Abstract
Objective
CYP2D6 is a polymorphic gene. It has been observed to be deleted, to be duplicated and to undergo recombination events involving the CYP2D7 pseudogene and surrounding sequences. The objective of this study was to discover the genomic structure of CYP2D6 recombinants that interfere with clinical genotyping platforms that are available today.
Methods
Clinical samples containing rare homozygous CYP2D6 alleles, ambiguous readouts, and those with duplication signals and two different alleles were analyzed by long-range PCR amplification of individual genes, PCR fragment analysis, allele-specific primer extension assay, and DNA sequencing to characterize alleles and genomic structure.
Results
Novel alleles, genomic structures, and the DNA sequence of these structures are described. Interestingly, in 49 of 50 DNA samples that had CYP2D6 gene duplications or multiplications where two alleles were detected, the chromosome containing the duplication or multiplication had identical tandem alleles.
Conclusion
Several new CYP2D6 alleles and genomic structures are described which will be useful for CYP2D6 genotyping. The findings suggest that the recombination events responsible for CYP2D6 duplications and multiplications are because of mechanisms other than interchromosomal crossover during meiosis.
Keywords: allele, CYP2D6, CYP2D7, CYP2D8, cytochrome P450, drug metabolism, pharmacogenetics, polymorphism, recombinant
Introduction
Cytochrome P450 2D6 (CYP2D6) is involved in phase I drug metabolism of many medications including several psychotropic medications [1,2] and it is involved in the bioactivation of tamoxifen [3]. Many clinical laboratories perform CYP2D6 genotyping for phenotype prediction, so it is important that all alleles and genomic structures be identified for this testing. CYP2D6 is a highly polymorphic gene involving the pseudogene CYP2D7, in part, because of recombinant events that generate duplications, multiplications, deletions, and gene conversions. There are also a large number of single-nucleotide polymorphisms that have been described (http://www.cypalleles.ki.se/cyp2d6.htm). This diversity of alleles and the incidence of phenotypes (poor, intermediate, extensive and ultrarapid) vary among ethnic groups [4].
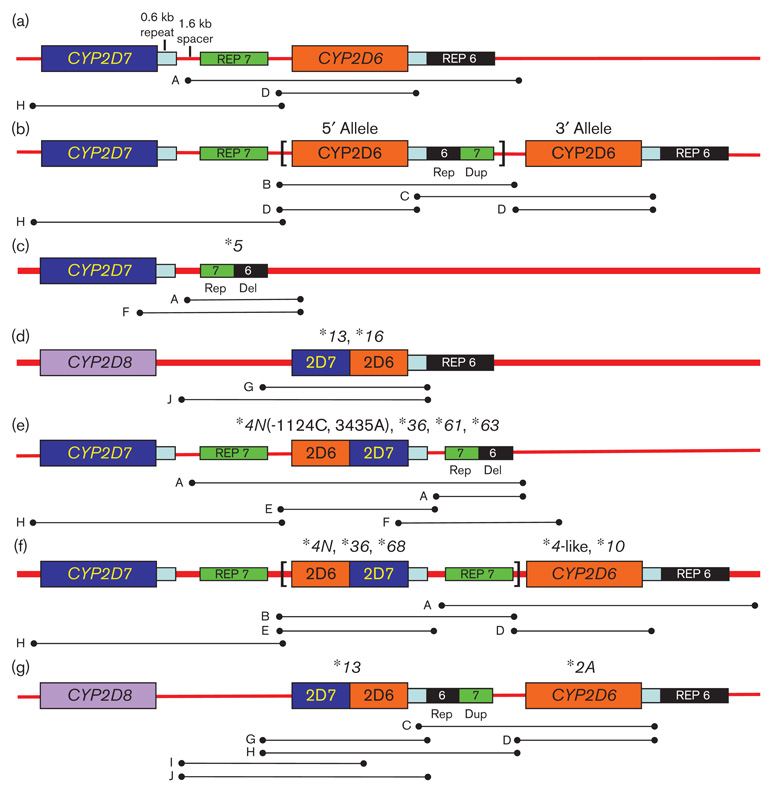
To understand CYP2D6 gene complexity, it is essential to understand the genomic structure of the gene and its surrounding sequences. Figure 1a–c shows the most commonly encountered CYP2D6 and CYP2D7 gene arrangements [5–10]. CYP2D6 is most commonly found in a nonduplicated arrangement (Fig. 1a), although duplications (Fig. 1b) and deletions (Fig. 1c) are also encountered. Gene multiplications resemble Fig. 1b except that they have more than two CYP2D6 genes in tandem on a single chromosome. Other arrangements have been described where recombination between the CYP2D6 and CYP2D7 genes results in CYP2D7/CYP2D6 hybrids, for example *13 and *16 (Fig. 1d), or CYP2D6/CYP2D7 hybrids, namely *4N and *36 (Fig. 1e and f). *36 has been described to occur as a single gene, a duplication arrangement and a tandem arrangement with *10B (e.g. *36+*10B) (Fig. 1f) [10–14]. *4N has only been reported in a *4N X N configuration to date [10].
Fig. 1.
CYP2D6, CYP2D7, and CYP2D8 gene arrangements and PCR amplicons used in PCR fragment analysis. Lettered lines under the structures depict PCR fragments used in analysis (Fig. 2, Table 1). Note that CYP2D8 is thought to be present in all of the structures shown but is only depicted in structure D and G for simplicity. (a) Nonduplicated CYP2D6 arrangement. The CYP2D7 pseudogene is followed by a 0.6-kb repeat, a unique 1.6-kb ‘spacer’ sequence, and rep 7. The CYP2D6 gene is next which is followed by a 0.6-kb repeat region and rep 6. (b) Typical CYP2D6 duplication arrangement. Rep dup is a hybrid containing a 5′ rep 6 sequence and a 3′ rep 7 sequence [5,6]. A multiplication structure contains two or more copies of the sequence found between the brackets. (c) The deletion arrangement lacks the coding and surrounding sequences for CYP2D6 but has CYP2D7 with the usual trailing sequence up to the rep del sequence. Rep del is a hybrid between a 5′ rep 7 and 3′ rep 6. (d) CYP2D7/CYP2D6 genes lack the CYP2D7 pseudogene and are followed by CYP2D6-related trailing sequences. (e) Single CYP2D6/CYP2D7 genes are trailed by the CYP2D7-specific sequences before transitioning into rep del. (f) CYP2D6/CYP2D7 genes found in a duplication arrangement have a 5′ recombinant gene that was trailed by a CYP2D7-specific spacer sequence and rep 7 before the 3′ CYP2D6 gene. Multiplications (not detected in this research) have multiples of the sequence shown between the brackets. (g) A CYP2D7/CYP2D6 gene in a duplication arrangement has a 5′ recombinant gene that is trailed by rep dup before the 3′ CYP2D6 gene. The chromosome lacks the CYP2D7 pseudogene.
