Abstract
Although it is well established that the circadian clock regulates mammalian reproductive physiology, the molecular mechanisms by which this regulation occurs are not clear. The authors investigated the reproductive capacity of mice lacking Bmal1 (Arntl, Mop3), one of the central circadian clock genes. They found that both male and female Bmal1 knockout (KO) mice are infertile. Gross and microscopic inspection of the reproductive anatomy of both sexes suggested deficiencies in steroidogenesis. Male Bmal1 KO mice had low testosterone and high luteinizing hormone serum concentrations, suggesting a defect in testicular Leydig cells. Importantly, Leydig cells rhythmically express BMAL1 protein, suggesting peripheral control of testosterone production by this clock protein. Expression of steroidogenic genes was reduced in testes and other steroidogenic tissues of Bmal1 KO mice. In particular, expression of the steroidogenic acute regulatory protein (StAR) gene and protein, which regulates the rate-limiting step of steroidogenesis, was decreased in testes from Bmal1 KO mice. A direct effect of BMAL1 on StAR expression in Leydig cells was indicated by in vitro experiments showing enhancement of StAR transcription by BMAL1. Other hormonal defects in male Bmal1 KO mice suggest that BMAL1 also has functions in reproductive physiology outside of the testis. These results enhance understanding of how the circadian clock regulates reproduction.
Keywords: circadian rhythms, fertility, testosterone, testes, sperm, StAR, mice
Disruption of circadian rhythms results in a variety of pathophysiologic states (Hastings et al., 2003). Reproductive physiology, in particular, is profoundly influenced by circadian rhythms (Boden and Kennaway, 2006). In various insect species, the circadian clock is necessary for proper ovulation, sperm production, and fertility (Giebultowicz et al., 1989; Beaver et al., 2002; Beaver et al., 2003; Beaver and Giebultowicz, 2004). In mammals, evidence indicates that the circadian clock regulates the serum concentrations of many reproductive hormones (Lucas and Eleftheriou, 1980; Clair et al., 1985; Chappell et al., 2003; Miller et al., 2004). For example, the surge of luteinizing hormone (LH) necessary for ovulation in rodents, which occurs at the same time of day during each estrous cycle, requires a functional circadian clock (Barbacka-Surowiak et al., 2003). In addition, at the onset of puberty, a clear diurnal rhythm of gonadotropin serum levels is established in both mice and humans (Andrews and Ojeda, 1981; Jean-Faucher et al., 1986; Dunkel et al., 1992; Apter et al., 1993). It is unclear whether this diurnal rhythm continues into adulthood, but testosterone serum concentration shows daily oscillations in adult male mice and humans (Lucas and Eleftheriou, 1980; Clair et al., 1985). Although the association between circadian rhythms and testosterone is a long-established phenomenon, the molecular mechanisms by which the circadian clock regulates testosterone production are unknown.
The circadian clock is based on a transcription translation feedback loop that results in the cyclic expression of genes and proteins over a 24-h period. At the core of the loop are 2 transcription factors, CLOCK and BMAL1 (ARNTL, MOP3), which bind to DNA as a heterodimer to activate transcription of 2 other circadian clock genes, called Period (Per) and Cryptochrome (Cry). At high concentrations, the PER and CRY proteins repress their own transcription by inhibiting CLOCK and BMAL1, resulting in decreased protein production. As the protein concentration falls, the repression is relieved, and the cycle begins anew. Mammals have 3 homologs of the Period gene—Per1, Per2, and Per3—and 2 homologs of the Cryptochrome gene, Cry1 and Cry2.
Of the circadian clock genes that make up the core oscillator, Bmal1 is indispensable. In contrast to other circadian clock gene knockout (KO) mice, mice without Bmal1 (Bmal1 KO) completely lack circadian rhythms (Bunger et al., 2000). This effect may be due to a lack of redundancy in gene function. Unlike other circadian clock gene knockout mice, Bmal1 KO mice also have a discernible phenotype. These mice develop signs of early aging, including a progressive arthropathy (Bunger et al., 2005; Kondratov et al., 2006). In addition, it has been suggested that homozygous Bmal1 KO mice are infertile, but no data have been published (Boden and Kennaway, 2006; Kondratov et al., 2006). Therefore, we examined the function of Bmal1 in mouse reproduction.
MATERIALS AND METHODS
Experimental Animals
Bmal1 KO mice were established in our mouse colony by mating heterozygous Bmal1 KO mice to C57BL6/J mice (Jackson Laboratories, Bar Harbor, ME). These mice were backcrossed at least 5 generations onto a C57BL6/J background prior to use. Homozygote animals were produced by breeding heterozygous pairs. Per2 KO mice were purchased from Jackson Laboratories. Colonies of Clock mutant mice, Per1 KO mice, and Per2 KO mice were established on a C57BL6/J background (12 generations). A colony of Per1 KO/Per2 KO double-knockout animals was established by breeding Per1 KO and Per2 KO mice. All mice were group housed in a 12L:12D light-dark cycle (lights on at 0600 h; lights off at 1800 h), except those used for hormone measurements. These mice were individually housed under the same 12L:12D light-dark cycle for 2 weeks prior to sacrifice and serum collection. Food and water were supplied ad libitum. For all experiments, mice were sacrificed by asphyxiation with CO2 gas. Care was in accordance with Institutional Animal Care and Use Committee guidelines at the University of Pennsylvania and at Oregon State University.
