the brain structure (Hendricks et al., 2000). The fruit fly, Drosophila melanogaster, on the other hand, has a relatively simple brain, making it a better model to address the cellular circuitry underlying sleep (Hendricks and Sehgal, 2004). Similarities between mammalian and fly sleep have been established and extend to the neurotransmitters underlying the regulation of sleep (Agosto et al., 2008; Andretic et al., 2005; Hendricks et al., 2000a; Shaw et al., 2000; Yuan et al., 2006).
We demonstrated recently that octopamine, the Drosophila equivalent of norepinephrine, is a potent wake promoting signal (Crocker and Sehgal, 2008). Octopamine is synthesized from tyramine by the action of tyramine beta-hydroxylase (Tβh) and tyramine is synthesized from tyrosine by tyrosine decarboxylase (dTdc) (Roeder, 2005). Mutations in both Tβh and Tdc result in an increased amount of sleep, and electrical manipulation of Tdc-expressing neurons also has predictable effects on sleep (Crocker and Sehgal, 2008). Likewise in mammals, norepinephrine promotes wakefulness. However, the many targets of norepinephrine-producing cells and the existence of multiple receptors has made it difficult to dissect the function of this pathway in sleep (Insel, 1996; Sara, 2009). There are five different classes of adrenergic receptors, with a total of nine receptors, both alpha and beta, and conflicting data on the roles these different receptors may play in sleep (Insel, 1996). Octopamine-producing neurons in Drosophila also project all over the brain and octopamine can signal through several different receptors (Evans and Robb, 1993). Nevertheless, the system is still simpler than in mammals with a total of 4 known genes for octopamine receptors: OAMB, Octβ1R (OA2), Octβ2R and Octβ3R (Evans and Robb, 1993; Han et al., 1998). In addition the gene for OAMB encodes two isoforms, AS and K3, resulting in five possible receptor types (Han et al., 1998; Lee et al., 2009).
Other neurotransmitters shown to affect Drosophila sleep are dopamine, serotonin and GABA (Agosto et al., 2008; Andretic et al., 2005; Kume et al., 2005; Yuan et al., 2006). However, each of these is produced by many neurons and the specific sub-group of neurons responsible for the effect on sleep is not known for any neurotransmitter. Dopamine and serotonin exert at least some of their effects through receptors expressed in the mushroom bodies, a region previously implicated in the regulation of sleep (Andretic et al., 2005; Joiner et al., 2006; Pitman et al., 2006; Yuan et al., 2006). However, the MBs are a complex structure (Tanaka et al., 2008) and the specific neurons relevant for the serotonin/dopamine phenotype were not identified The circadian large ventral lateral neurons mediate effects of light and GABA on sleep, but the source of the GABA is not known (Chung et al., 2009; Parisky et al., 2008; Sheeba et al., 2008). Finally, neurons in the pars intercerebralis (PI) region promote sleep, but their neurochemical identity remains to be determined (Foltenyi et al., 2007). Thus, little is known about the cellular basis of sleep in Drosophila and there has been no concerted effort to dissect the cellular and molecular circuit underlying the effect of any particular neurotransmitter.
In this study we set out to map the octopamine pathway important for sleep. Using a technique known as Mosaic Analysis with a Repressible Cell Marker (MARCM) we were able to map the wake-relevant octopamine-producing cells to specific cells in the medial protocerebrum (Wu and Luo, 2006). We also used an unbiased approach to map the octopamine target neurons that mediate the effects on sleep:wake. Building upon the observation that the effects of octopamine on sleep are protein kinase A (PKA)-dependent, we blocked PKA signaling in various parts of the fly brain and assayed the behavioral response to orally ingested octopamine. We find that a subset of neurons in the pars intercerebralis (PI) mediates effects of octopamine on sleep. Consistent with their identification as octopamine-responsive cells, these neurons in the PI demonstrate changes in electrical activity, as well as in cAMP levels, in response to exogenous octopamine. Acting through the OAMB receptor on these PI neurons, octopamine signals through cAMP and PKA to promote wakefulness. Our data provide a molecular pathway as well as the cellular circuitry that mediates the wake-promoting effects of octopamine. Since the PI neurons implicated here are an integral part of the neuroendocrine PI, we believe these findings highlight the similarity between the PI and the mammalian hypothalamus. These findings may also provide insight into how the adrenergic system works to influence sleep in mammals.
Results
Identifying the octopaminergic cells that regulate sleep
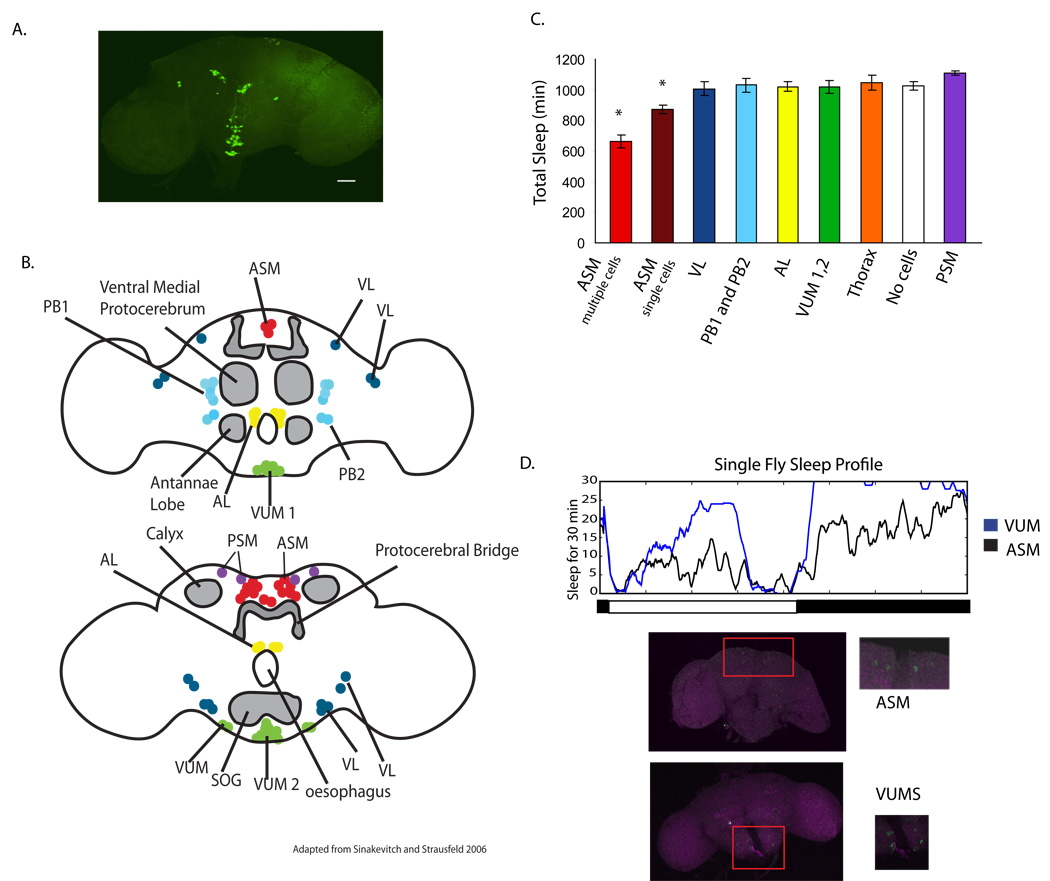
In mammals, the release of norepinephrine from a specific group of cells in the Locus Coeruleus (LC) is known to regulate sleep (Sara, 2009). We set out to determine if a specific cell group in the fly is responsible for the effect of octopamine on sleep. To address this question we employed the MARCM technique which allows one to restrict the expression of a transgene to a subset of its normal pattern (Wu and Luo, 2006); (Agosto et al., 2008). We showed previously that expression of a sodium channel (NaChBac) by a tyrosine decarboxylase 2 (Tdc2) GAL4 driver, which expresses in tyramine and octopamine producing cells, decreases sleep (Cole et al., 2005; Crocker and Sehgal, 2008). The NaChBac transgene is derived from a gene encoding a bacterial Na+ channel which has the characteristics of high open probability and low inactivation, thus driving membrane voltage to a more depolarized and easily excited state (Luan et al., 2006; Nitabach et al., 2006) Thus, excitation of Tdc2-positive neurons results in decreased sleep. However, Tdc2-GAL4 is expressed in a number of cells in the fly brain (Figure 1a), any of which could be responsible for the decrease in sleep. In the MARCM technique, a repressor of GAL4 activity, GAL80, is co-expressed and then is excised in a random fashion from groups of cells (see Supplemental Figure 1). By driving this excision in populations of developing Drosophila, one can generate large numbers of flies, each of which expresses GAL4 in a different subset of the original pattern. Using MARCM we were able to reduce the number of octopamine cells expressing the Na+ channel down to a single cell in some cases. We chose the Na+ channel in part because it has a robust effect on sleep and also because it is tagged with green fluorescent protein (GFP). The GFP marker allowed us to identify the cells expressing the sodium channel.