The intent of this study was to look for genotypes which could not be detected by a commercially available allele-specific primer extension assay (ASPE), commonly used for genotyping of clinical samples. The samples studied included (i) samples with a duplication signal and heterozygosity for CYP2D6 alleles, as the existing platform could not predict which alleles were duplicated in these situations, (ii) homozygous genotypes of rare alleles suggesting failure of the ASPE assay to detect one allele, for example, because of recombinations, and (iii) samples that yielded raw data that could not be interpreted for heretofore unknown reasons.
Methods
Samples
De-identified clinical samples submitted to the Nucleotide Polymorphism Laboratory of the Department of Laboratory Medicine and Pathology at the Mayo Clinic for CYP2D6 genotyping were studied with Mayo Clinic Institutional Review Board approval. From these samples we selected the following samples: (i) 50 samples yielding a duplication signal and heterozygosity for CYP2D6 alleles, (ii) two samples which yielded genotypes of homozygous alleles that were seen only once in our practice, (iii) five samples that yielded uninterpretable readouts using the CYP2D6 ASPE kit.
Genotyping by allele-specific primer extension assay and sequencing
Genotyping of genomic DNA and amplicons of individual genes was performed with the Luminex-based Tag-It Mutation Detection Kit P450-CYP2D6 version 1 (CYP2D6 ASPE kit) (Austin, Texas, USA). This kit incorporates multiplex PCR and multiplex ASPE with a proprietary tag sorting system on the Luminex100 xMAP platform and is capable of detecting the following: gene duplication, gene deletion (*5), − 1584 C>G (*2A), 100 C>T (*4A–L, *10A, B), 124G>A (*12), 883G>C (*11), 1023C>T (*17), 1707T>del (*6A–D), 1758G>T (*8), 1846G>A (*4A–L), 2549A>del (*3A, B), 2613–2615 del AGA (*9), 2850C>T (*2, *17, *41), 2935A>C (*7). All assays were read on the Luminex 100 IS device using IS 2.3 software (Austin, Texas, USA). Genotyping of genomic DNA was carried out as described in the product manual. However, when individual genes were amplified as described below, an optimized procedure using the CYP2D6 ASPE kit was done. In addition, the 2988G>A (*41) polymorphism was assessed by PCR amplification of the region of interest followed by direct sequencing as described below.
TaqMan CYP-2D6 gene copy number assay
The gene copy number was determined on samples with duplication signals by the CYP2D6 ASPE kit by using the TaqMan CYP-2D6 Gene Copy Number Assay (Applied Biosystems, Foster City, California, USA, Assay ID Hs00010001_cn) per product literature. This assay can detect high copy numbers according to the product manufacturer. Genomic DNA (5 ng) was combined with the TaqMan Universal PCR Master Mix, FAM-MGB CYP-2D6 specific probe, and primers. RNase P gene (VIC/Tamra Probe) was used as an endogenous control. Samples were run in triplicate on an ABI 7900 HT Fast Real-Time PCR System using absolute quantification and the copy number was determined by relative quantification (ΔΔCt).
PCR and amplicon verification
The PCR primers used in this study were selected using Oligo Primer Analysis Software version 6.71 (Molecular Biology Insights Inc., Cascade, Colorado, USA) and are presented in Table 1. PCR amplicon locations are shown in Fig. 1a–g as black bars under each structure. Primers were designed to prevent cross-reactivity with the incorrect gene or pseudogene which is important on account of the high degree of sequence homology existing among CYP2D6, CYP2D7, and CYP2D8.
Table 1.
PCR primers and amplicon characteristics
| PCR product |
Primer | Sequence | Arrangement amplified | Size (bp) |
|---|---|---|---|---|
| A | Seq B1 | GTCCCACACCAGGCACCTGTACT | Nonduplicated allele and deletion arrangement |
15 629, 3471 |
| B | Rep 7R3 | GAATTAGTGGTGGTGGGTGTTTG | 5′ allele of duplication arrangement | 12 052 |
| Prealpha F | TCACCCCCAGCGGACTTATCA | |||
| 2D63- | CCACAGCCCTCAATAAGTGAA | |||
| C | A‘F | CCCTGGGAAGGCCCCATGGAAG | 3′ allele of duplication arrangement | 12 103 |
| Prenest R2 | TAGGTAGCCCTGGCCTATAGCTCCCTGACGCC | |||
| D | Prenest F | TTGCCACATTATCGCCCGTGAAA | Nonduplicated allele & 3′ allele of duplication arrangement |
8433 |
| E | Prenest R2 | TAGGTAGCCCTGGCCTATAGCTCCCTGACGCC | CYP2D6/CYP2D7 recombinant allele | 6714 |
| Prealpha F | TCACCCCCAGCGGACTTATCA | |||
| Rep 7B rev | TACGGTGGGCTCCCTGCGAG | |||
| F | G2D7ex9F | AGCCACTCTCGTGTCGTCAGCTT | PCR fragment from CYP2D7 exon 9 to unique region 30 of CYP2D6 and rep 6 |
5173 |
| G | GER | CAGGCATGAGCTAAGGCACCCAGA | CYP2D7/CYP2D6 recombinant allele | 5742 |
| *16F | CCTGTGTGGGCTTGGGGAGCTTG | |||
| *16R5 | TGTGGTGAGGTGACGAGGCTGA | |||
| H | *16F | CCTGTGTGGGCTTGGGGAGCTTG | CYP2D7 and CYP2D7/CYP2D6 Recombinants | 119 98, 104 42 |
| 2D63- | CCACAGCCCTCAATAAGTGAA | |||
| I | 2D8F | ACCTGGACGCCTGACTTTA | PCR fragment from CYP2D8-unique 30 region to CYP2D6 intron 3 |
5815 |
| J | LXR2- | ACCGGATTCCAGCTGGGAAATG | PCR fragment from CYP2D8-unique 30 region to CYP2D6 3′ region |
9525 |
| 2D8F | ACCTGGACGCCTGACTTTA | |||
| Prenest R2 | TAGGTAGCCCTGGCCTATAGCTCCCTGACGCC |
The primers listed here were used to amplify genes for testing by allele-specific primer extension assay, DNA sequencing, or for PCR fragment analysis. Refer to Fig. 1 for approximate annealing sites of these primers on the various genomic structures.
