Abstract
Cadherins and protocadherins are cell adhesion proteins that play an important role in neuronal migration, differentiation and synaptogenesis, properties that make them targets to consider in schizophrenia (SZ) and bipolar disorder (BD) pathogenesis. Consequently, allelic variation occurring in protocadherin and cadherin encoding genes that map to regions of the genome mapped in SZ and BD linkage studies are particularly strong candidates to consider. One such set of candidate genes is the 5q31-linked PCDH family, which consists of more than 50 exons encoding three related, though distinct family members – α, β, and γ – which can generate thousands of different protocadherin proteins through alternative promoter usage and cis-alternative splicing. In this study, we focused on a SNP, rs31745, which is located in a putative PCDHα enhancer mapped by ChIP-chip using antibodies to covalently modified histone H3. A striking increase in homozygotes for the minor allele at this locus was detected in patients with BD. Molecular analysis revealed that the SNP causes allele-specific changes in binding to a brain protein. The findings suggest that the 5q31-linked PCDH locus should be more thoroughly considered as a disease-susceptibility locus in psychiatric disorders.
1. Introduction
Cadherins are transmembrane proteins with extensive extracellular domains that exhibit adhesion properties by homophilic and heterophilic protein-protein interactions, through which they guide neuronal migration and positioning during development (reviewed by Yagi and Takeichi, 2007). They also play a role in neuronal differentiation and synaptogenesis, processes that are believed to underlie the development of schizophrenia (SZ) and bipolar disorder (BD). Thus, genetic variation occurring in cadherin-encoding genes, especially those that map to regions of the genome implicated in SZ and BD by linkage analysis, should be viewed as candidates underlying disease susceptibility.
The cadherin family consists of nearly 100 different genes scattered throughout the genome either as separate entities or as members of tandem clusters that arose through gene duplication. The largest such cluster is the PCDHαβγ multigene family of protocadherins on chromosome 5q31 (Sano et al., 1993; Wu and Maniatis, 1999; Frank and Kemler, 2002; Tasic et al., 2002; Hirayama and Yagi, 2006; Zou et al., 2007). The organization and regulation of the PCDHαβγ cluster is consistent with an underlying innate mechanism for generating protein diversity. An array of PCDHα, and PCDHγ proteins is generated from a series of N-terminal encoding variable exons, which are transcribed via alternative promoter usage, and cis-alternative splicing to one of several different genes coding for C-terminal constant domains (Wu and Maniatis, 1999; Tasic et al., 2002; Wang et al., 2002; Hirayama and Yagi, 2006; Kanecko et al., 2006). There are 13 variable PCDHα exon domains, which are paired with one of two constant genes encoding class-specific C-termini. PCDHγ is configured in a similar manner with 19 variable exons and 3 constant genes. The PCDHβ gene locus contains 18 variable exons, but lacks a constant region. The repertoire of diverse isoforms encoded by this locus is increased by the presence of an unusually large number of polymorphic, nonsynonymous SNPs. With respect to the genetic diversity generated from a relatively small number of subunits, the 5q31-linked PCDH locus displays characteristics similar to the immunoglobulin and T-cell receptor loci, except that genetic diversity in B and T-lymphocytes is generated by somatic rearrangement.
PCDHα is expressed in the central nervous system during development, primarily in the postsynaptic density fractions, whereas PCDHγ is more ubiquitous (Bonn et al., 2007). Interestingly, RT-PCR analysis and DNA sequencing carried out in single Purkinje cells indicate that only one or two PCDHα and γ variable exons are expressed in individual cells, and that expression occurs in an allele-specific manner (Kohmura et al., 1998; Esumi et al, 2005; Kaneko et al., 2006). Restricting protocadherin expression irrespective of the isoform diversity capable of being generated is consistent with the notion that these proteins provide specific instructions and addresses to individual cells in their migration paths during development. Cell-cell contact is accomplished through homophilic interaction of protocadherin variable subunits. However, protocadherins contain specific disulfide-bonded Cys-X(5)-Cys motifs not found in classical cadherins, which suggest heterophilic cell adhesion properties as well, possibly through beta1 integrin or between PCDH α and γ proteins (Morishita et al., 2006; Bonn et al., 2007).