Figure 1. Cells in the medial protocerebrum mediate wake-promoting effects of octopamine.
A. Expression pattern of the Tdc2–Gal4 line as visualized with a GFP reporter. Octopamine is produced in a subset of neurons in the brain. Expression was characterized by crossing Tdc2–Gal4 with UAS–GFPnls. B. A schematic of the different octopamine neurons, labeled according to Sinakevitch and Strausfeld (Sinakevitch and Strausfeld, 2006). Cell groups are clustered according to color, with similar colors indicating cells that could not be distinguished from each other in our analysis. C. Total sleep amounts for each cell population indicated in B. The colors of the graph correspond to clusters of the same color in B. The ASMs were the only cell group that showed a significant decrease in sleep compared to the other cell groups. The graph depicts mean sleep ± S.E. and * is p≤0.01 using 2 way ANOVA. Values are found in Supplemental Table 1. D. Sleep data for single flies. Sleep over 24 hours is shown for two flies, one expressing Na+ channel in 4 cells in the ASM region (Black line) and one for a fly expressing the Na+ channel in about 8 VUMS (Blue line). Day and night are depicted by the black and white lines respectively. Pictures below are of those individual fly brains stained for nc82 (pink) and GFP (green). The generation of the MARCM flies is outlined in Supplemental Figure 1.
Since each fly generated through MARCM expresses the relevant transgene in a distinct pattern, we assayed each individual fly for sleep and then for expression of GFP. Figure 1b shows the different cell populations labeled by Tdc2-GAL4 in a schematic of the anterior and posterior sections of the fly brain. The schematic is derived from the anatomical map generated by Sinakevitch and Strausfeld (Sinakevitch and Strausfeld, 2006), while the nomenclature is based upon the studies of Busch and colleagues (Busch et al., 2009). We were unable to discriminate some cell clusters as anterior or posterior and so grouped them together, such as the ASM cells. We found that expression of the Na+ channel in the ASM group of cells produced a decrease in sleep similar to that seen with the Tdc2-GAL4 driver expressing the Na+ channel in all neurons (Figure 1c,d, Supplemental Table 1). The sleep loss was approximately 320 minutes when the Na+ channel was expressed in multiple cells of the ASM group, but was less when it was expressed in only one cell in this group. Thus, the effect of this neuronal group on sleep appears to reflect the cumulative contribution of multiple cells. Expression of the sodium channel in other areas of the brain and thorax did not result in a significant drop in sleep (Figure 1c).
Mapping Brain Regions that mediate Post Synaptic Effects of octopamine
We showed previously that protein kinase A (PKA) is necessary for the relay of the wake-promoting octopamine signal (Crocker and Sehgal, 2008). In this study we attempted to localize the areas of the fly brain that respond to this signal. We overexpressed the regulatory subunit of PKA (PkaR), which acts to block PKA signaling, using a number of different GAL4 drivers (Figure 2a)(Rodan et al., 2002). We found that many of the resulting lines showed increases in total sleep as compared to the control line Iso31 (Isogenic w1118 line). This effect is consistent with our previous work showing that an increase in PKA signaling reduces sleep (Hendricks et al., 2001). In addition, a subset of the GAL4 lines expressing PkaR were no longer sensitive to the decrease in sleep caused by feeding octopamine (Figure 2a, Supplemental Table 2). While some of these lines express GAL4 in many cell types, the one region of the brain where the expression overlaps is the PI (Figure 2b). In fact, the Dilp2 GAL4 driver, which expresses GAL4 in cells producing Drosophila insulin like peptide 2 (Dilp2), is expressed in only a subset of PI neurons (Rulifson et al., 2002) and expression of PkaR by this driver has as robust an effect as any other. While this line did not show a significant increase in nighttime sleep relative to wildtype, it did show a significant increase in sleep consolidation- decreased number of sleep bouts with a corresponding increase in sleep bout length (Supplemental Figure 4). Interestingly, expression of PkaR in the mushroom body, a region previously implicated in the regulation of sleep, does not affect the response to octopamine.
Figure 2. Octopamine signals through cells in the pars intercerebralis (PI).
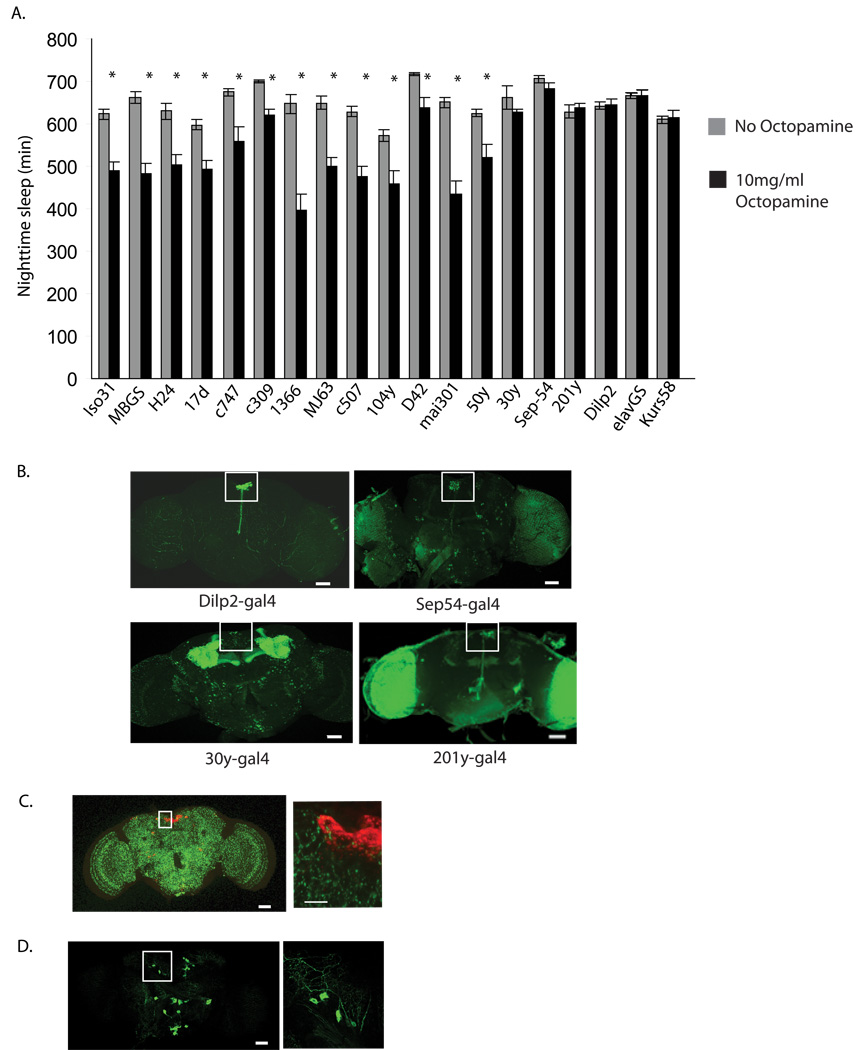
A. Sleep in flies expressing the regulatory subunit of protein kinase A (PkaR) in different regions of the fly brain. Progeny obtained by crossing each of the 18 GAL4 driver lines indicated to w1118;UAS-BDK33 (PkaR) were analyzed for sleep. w1118 (Iso31), the background for all the lines, was used as control line. Sleep values were determined for the night prior to the initiation of 10mg/ml octopamine feeding and then the third night after providing octopamine. Grey bars represent sleep prior to octopamine; black bars, sleep following 10mg/ml octopamine. Sleep values are found in Supplemental Table 2 and are plotted as mean ± s.e.m. Nighttime sleep values for each line is depicted in Supplemental Table 2 and sleep parameters during nighttime sleep for expression of PkaR in Dilp2 producing neurons is shown in Supplemental Figure 4. B. Expression of a nuclear-targeted GFP (GFPn) under the control of the following GAL4 drivers; Dilp2-GAL4, Sep54- GAL4, 30y-GAL4, 201y-GAL4. The PI region is denoted by the white square in each image. C. Expression of a synaptically targeted GFP (syt-GFP) under the control of Tdc2-GAL4. PI neurons were stained with anti-Dilp2 and are shown in red. An enlarged image of the PI region is shown in the right hand panel. D. Expression of a membrane bound GFP (CD8-GFP) Tdc2 producing cells using the Tdc2-GAL4 driver. A single confocal image through the ASM neurons is shown on the left. On the right is an enlarged image of a 5υm thick stack through the ASM neurons, which appear to show dorsal as well as lateral projections.