The PCR master mix was composed of 12 µl reactions containing 0.12 µl TaKaRa LA Taq HS (5 U/µl, TaKaRa Bio. Inc., Otsu, Shiga, Japan), 1.2 µl 10X LA PCR Buffer II (25mmol/l Mg2+), 2 µl dNTP mixture (2.5 mmol/l each), 6.48 µl water and 1 µl betaine monohydrate (16.25 mol/l, Fluka Biochemical, Steinheim, Germany), 0.5 µl of each primer and 0.2 µl genomic DNA (250 ng/µl).
To determine which genes were present in a duplication or multiplication arrangement, thermocycler parameters for fragment A, fragment B, and fragment C were as follows: 94°C for 1 min followed by 96°C for 10 s, 64°C for 30 s, 68°C for 11 min for 30 cycles, and a final extension at 72°C for 10 min and a 4°C hold. Note that placement of the amplicon B primers (Fig. 1, Table 1) permitted each CYP2D6 gene in a duplication or multiplication structure to yield a PCR product. Thus, if nonidentical tandem alleles were present, heterozygous single-nucleotide polymorphisms would be identified by either the CYP2D6 ASPE kit or DNA sequencing.
To characterize samples for which the CYP2D6 ASPE kit yielded raw data suggesting a deletion signal as well as a partial allele and another full allele, and to test for 2D6/2D7 recombinants, PCR conditions for fragment D and fragment E were as follows: 94°C for 1 min followed by 96°C for 10 s and 68°C for 9 min for 30 cycles before a final extension step of 72°C for 10min and a 4°C hold temperature. Parameters for fragment F were 94°C for 1 min followed by 96°C for 10 s and 68°C for 4 min for 30 cycles, before a final extension step of 72°C for 10 min and a 4°C hold temperature.
For the detection of 2D7/2D6 recombinants using fragment G, thermocycler parameters were 94°C for 1 min followed by 96°C for 10 s and 72°C for 8 min for 28 cycles before a final extension step of 72°C for 10 min and a 4°C hold temperature. For fragment H, thermocycler parameters were 94°C for 1 min followed by 96°C for 10 s and 68°C for 15 min for 30 cycles before a final extension step of 72°C for 10 min and a 4°C hold temperature.
To determine whether CYP2D7 was present upstream of *13 and *16 (e.g. CYP2D7/CYP2D6 recombinants), PCR parameters for fragments I and J were as follows: 94°C for 1 min followed by 96°C for 10 s, 64°C for 30 s, 68°C for 9 min for 28 cycles and a final extension at 72°C for 10 min and a 4°C hold.
PCR fragments were analyzed using the Agilent Technologies DNA 7500 Kit (Agilent Technologies, Waldbronn, Germany) as per manufacturer’s protocol.
DNA sequencing
DNA sequencing was performed using an Applied Biosystems 3730xl DNA Analyzer and the ABI PRISM dRhodamine Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA Polymerase, FS. Sequencing primer details are available upon request.
Bioinformatics tools
DNA sequence data was compiled using Sequencher software, version 4.9, (Gene Codes Corp., Ann Arbor, Michigan, USA). Sequences were compared with the CYP2D6 alleles listed by the CYP2D6 Nomenclature Committee (http://www.cypalleles.ki.se/cyp2d6.htm). GenBank entry M33388 (with modifications per AY515216) served as a reference sequence for CYP2D6 and GenBank entry M33387 served as reference sequences for CYP2D7 and CYP2D8. Comparisons were performed using the BLAST 2 SEQUENCES program (bl2seq; http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi) and CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw/index.html) at default settings.
Mapping of recombinant regions
The high degree of sequence similarity between the CYP2D6 and CYP2D7 genes makes it difficult to determine exact recombination regions in recombinant genes. A conservative rule for determining the region involved in the recombination event was used. The recombination region was said to begin and end when the nucleotide sequence was consistent with an uninterrupted string of five nucleotide differences according to each gene’s reference sequence.
Phenotype prediction
Phenotype prediction was based on the genotype. Phenotype prediction was binned into four categories in a manner similar to those reviewed by Ingelman-Sundberg [15] and Kirchheiner et al. [2] to describe the relevance of CYP2D6 allelic variability in clinical settings. An ultrarapid metabolizer had more than two functioning alleles, extensive metabolizer had two functioning alleles, an intermediate metabolizer had one functioning allele, and a poor metabolizer had no normally functioning alleles. Functionality of alleles was determined by review of the CYP2D6 Nomenclature Committee web page (http://www.cypalleles.ki.se/cyp2d6.htm).
Results
DNA samples
Fifty samples that had duplication signals and heterozygosity for CYP2D6 alleles were studied. These were found in approximately 2000 samples processed by the Nucleotide Polymorphism Laboratory of the Department of Laboratory Medicine and Pathology at the Mayo Clinic over a 4-year period. These samples were of interest because the CYP2D6 ASPE kit is unable to determine which allele was duplicated thus introducing uncertainty to the prediction of phenotype.
Two samples that were initially clinically genotyped as *3/*3 and *14B/*14B were also studied. These were rare homozygous alleles and we hypothesized that they were incorrectly genotyped because of allelic drop out, for example, because of recombination. In the case of the *14B homozygote, the CYP2D6 ASPE kit failed to produce the 1758G>Toutput, hence sequencing of the sample was carried out which revealed 1758G>A which is characteristic of CYP2D6*14B.