The 5q31-linked PCDH family is also a positional candidate locus in SZ and BD since linkage to the region has been reported in several studies in both conditions, although the most positive findings are somewhat telomeric (Schwab et al., 1997; Straub et al., 1997,2002; Kendler et al., 2000; Gurling et al., 2001; Paunio et al., 2001; DeLisi et al., 2002; Devlin et al., 2002; Lewis et al., 2003; Sklar et al., 2004; Hong et al., 2004; Herzberg et al., 2006; Kerner et al., 2007).
Several candidate genes in the region have been considered including epsin 4 (CLINT1), and the GABAA locus between 160–170Mb, although the results are equivocal (Ikeda et al., 2005; Liu et al., 2005; Petryshen 2005; Pimm et al., 2005; Tang et al., 2006; Jamra et al., 2007; Lo et al., 2007). Recently, Fanous et al., (2007) presented data showing a modest association in SZ to a haplotype in the neurogenin 1 gene, which maps to ~134.9Mb. Kirov et al (2003) found a significant difference in the distribution of a frameshift mutation in the PCDHγA8 gene in SZ and controls. However, the frequency of the variant was higher in controls than patients and no selective transmission of the allele was found in family triads. In a previous study, we analyzed a polymorphic copy number variation (CNV), a16.7 kilobase (kb) deletion affecting PCDH exons α8–10, initially characterized by Noonan, et al., (2003). However, no differences were detected in patients with BD and SZ compared with controls (in press).
We have now extended our analysis of this interesting locus by examining a SNP located in an enhancer element located 3’ to the PCDHα locus. A significant increase in homozygosity for the minor allele was found in patients with BD.
2. Methods and Materials
2.1. Subjects
Individuals with BD from the Czech Republic were unrelated subjects recruited from in-patient and out-patient units at the Prague Psychiatric Center, Psychiatric Hospital Bohnice, Psychiatric Clinic (n=167). Diagnosis was made on the basis of either a Schedule for Affective Disorders and Schizophrenia-Lifetime (SADS-L; Endicott and Spitzer, 1978) interview (N=68) or by unstructured clinical interview modified from SADS-L using Research Diagnostic Criteria (RDC) criteria for diagnosis of either bipolar disorder I or bipolar disorder II (Spitzer et al, 1978) (N=99). Control subjects from the Czech Republic were blood-bank donors and individuals hospitalized for medical reasons (n=211). Seventy-one control subjects did not have underlying psychiatric illness based on a brief psychiatric clinical interview. In the remaining controls, all from blood bank donors, no formal testing procedure was used to screen for personal history of mental illness. However, the blood bank only accepted subjects who were not being treated for a psychiatric illness and had no family history of mental illness.
Individuals with SZ (n=176) were recruited from Rockland State Hospital. Diagnosis was established by Research Diagnostic Criteria (RDC) using SCID or clinical interview. U.S. controls (n=175) were blood bank donors. No formal testing procedure was used to screen these subjects to rule out individuals who had a personal or family history of mental illness, although the frequency of BD and SZ in a population of blood donors would be expected to be 1% or less for each. All subjects were Caucasians. They each signed an informed consent approved by the Ethical Committee on Clinical Investigation (Czech samples) and the AECOM IRB (U.S. samples).
2.2 ChIP-chip
A custom tiled microarray was designed containing the entire PCDHαβγ cluster including extensive flanking domains (chromosome 5, 138,700,000–141,500,000). 50-mer oligonucleotide probes were tiled every 38 bp across the region, minus repetitive sequences (NimbleGen Systems [NGS]; Madison WI and Reykjavik, Iceland). The array included a number of other SZ and BD candidate genes, the results of which will be reported elsewhere. Coordinates are from the hg18 assembly, NCBI build 36.1 (genome.ucsc.edu). Chromatin immunoprecipation was carried on brain tissue from a 20-week old aborted fetus using procedures described in Oberley et al., (2004) and Xu et al., (2007). Tissue was obtained from the Human Fetal Tissue Repository at the Albert Einstein College of Medicine under an IRB-approved protocol. IgG antibody to acetylated histone H3K9/14 (H3K9/14Ac) (Upstate Cell Signaling, now part of Millipore, Billerica, MA USA), and monomethylated histone H3K4 (H3K4me1) (Abcam ab8895) were used for the immunoprecipitation. Sheared input chromatin was used as a control. The entire procedure for preparing ligation mediated PCR amplified DNA from immunoprecipitated chromatin can be found in supplementary table 1. H3K4me1 ChIP-chip was carried out twice, while the H3K9/14Ac experiment was performed only once since results from this and other arrays showed that there was excellent overlap with the monomethylation data. Arrays were hybridized with Cy5 and Cy3-labeled DNA. The samples were labeled and hybridized by NGS using their standard in-house protocols. GFF files generated from the hybridization signals were imported for data analysis. Significant differences in the log2 ratio signal between modified histone immunoprecipitates and control DNA and an estimate of the false discovery rate (FDR) were determined using NimbleScan (NimbleScan user’s guide). An FDR <0.05 (based on randomizing the data 20 times) provides the highest confidence level that the peak corresponds to a true protein binding site (red peaks). FDR rates between 0.05 and 0.2 are also indicative of a protein-binding site (orange peaks, 0.05–0.1; yellow peaks, 0.1–0.2. Grey peaks (FDR>0.2) were viewed as false positives.