We were interested in determining whether wake-promoting octopaminergic cells project to the PI. To this end, we expressed a synaptically tagged GFP under the control of the Tdc2-GAL4 and co-labeled the brains with antibodies specific for the Dilp2 neurons. As shown in Figure 2c, synaptic boutons of Tdc2 neurons are found in the vicinity of the Dilp2 neurons. In addition, expression of a membrane bound GFP (CD8-GFP) in TDC2 producing cells indicates dorsal as well as lateral projections of ASM neurons. This is similar to the findings of Busch et al, who examined whole mount projections of the octopamine producing ASM neurons (Busch et al., 2009). Together these data indicate that the Dilp2 neurons likely receive octopaminergic signals and mediate wake-promoting effects of octopamine.
Modulation of Dilp2 producing neurons alters sleep
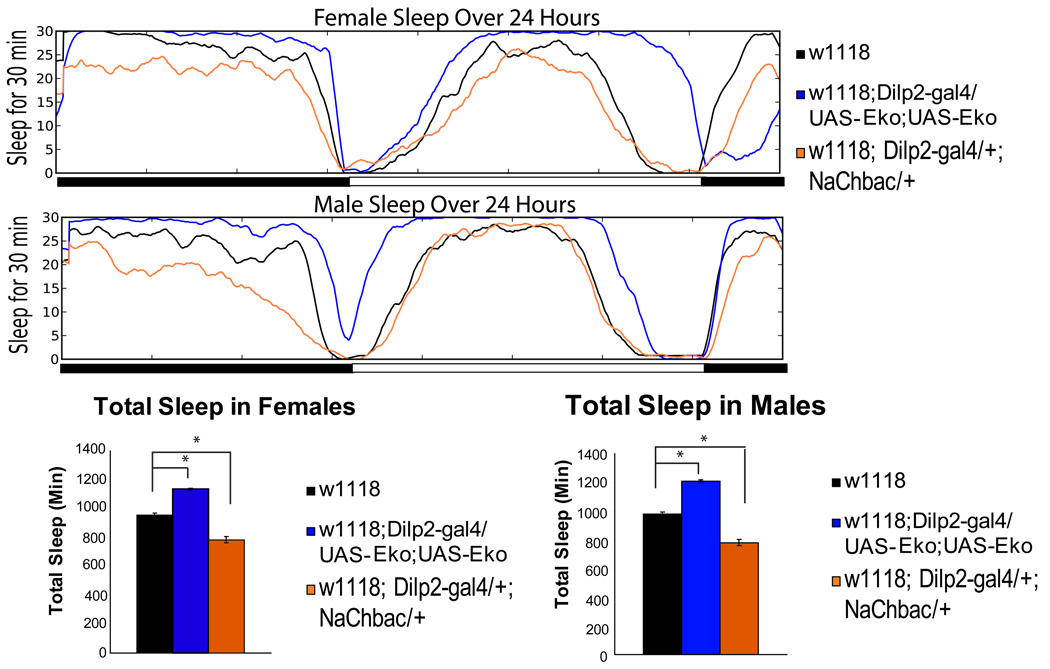
If octopamine signals through the PI to regulate sleep, then modulating the firing of the relevant PI neurons should also affect sleep. Thus, we used the Dilp2–GAL4 driver to express either a depolarizing or a hyperpolarizing ion channel (UAS–NaChBac or UAS–EkoIII) (Luan et al., 2006; White et al., 2001) and examined effects on sleep. Expression of the Na+ channel in Dilp2-positive cells resulted in an average decrease in daily sleep of 172 min (Figure 3) (Supplemental Table 3). The loss of sleep was more specific to the nighttime, similar to what we see when Tdc2 producing cells are depolarized (Crocker and Sehgal, 2008). A similar decrease in sleep was seen in male flies expressing the Na+ channel in the Dilp2 neurons (Figure 3)(Supplemental Table 3).
Figure 3. Sleep is altered by manipulations of the electrical activity of PI neurons.
A 24 hour profile of sleep in w1118 flies (black line), and Dilp2-GAL4 flies expressing either 3xUAS-Eko (Blue line) or UAS-NaChBac (orange line). Top panel shows female data and bottom panel male data. Total sleep amount for females and males of each genotype is shown below the profiles. Actual values are in Supplemental Table 3. Total sleep is plotted as mean ± s.e.m. Asterisk= P < 0.01 by 2-way Anova.
When UAS-EkoIII, a modified Shaker K+ channel, was expressed in Dilp2 neurons, there was an increase in sleep during both the night and the day (Figure 3a, b) (Supplemental Table 3). These data support the idea that excitation of the Dilp2 neurons promotes wakefulness while silencing these neurons increases sleep.
Octopamine modulates a potassium current in PI neurons
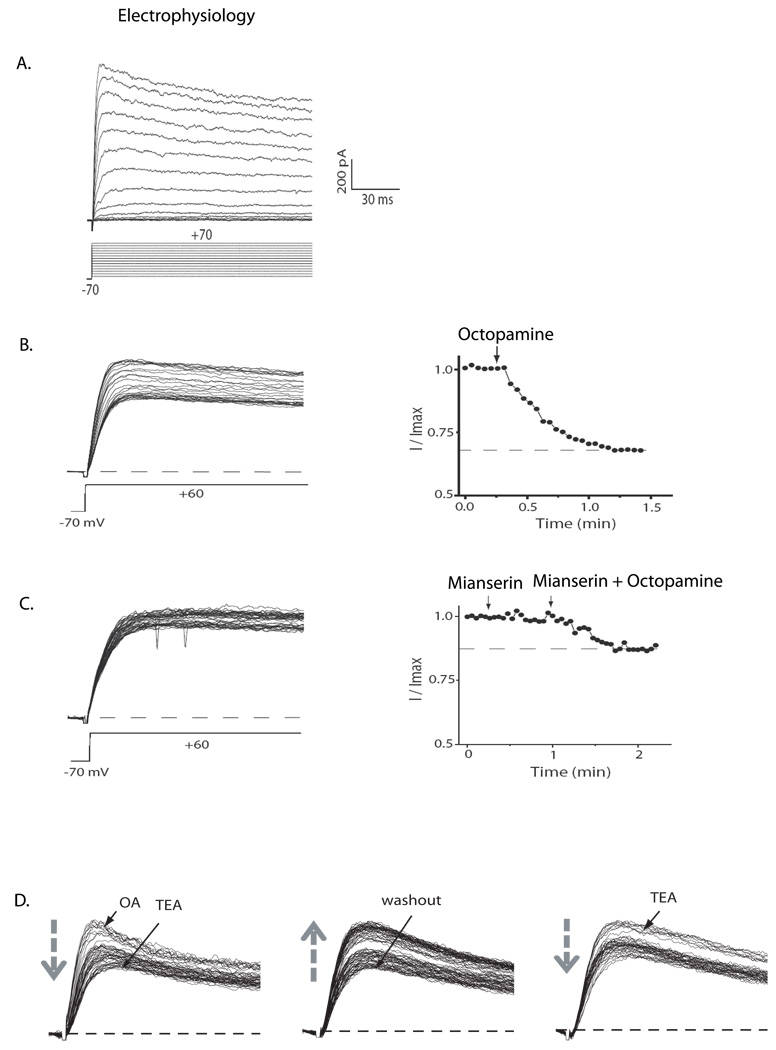
Since PI neurons are implicated in the wake-promoting effects of octopamine, we asked whether octopamine application could influence their electrical excitability. Accordingly we generated flies in which the PI neurons were labeled with green fluorescent protein (GFP) under the control of the Dilp2 GAL4 driver. The PI neurons are large and located close to the surface of the brain, and so we were able to measure whole-cell currents by patch clamp analysis, in the brains of living flies (Shahidullah et al., 2009). Voltage-dependent outward currents were evoked by depolarizing voltage steps in the whole cell recording mode (Figure 4a). In order to examine the outward current in the presence of octopamine, we evoked currents by applying a repetitive depolarizing pulse to +60 mV from a holding potential of −70 mV, and then added 1 mM octopamine to the bath solution. As shown in Figure 4b, 1 mM octopamine reduces the total outward current by approximately 30%. Since mianserin antagonizes effects of ingested octopamine on sleep:wake (Crocker and Sehgal, 2008), we determined whether this antagonist (Maqueira et al., 2005) could block the effects of octopamine on the whole cell outward current. We applied the same protocol described above to evoke outward current, and then added 0.25 mM mianserin to the bath solution followed by 1mM octopamine (Figure 4c). Indeed, mianserin greatly reduces the effect of octopamine on the outward current.
Figure 4. Octopamine modulates outward potassium current in PI neurons.