Histology and Immunohistochemistry
For light microscopy, testis sections were prepared as described (Alvarez et al., 2003) and stained with hematoxylin and eosin. For immunohistochemistry, C57BL6 mice (10-15 weeks old) were anesthetized by intraperitoneal injection of sodium pentobarbital (6.5 mg per 100 g of body weight) and perfused through the right ventricle with 100 mL of heparin-supplemented phosphate-buffered saline (PBS; 100 USP units/10 mL) followed by 150 mL of Bouin’s fixative. Testes were dissected, postfixed overnight at 4 °C in Bouin’s, washed in 70% ethanol for 24 h, dehydrated, and embedded in paraffin. Sections (10 μm) were spread on gelatin-coated glass slides, deparaffinized, hydrated, washed with PBS, and blocked for 1 h at room temperature (RT) in PBS containing 0.1% bovine serum albumin (BSA), 5% normal goat serum (NGS), and 0.3% Triton X-100 (TX-100). Sections were incubated in chicken antihuman MOP3 polyclonal antibody (Alpha Diagnostics, Inc., San Antonio, TX) diluted 1:1000 in PBS supplemented with 0.1% BSA and 0.03% TX-100 (PBST). For control sections, antibody was preadsorbed overnight at 4 °C with 50 μg/μL of antigenic peptide (Alpha Diagnostics, Inc.). Sections were then washed in PBST, incubated for 1 h at RT in AlexaFluor 488 goat antichicken IgG (Molecular Probes, Carlsbad, CA) diluted 1:1000 in PBST supplemented with 3% NGS, and rewashed with PBST; nuclei were stained with Hoechst 33258 diluted 1:10,000 in PBS for 5 min and mounted in Fluoromount G (Southern Biotech Associates, Birmingham, AL). Analysis used a Leica DMBR fluorescent microscope and a SPOT digital camera and ImageJ software (Diagnostic Instruments, Waltham, MA). The relative scale for densitometric analysis was from 0 for black to 255 for maximum green opacity. Measurements of the average density were taken from a circular area of the same size on photomicrographs of the Leydig cells. Five measurements were taken separately for cytoplasm and nuclei from different cells on 1 section. Four sections for each of the 5 animals were analyzed per time point.
Capacitation Analysis
Analysis was performed essentially as described previously (Travis et al., 2001). Briefly, sperm were collected from cauda epididymides of Bmal1 KO, Bmal1 heterozygous, and wild-type littermates and placed into capacitating or noncapacitating media for 3 h. Protein extract was collected and separated by electrophoresis on 10% sodium dodecyl sulfate (SDS)–polyacrylamide gels, transferred to nylon membranes, and probed with antiphosphotyrosine antibodies (α-PY, clone 4G10, Upstate Biotechnology, Lake Placid, NY) at a dilution of 1:10,000.
In Vitro Fertilization
Six-week-old CF-1 female mice (Harlan, Indianapolis, IN) were superovulated by intraperitoneal injection with 5 IU equine chorionic gonadotropin (eCG) followed 48 h later by 5 IU human chorionic gonadotropin (hCG). Cumulus-intact eggs were released from the oviducts 14 h after hCG injection and transferred to 50-μL drops of TYH medium under light mineral oil (Toyoda et al., 1971). Sperm were obtained from the cauda epididymides of 12- to 16-week-old Bmal1 KO, Bmal1 heterozygous, or wild-type littermates and capacitated for 3 h in 400-μL drops of TYH medium under light mineral oil. Sperm (5 × 105) were added to the 50-μL fertilization drop, and fertilization was allowed to proceed for 3 h. The eggs were removed, washed free of unbound sperm, and incubated at 37 °C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2. The inseminated eggs were examined for progression to the 2-cell stage, which was considered successful fertilization. Five separate experiments were performed. The reported data are an average of the 5 experiments with standard error of the mean.
Hormone Measurements
Mice were sacrificed at ZT3. Immediately upon death, the chest cavity was opened and blood was collected by cardiac puncture, transferred to 1.5-mL microfuge tubes, placed on ice for 1 h, and then stored at 4 °C for 6 to 8 h to allow clotting. The serum was separated from the blood clot by centrifugation at 4 °C for 15 min at 5000 rpm, collected into a fresh 1.5-mL microfuge tube, and stored at −70 °C. Hormone measurements were carried out by the National Hormone and Peptide Program (Harbor-UCLA Medical Center).
RNA Preparation and Quantitative Polymerase Chain Reaction
RNA was collected and prepared, and quantitative polymerase chain reaction (qPCR) was performed as described (Alvarez and Sehgal, 2005). Taqman assays for all genes examined were purchased from Applied Biosystems (Foster City, CA).
Immunoblot Analysis
Testes were collected, decapsulated, and homogenized in a buffer containing 50 mM Tris-HCl (pH 8.0), 250 mM NaCL, 2 mM EDTA, 1% NP-40, complete protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), 50 mM NaF, and 1 mM Na vanadate. Cells were lysed on ice for 1 h. Protein concentration was determined by the Dc Protein Assay (Bio-Rad, Hercules, CA). Then, 25 μg of protein extract was run on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon, Millipore Corporation, Billerica, MA) by electroblotting overnight. The membrane was washed in 1× PBS/0.3% Tween-20, blocked with buffer containing 1% BSA and 5% milk, and incubated with an anti–steroidogenic acute regulatory protein (StAR) antibody at a concentration of 1:200 (Devoto et al., 2001) for 1.5 h at room temperature. The blot was then washed with buffer, and a secondary antibody of horseradish peroxidase-conjugated donkey antirabbit IgG (Amersham Biosciences, Piscataway, NJ) at 1:1000 diluted in 1× PBS/0.3% Tween-20/1% BSA/5% milk was incubated for 1.5 h at room temperature. The blot was washed and visualized using the ECL reagent (Amersham) according to the manufacturer’s instructions.
Cell Culture, Transient Transfection, and Luciferase Assays
The 2.5-kb StAR promoter luciferase reporter construct was constructed using a construct containing 3.6 kb of murine StAR promoter (pBS-StAR GH; Dr. Barbara Clark, University of Louisville). A 2.5-kb fragment of promoter was isolated by digestion with Xba I, Hind III, and Sca I. This was ligated into a pGL2 luciferase reporter vector (Promega, Madison, WI) digested with Nhe I and Sma I. The resultant construct was labeled pGL2-StAR2.5Bl-luc.
MA-10 cells were cultured in Waymouth’s MB752 medium containing 15% horse serum, 20 mM HEPES, and 50 μg/mL gentamicin at 37 °C under 5% CO2. All culture reagents were purchased from Invitrogen (Carlsbad, CA). The night before transfection, cells were split into 12-well cell culture plates at a concentration of 2.5 × 105 cells per well. At 3 h prior to transfection, media were replaced with 1 mL of fresh media. Transfection was carried out in individual wells according to the manufacturer’s instructions with 1.5 μL of FUGENE 6 transfection reagent (Roche Diagnostics) diluted into 50 μL of OptiMEM media (Invitrogen) containing the appropriate amounts of DNA constructs. DNA constructs used (with amounts per well) were as follows: pGL2-StAR2.5Bl-luc (500 ng), pGL2-StAR0.9 (500 ng; generously provided by Dr. Barbara Clark; Caron et al., 1997), pGL2 (500 ng; Promega), pRL-TK (20 ng; Promega), Bmal1 expression construct in pcDNA3.1 (100 ng; generously provided by Dr. Garrett FitzGerald), pcDNA3.1 (100 ng; Invitrogen). An additional 1 mL of media was added 12 h after transfection. At 48 h following transfection, luciferase activity was assayed using the Dual Glo luciferase assay (Promega) according to the manufacturer’s instructions.