2.2. Genotyping
Genotyping was carried out using the TaqMan® Allelic Discrimination Technique according to the manufacturer’s instructions. Samples were amplified by PCR in 384 well plates using an Applied Biosystems Model 7900HT thermal cycler and SDS 2.1 software (Applied Biosystems). A total of six SNPS were analyzed in the PCDHα gene locus, which spans ~260kb (140,146,060–140,372,113): rs10036519 (140,131,885 upstream of start site), rs3756337 (140,166,648 in PCDHα4), rs59479 (140,241,887 in PCDHα13), rs6876364 (140,387,178), rs31745 (140,400,408 in PCDHα 3’ enhancer) and rs17119385. (140,402,321). All SNPs were available through the ABI pre-designed assays except rs59479, which was designed from a ~600bp sequence using File Builder v3.1 software (Applied Biosystems) (supplementary table 2).
2.3. Electromobility Gel Shift Assay (EMSA)
EMSA was performed according to published procedures (Hope et al., 1994). Briefly, double-stranded oligonucleotide probes containing the polymorphic variants were constructed. These were annealed and end-labeled with 32P deoxynucleotides using Klenow polymerase to fill in 5’ overhangs, which generated double stranded probes that were used for protein binding experiments (probe size 32bp after filling in: primers used to generate allele specific probes are shown in supplemental table 2). Nuclear protein extract was isolated from fetal (whole brain) and adult brains (parietal lobe). Protein (10 micrograms) was mixed with probe (1 nanogram, ~106 counts) and incubated for 20 minutes at room temperature. Probes containing the different alleles were labeled simultaneously using the same amount of DNA and radioactive nucleotides. After purifying the probes, an aliquot was analyzed by scintillation counting to ensure that the labeling efficiency was greater than 108 cpm/µg. DNA-protein complexes were resolved by electrophoresis in a non-denaturing gel system containing 6% acrylamide and 1.6% glycerol. Specificity of the resulting binding activity was demonstrated by competition with non-radioactive probe added in 100-fold excess. Autoradiograms were scanned and quantified by normalizing against unused probe. Differences between the two alleles for each sample were analyzed by using a paired t-test.
2.4. Analysis of copy loss by Quantitative PCR
Quantitative real-time PCR (qPCR) was used to detect copy differences applying the technique described by Meijerink et al (2001), Ponchel et al (2003), and Weksberg et al (2005). The fluorescent signal from the TaqMan® assay was used to assess the test region, while for the control gene, H6PD (hexose-6-phosphate dehydrogenase — the autosomal or H-form of glucose 6-phosphate dehydrogenase PCR was carried out in the presence of SYBR® Green 1 (SYBR® Green 1 Master Mix, Applied Biosystems). The complete procedure is shown in supplementary table 3.
2.5. Statistical analysis
Tests for Hardy-Weinberg equilibrium (HWE) were conducted for each of the SNPs separately for controls and cases. The exact p-value for the HWE test was approximated by 10,000 permutations. A statistical program, StatXACT_5 (Cytel Software Corporation, Cambridge MA) was used to compute chi square statistics for differences in allele and genotype frequencies. The level of significance was set at p<.05.