The Dilp2 neurons were identified by expressing GFP under the control of a Dilp2-Gal4 driver, and voltage-dependent outward current was recorded in the whole cell patch recording mode. A. Whole-cell outward currents evoked by 150 ms depolarizing voltage steps from −60 mV to +70 mV in 10 mV increments, from a holding potential of −70 mV. The scale bars apply to all portions of this figure. B. Repetitive pulses to a single voltage, +60 mV, were used to examine the effects of octopamine (OA) on the outward current. 1 mM OA was added to the bath solution during the recording after which the outward current at +60 mV decreases as a function of time (B, left). Peak current amplitudes are plotted against time and the application of octopamine is shown by an arrow (B, right). C. The same voltage protocol as in B was used to evoke outward current, and 0.5 mM mianserin (MI) was added to the bath solution followed by the application of 1 mM OA (C, left). Peak current amplitudes are plotted against time; the applications of mianserin and OA are shown by arrows (C, right). D. The same voltage protocol was used to evoke outward current, and 1 mM OA was added to the bath solution. After the modulation by octopamine, subsequent addition of TEA does not block the residual outward current (D, left). Following washout of the octopamine and TEA, the current returns to baseline levels (D, middle). Subsequent addition of TEA blocks about 30% of the outward current (D, right), suggesting that octopamine selectively modulates the TEA-sensitive component of the current. The arrows illustrate the direction of current change over time after drug addition or withdrawal.
We next asked which component of the outward potassium current is modulated by octopamine application. At low concentrations, the potassium channel blocker tetraethylammonium (TEA) is highly selective for the calcium-dependent potassium current carried by Slowpoke channels (Shen et al., 1994), and we have demonstrated that 1 mM TEA selectively blocks Slowpoke current in the PI neurons in vivo (Shahidullah et al., 2009). As shown in Figure 4d, 1 mM TEA has little effect on the outward current following octopamine application, suggesting that octopamine inhibits the TEA-sensitive component of outward current attributable to Slowpoke. Following washout of octopamine, the TEA-sensitive current returns. These data demonstrate that octopamine selectively inhibits the Slowpoke calcium-dependent potassium current in the Dilp2 neurons, thereby increasing their excitability.
Octopamine affects cAMP signaling in PI Neurons
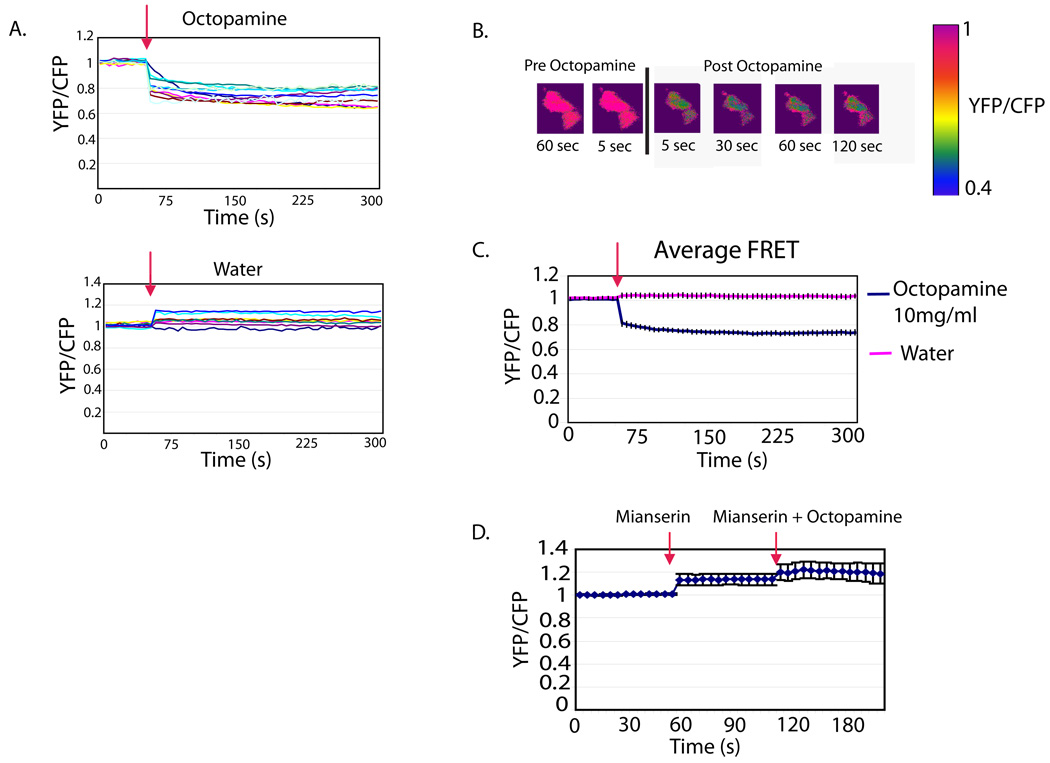
As mentioned above, we found that altering PKA levels in the PI neurons blocks the flies’ response to octopamine, suggesting that octopamine increases cAMP. Thus, we asked whether there is a change in cAMP signaling in Dilp2 neurons following application of octopamine. To monitor cAMP activity, we used a recently described Epac-cAMP construct which reports the activation of Epac by cAMP through a FRET-based assay (Shafer et al., 2008). An increase in cAMP produces a decrease in the FRET associated with this construct. We drove the UAS-Epac construct with a Dilp2 driver, dissected brains from the resulting flies and measured FRET in response to octopamine. Figure 5a shows that the FRET signal decreases following application of 10mg/ml octopamine in the bath. The response is fairly rapid, similar to that seen when Gs coupled receptors in other parts of the brain are stimulated (Shafer et al., 2008). As one might expect, most of the changes in FRET are intracellular (Figure 5b). We also determined if 0.25mM mianserin, a blocker of octopamine receptors, could block the effects of octopamine application. The addition of mianserin completely blocks the increase in cAMP activity seen with octopamine administration alone (Figure 5d). Interestingly mianserin alone produces a small decrease in cAMP activity. Together these studies show that octopamine both depolarizes Dilp2 neurons and activates cAMP signaling in them, with a similar time course.
Figure 5. Octopamine increases cAMP signaling in PI neurons.
A. The response of Dilp2 neurons to 1 mM Octopamine. A FRET-based cAMP sensor was expressed in Dilp2 neurons and the FRET signal was measured in dissected brains following the application of octopamine. In all graphs the red arrow indicates the start of bath application of octopamine or vehicle. The lower panel shows the response of Dilp2 neurons from 8 brains to vehicle (water). B. A pseudocolored time course of Epac1-camps FRET loss in a cluster of 3 Dilp2 cell bodies in response to 1 mM octopamine. The black bar represents the addition of octopamine. The time course is outlined below the images. The table values on the right represent raw YFP/CFP ratios. C. Quantification of the data in A showing the average YFP/CFP value for each time point over 300 seconds. Error bars indicate the standard error of the mean. Through one-way repeated measures ANOVA, all time points following octopamine administration were significantly different from vehicle with p<.001; prior to octopamine administration there was no difference between vehicle and octopamine groups. D. Effects of mianserin on the response to octopamine. The plot shows the average YFP/CFP value for each time point over 300 seconds following application of 0.25mM mianserin and then 0.25mM mianserin plus 10mg/ml octopamine. Error bars indicate the standard error of the mean. Through one-way repeated measures ANOVA, all time points following octopamine administration were not significantly different from previous time points, despite there being a trend towards an increased ratio.
The OAMB receptor acts in PI neurons to regulate sleep
Mammals express many different norepinephrine receptors and it has been difficult to pinpoint those relevant for sleep. Since our previous work indicated that effects of octopamine are mediated by PKA, we focused on the octopamine beta and OAMB receptors which signal through cAMP and are homologous to the beta adrenergic receptors. As noted above, the OAMB receptor has two isoforms, K3 and AS. Activation of the K3 isoform results in increased cAMP signaling and Ca++ signaling whereas activation of the AS isoform only increases Ca++ (Lee et al., 2009).
OctB2R is an octopamine beta receptor that is known to signal through cAMP (Evans and Robb, 1993), but has not been characterized genetically in the fly. We obtained flies carrying a P-element insertion in the 5th exon of the OctB2R gene and analyzed them on molecular and behavioral levels. This P-element disrupts the conserved 7th transmembrane domain of the predicted protein and results in a loss of mRNA signal, as seen in Supplemental Figure 3. To assay the effect of the P-induced mutation on sleep, we first outcrossed it into an Iso31 background. We found that disruption of the OctB2R gene did not affect nighttime sleep, but produced a significant decrease in daytime sleep resulting in an overall decrease in sleep (Supplemental Table 3). Since this P-element disrupts expression of the OctB2R receptor and yet affects sleep in the opposite direction from octopamine, we did not consider it further as a candidate for mediating effects of octopamine (Figure 6b)
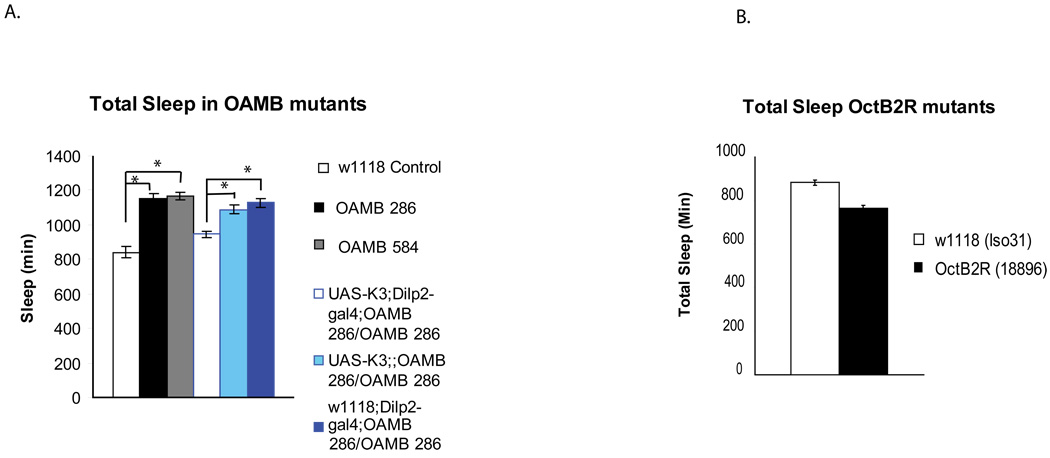
Figure 6. Mutations in the OAMB receptor affect sleep levels.