RESULTS
Bmal1 KO mice Are Infertile
Bmal1 KO mice have multiple anatomic abnormalities that develop with age (becoming microscopically apparent at approximately 12 weeks of age). These include hind limb cartilage ossification, sarcopenia, cataract formation, and a reduction in subcutaneous body fat (Bunger et al., 2005; Kondratov et al., 2006). However, we found that homozygous Bmal1 KO mice were unable to breed with each other even at young, as yet unaffected, ages. We directly tested whether homozygous Bmal1 KO mice were fertile by mating to wild-type mice. Ten cages containing a male Bmal1 KO mouse (aged between 6 and 10 weeks) and 2 wild-type female mice were established. Similarly, 10 cages containing a single Bmal1 KO female mouse (aged between 6 and 10 weeks) and a wild-type male mouse were established. Over a 10-week period of mating, no litters were produced in any of the cages. However, after 10 weeks, the wild-type mice were mated to other wild-type mice, and normal-sized litters were produced, indicating that homozygous male and female Bmal1 KO mice are infertile. Notably, we never observed successful fertility in either male or female Bmal1 KO mice even up to 6 months of age.
Bmal1 KO Mice Have Low Sperm Counts
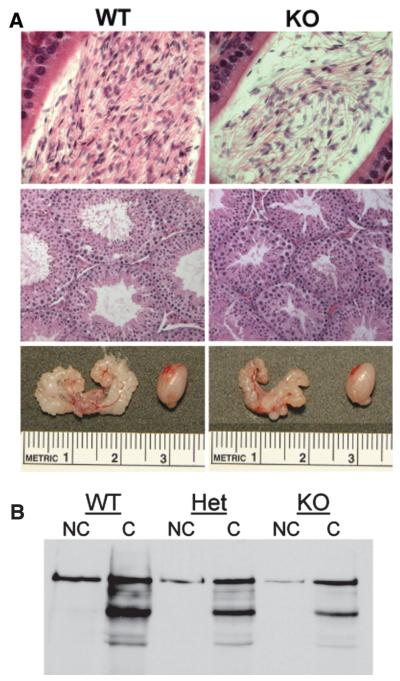
Gross dissection of male and female Bmal1 KO mice revealed that the reproductive anatomy was normal (data not shown). However, although body weight of male Bmal1 KO mice did not differ from wild-type mice between the ages of 10 and 14 weeks, the testes were slightly smaller and the seminal vesicles were markedly shrunken (Table 1 and Fig. 1). Microscopically, testes from Bmal1 KO mice were normal except that the average seminiferous tubule diameter was reduced by approximately 20% (Table 1 and Fig. 1A). Strikingly, the sperm counts of male Bmal1 KO mice were reduced by approximately 70% (Fig. 1A and Table 1).
Table 1.
Reproductive measurements.
| Genotype | Body Weight, g | Testes Weight, mg | Sperm Count, × 106 | Seminiferous Tubule, μm |
Seminal Vesicle, mg |
|---|---|---|---|---|---|
| Wild type | 24.4 ± 0.29 (31) | 181.9 ± 5.3 (36) | 11.2 ± 0.95 (31) | 179.9 ± 18.2 (11) | 179.0 ± 6.8 (34) |
| Heterozygous | 25.2 ± 0.28 (50) | 175.4 ± 4.7 (50) | 9.37 ± 0.69 (50) | ND | 171.4 ± 4.8 (50) |
| Homozygous | 23.7 ± 0.34 (48) | 142.9 ± 4.6 (47)** | 3.86 ± 0.35 (43)** | 146.7 ± 13.0 (11)* | 102.7 ± 4.6 (48)** |
Error represents standard error of the mean, except for seminiferous tubule diameter, where error represents the standard deviation.
Number is in parentheses. ND, not determined.
p = 0.0001 by Student t test.
p < 0.0001 by Student t test.
Figure 1.
Bmal1 knockout (KO) male mice have reproductive deficiencies but functional sperm. (A) Photos are of organs collected from 12-week-old littermates. Genotypes are indicated. Top panel: Sections of caput epididymi showing fewer sperm in the Bmal1 KO animal in comparison to its wild-type littermate. Exact counts are provided in Table 1. Magnification is ×40. Middle panel: Sections of testes showing smaller seminiferous tubules in the Bmal1 KO mice in comparison to the wild-type littermate. Magnification is ×20. Bottom panel: Photos comparing the sizes of the seminal vesicles and a testis from wild-type and Bmal1 KO littermates. (B) There is no difference in capacitation of sperm from Bmal1 KO, heterozygous, and wild-type mice. Sperm were collected from the caudal epididymi of 12-week-old littermates and subjected to capacitation analysis by detection of proteins containing phosphotyrosine. Fewer sperm were collected from Het and KO mice (due to the lower sperm counts in these animals), resulting in lighter bands. However, the pattern of phosphorylated proteins is the same.
Although a low sperm count can cause infertility in mice, this is generally observed only in mice with a greater than 90% reduction in sperm compared to wild type, which is a far greater reduction than observed in Bmal1 KO homozygotes (Cooke and Saunders, 2002; Matzuk and Lamb, 2002). To investigate the possibility of a functional defect in the sperm of Bmal1 KO mice, we isolated sperm and analyzed motility. Visually, the sperm were motile with no apparent difference between wild-type and Bmal1 KO sperm. In addition, computer-assisted semen analysis (CASA) indicated that there were no differences in sperm motility between wild-type and Bmal1 KO mice (data not shown). We also assessed whether these sperm were able to undergo capacitation, an obligatory process that precedes fertilization, as measured by the appearance of proteins that are phosphorylated on tyrosines (Visconti and Kopf, 1998). There was no difference in the appearance of tyrosine-phosphorylated proteins when wild-type and Bmal1 KO sperm were incubated under capacitation conditions (Fig. 1B). To determine if sperm from Bmal1 KO mice were fully functional, we performed in vitro fertilization. Although the percentage of eggs fertilized by the Bmal1 KO sperm was less than that of wild-type littermates (52.6% ± 14.8% vs. 34.0% ± 11.1%), the difference was not significant (paired Student t test).