Finally, five additional samples were studied because the CYP2D6 ASPE kit failed to yield raw data that could be interpreted. In four of these samples, the raw data yielded a deletion signal and the presence of a partial CYP2D6 allele and another complete allele. Another sample had raw data suggesting the presence of a duplication, a deletion, and a *2A allele.
Naming of novel alleles
All novel alleles and genomic structures were submitted to the CYP2D6 Allele Nomenclature Committee and all alleles were submitted to GenBank and were assigned unique accession numbers. Three alleles were named by the CYP2D6 Allele Nomenclature Committee and were assigned *61 (EU530607), *63 (EU530608), and *68 (EU530606) (Table 2). Other alleles did not meet the Committee’s criteria for naming, in that they were not expected to affect gene function, so it was recommended that these alleles be given descriptive names and that they be submitted to GenBank. Therefore the following descriptive names and accession numbers were assigned: ‘CYP2D6*1 (270 T)’-EU530602, ‘CYP2D6*2A-*1A’-EU530603, ‘CYP2D6*4N(-1124C, 3435A)’-EU530604, and ‘CYP2D6*4-like’-EU530605. ‘CYP2D6*13’-EU530609 and ‘CYP2D6* 16’-EU530610 are not new alleles as they have been described earlier, but data on their exact sequence was not present in GenBank so these were also submitted [12,16]. The characteristics of these alleles are found in Table 2.
Table 2.
Polymorphisms and crossover regions of selected alleles
 |
This table lists the polymorphisms with dbSNP rs#, where known. The sequences surrounding the SNPs, and their locations on the reference sequences for CYP2D6, M33388 and AY545216 (in brackets) are shown. The relative locations of the recombination regions are crosshatched. Gray boxes are regions consistent with CYP2D7 sequence and the SNPs in those regions are not specified. The black boxes depict the presence of a given SNP in an allele. Thick black horizontal lines separate the results for a given sample of interest. For duplicated samples of any configuration, the first allele listed represents the 5′ gene sequence and the second is the 3′ gene. All of the alleles shown had full DNA sequencing from − 1770 through + 4401.
SNPs, single-nucleotide polymorphisms.
Characterization of gene duplication and multiplication arrangements
The analysis of 50 DNA samples with CYP2D6 duplication signals and heterozygosity for alleles revealed a total of 54 duplication and multiplication structures. Of the 50 DNA samples studied, 48 had duplications (Fig. 1b), two had multiplications (Fig. 1b), and four had duplication structures containing a hybrid gene (Fig. 1f). A total of 54 duplication and multiplication structures were present because four samples were discovered to have duplications on both chromosomes as described below. In all of the instances of typical duplications and multiplications, except one, the tandem genes were identical to the level of detection of the CYP2D6 ASPE kit. In the one duplication arrangement where different tandem alleles were detected, the 5′ gene had a CYP2D6*2A sequence from the promoter region through intron 1 before reverting to *1A sequence. This gene has been described once earlier [17] and it is referred to as *2A-*1A (EU530603) here (Table 2). The 3′ gene in this structure also differed from CYP2D6*1A because of the presence of a 270T polymorphism (rs29001678). This CYP2D6*1 variant is described here for the first time and is named *1 (270 T) (EU530602). For the remaining 49 chromosomes with typical CYP2D6 duplication or multiplication arrangements, we found the following frequencies: *2A+*2A (n=22), *1A+*1A (n=10), *2+*2 (n=8), and *4+*4 (n=3), *3+*3 (n=1), *6+*6 (n=1), *9+*9 (n=1), *41+*41 (n=1) *1A X 5 (n=1), and *1A X 3 (n=1).
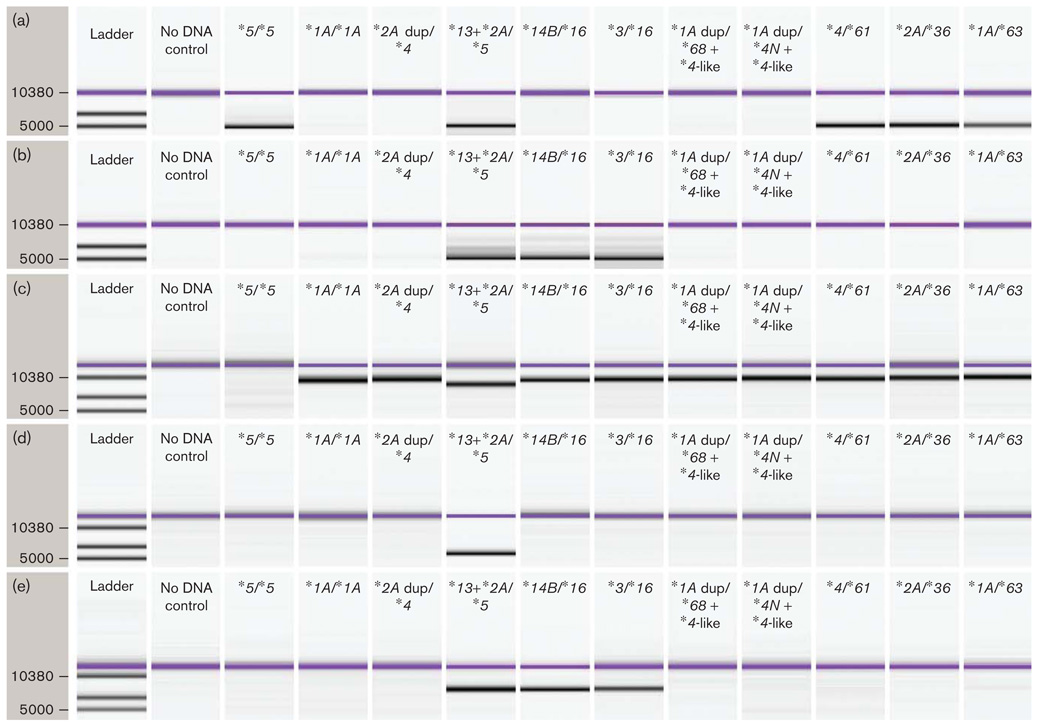
As mentioned, four of the 50 samples were discovered to have a second duplication structure on the other chromosome. These four samples contained a CYP2D6/CYP2D7+CYP2D6 arrangement (Fig. 1f). Amplicons of the individual genes from these samples were sequenced. Two *4N+*4-like and two *68+*4-like arrangements were found (Table 2). *4N and *68 differ only by their recombination regions and the CYP2D6 portion of these recombinant genes were identical to that found in the trailing *4-like allele. The *4-like allele (EU530605), described here for the first time, is 100% identical to *4N except it lacks the conversion to CYP2D7 in exon 9. This genomic structure is distinct from the CYP2D6*4N X N described by Gaedigk et al. [10], because the 3′ gene did not contain the exon 9 conversion, as determined by DNA sequencing and further confirmed by testing for amplicon F, described by these authors (Fig. 2a).