A. Sleep amounts for females of the following genotypes- w1118 (white), w1118;;OAMB286/OAMB 286 (black), w1118;;OAMB 584/OAMB 584 (grey), UAS-K3;Dilp2-GAL4;OAMB 286/OAMB286 (white), UAS-K3;;OAMB 286/OAMB286 (light blue), and w1118;Dilp2-GAL4;OAMB 286/OAMB286 (blue). Total sleep is plotted as mean ± s.e.m. Asterisk= P < 0.01 by 2-way Anova. Actual values and male data are in Supplemental Table 3. B. Sleep amounts for the OctB2R (18896) outcrossed into the w1118 (Iso31) background. Total sleep is shown as in part A for Iso31 (control) in white and 18896 in black. Actual values and male data are in Supplemental Table 3. Verification of loss of OAMB transcript in OAMB 286 mutant and OctB2R transcript in the OctB2R mutant is shown in Supplemental Figure 2. Expression of OAMB transcript in Dilp2 producing neurons is shown in Supplemental Figure 3.
We examined two previously characterized OAMB mutants, the OAMB 286 mutant and the OAMB 584 mutant (Lee et al., 2003), for effects on sleep:wake. The OAMB 286 mutant is a known null (Supplemental Figure 2) and the OAMB 584 mutant deletes the first 3 exons of the OAMB gene. Both OAMB 286 and OAMB 584 disrupt the K3 and AS isoforms. Both mutants show increases in sleep (Figure 6a), which manifest as longer sleep bouts at night and greater numbers of sleep bouts during the day (Supplemental Table 3). An effect on number and duration of sleep bouts suggests that the mutants affect the initiation as well as the maintenance of sleep. For the rest of our studies we focused on the null OAMB 286.
OAMB is expressed in the mushroom body, thereby its name (octopamine mushroom body receptor), but was not known to express in the PI neurons (Han et al., 1998). To determine if OAMB is expressed in the PI, we collected extracts from single Dilp2 neurons via the whole cell recording electrode, prepared RNA and ran rtPCR experiments. We found that the K3 isoform of OAMB, which couples to both cAMP and calcium, is expressed in the Dilp2 neurons. Indeed, sequence analysis of the single band observed on the gel confirmed its identity as the K3 isoform (data not shown). We did not detect the Ca++ only isoform, A3 (Supplemental Figure 3). To confirm the specificity of the rtPCR experiments, we also ran them on OAMB mutants. The Dilp2 neurons of these mutants did not express the K3 isoform of OAMB (Supplemental Figure 3)
We then asked whether restoring OAMB to the PI neurons would rescue the sleep phenotype of the OAMB mutants. To address this question, we drove expression of the K3 isoform in the Dilp2 neurons of OAMB 286 mutants. Expression of K3, under the control of the Dilp2 driver, resulted in a significant reduction in sleep in the 286 background, thus effectively rescuing the phenotype (Figure 6a). These data support the conclusion that octopamine acts through OAMB mediated cAMP signaling in Dilp2 neurons to modulate sleep.
The OAMB receptor mutants act in PI neurons to mediate wake-promoting effects of octopamine
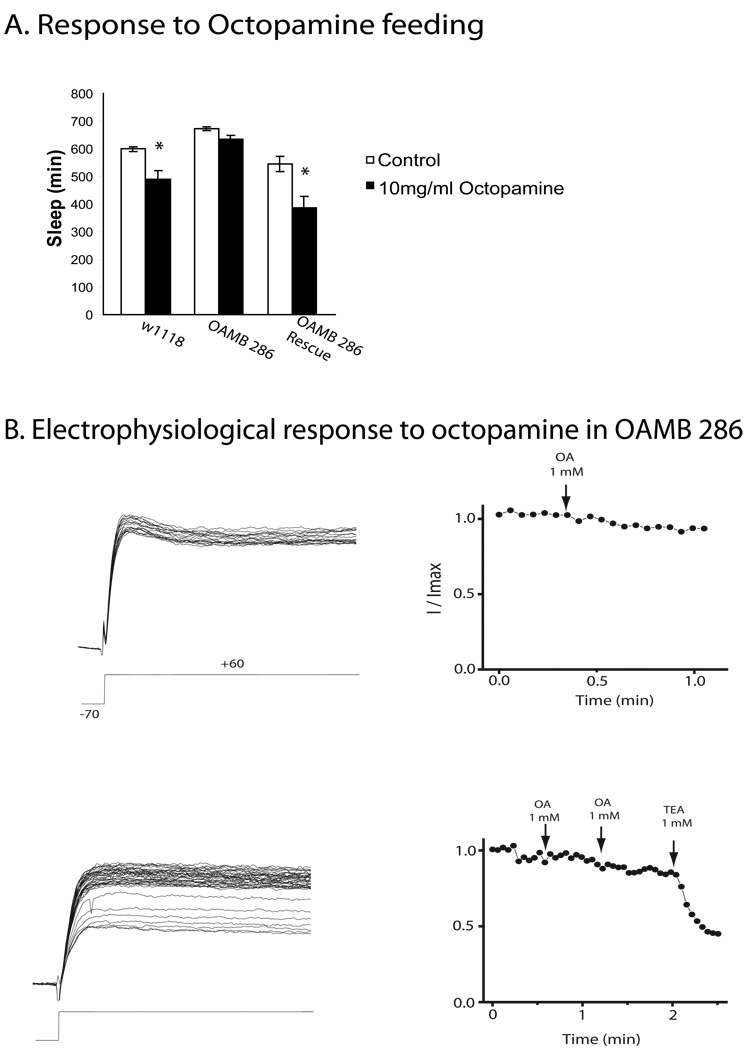
We then asked whether OAMB mutants are responsive to the wake promoting effects of octopamine. Feeding 10mg/ml octopamine to OAMB 286 flies did not produce the decrease in nighttime sleep seen in wildtype controls (Figure 7a). However, consistent with a role for the Dilp2 neurons in mediating effects of OAMB on sleep:wake, the response of 286 flies to octopamine was rescued by Dilp2-Gal4-driven expression of the K3 isoform (Figure 7a).
Figure 7. The OAMB mutant shows an attenuated response to octopamine.
A. Sleep amounts prior to treatment with 10mg/ml octopamine (white bar) and post octopamine (black bar) for females of the genotypes w1118, w1118;;OAMB286/OAMB286, and UAS-K3;Dilp2-GAL4;OAMB286/OAMB286. Sleep is plotted as mean ± s.e.m. Asterisk= P < 0.01 by 2-way Anova. Nighttime sleep values are shown in Supplemental Figure 2. B. Whole-cell outward current in PI neurons in OAMB 286 flies. Current at +60 mV (as in Figure 4) is modulated only slightly by two successive applications of 1 mM octopamine (OA). Subsequent application of 1 mM TEA blocks about 30% of the outward current (B, left). Peak current amplitudes are plotted against time, and the applications of octopamine and TEA are shown by arrows (B, right).
We also recorded in vivo from Dilp2 neurons in the OAMB 286 flies and found they are virtually unresponsive to octopamine (Figure 7b). To determine whether the Slowpoke calcium-dependent potassium current is present in Dilp2 neurons in the OAMB 286 mutant flies, we applied 1 mM TEA after several attempts to modulate the current with octopamine (Figure 7b). TEA blocks about 30% of the outward current, as it does in wild type flies (Figure 4), confirming that octopamine receptor deletion does not directly affect the Slowpoke current, but eliminates its modulation by octopamine.