Gonadotropin Levels Are Altered in Bmal1 KO Mice
The low sperm counts and shrunken seminal vesicles observed in male Bmal1 KO mice are indicative of low serum testosterone concentration (Bartke, 1974). Indeed, testosterone concentrations in male Bmal1 KO mice were reduced approximately 70% in comparison to wild-type and heterozygous littermate controls (Table 2). Although testosterone serum concentration varies over the course of the day, we assayed sera collected in the morning, which is the time when it should be highest (Lucas and Eleftheriou, 1980).
Table 2.
Hormone concentrations.
| Genotype | Testosterone, ng/mL | Luteinizing Hormone, ng/mL |
Follicle-Stimulating Hormone, ng/mL |
|---|---|---|---|
| Wild type and heterozygous (n = 14) | 3.12 ± 0.93 | 9.6 ± 1.5 | 10.6 ± 4.02 |
| Homozygous (n = 10) | 1.00 ± 0.47* | 4.2 ± 0.6** | 35.9 ± 3.85** |
Error represents standard error of the mean.
p = 0.015 by Wilcoxon rank sum test.
p ≤ 0.001 by Wilcoxon rank sum test.
Testosterone is produced by Leydig cells in the testis under the influence of LH from the pituitary, which in turn is negatively regulated by testosterone in a feedback loop. To determine if the low testosterone concentration in Bmal1 KO mice is caused by low levels of LH, we measured the concentration of LH in the serum of these mice. LH serum levels in Bmal1 KO mice were actually 3-fold higher than those of wild-type and heterozygous controls (Table 2), indicating that a defect in LH production does not underlie the low concentration of testosterone and rather suggests a defect in the ability of Leydig cells to produce testosterone. We also measured the serum concentration of follicle-stimulating hormone (FSH), which is produced by the pituitary and is necessary for high-quantity spermatogenesis (Kumar et al., 1997). FSH serum concentration was reduced in Bmal1 KO mice as compared with wild-type and heterozygous littermates, suggesting that BMAL1 has multiple functions within the hypothalamus-pituitary-testis (HPT) axis (Table 2).
Steroidogenic Gene Expression Is Reduced in Bmal1 KO Mice
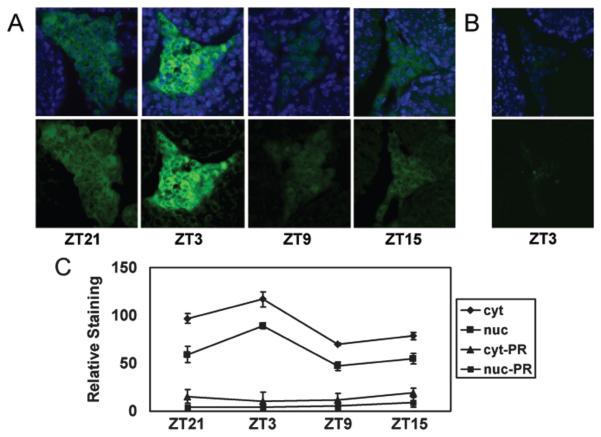
Our data suggest that the diminished testosterone concentration in Bmal1 KO mice is due, at least in part, to a failure by Leydig cells to produce testosterone. To examine whether this is possibly a cell autonomous effect of BMAL1 in Leydig cells, we assessed expression of BMAL1 in the testis by immunohistochemistry. BMAL1 was cyclically expressed in Leydig cells, with strongest expression being noted at ZT3 (3 h after lights-on; Fig. 2). We also observed minimal expression in seminiferous tubules, although it is currently unclear which specific cells in the tubules express BMAL1. These data strongly suggest that BMAL1 functions within Leydig cells facilitate testosterone production.
Figure 2.
BMAL1 expression oscillates in Leydig cells. (A) Immunohistochemical analysis of sectioned testes detected BMAL1 almost exclusively in the Leydig cells. Top panel: Merged images of BMAL1 (green) and nuclear dye Hoechst 33258 (blue). Bottom panel: Staining for BMAL1 only. BMAL1 staining intensity oscillates over the course of day, with a peak at ZT3 and a trough at ZT9. Images shown are representative of 5 testes processed at each time point. (B) Preadsorption of antibody with BMAL1 peptide dramatically reduces staining. Magnification in all pictures is ×40. (C) BMAL1 oscillates in both cytoplasm (cyt) and nuclei (nuc), as measured by densitometry. Data represent the average (± SEM) level of staining in 20 sections derived from 5 animals per time point. The vertical axis represents relative staining intensity, where 0 represents no staining (black) and 255 represents maximum saturated staining (green) assigned by ImageJ software. Significant differences (p < 0.05) in BMAL1 signal in both cytoplasmic and nuclear staining were observed between all time points, except between ZT21 and ZT15 (paired Student t test). Minimal staining using antibody preadsorbed with peptide was observed in cytoplasm (cyt-PR) and nuclei (nuc-PR).