Fig. 2.
PCR fragment analysis. The PCR products for amplicons F–J were used in fragment analysis on the Agilent Technologies DNA 7500 Kit to show the presence or absence of the CYP2D7 gene, CYP2D6 genes, or recombinant allele. Fragments A–E are not shown as these were primarily useful for the generation of amplicons used in allele-specific primer extension assay reactions and sequencing reactions. Genotypes tested are shown at the top of each lane. The ladder in the left column shows the size in base pairs of the molecular weight marker. The uppermost purple band in all sample lanes represents a marker depicting the maximum size that the kit can detect reliably. (a) PCR fragment F analysis detects CYP2D7 exon 9 through the region 3′ of rep 6. The *68 +*4-like and *4N+*4-like arrangements did not produce a product because the *4-like allele did not have the exon 9 conversion present. The sample containing the *5/*5 genotype and those with single *36, *61, and *63 alleles generated an amplicon. The size of the product is consistent with the presence of the CYP2D7-derived spacer sequence which was confirmed by DNA sequencing. (b) PCR fragment G analysis that detects CYP2D7/CYP2D6 genes. Only samples with *13 and *16 generated an amplicon. (c) PCR fragment H analysis detects DNA spanning from the CYP2D7 promoter to CYP2D6 promoter regions. The presence or absence of 1.6 kb spacer sequence can be visualized based on the amplicon size. Genotype *5/*5 lacks the CYP2D6 promoter region, so does not generate an amplicon. Genotype *13 +*2A/*5 lacks the 1.6 kb spacer sequence, so the amplicon is smaller than the products generated by the other genotypes tested. (d) PCR fragment I analysis generates an amplicon spanning from CYP2D8 3′region to a unique region in CYP2D6 intron 3. The *13+*2A-containing sample generated an amplicon because CYP2D7 was absent. No amplicon was detected in *16 samples because the crossover region in *16 occurs after exon 7, which eliminates the reverse primer binding site for the amplicons. Normal gene arrangements and typical duplication arrangements do not generate a product because the size of the amplicons is too great. (e) PCR fragment J analysis generates amplicons spanning from the CYP2D8 3′ region to the CYP2D6-specific trailing sequences. Amplicons are generated for the *13 +*2A-containing and *16-containing samples because CYP2D7 is absent in these samples. Normal gene arrangements and typical duplication arrangements do not generate a product because the size of the amplicons is too great.
The detailed analysis resulted in more informative genotypes which should allow more accurate phenotype prediction in 28 of the 50 samples that we tested. Specifically, the testing allowed distinction between an intermediate metabolizer versus extensive metabolizer in 27 cases (e.g. *1A/*4 with duplication) and an intermediate metabolizer versus intermediate to extensive metabolizer in one case (*4/*9 with duplication), because the *9 allele is a functionally impaired allele. The testing had no impact on phenotype prediction in 22 cases.
Characterization of samples causing allelic drop out on the allele-specific primer extension assay
The samples initially genotyped as *3/*3 and *14B/*14B, when further studied using PCR fragment analysis and DNA sequencing, had a CYP2D6*16 allele (EU530610) on one chromosome (Fig. 2b). These samples did not contain a duplication arrangement. Interestingly, the chromosome containing the *16 allele lacked a 5′ CYP2D7 pseudogene (Fig. 2e) as well as a normal CYP2D6 allele as determined by PCR fragment analysis and DNA sequencing [12]. Detailed genotyping of the *14B-containing samples allowed estimation of an intermediate metabolizer status (e.g. *14B/*16) compared with intermediate to extensive metabolizer status (*14B/*14B) noting that *14B is an allele with reduced activity in vitro [18]. Thus, allelic dropout had occurred in these samples due to limitations of the CYP2D6 ASPE kit and the correct genotype was found to be *3/*16 and *14B/*16, respectively.
Characterization of samples yielding ambiguous raw data on the allele-specific primer extension assay
Four DNA samples yielding ambiguous data suggested a partial allele, a complete allele, and a CYP2D6 gene deletion using the CYP2D6 ASPE kit. All of these samples contained a single CYP2D6/CYP2D7 hybrid gene followed by the 1.6 kb CYP2D7-derived spacer sequence and rep del sequences as shown in Fig. 1e. One sample contained a *36 allele. Three novel alleles were detected and two were named *61 and *63 by the CYP2D6 Nomenclature Committee. The third novel allele resembled *4N but had − 1124C and 3435A polymorphisms and it also lacked the gene conversion to CYP2D7 in exon 9. It is named *4N ( − 1124C, 3435A) (EU530604) here (Table 2). The *4N ( − 1124C, 3435A) was unique as it had a recombination region in the 0.6 kb repeat located beyond the stop codon of the gene. The absence of a duplication arrangement in these four samples was determined as all attempts to generate PCR fragment B failed (data not shown). PCR fragment F was generated from the CYP2D6*36, CYP2D6*61, and CYP2D6*63-containing samples and its length was approximately 5742 bp (Fig. 2a). The sample containing *4N ( − 1124C, 3435A) did not produce PCR fragment F as the forward primer is specific for CYP2D7 exon 9, which was absent in this allele (data not shown). These four samples have gene arrangements that are similar to that described by Fukuda et al. [19]. Phenotype estimation was possible for only one of these samples (*2A/*36, intermediate metabolizer) as the other samples contained alleles whereby phenotype has not been assigned.