Discussion
Most animals have evolved mechanisms to maintain alertness as well as sleep states (Tobler, 2005). Here we identify a pathway through which octopamine regulates arousal in the insect brain. This study is the first to map the sleep:wake-regulating effect of a particular neurotransmitter to specific cells in the fly brain. We show that a specific cell group, known as the ASM cell group, comprises the octopamine-producing cells important for the effects of octopamine on arousal. These cells are found in the lateral protocerebrum region and are also known as the G1/G4a cells (Sinakevitch and Strausfeld, 2006). Before this report there was no known function of these cells. Using MARCM to specifically express a Na+ channel in ASM cells we were able to mimic the sleep phenotype seen with Na+ channel expression in all octopamine-producing cells. We find that the magnitude of the decrease in sleep depends upon the number of ASM cells activated, although even the activation of a single cell produces a phenotype. Thus, as in mammals, discrete cell populations encoding a specific neurotransmitter regulate sleep and arousal in the fly.
We also demonstrate that the Pars Intercerebralis (PI) is an important region of the fly brain for relaying the octopamine arousal-promoting signal. Inhibiting PKA, a downstream signal of octopamine receptors, in specific PI neurons blocks the effect of ingested octopamine on sleep. Modulating the electrical activity of these PI neurons also affects sleep. While we have not demonstrated actual synaptic contact between the ASM neurons discussed above and the relevant PI neurons, there is reason to believe that these neuronal groups are directly connected. Our work, and that of Busch et al, suggests that Tdc2 neurons, including the ASM group, extend projections towards the dorsal region of the brain (Busch et al., 2009). In fact, we show termini of Tdc2 projections in the vicinity of the PI (Figure 2).
Previous work identified sleep-promoting cells in the region of the PI, but those appear to be distinct from the ones that mediate the effects of octopamine (Foltenyi et al., 2007). When we decreased PKA signaling in cells labeled by 50y-GAL4 (which targets the sleep promoting cells identified in the other study) we observed a normal response to octopamine (Figure 2). Conversely, none of the drivers that were effective in our studies were reported in the study that identified the sleep-promoting neurons (Foltenyi et al., 2007). Also, the sleep-promoting neurons appear to be morphologically distinct from many of the other PI neurons, in that they do not show long projections to the tritocerebrum. It should be noted that the PI is a very heterogeneous structure composed of many different cell types (Siegmund and Korge, 2001). Functionally, given that it contains many neuroendocrine cells, it appears to be most analogous to the mammalian hypothalamus (Toivonen and Partridge, 2009). The fact that both sleep-promoting and arousal-promoting neurons can be found in the PI, as in the hypothalamus, lends credence to this analogy. Interestingly, we have found that the PI is also important for circadian output, which suggests the possibility that it integrates circadian and homeostatic signals (Jaramillo et al., 2004).
The PI neurons that mediate the effects of octopamine are the major insulin secreting cells in the fly (Rulifson et al., 2002). This may have important implications for the much-hypothesized link between metabolism and sleep (Trenell et al., 2007). Recent work on human sleep indicates that sleep deprivation modulates the insulin signaling pathway (Knutson and Van Cauter, 2008; Spiegel et al., 2009). On the other hand, disruptions in metabolism may also cause changes in sleep (Laposky et al., 2008; Laposky et al., 2006; Trenell et al., 2007). It is reasonable to assume that if sleep is conserved across species then its most basic functions may also be conserved. We find that an arousal pathway in flies includes the major neurosecretory cells in the PI (Toivonen and Partridge, 2009). In mammals, growth hormone regulation more closely follows the sleep wake cycle than the circadian cycle (Sassin et al., 1969; Takahashi et al., 1968). It may turn out that similar hormones released from the PI or PI projections closely match the sleep wake cycle. Indeed, insulin, produced by the Dilp2 neurons, has growth-promoting effects in Drosophila (Brogiolo et al., 2001).
In mammals, it has been difficult to tease apart the adrenergic receptors important for the arousal-promoting effects of norepinephrine. In this study we show that the OAMB receptor coupled to Ca++ and cAMP regulates arousal in response to octopamine. Specifically we demonstrate that the cAMP-stimulating action of this receptor is required. In mammals, the Alpha-1 receptors are coupled to Ca++ or PKC, the Alpha2 receptors inhibit cAMP and the Beta receptors stimulate cAMP (Insel, 1996). Given that cAMP also regulates sleep in mammals (Cirelli et al., 1996; Kanyshkova et al., 2009; Zamboni et al., 1999) and CREB appears to be a wake-promoting signal (Graves et al., 2003), it is likely that mammalian beta receptors are responsible for the wake-promoting effects of norepinephrine. Thus, the prediction is that A2 adrenergic receptors will be found to increase sleep while beta receptors decrease sleep. In mammals, the agonists of these receptors, when focally applied to the medial preoptic area, do seem to follow the hypothesis above such that the A2 agonists promote sleep and the beta agonist promotes wakefulness (Mallick and Alam, 1992). Finer injections of agonists and antagonists specifically into the ventral lateral preoptic area (the major mammalian sleep promoting area (Saper et al., 2005), will likely provide stronger support for this idea.
In the PI neurons identified here, the activation of cAMP by octopamine is accompanied by an inhibition of the outward potassium current, measured by whole cell patch clamp in vivo. By using low concentrations of the highly selective potassium channel blocker TEA, we identified the Slowpoke calcium-dependent potassium current as the specific target of octopamine. Because Slowpoke current contributes significantly to action potential repolarization (Shao et al., 1999), a decrease in this current will lead to an increase in action potential duration in the PI neurons (Shahidullah et al., 2009), thereby increasing the activity of neural circuits in which these neurons participate. It is noteworthy that calcium-dependent potassium channels have long been known to be modulated by PKA-dependent phosphorylation and dephosphorylation (DePeyer et al., 1982) (Ewald et al., 1985) (Chung et al., 1991) (Zhou et al., 2002). The present findings provide an intriguing physiological and behavioral context for these earlier cellular and molecular studies.
To map the octopamine target neurons relevant for sleep:wake, we genetically decreased PKA levels in different brain regions and then monitored the response to food supplemented with octopamine. In many subgroups of neurons, dropping the level of PKA leads to death or very sick flies, making it difficult to address their role in sleep behavior. Thus, there may be areas in addition to the PI that are important for the regulation of sleep by octopamine. Likewise, there are other brain regions that mediate effects of PKA on sleep and wake. For instance, a screen to map the sleep-relevant site of PKA action identified the mushroom body as an important sleep-regulating structure (Joiner et al., 2006; Pitman et al., 2006). We found that over-expressing PKA in Dilp2 cells results in pupal lethality, which may explain why the PI neurons did not show up in the screen that identified the MBs. In addition, since the octopamine signal did not map to the mushroom body, there are as yet unknown signals that increase PKA in the MB.
Clearly, the regulation of sleep in Drosophila is complex and likely involves many sites and signaling molecules. As noted earlier, in addition to the MBs and the PI, the circadian large ventral lateral neurons have a role in the regulation of sleep (Parisky et al., 2008). How these different sites communicate with each other and are ordered in a sleep-regulating circuit will be an important question to address. We propose that the PI lies downstream of these other sites and integrates circadian and homeostatic signals to control sleep:wake. Consistent with this idea, the PI is known to be a major output of the fly brain (Toivonen and Partridge, 2009). In addition, we know that some projections of the central clock neurons are in the vicinity of the PI (Jaramillo et al., 2004) and that the PI contains wake and sleep regulating cells. We hypothesize that the decrease in sleep produced by the ablation of mushroom bodies (Joiner et al., 2006; Pitman et al., 2006) is due to the imbalance of inputs to the PI neurons, resulting in an increase of wake promoting signals over sleep promoting signals. This is supported by our data showing that flies lacking MBs are more sensitive to octopamine feeding (Crocker and Sehgal, 2008), implying that the MBs normally exert a moderating influence.
Experimental Procedures
Fly Stocks
The following lines were ordered from the Bloomington Stock Center: Tdc2–GAL4 (9313), UAS–NaChBac (9466), UAS–GFP syt(9313), UAS-mCD8::GFP (5137), UAS–GFPnls (7032), P{neoFRT}19A, P{tubP-GAL80}LL1, P{hsFLP}1, w[*]; Pin[Yt]/CyO (5133), y[1] w[1118] P{neoFRT}19A (1744), 18896 (OctB2R),and Iso31 (5905). MBGS, H24-GAL4, 17d-GAL4, c747-GAL4, c309-GAL4, 1366-GAL4, MJ63-GAL4, c507-GAL4, 104y-GAL4, D42-GAL4, mai301-GAL4, ElavGS, 30y-GAL4, Sep54-GAL4, 201y-GAL4, Dilp2-GAL4, UAS-CD8-GFP and UAS-BDK33(PkaR) were used previously in the laboratory (Crocker and Sehgal, 2008; Joiner et al., 2006; Zheng et al., 2007). OAMB 286, OAMB 584 and UAS-K3;OAMB286 were a gift from Dr. K. Han, 50y-GAL4 was a gift from Dr. R. Greenspan and Kurs58-GAL4 was a gift from Dr. U. Heberlein. UAS-Epac1-cAMP50 was a kind gift from Dr. P. Taghert.