3. Results
3.1. ChIP-chip
The <50 different PCDHαβγ exons are heterogeneously expressed throughout the central nervous system. We reasoned, therefore, that if this locus is involved in SZ or BD pathogenesis, which appear to affect a variety of cortical and subcortical regions, regulatory factors that influence the expression of many components, such as an enhancer element, would be the most reasonable targets to analyze for disease involvement. To identify putative regulatory domains in the PCDH locus in human tissue, ChIP-chip was used. Chromatin from fetal brain was immunoprecipitated with antibodies to histone H3 monomethylated at lysine 4 (H3K4me1), which is associated with expressed genes, active promoters and some enhancers, and histone H3 acetylated at lysines 9 and 14 (H3K9/14Ac), which is enriched in promoters, enhancers and putative regulatory domains of unknown function (Bernstein 2005; Roh et al., 2005, 2007; Heintzman et al., 2007). Immunoprecipitates were hybridized to tiled microarrays containing the entire PCDHαβγ locus. Hybridization peaks from representative H3K9/14Ac and H3K4me1 ChIP-chip experiments, indicative of enrichment of DNA in the chromatin immunoprecipitates generated using antibodies to modified histones compared to control chromatin, are shown in supplementary figure 1 and supplementary figure 2; a summary of the findings is shown on table 1. A total of 13 peaks or peak clusters were found in the H3K9/14Ac ChIP-chip, and 12 were found in the H3K4me1 ChIP-chip experiments. Of these, 11 overlapped. A number of peaks are near the transcription start sites (TSS) for several genes, corresponding to enrichment of their respective promoter regions in the immunoprecipitates, including PCDHαC1, PCDHαC2, PCDHγA1, PCDHγC3, PCDHγC4, PCDHγC5, and TAF7, a closely linked gene. The ability to detect promoters with the ChIP-chip strategy we used is consistent with previous observations (Bernstein et al., 2005), Roh et al., 2005) and Heintzman et al., 2007; Lachman et al, submitted). Of note, however, is the absence of significant peaks at all but one of the PCDHαβγ variable exon promoters; the exception is PCDHγA1 (H3K9/14Ac peak 6). The absence of other variable exon promoter peaks is most likely due to their heterogeneous activation throughout the CNS. This is consistent with data reported showing DNA hypomethylation at an active PCDHα promoter in a cell line in contrast to the heterogeneous DNA methylation pattern found in brain tissue (Kawaguchi et al., 2008).
TABLE 1. Summary OF ChIP-chip findings in PCDHαβγ locus.
Summary of ChIP-chip peaks shown in supplemental figure 1 and supplemental figure 2 for H3K9/14Ac and H3K4me1 immunoprecipitates. Map distances on chromosome 5 are from NCBI build 36.1, hg18.
| H3K9/14 Ac Peaks | H3K4me1 Peaks | Notes |
|---|---|---|
| 1. 140,286,433–140,287,582 | 1. 140,287,298–140,287,602 | PCDHαC1 promoter (TSS 140,286,486) |
| 2. 140,326,988–140,327,157 | 2. 140,326,798–140,328,277 | PCDHαC2 promoter (TSS 140,326,296) |
| 3. 140,361,419–140,362,779 | 3. 140,361,319–140,362,458 | corresponds to HS 7 |
| 4. 140,400,198–140,400,868 | 4. 140,399,758–140,400,086 | corresponds to HS 5-1 |
| 5. 140,680,813–140,681,002 | 5. 140,679,488–140,679,732 | TAF7 promoter (TSS 140,678,241) |
| 6. 140,686,031–140,686,385 | PCDHγA1 (TSS 140,690,436) | |
| 7. 140,836,076–140,838,601 | 6. 140,835,361–140,839,748 | PCDHγC3 (TSS 140,835,753) |
| 8. 140,844,072--40,845,334 | 7. 140,843,692–140,847,314 | PCDHγC4 (TSS 140,844,925) |
| 8. 140,848,685–140,851,154 | PCDHγC5 (TSS 140,848,992) and exon | |
| 9. 140,852,520–140,855,580 | 9. 140,853,196–140,856,220 | common constant region exon |
| 10. 140,868,194–140,869,553 | 10. 140,867,949–140,869,553 | ~1kb 5’ to PCDHγ constant region terminal exon |
| 11. 140,872,814–140,874,481 | 11. 140,872,879–140,874,516 | 3’ to PCDHγ locus |
| 12. 140,883,968–140,884,963 | 12. 140,884,193–140,884,922 | 3’ to PCDHγ locus, 5’ end of EST AK094264 |
| 13. 140,917,578–140,919,962 | EST BC041908 at 140,918,062; DIAPH1 intron |
ChIP-chip peaks were also found near constant gene exons (H3K4me1 peaks 8–10; H3K9/14Ac peaks 9 and 10), suggestive of a role in alternative splicing. Finally, most relevant to this study, two peaks (peaks 3 and 4 in both ChIP experiments) were detected in regions corresponding to two PCDH enhancers identified in the murine PCDHα locus by Ribich et al., referred to as HS7 and HS5–1, respectively. Of these, HS5–1 (peak 4) was found to increase the expression of PCDHαC1, which codes for one of the constant genes, and all PCDHα variable exons, in the CNS of transgenic mice (Ribich et al., 2006). Based on our hypothesis that involvement of PCDHαβγ in SZ and BD would more likely involve multiple genes in the locus, we decided to focus on this region for further investigation.