The remaining samples studied had a duplication signal, a deletion signal, and a CYP2D6*2A allele. Amplification of individual genes and DNA sequencing revealed that one chromosome had a CYP2D6*5 arrangement, whereas the other chromosome carried a *13+*2A arrangement. The CYP2D6 part of the *13 allele (EU530609) was identical to the corresponding portion of the trailing *2A allele (Table 2) [16]. Analysis of PCR fragment H yielded a single band that was approximately 1.6 kb smaller compared with that derived from CYP2D7 (Fig. 2c). This is consistent with a CYP2D7/CYP2D6 hybrid gene that possesses a downstream rep region without the CYP2D7-derived 1.6 kb spacer (Fig. 2c). The chromosome containing *13 lacked the CYP2D7 pseudogene as evidenced by the presence of PCR fragments H and J (Fig. 2d and e).
Identification of recombination regions
The high degree of similarity between the CYP2D6 and CYP2D7 sequences prevented the precise determination of the recombination sites in the hybrid genes. Estimated recombination regions are shown in Table 2.
Discussion
CYP2D6 is widely genotyped for pharmacogenomic purposes. The enzyme is involved in the metabolism of a large number of medications including psychotropics and the anticancer agent, tamoxifen. Accurate genotyping is essential because all pharmacogenomics-based treatment decisions are based on genotype–phenotype correlation.
The main findings of this research are as follows: (i) we found that typical CYP2D6 duplications and multiplications where two alleles were detected had the same alleles in the tandem in 49 of 50 samples studied. This is critical information as some genotyping platforms in existence do not predict which allele is duplicated or multiplied. In cases where alleles encoding an active and an inactive enzyme are detected, this uncertainty can interfere with phenotype prediction. For example, at *1/*4 with duplication, readout could be either an intermediate or an extensive metabolizer phenotype but this cannot be accurately predicted unless the phase of duplication is known. Complete genotyping had an impact on phenotype prediction in 28 of 50 samples studied here. (ii) Genomic structures which contain a recombinant gene followed by a CYP2D6 gene (Fig. 1f and g) had identical corresponding CYP2D6 DNA sequences. In other words, the CYP2D6 portion of *4N, *68, and *13 alleles were identical to the corresponding sequence of the trailing *4-like or *2A alleles in these genomic arrangements. This observation is essential when designing testing platforms. (iii) In two samples that appeared to have rare homozygous alleles on initial genotyping (e.g. *3/*3 and *14B/*14B), allelic drop out had actually occurred. This had an impact on predicted phenotype for the *14-containing sample. This finding emphasizes that when alleles are not detected by a particular platform, accurate phenotype prediction is not possible. The frequency of undetected *13 and *16 alleles in clinical samples is an ongoing focus of our research. Although these alleles are rare, more accurate genotyping will have an impact on the care of patients who have them, especially in cases where activation of life-preserving medications (e.g. tamoxifen) are involved. (iv) Chromosomes with CYP2D7/CYP2D6 genes either in unison or in a duplication arrangement lack CYP2D7 5′ to the recombinant gene (Fig. 1d and g). (v) Finally, all structures containing a single CYP2D6/CYP2D7 gene studied here were followed by a rep del sequence (Fig. 1e). These samples produced raw data outputs using the CYP2D6 ASPE kit, which could not be interpreted before this research.
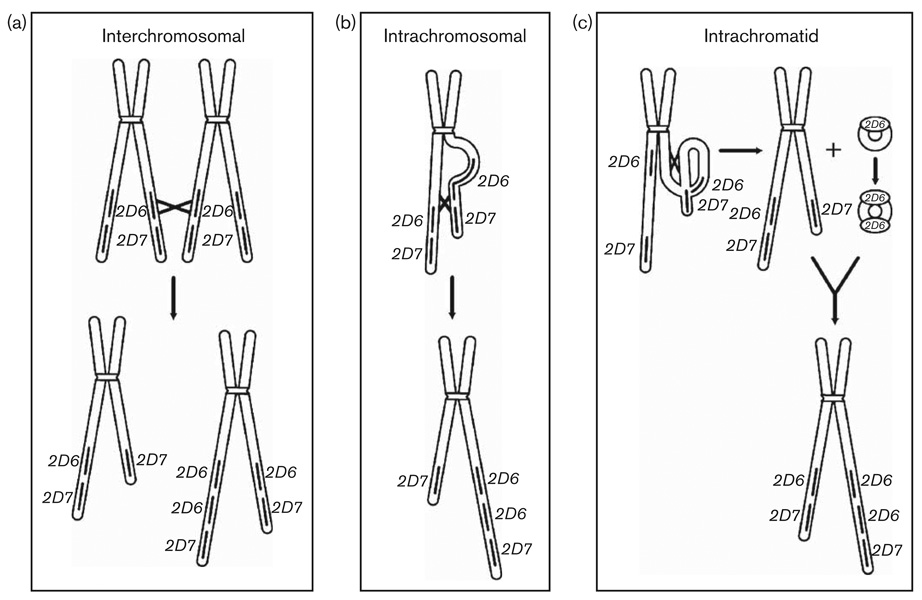
These findings lead us to consider the molecular mechanism whereby the observed genomic structures were generated. For this reason, we used these structures to model fit for interchromosomal, intrachromosomal, and intrachromatidic recombination mechanisms (Fig. 3) in a manner similar to that first proposed by Lundqvist et al. [21].
Fig. 3.
Interchromosomal, intrachromosomal, and intrachromatidic recombination. This diagram depicts interchromosomal, intrachromosomal, and intrachromatidic recombination using the descriptions as per Stankiewicz and Lupski [20]. Chromosomes are shown in black. Blocks in the chromosomes depict CYP2D6 and CYP2D7. (a) Interchromosomal recombination occurs between two independent homologous chromosomes during meiosis. (b) Intrachromosomal recombination occurs between sister chromatids of the same chromosome. (c) Intrachromatidic recombination involves recombination between low copy repeats on the same chromatid. Episomal DNA circles can undergo reinsertion after DNA replication at the excision site or at different loci on the same chromatid.