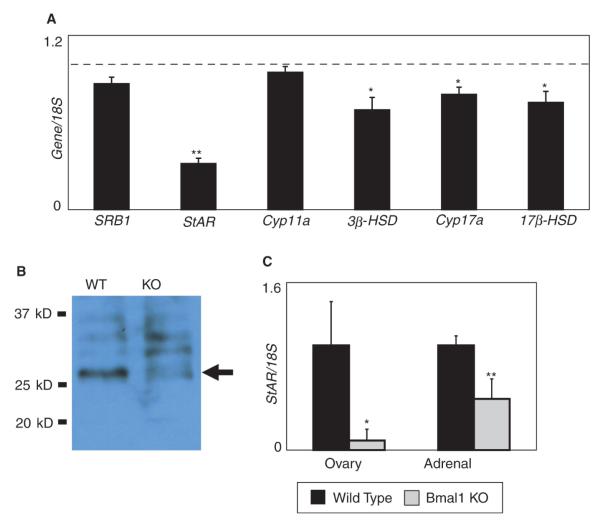
Given its function as a transcription factor, we hypothesized that BMAL1 is necessary for proper expression of steroidogenic genes involved in testosterone production. We compared the expression of the following genes in testes from Bmal1 KO mice and from wild-type littermates: scavenger receptor class B-1 (SRB1), StAR, p450 side chain cleavage enzyme (Cyp11a), 17α-hydroxylase (Cyp17a), 3β-hydroxysteroid dehydrogenase (3β-HSD), and 17β-hydroxysteroid dehydrogenase (17β-HSD). Expression of most of these genes was reduced in the testes of Bmal1 KO mice (Fig. 3A). Notably, StAR expression was affected the greatest, being reduced to approximately 30% of the wild-type value (Fig. 3A). StAR regulates the transport of cholesterol from the outer leaflet to the inner leaflet of the mitochondrial membrane, which is the rate-limiting step of steroidogenesis (Privalle et al., 1983; Stocco and Clark, 1996). StAR protein expression in the testis was also dramatically reduced in Bmal1 KO testis as compared with wild-type littermates (Fig. 3B). In addition, StAR expression in other steroidogenic tissues (adrenal and ovary) was low in Bmal1 KO mice, indicating that the effect of Bmal1 absence on StAR expression is not restricted to the testis (Fig. 3C).
Figure 3.
Steroidogenic acute regulatory protein (StAR) expression is reduced in the testis of Bmal1 knockout (KO) mice. (A) Steroidogenic gene expression is reduced in the testis of Bmal1 KO mice. Testis RNA from Bmal1 KO mice and wild-type littermates (8 pairs) was analyzed by quantitative polymerase chain reaction (qPCR). Expression is shown relative to wild-type expression (dashed line). Error bars represent SEM. A single asterisk represents p < 0.05, and a double asterisk represents p < 0.005 (paired Student t test). (B) StAR protein expression is reduced in the testes of Bmal1 KO mice. The arrow indicates the StAR protein. Upper bands represent preprocessed forms of the protein. Data shown are representative of experiments repeated on a total of 3 littermate pairs. (C) StAR expression is reduced in both adrenal glands and in ovaries of Bmal1 KO mice. Adrenal glands were collected from male Bmal1 KO mice and wild-type littermates (2 pairs), and adrenal glands and ovaries were collected from female Bmal1 KO mice and wild-type littermates (3 pairs). Error bars represent standard deviation. Data were normalized such that wild-type expression was set at 1. Data are significant to p = 0.02 (ovary) and <0.001 (adrenal).
BMAL1 Directly Regulates StAR Expression
Low StAR expression in the testis of Bmal1 KO mice may result from a direct effect of BMAL1 on the StAR promoter, from an indirect effect of BMAL1 that stimulates StAR expression, or from a combination of these 2 possibilities. StAR expression is induced by LH signaling through the LH receptor (Clark et al., 1994). Expression of the LH receptor gene (Lhcgr) was decreased in the testis of Bmal1 KO mice; however, we do not believe that signaling by the LH receptor is completely impaired, if at all (Fig. 4A). LH receptor knockout mice have dramatically reduced expression of both StAR and Cyp11a (Zhang et al., 2001). In contrast, expression of Cyp11a was not affected in Bmal1 KO mice (Fig. 4A).
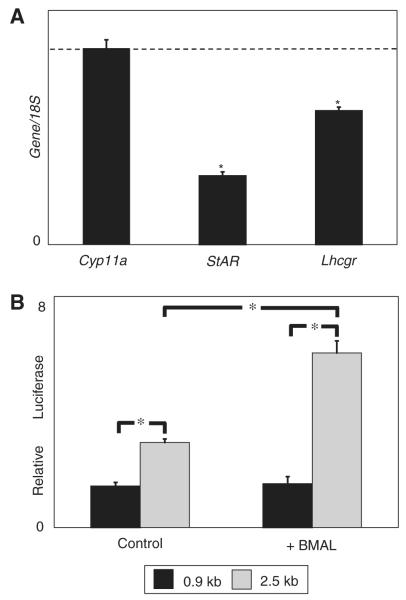
Figure 4.
BMAL1 directly enhances steroidogenic acute regulatory protein (StAR) expression. (A) Reduced expression of the luteinizing hormone (LH) receptor does not account for the effect on StAR expression observed in Bmal1 knockout (KO) mice. Testis RNA was collected from 5 pairs of Bmal1 KO mice and wild-type littermates and analyzed by quantitative polymerase chain reaction (qPCR). Expression is shown relative to wild-type expression level (dashed line). Error bars represent SEM. Asterisk represents a significant reduction (p < 0.05; paired Student t test). (B) BMAL1 enhances StAR expression in vitro. Expression constructs containing either 966 bp or 2.5 kb of the StAR promoter were transfected into the MA-10 Leydig tumor cell line with either a BMAL1 expression construct or an empty expression construct (pcDNA3.1). After 48 h, the cells were collected and luciferase activity was measured. Luciferase activity is higher with the 2.5-kb promoter sequence and is increased in the presence of BMAL1. Activity from the 966-bp promoter is unaffected by BMAL1. Shown is a single representative experiment with data points collected in triplicate. Differences are significant with p < 0.001 (unpaired Student t test). This experiment has been repeated once with similar results.
We investigated whether BMAL1 can directly activate StAR expression. StAR expression in vitro requires only the first 966 bp of DNA sequence upstream of the transcription start site (Caron et al., 1997). Although a number of transcription factors bind to this region, there are no canonical BMAL1 binding sites, called E-boxes (CACGTG), in this region (Manna et al., 2003). However, inspection of DNA sequence upstream of this region revealed the presence of a noncanonical E-box (CACGTT) at −1.5 kb and 2 E-boxes at −2.0 kb relative to the transcription start site. These sites are conserved among mouse, rat, and human StAR promoters, although only 1 E-box is present at −2.0 kb in the human promoter. To test whether BMAL1 directly affects StAR transcription, we used constructs containing either 966 bp or 2.5 kb of the murine StAR promoter ligated to a luciferase reporter gene. We transfected these constructs into the MA-10 Leydig cell line (Ascoli, 1981). MA-10 cells express Bmal1 at a low but detectable level (data not shown). The 2.5-kb promoter construct yielded higher baseline StAR expression than the 966-bp promoter construct (Fig. 4B). When cotransfected with a BMAL1 expression vector, we observed a marked increase in StAR expression from the 2.5-kb luciferase construct but no increase from the 966-bp promoter construct (Fig. 4B). These results indicate that BMAL1 directly increases StAR expression and suggest that the upstream E-boxes are responsible.