Generating flies for MARCM
The MARCM method was used to generate flies in which different, random subsets of Tdc2-expressing neurons expressed the sodium channel, NachBac (Lee and Luo, 1999). The crosses that created the MARCM flies are shown in Supplemental Fig 1. The original lines are: P{neoFRT}19A, P{tubP-GAL80}LL1, P{hsFLP}1, w[*]; Pin[Yt]/CyO (5133)of the Bloomington stock Center (donated by L. Luo) UAS–NaChBac(9466) line of the Bloomington stock Center (donated by B. White) y[1] w[1118] P{neoFRT}19A (1744) line Bloomington Stock Center (donated by L. Luo) Tdc2–GAL4 (9313) line Bloomington Stock Center (donated by J. Hirsh) The final MARCM flies contain a Tdc2-Gal4, a UAS-NachBac (B16B) a tubulin-Gal80 and a heat shock promoter-FLP recombinase transgene. In the presence of the Gal80, Gal4 activity is suppressed and so NachBac is not expressed in any cell. However, the Gal80 can be randomly excised from subsets of cells by the heat shock activated FLP recombinase. Crosses were raised at 18°C and the parental generations were transferred to new vials following egg laying for the times indicated in Supplemental Fig. 1. To remove the GAL80, animals received a heat shock at 37°C for 20–30 minutes between 1 and 2 days of development.. P{neoFRT}19A, P{tubP-GAL80}LL1, P{hsFLP}1, w[*]/P{neoFRT}19A ; UAS-NaChBac-EGFP/TDC2-Gal4 female flies were selected for sleep and mapping analysis.
Sleep Analysis
Sleep analysis was performed as described previously (Gilestro and Cirelli, 2009; Joiner et al., 2006). All flies were kept on a 12 h light/dark (LD) cycle at 25°C schedule. Female MARCM flies, 6–8 d old, were placed in 65 × 5 mm tubes containing 5% sucrose and 2% agar and entrained for 24–36 h before the sleep recording. Baseline sleep was determined by monitoring activity for at least 3 d with no disruptions in an LD cycle. Locomotor activity was monitored using the DAMS/Trikinetics system as described previously (Gilestro and Cirelli, 2009; Joiner et al., 2006). Sleep was defined as a 5 min bout of inactivity as described previously (Shaw, 2003). Latency to sleep was determined for the octopamine receptor mutants and is defined as the time in minutes from the moment lights are turned off to the first bout of sleep.
Immunohistochemistry
Brains of adult female flies were dissected in cold PBS and fixed in 4% paraformaldehyde in PBS at 4°C overnight. After four 10-min washes in 0.1% Triton X-100 in PBS samples were permeabilized with 3% normal goat serum (NGS) 2 h at room temperature. Samples were then incubated in a primary antibody solution containing mouse anti-nc82 (1:50; Developmental Studies Hybridoma Bank) and rabbit anti-GFP IgG (1:1000; no. A11122, Invitrogen) in 3% NGS at 4°C overnight. After four 10-min washes in 0.1% Triton X-100 in PBS, brains were incubated at 4°C overnight in a secondary antibody solution containing Alexa Fluor 568 goat anti-mouse IgG (1:1000, Invitrogen) and Alexa Fluor 488 goat anti-rabbit IgG (1:1000, Invitrogen) in 3% NGS. After 4 10-minute washes in PBS brains were mounted using Vectashield mounting medium (Vector Labs) and covered with a no. 1 glass coverslip before imaging. Immunolabeled adult brains were imaged with a 488 and 510 laser-scanning confocal microscope (Leica) under 20x magnification. z stack images were scanned at 1micron section intervals with a resolution of 1024 × 1024 pixels.
Analysis of MARCM data
Following sleep analysis, and immunohistochemistry to detect GFP, individual brains were analyzed to correlate the sleep phenotype with the cellular expression of NachBac-GFP. All mapping analysis was performed blind to the sleep analysis. Each group contains flies with expression only in that subgroup, with the exception of the ASM flies and PSM flies. Due to the low numbers of flies that expressed NachBac-GFP in these regions alone we included flies that also expressed in other areas. We had four flies that had expression only in ASM neurons and an additional 8 flies which had expression in other areas in addition to ASM, bringing the total number to 12. These 8 flies were not counted in any other category. In the group showing expression in only a single ASM neuron, we had 3 flies with only one such neuron labeled and another 8 lines which had expression elsewhere, in addition to the single ASM neuron, to bring the number to 11. These lines were not counted in any other category. The number of flies indicated in the “no cells” category is probably an underestimate since we stopped counting these flies at some point. Over 1,000 flies were screened in this study, similar to the number Busch and colleagues screened to map octopamine neurons (Busch et al., 2009).
Mapping the site of PKA action
To map the site of PKA action, we crossed each of the following drivers- MBGS, H24-GAL4, 17d-GAL4, c747-GAL4, c309-GAL4, 1366-GAL4, MJ63-GAL4, c507-GAL4, 104y-GAL4, D42-GAL4, mai301-GAL4, 50y-GAL4, ElavGS, 30y-GAL4, Sep54-GAl4, 201y-GAL4, Dilp2-GAL4, Kurs58-GAl4- into UAS–BDK33 flies (Rodan et al., 2002). Female flies were used from these crosses for all sleep and octopamine analysis. Octopamine at 10 mg/ml as previously described was fed to each fly for 3 days following a 3 day baseline(Crocker and Sehgal, 2008). The GeneSwitch construct in elavGS and MBGS can be turned on during adulthood using the drug RU486 (11β-(4-dimethylamino)phenyl-17β-hydroxy-17-(1-propynyl)estra-4,9-dien-3-one). We placed the animals on 5% sucrose/2% agar tubes containing either 500 µm RU486 dissolved in ethanol or ethanol alone (1%) for 3 d. Half of each group was then transferred to octopamine-containing food with either 5% sucrose/2% agar plus 500 µm RU486 plus octopamine or 5% sucrose/2% agar plus ethanol (1%) plus octopamine. Both groups were also simultaneously fed 10 mg/ml octopamine for 3 d. At the lights-on transition at the end of this period, animals were transferred off of octopamine onto 5% sucrose/2% agar containing either 500 µm RU486 or ethanol (1%). Sleep analysis was performed as described above.
Electrophysiology
For in vivo patch recording from PI neurons, flies were anesthetized with CO2 and glued ventral side down to a glass cover slip. The cover slip was placed in a chamber containing extracellular solution (NaCl 101, KCl 3, MgCl2 4, CaCl2 1, NaH2PO4 1.25, NaHCO3 20.7, Glucose 5 mM with pH 7.2) and then the cuticle was peeled off using fine forceps to expose the surface of the brain. The chamber was placed on the stage of an Olympus BX51 fluorescent microscope, and PI neurons were identified by their location and bright green fluorescence. Patch recording electrodes (WPI, Inc) were fire-polished, and had resistances from 3 – 4 MΩ when filled with intracellular solution (K-gluconate 102, NaCl 17, CaCl2 0.085, Mg-ATP 4, Na-GTP 0.5, EGTA 0.94, HEPES 8.5 mM with pH 7.2). Standard technique was used to record macroscopic currents in the whole cell voltage clamp mode with an Axopatch 200A amplifier (Molecular Devices, Union City, CA). Data were digitized with a Digidata 1322A interface (Molecular Devices, Union City, CA) and stored on a PC hard drive for further analysis with pClamp9 software (Molecular Devices, Union City, CA).
FRET Imaging
The TDC2-Gal4 flies were crossed to flies carrying a UAS-Epac1-cAMPs(50A) transgene (Shafer et al., 2008). Brains were dissected under ice-cold calcium-free fly hemolymph-like saline (HL3) containing 70 mM NaCl, 5 mM KCl, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, and 5 mM HEPES (pH 7.1). The brains were then laid at the bottom of a 35 × 10 mm plastic FALCON Petri dish (Becton Dickenson Labware), given a few seconds to adhere and covered with 1.6 ml HL-3 containing 1.5 mM CaCl2 (Shafer et al., 2008).
Time course FRET imaging of Epac1-camps was performed on individual brains using the Leica TCS SP5 confocal microscope using a HCX APO L 40x/0.80 dipping objective. We performed octopamine experiments by administering 140ul of 125mg/ml of octopamine in dH2O into the dish making the final concentration of octopamine 10mg/ml octopamine. In the water control, 140 ul of water was added to the well. For the mianserin experiments, initially mianserin alone was added at a concentration of 0.25mM, followed by a second application of 0.25mM mianserin along with 140 ul of the 125 mg/ml stock of octopamine. Analysis of FRET was done using the Sensitized Emission FRET wizard built into the Leica Application Suite for the TCS SP5 using Method 1 for FRET analysis. Images were taken every 5 seconds at 512 × 512 and line averaged 3 times. A time series was taken for 5 minutes of baseline, followed by octopamine administration and 15 minutes post octopamine administration. For the mianserin experiments, the 5 minutes of baseline preceded the first application of mianserin alone; this was followed 1 minute later by mianserin plus octopamine. We then outlined the PI neurons and selected them as our region of interest for analysis. Results are plotted as the minute prior to octopamine administration followed by 240 seconds post octopamine administration. Longer periods of time did not change the FRET levels. 12 brains were plotted for octopamine administration, 8 for the water control and 9 brains for the mianserin treatment.