Interchromosomal recombination could generate the typical duplication structure defined by Fig. 1b when there are different alleles in tandem, which was observed in the case of genotype, *2A-*1A+*1 (270 T). The structure found in Fig. 1e could only be generated by interchromosomal recombination. Intrachromosomal and intrachromatidic recombination mechanisms cannot yield the *2A-*1A+*1 (270T) structure or structures depicted by Fig. 1e.
All other arrangements depicted in Fig. 1 can be generated by all three recombination mechanisms considered here and they are not informative about the type of recombination that has generated them unless sequence data of tandem genes in duplication or multiplication arrangements is taken into consideration. When this is done, the results presented here and by other researchers provide compelling evidence for intrachromosomal or intrachromatidic recombination. The finding that all but one of the duplication and multiplication structures studied here had the same 5′ and 3′ genes strongly supports these mechanisms. The duplication, multiplication, *4N+*4-like, *68+*4-like, and *13+*2A arrangements fit this model as well. Furthermore, the *36+*10B arrangement fits this model as the CYP2D6 nucleotide sequence found in *36 matches the corresponding CYP2D6 portion of *10B [11,14]. It should be noted that although interchromosomal recombination, also known as sister chromatid exchange, can generate these structures, multiple intrachromosomal exchanges would be necessary to generate the high copy number repeats that have been reported by others [7].
Intrachromatidic recombination represents a plausible mechanism for all of the recombinants that we observed except the *2A-*1A+*1 (270T) structure and the structure depicted by Fig. 1e. This involves the excision and possible reinsertion of an episomal DNA intermediate analogous to the double minute model of extra-chromosomal and genomic gene amplification [22–24]. These episomes have been observed in developmental stages of insect [25–27], frog [28,29] and mouse cell lines [30], and in mammalian cancer cells [31–35]. Carroll et al. [22] showed that mitotic gene amplification of the carbamoyl-phosphate synthetase/aspartate transcarbamoylase/dihydroorotase gene complex was mediated by episomal DNA circles in a Chinese Hamster Ovary cell line grown in the presence of the carbamoyl-phosphate synthetase/aspartate transcarbamoylase/dihydroorotase inhibitor N-phosphonacetyl-l-aspartate. Cells containing episomes had lost the corresponding chromosomal sequences. The episomes underwent amplification of their sequences to form larger episomes known as double minutes; these episomes were observed to reintegrate into the chromosome thereby amplifying the gene involved. Similarly, Von Hoff et al. [31] studied the location of amplified c-myc oncogene sequences in HL-60 promyelocytic leukemia cells through several cell passages and were able to follow the presence of episomal DNA through a double minute stage before they integrated into a chromosomal site as an amplified sequence. Amplification of drug resistance genes through this mechanism has been associated with drug resistance of patients’ tumors to antineoplastic agent [36–39].
The intrachromatidic mechanism can also generate recombinant arrangements (i.e. *4N+*4, *36+10B, *13+2A, and *16), large copy number repeats of identical alleles [7], and gene deletions (CYP2D6*5).
Future research should focus on determining the frequency of the genomic structures and alleles that we have found in larger clinical samples. Furthermore, a more comprehensive genotyping test should be designed to detect these structures and alleles. Finally, additional research to determine the actual mechanisms behind gene amplification and other recombinant events is warranted.
Acknowledgements
The authors thank Andrea Gaedigk PhD, David Flockhart MD PhD, and Richard Weinshilboum MD for reviewing this manuscript and providing constructive comments. Department of Psychiatry and Psychology, Department of Laboratory Medicine and Pathology and the Mayo Clinic Center for Translational Science Activities Program, Mayo Clinic, Rochester, MN, USA are the funding sources. Drs Black, O’Kane, and Mrazek have filed a patent disclosure for a psychiatric drug treatment algorithm involving CYP2D6 and have a licensing agreement with AssureRx for which they have received no fees to date. The authors have filed a patent disclosure for some information contained in this article. GenBank accession numbers: CYP2D6*1 (270T)-EU530602, CYP2D6*2A-*1A-EU530603, CYP2D6*4N(−1124C, 3435A)-EU530604, CYP2D6*4-like-EU530605, CYP2D6*68-EU530606, CYP 2D6*61-EU530607, CYP2D6*63-EU530608, CYP2D6*13-EU530609, CYP2D6*16-EU530610.
References
- 1.Black J, III, O’Kane D, Mrazek D. The impact of CYP2D6 allelic variation on antidepressant drug metabolism: a review. Expert Opin Drug Metabol Toxicol. 2007;3:21–31. doi: 10.1517/17425255.3.1.21. [DOI] [PubMed] [Google Scholar]
- 2.Kirchheiner J, Nickchen K, Bauer M, Wong M-L, Licinio J, Roots I, et al. Pharmacogenomics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiat. 2004;9:442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 3.Goetz M, Knox S, Suman V, Rae J, Safgren S, Ames M, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 4.Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenetics Genomics. 2007;17:93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 5.Lovlie R, Daly A, Molven A, Idle J, Steen V. Ultrarapid metabolizers of debrisoquine: characterization and PCR-based detection of alleles with duplication of the CYP2D6 gene. FEBS Lett. 1996;392:30–34. doi: 10.1016/0014-5793(96)00779-x. [DOI] [PubMed] [Google Scholar]
- 6.Gaedigk A, Blum M, Gaedigk R, Eichelbaum M, Meyer U. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am J Hum Genet. 1991;48:943–950. [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson I, Lundqvist E, Bertilsson L, Dahl M-L, Sjoqvist F. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci U S A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen V, Andreassen O, Daly A, Tefre T, Borresen A, Idle J, Gulbrandsen A. Detection of the poor metabolizer-associated CYP2D6(D) gene deletion allele by long-PCR technology. Pharmacogenetics. 1995;5:215–223. doi: 10.1097/00008571-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Steen V, Molven A, Aarskog N, Gulbrandsen A-K. Homologous unequal cross-over involving a 2.8 kb direct repeat as a mechanism for the generation of allelic variants of the human cytochrome P450 CYP2D6 gene. Hum Mol Genet. 1995;4:2251–2257. doi: 10.1093/hmg/4.12.2251. [DOI] [PubMed] [Google Scholar]