Other Circadian Clock Genes Also Affect StAR Expression
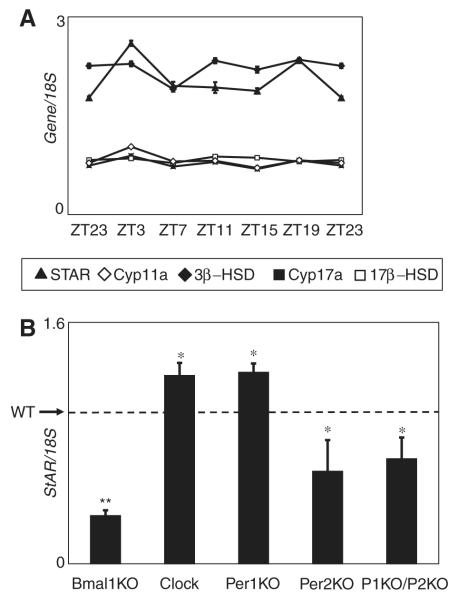
Although BMAL1 is central to the molecular workings of the circadian clock, it is unclear whether BMAL1 regulation of StAR expression is due to its circadian function. If BMAL1 regulation of StAR expression is circadian, we would expect to find cyclic expression of StAR over the course of the day. We found that StAR expression is nonrhythmic in the testis over a 24-h period, although there is a minimal but reproducible increase at ZT3, which coincides with the peak of BMAL1 expression in Leydig cells (Fig. 5A). The expression of a number of other steroidogenic genes that are affected by the absence of Bmal1 is also nonrhythmic over a 24-h period (Fig. 5A).
Figure 5.
Expression of steroidogenic genes in testes is not circadian. Expression of the steroidogenic genes does not oscillate over the course of a day. (A) Testis RNA collected from 12-week-old C57BL6/J individually housed male mice over a continuous 24-h period was examined by quantitative polymerase chain reaction (qPCR). Data shown are from 1 representative experiment. This was repeated 3 times with similar results. (B) Expression of steroidogenic acute regulatory protein (StAR) is affected by mutations of the circadian clock genes. StAR expression in testes from the indicated mutant mice and wild-type mice was measured by qPCR. Expression of the genes is shown relative to wild-type expression level (dashed line). Error bars represent SEM. Clock mutant mice (9 pairs) and Bmal1 knockout (KO) mice (8 pairs) are compared with wild-type littermates. Per1 KO (3 mice), Per2 KO (3 mice), and Per1 KO/Per2 KO mice (4 mice) are compared with age-matched C57BL6/J male mice (4 mice). All data are significant to p < 0.05 (single asterisk), except for the Bmal1 KO mice, which are significant to p = 0.002 (unpaired Student t test).
In the circadian clock, BMAL1 partners with CLOCK or NPAS2 to activate transcription while the PER and CRY proteins negatively regulate the function of the CLOCK/BMAL1 heterodimer. If BMAL1 functions as a circadian molecule to regulate StAR expression, then we would expect disruption of other circadian clock components to also affect StAR expression. For example, if CLOCK partners with BMAL1 in Leydig cells to regulate StAR expression, we would expect that StAR expression would be low in mice with a disrupted Clock gene. However, in Clock mutant mice, which express a dominant negative form of CLOCK, StAR expression was increased in comparison to wild-type littermates, suggesting that CLOCK is not the partner of BMAL1 in stimulating StAR expression (Fig. 5B). Examination of testes of mice lacking Per1, Per2, or both genes revealed distinct effects on StAR expression. In Per1 KO mice, StAR expression was increased; however, in Per2 KO mice and in Per1 KO/Per2 KO mice, StAR expression was decreased (Fig. 5B). These data show that although expression of StAR is not rhythmic, it is affected by the other circadian clock proteins.
DISCUSSION
In the current study, we establish that mice lacking the circadian clock protein, BMAL1, are infertile. The mechanism leading to infertility probably involves a combination of factors. The normal hormonal milieu of male Bmal1 KO mice is disrupted (low testosterone, high LH, low FSH), which undoubtedly leads to the physical phenotype observed, including small testes, small seminal vesicles, and a low sperm count. However, confirmation of this hypothesis will require hormone replacement experiments. Because the sperm of Bmal1 KO mice are functional in vitro, there is probably also a behavioral component to the infertility. Male Bmal1 KO mice attempt mating, but we have not observed successful copulation as assessed by the appearance of vaginal plugs in mating partners. Whether this is due to an inability to mate or due to disturbed mating behavior requires further investigation. Because prenatal exposure to testosterone is necessary for the development of normal sexual behavior later in life, low testosterone in Bmal1 KO mice may result in abnormal sexual behavior (MacLusky and Naftolin, 1981). Notably, female Bmal1 KO mice mate, as assessed by the appearance of vaginal plugs, so presumably female mating behavior in these mice is essentially normal (our unpublished data). Therefore, female sterility most likely results from altered levels of reproductive hormones.
Although BMAL1 probably functions in multiple aspects of reproduction, we propose that a defect in Leydig cells contributes to the infertility. BMAL1 presumably activates transcription in Leydig cells, as there is reduced expression of a number of steroidogenic genes in the testes of Bmal1 KO mice. We hypothesize that low levels of StAR in Bmal1 KO mice underlie the low testosterone state. BMAL1 is clearly necessary for full StAR expression, and our data strongly suggest that this is a direct effect of BMAL1 on the StAR promoter. Complete StAR expression reportedly requires only the first 966 bp of promoter sequence; however, we showed that BMAL1 enhances StAR expression in vitro when using 2.5 kb of promoter sequence. The original study defining the minimal 966-bp sequence showed that there was no significant difference with respect to basal- or hormone-induced StAR expression in comparison to a 3.6-kb fragment (Caron et al., 1997). Possibly additional sequences exist between −2.5 kb and −3.6 kb of the StAR promoter, which serve to reduce StAR expression. Future efforts will focus on characterizing the exact nature of the interaction of BMAL1 with the StAR promoter.