Receptor Characterization
RT-PCR was run according to previously published protocols.(Ousley et al., 1998) rtPCR primers: OAMB primers were previously described (Han papers); we also generated new primers for the K3 and AS isoforms. New primers are for K3: 5’-3’ forward CTGCCGTGAGAACGACGAG 5’-3’ reverse GCGCAATATGAGCTGGGACT. Primers for AS were: 5’-3’ Forward CTGCCGTGAGAACGACGAG 5’-3’ reverse ATGTATGCGCAATGTGAGGC OctB2R receptor primers were: 5’-3’Forward ATGCTGATGCACCGACCAT 5’-3’ reverse AAGGCAGCCAGCAGAGGAT
Single cell rt-PCR
Whole cell extracts were taken from Dilp2 neurons following electrical recording. 10 cells were pooled and then rt-pcr was run on them with the following modifications: 10ul of buffer (150 mM sodium acetate, 50 mM Tris, pH 9.0, 5 mM EDTA, pH 8.0, 1% SDS containing 1/100th volume diethyl pyrocarbonate) was used to place each recording pipette tip into, than those were pooled to equal 100ul. Following PCR with K3 and AS primers, the K3 band was cut out of the gel and sent for sequencing to verify K3 band. We did not see an AS band so that was not sequenced.
Supplementary Material
Acknowledgements
This work was partially supported by a program project grant from the National Institutes on Aging. A.C. was supported by a training grant to the Center for Sleep and Respiratory Neurobiology at the University of Pennsylvania and by a National Research Service Award from the National Institute of Mental Health. M.S. is supported from a grant to I.B.L. from the National Institutes of Health. We thank Drs. Kyung-An Han, Ralph Greenspan, Ulrike Heberlein, Ben White, Bill Joiner and Paul Taghert for providing flies used in this study. We thank Eric Rulifson for the Dilp2 antibody. We also thank K. Luu, Z. Yue and A. Joseph for assisting with animal maintenance. We also would like to thank Andrea Stout for comments and advice on confocal and FRET experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Busch S, Selcho M, Ito K, Tanimoto H. A map of octopaminergic neurons in the Drosophila brain. J Comp Neurol. 2009;513:643–667. doi: 10.1002/cne.21966. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19:386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SK, Reinhart PH, Martin BL, Brautigan D, Levitan IB. Protein kinase activity closely associated with a reconstituted calcium-activated potassium channel. Science. 1991;253:560–562. doi: 10.1126/science.1857986. [DOI] [PubMed] [Google Scholar]
- Cole SH, Carney GE, McClung CA, Willard SS, Taylor BJ, Hirsh J. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J Biol Chem. 2005;280:14948–14955. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePeyer JE, Cachelin AB, Levitan IB, Reuter H. Ca2+-activated K+ conductance in internally perfused snail neurons is enhanced by protein phosphorylation. Proc Natl Acad Sci U S A. 1982;79:4207–4211. doi: 10.1073/pnas.79.13.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Robb S. Octopamine receptor subtypes and their modes of action. Neurochem Res. 1993;18:869–874. doi: 10.1007/BF00998270. [DOI] [PubMed] [Google Scholar]
- Ewald D, Williams A, Levitan IB. Modulation of single Ca2+-dependent K+ channel activity by protein phosphorylation. Nature. 1985;315:503–506. doi: 10.1038/315503a0. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Gilestro GF, Cirelli C. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics. 2009;25:1466–1467. doi: 10.1093/bioinformatics/btp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;90:1152–1159. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- Han KA, Millar NS, Davis RL. A novel octopamine receptor with preferential expression in Drosophila mushroom bodies. J Neurosci. 1998;18:3650–3658. doi: 10.1523/JNEUROSCI.18-10-03650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Sehgal A. Why a fly? Using Drosophila to understand the genetics of circadian rhythms and sleep. Sleep. 2004;27:334–342. doi: 10.1093/sleep/27.2.334. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Sehgal A, Pack AI. The need for a simple animal model to understand sleep. Prog Neurobiol. 2000;61:339–351. doi: 10.1016/s0301-0082(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Insel PA. Seminars in medicine of the Beth Israel Hospital, Boston. Adrenergic receptors--evolving concepts and clinical implications. N Engl J Med. 1996;334:580–585. doi: 10.1056/NEJM199602293340907. [DOI] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 2004;5:3. doi: 10.1186/1471-2202-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Knutson KL, Van Cauter E. Associations between sleep loss and increased risk of obesity and diabetes. Ann N Y Acad Sci. 2008;1129:287–304. doi: 10.1196/annals.1417.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryger MH, Roth T, Dement WC. Principles and practice of sleep medicine. 4th edn. Philadelphia, PA: Elsevier/Saunders; 2005. [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Rohila S, Han KA. The octopamine receptor OAMB mediates ovulation via Ca2+/calmodulin-dependent protein kinase II in the Drosophila oviduct epithelium. PLoS One. 2009;4:e4716. doi: 10.1371/journal.pone.0004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seong CS, Kim YC, Davis RL, Han KA. Octopamine receptor OAMB is required for ovulation in Drosophila melanogaster. Dev Biol. 2003;264:179–190. doi: 10.1016/j.ydbio.2003.07.018. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick BN, Alam MN. Different types of norepinephrinergic receptors are involved in preoptic area mediated independent modulation of sleep-wakefulness and body temperature. Brain Res. 1992;591:8–19. doi: 10.1016/0006-8993(92)90972-c. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousley A, Zafarullah K, Chen Y, Emerson M, Hickman L, Sehgal A. Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics. 1998;148:815–825. doi: 10.1093/genetics/148.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annu Rev Entomol. 2005;50:447–477. doi: 10.1146/annurev.ento.50.071803.130404. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Sara SJ. The locus coeruleus and noradrenergic modulation of cognition. Nat Rev Neurosci. 2009;10:211–223. doi: 10.1038/nrn2573. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidullah MSR, Fei H, Levitan IB. In Vivo Role of a Potassium Channel-binding Protein in Regulating Neuronal Excitability and Behavior. Journal of Neuroscience. 2009 doi: 10.1523/JNEUROSCI.3024-09.2009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao LR, Halvorsrud R, Borg-Graham L, Storm JF. The role of BK-type Ca2+-dependent K+ channels in spike broadening during repetitive firing in rat hippocampal pyramidal cells. J Physiol 521 Pt. 1999;1:135–146. doi: 10.1111/j.1469-7793.1999.00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. Awakening to the behavioral analysis of sleep in Drosophila. J Biol Rhythms. 2003;18:4–11. doi: 10.1177/0748730402239672. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KZ, Lagrutta A, Davies NW, Standen NB, Adelman JP, North RA. Tetraethylammonium block of Slowpoke calcium-activated potassium channels expressed in Xenopus oocytes: evidence for tetrameric channel formation. Pflugers Arch. 1994;426:440–445. doi: 10.1007/BF00388308. [DOI] [PubMed] [Google Scholar]
- Sinakevitch I, Strausfeld NJ. Comparison of octopamine-like immunoreactivity in the brains of the fruit fly and blow fly. J Comp Neurol. 2006;494:460–475. doi: 10.1002/cne.20799. [DOI] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009;5:253–261. doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Tobler I. In: Phylogeny of sleep regulation. In Principles and practice of sleep medicine. Kryger MH, Roth T, Dement WC, editors. Philadelphia: Elsevier/Saunders; 2005. pp. 77–90. [Google Scholar]
- Toivonen JM, Partridge L. Endocrine regulation of aging and reproduction in Drosophila. Mol Cell Endocrinol. 2009;299:39–50. doi: 10.1016/j.mce.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Trenell MI, Marshall NS, Rogers NL. Sleep and metabolic control: waking to a problem? Clin Exp Pharmacol Physiol. 2007;34:1–9. doi: 10.1111/j.1440-1681.2007.04541.x. [DOI] [PubMed] [Google Scholar]
- White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, Keshishian H. Targeted attenuation of electrical activity in Drosophila using a genetically modified K(+) channel. Neuron. 2001;31:699–711. doi: 10.1016/s0896-6273(01)00415-9. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang J, Wen H, Kucherovsky O, Levitan IB. Modulation of Drosophila slowpoke calcium-dependent potassium channel activity by bound protein kinase a catalytic subunit. J Neurosci. 2002;22:3855–3863. doi: 10.1523/JNEUROSCI.22-10-03855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.