- 10.Gaedigk A, Ndjountche L, Divakaran K, DiAnne Bradford L, Zineh I, Oberlander T, et al. Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clinical Pharmacol Therap. 2007;81:242–251. doi: 10.1038/sj.clpt.6100033. [DOI] [PubMed] [Google Scholar]
- 11.Chida M, Ariyoshi N, Yokoi T, Nemoto N, Inaba T, Kinoshita M, et al. New allelic arrangement CYP2D6*36 × 2 found in a Japanese poor metabolizer of debrisoquine. Pharmacogenetics. 2002;12:659–662. doi: 10.1097/00008571-200211000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Daly A, Fairbrother K, Andreassen O, London S, Idle J, Steen V. Characterization and PCR-based detection of two different hybrid CYP2D7P/CYP2D6 alleles associated with the poor metabolizer phenotype. Pharmacogenetics. 1996;6:319–328. doi: 10.1097/00008571-199608000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Johansson I, Oscarson M, Yue Q-Y, Bertilsson L, Sjoqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46:452–459. [PubMed] [Google Scholar]
- 14.Gaedigk A, DiAnne Bradford L, Alander SW, Steven Leeder J. CYP2D6*36 gene arrangements within the CYP2D6 locus: associations of CYP2D6*36 with poor metabolizer status. Drug Metab Dispos. 2006;34:563–569. doi: 10.1124/dmd.105.008292. [DOI] [PubMed] [Google Scholar]
- 15.Ingelman-Sundberg M. Genetic Polymorphisms of cytochrome p450 2D6 (CYP2D6) clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 16.Panserat S, Mura C, Gerard N, Vincent-Viry M, Galteau M, Jacqz-Aigrain E, et al. An unequal cross-over event within the CYP2D gene cluster generates a chimeric CYP2D7/CYP2D6 gene which is associated with the poor metabolizer phenotype. Br J Clin Pharmacol. 1995;40:361–367. doi: 10.1111/j.1365-2125.1995.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanger U, Fischer J, Raimundo S, Stuven T, Evert B, Schwab M, et al. Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics. 2001;11:573–585. doi: 10.1097/00008571-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Sakuyama K, Sasaki T, Ujiie S, Obata K, Mizugaki M, Ishikawa M, et al. Functional characterization of 17 CYP2D6 allelic variants (CYP2D6.2, 10, 14A–B, 18, 27, 36, 39, 47–51, 53–55, and 57) Drug Metab Dispos. 2008;36:2460–2467. doi: 10.1124/dmd.108.023242. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda T, Maune H, Ikenaga Y, Naohara M, Fukuda K, Azuma J. Novel structure of the CYP2D6 gene that confuses genotyping for the CYP2D6*5 allele. Drug Metabol Pharmacokinet. 2005;20:345–350. doi: 10.2133/dmpk.20.345. [DOI] [PubMed] [Google Scholar]
- 20.Stankiewicz P, Lupski J. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 21.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226:327–338. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 22.Carroll S, DeRose M, Gaudray P, Moore C, Needham-Vandevanter D, Von Hoff D, et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol. 1988;8:1525–1533. doi: 10.1128/mcb.8.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll S, Gaudray P, DeRose M, Emery J, Meinkoth J, Nakkim E, et al. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: functional evidence for a mammalian replication origin. Mol Cell Biol. 1987;7:1740–1750. doi: 10.1128/mcb.7.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn P. Molecular biology of double-minute chromosomes. BioEssays. 1993;15:477–484. doi: 10.1002/bies.950150707. [DOI] [PubMed] [Google Scholar]
- 25.Stanfield S, Helinski D. Small circular DNA in Drosophila melanogaster. Cell. 1976;9:333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- 26.Cohen S, Agmon N, Yacobi K, Mislovati M, Segal D. Evidence for rolling circle replication of tandem genes in Drosophila. Nucleic Acids Res. 2005;33:4519–4526. doi: 10.1093/nar/gki764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in drosophila. Genome Res. 2003;13:1133–1145. doi: 10.1101/gr.907603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen S, Mechali M. A novel cell-free system reveals a mechanism of circular DNA formation from tandem repeats. Nucleic Acids Res. 2001;29:2542–2548. doi: 10.1093/nar/29.12.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen S, Mechali M. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 2002;3:1168–1174. doi: 10.1093/embo-reports/kvf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tlsty T, Brown P, Schimke R. UV radiation facilitates methotrexate resistance and amplification of the dihydrofolate reductase gene in cultured 3T6 mouse cells. Mol Cell Biol. 1984;4:1050–1056. doi: 10.1128/mcb.4.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Von Hoff D, Forseth B, Clare C, Hansen K, VanDevanter D. Double minutes arise from circular extrachromosomal DNA intermediates which integrate into chromosomal sites in human HL-60 leukemia cells. J Clin Invest. 1990;85:1887–1895. doi: 10.1172/JCI114650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beland J, Longo J, Hahn P. CpG island mapping of a mouse double-minute chromosome. Mol Cell Biol. 1993;13:4459–4464. doi: 10.1128/mcb.13.8.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll S, DeRose M, Kolman J, Nonet G, Kelly R, Wahl G. Localization of a bidirectional DNA replication origin in the native locus and in episomally amplified murine adenosine deaminase loci. Mol Cell Biol. 1993;13:2971–2981. doi: 10.1128/mcb.13.5.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen S, Lavi S. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol Cell Biol. 1996;16:2002–2014. doi: 10.1128/mcb.16.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Loon N, Miller D, Murnane J. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 1994;22:2447–2452. doi: 10.1093/nar/22.13.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaeffeler E, Schwab M, Eichelbaum M, Zanger U. CYP2D6 genotyping strategy based on gene copy number determination by TaqMan real-time PCR. Hum Mutat. 2003;22:476–485. doi: 10.1002/humu.10280. [DOI] [PubMed] [Google Scholar]
- 37.Schimke R. Gene amplification, drug resistance and cancer. Cancer Res. 1984;44:1735–1742. [PubMed] [Google Scholar]
- 38.Stark G. DNA amplification in drug resistant cells and in tumors. Cancer Surv. 1986;5:1–5. [PubMed] [Google Scholar]
- 39.Slamon D, Clark G, Wong S, Levin W, Ullrich A, McGuire W. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]