Importantly, we found cyclic expression of BMAL1 in Leydig cells. This was somewhat unexpected given our previous findings that expression of Bmal1 is noncyclic in whole testis (Alvarez et al., 2003; Alvarez and Sehgal, 2005). There are a number of possible explanations for this discrepancy. For one, Bmal1 mRNA may be statically expressed but not translated to a high level in the spermatogenic compartment of the testis, which masks cyclic expression in the Leydig cells. It is also possible that Bmal1 mRNA expression is noncyclic within Leydig cells, but translation or posttranslational modification and/or degradation of the protein is cyclic. Finally, it is also possible that our methods to measure mRNA concentration only detected alternative spliced forms of Bmal1 mRNA that do not cycle (Yu et al., 1999).
Despite the finding of cyclic BMAL1 expression in Leydig cells and our data suggesting a direct function of BMAL1 on the StAR promoter, we did not observe cyclic expression of StAR in whole testis. This finding is consistent with recent microarray data demonstrating constant expression of StAR in the adrenal gland; however, these data are inconsistent with a recent report of cyclic expression of StAR in F1 follicles of the chicken ovary (Oster et al., 2006; Nakao et al., 2007). The chicken StAR promoter has an E-box that is close to the transcription start site, which may account for the difference (Nakao et al., 2007). Alternatively, cyclic expression of StAR may occur in Leydig cells that is masked when whole testes are examined due to noncyclic expression in other cell types such as Sertoli cells (Gregory and DePhilip, 1998). It is also possible that StAR protein expression or activity cycles irrespective of the mRNA expression pattern. Notably, we find a consistent increase of StAR expression at ZT3, which coincides with the peak of BMAL1 expression in Leydig cells. Although it is possible that this increase accounts for the testosterone rhythm, it is also possible that rhythmic release of testosterone is not regulated at the level of the Leydig cell but rather may result from diurnal variations in LH release from the pituitary (Jean-Faucher et al., 1986; Dunkel et al., 1992).
We cannot yet conclude that BMAL1 is functioning within the well-known circadian clock mechanism to regulate StAR expression. The effect of deletion or mutation of other circadian clock genes on StAR expression does not follow predicted patterns if BMAL1 were functioning within a canonical circadian clock mechanism to regulate StAR expression. Since PER2 indirectly activates Bmal1 expression by repressing REV-ERBα transcription (Preitner et al., 2002), the reduced levels of StAR in Per2 KO mice are consistent with the circadian regulation of StAR. However, the phenotype of the Clock mutant mouse is not: StAR levels are increased in Clock mutant mice. If CLOCK were acting as the sole BMAL1 binding partner to activate StAR expression, we would expect a decrease in expression. Although NPAS2 may act redundantly for CLOCK within Leydig cells, it is not clear that this occurs in peripheral tissues (DeBruyne et al., 2007). Thus, the role of other clock genes in regulating StAR expression is still unclear, but the effects of BMAL1 on StAR must, at least partly, underlie its effects on fertility. Future work using the established cell line model system will allow a better understanding of how the various circadian clock proteins function in regulating StAR expression.
ACKNOWLEDGMENTS
We thank Dr. Chris Bradfield (University of Wisconsin), Dr. Celeste Simon (University of Pennsylvania), and Dr. Garrett FitzGerald (University of Pennsylvania) for supplying Bmal1 KO mice; Dr. Joseph Takahashi (Northwestern University, Evanston, IL) for supplying Clock mutant mice; Dr. Cheng-Chi Lee (University of Texas–Houston) for supplying Per1 KO mice; Dr. Mario Ascoli (University of Iowa) and Dr. Jerome Strauss (University of Pennsylvania) for supplying the murine Leydig tumor cell line MA-10; and Dr. Barbara Clark (University of Louisville) for supplying various StAR reporter plasmids. We thank Jen Wood and Lane Christianson for scientific discussions and Lisa Haig-Ladewig, Brian Jones, and Dechun Chen for technical assistance. This work was supported by grants HD040297 (to JDA), HD044740 (to CW), and HD06427 (to SM). AS is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Alvarez JD, Chen D, Storer E, Sehgal A. Non-cyclic and developmental stage-specific expression of circadian clock proteins during murine spermatogenesis. Biol Reprod. 2003;69:81–91. doi: 10.1095/biolreprod.102.011833. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Sehgal A. The thymus is similar to the testis in its pattern of circadian clock gene expression. J Biol Rhythms. 2005;20:111–121. doi: 10.1177/0748730404274078. [DOI] [PubMed] [Google Scholar]
- Andrews WW, Ojeda SR. A detailed analysis of the serum luteinizing hormone secretory profile in conscious, free-moving female rats during the time of puberty. Endocrinology. 1981;109:2032–2039. doi: 10.1210/endo-109-6-2032. [DOI] [PubMed] [Google Scholar]
- Apter D, Butzow T, Laughlin G, Yen S. Gonadotropin-releasing hormone pulse generator activity during pubertal transition in girls: Pulsatile and diurnal patterns of circulating gonadotropins. J Clin Endocrinol Metab. 1993;76:940–949. doi: 10.1210/jcem.76.4.8473410. [DOI] [PubMed] [Google Scholar]
- Ascoli M. Characterization of several clonal lines of cultured Leydig tumor cells: Gonadotropin receptors and steroidogenic responses. Endocrinology. 1981;108:88–95. doi: 10.1210/endo-108-1-88. [DOI] [PubMed] [Google Scholar]
- Barbacka-Surowiak G, Surowiak J, Stoklosowa S. The involvement of suprachiasmatic nuclei in the regulation of estrous cycles in rodents. Reprod Biol. 2003;3:99–129. [PubMed] [Google Scholar]
- Bartke A. Increased sensitivity of seminal vesicles to testosterone in a mouse strain with low plasma testosterone levels. J Endocrinol. 1974;60:145–148. doi: 10.1677/joe.0.0600145. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Giebultowicz JM. Regulation of copulation duration by period and timeless in Drosophila melanogaster. Curr Biol. 2004;14:1492–1497. doi: 10.1016/j.cub.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM. Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:2134–2139. doi: 10.1073/pnas.032426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver LM, Rush BL, Gvakharia BO, Giebultowicz JM. Noncircadian regulation and function of clock genes period and timeless in oogenesis of Drosophila melanogaster. J Biol Rhythms. 2003;18:463–472. doi: 10.1177/0748730403259108. [DOI] [PubMed] [Google Scholar]
- Boden MJ, Kennaway DJ. Circadian rhythms and reproduction. Reproduction. 2006;132:379–392. doi: 10.1530/rep.1.00614. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron KM, Ikeda Y, Soo S-C, Stocco DM, Parker KL, Clark BJ. Characterization of the promoter region of the mouse gene encoding the steroidogenic acute regulatory protein. Mol Endocrinol. 1997;11:138–147. doi: 10.1210/mend.11.2.9880. [DOI] [PubMed] [Google Scholar]
- Chappell PE, White RS, Mellon PL. Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1-7 cell line. J Neurosci. 2003;23:11202–11213. doi: 10.1523/JNEUROSCI.23-35-11202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clair P, Claustrat B, Jordan D, Dechaud H, Sassolas G. Daily variations of plasma sex hormone-binding globulin binding capacity, testosterone and luteinizing hormone concentrations in healthy rested adult males. Horm Res. 1985;21:220–223. doi: 10.1159/000180052. [DOI] [PubMed] [Google Scholar]
- Clark B, Wells J, King S, Stocco D. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells: Characterization of the steroidogenic acute regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Cooke HJ, Saunders PT. Mouse models of male infertility. Nat Rev Genet. 2002;3:790–801. doi: 10.1038/nrg911. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- Devoto L, Kohen P, Gonzalez RR, Castro O, Retamales I, Vega M, Carvallo P, Christenson LK, Strauss JF., III Expression of steroidogenic acute regulatory protein in the human corpus luteum throughout the luteal phase. J Clin Endocrinol Metab. 2001;86:5633–5639. doi: 10.1210/jcem.86.11.7982. [DOI] [PubMed] [Google Scholar]
- Dunkel L, Alfthan H, Stenman U, Selstam G, Rosberg S, Albertsson-Wikland K. Developmental changes in 24-hour profiles of luteinizing hormone and follicle-stimulating hormone from prepuberty to midstages of puberty in boys. J Clin Endocrinol Metab. 1992;74:890–897. doi: 10.1210/jcem.74.4.1548356. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM, Riemann JG, Raina AK, Ridgway RL. Circadian system controlling release of sperm in the insect testes. Science. 1989;245:1098–1100. doi: 10.1126/science.245.4922.1098. [DOI] [PubMed] [Google Scholar]
- Gregory C, DePhilip R. Detection of steroido-genic acute regulatory protein (stAR) in mitochondria of cultured rat Sertoli cells incubated with follicle-stimulating hormone. Biol Reprod. 1998;58:470–474. doi: 10.1095/biolreprod58.2.470. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Jean-Faucher C, Berger M, de Turckheim M, Veyssiere G, Jean C. Circadian variations in plasma LH and FSH in juvenile and adult male mice. Horm Res. 1986;23:185–192. doi: 10.1159/000180321. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Lucas LA, Eleftheriou BE. Circadian variation in concentrations of testosterone in the plasma of male mice: A difference between BALB/cBy and C57BL/6By inbred strains. J Endocrinol. 1980;87:37–46. doi: 10.1677/joe.0.0870037. [DOI] [PubMed] [Google Scholar]
- MacLusky N, Naftolin F. Sexual differentiation of the central nervous system. Science. 1981;211:1294–1302. doi: 10.1126/science.6163211. [DOI] [PubMed] [Google Scholar]
- Manna PR, Wang X-J, Stocco DM. Involvement of multiple transcription factors in the regulation of steroidogenic acute regulatory protein gene expression. Steroids. 2003;68:1125–1134. doi: 10.1016/j.steroids.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(suppl):S33–S40. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao N, Yasuo S, Nishimura A, Yamamura T, Watanabe T, Anraku T, Okano T, Fukada Y, Sharp PJ, Ebihara S, et al. Circadian clock gene regulation of steroidogenic acute regulatory protein gene expression in preovulatory ovarian follicles. Endocrinology. 2007;148:3031–3038. doi: 10.1210/en.2007-0044. [DOI] [PubMed] [Google Scholar]
- Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Privalle CT, Crivello JF, Jefcoate CR. Regulation of intramitochondrial cholesterol transfer to side-chain cleavage cytochrome P-450 in rat adrenal gland. Proc Natl Acad Sci USA. 1983;80:702–706. doi: 10.1073/pnas.80.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocco D, Clark B. Regulation of the acute production of steroids in steroidogenic cells. Endocrinol Rev. 1996;17:221–244. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Yokoyama M, Hosi T. Study on the fertilization of mouse eggs in vitro: I. In vitro fertilization of eggs by fresh epidiymal sperm. Jpn J Anim Reprod. 1971;16:147–151. [Google Scholar]
- Travis AJ, Jorgez CJ, Merdiushev T, Jones BH, Dess DM, Diaz-Cueto L, Storey BT, Kopf GS, Moss SB. Functional relationships between capacitation-dependent cell signaling and compartmentalized metabolic pathways in murine spermatozoa. J Biol Chem. 2001;276:7630–7636. doi: 10.1074/jbc.M006217200. [DOI] [PubMed] [Google Scholar]
- Visconti PE, Kopf GS. Regulation of protein phosphorylation during sperm capacitation. Biol Reprod. 1998;59:1–6. doi: 10.1095/biolreprod59.1.1. [DOI] [PubMed] [Google Scholar]
- Yu W, Ikeda M, Abe H, Honma S, Ebisawa T, Yamauchi T, Honma K-I, Nomura M. Characterization of three splice variants and genomic organization of the mouse BMAL1 gene. Biochem Biophys Res Commun. 1999;260:760–767. doi: 10.1006/bbrc.1999.0970. [DOI] [PubMed] [Google Scholar]
- Zhang F-P, Poutanen M, Wilbertz J, Huhtaniemi I. Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol Endocrinol. 2001;15:172–183. doi: 10.1210/mend.15.1.0582. [DOI] [PubMed] [Google Scholar]