3.2. Genetic analysis of rs31745
The region defined by peak 4 is highly conserved, contains two transcription factor binding sites (TFBS track, UCSC Browser), and has been immunoprecipitated with antibodies to H3K4me3 and c-myc in hes-3 ES cells and P493 cells, respectively, in other ChIP experiments (ChIP-PET track from Genome Institute of Singapore, UCSC Browser). These are features characteristic of enhancers. The two TFBS within the peak are LHX3, a LIM homeobox transcription factor, and the pro-apoptotic transcription factor CHOP/GADD153, which is a C/EBP homologue, a transcription factor targeted by the GSK3β signaling pathway, a target of lithium salts (Gould et al., 2007; Mennen et al., 2007; Mullen et al., 2007).
Four SNPs are posted in dbSNP in the region contained within the peak 4. We chose to analyze rs31745, which maps to 140,400,408, because it is found in the region conserved only in higher order mammals (table 2). In addition the major allele (C) appears to be exclusive to humans, while the minor allele (T) is ancestral, being found in chimp, rhesus, dog, mouse, horse and armadillo. Two data sets were analyzed using a case control association design; cohorts of European Caucasian patients with SZ from the U.S. and Caucasian patients with BD from the Czech Republic. As seen in table 3, a significant increase in homozygosity for the T allele was detected in the bipolar patients; 5% had this genotype, but no controls (χ2 test for independence of genotype distribution, Pearson χ2 statistic=12.63, exact p=0.001, 2 df, two-tailed). However, the distribution of alleles did not differ between patients and controls because of the smaller number of heterozygotes detected in the bipolar patients (allele frequency Pearson χ2 statistic=0.32, asymptotic p=0.57, 1df, two-tailed). An increase in homozygotes for the T allele was also seen in SZ patients, but the results were not statistically significant; a larger sample size will be needed to more accurately assess this marker in SZ.
Table 2. Sequence comparison near rs31745.
Sequence conservation around rs31745 for seven different species taken from UCSC browser. Major allele (C) highlighted in bold, caps and underlined.
| Human | ctcttCttgtctaattagtcgctaagcaactg |
| Chimp | ctctttttgtctaactagtcgctaggcaactg |
| Rhesus | ctctttttgtctaactagtcgctaggcaactg |
| Mouse | tttttgtctaactggtcgctaggcaactg |
| Dog | ctctttttgtctaatgcattgctaagcaaccg |
| Horse | ttctttttgtctaatttgttgctaggtaactg |
| Armadillo | ttctttttgt----------------------------- |
TABLE 3. Analysis of rs31745 in BD and SZ.
BD vs CONTROLS: allele frequency Pearson χ2 statistic=0.32, asymptotic p=0.57 (1df, two-tailed); genotype distribution χ2 test for independence, Pearson χ2 statistic=12.63, exact p=0.001 (2 df, two-tailed); HWE: CONTROLS, p=0.14; BD, p=0.0004. SZ vs CONTROLS allele frequency Pearson χ2 statistic=0.45, asymptotic p=0.50 (1df, two-tailed); genotype distribution χ2 test for independence, Pearson χ2 statistic=1.28, exact p=0.53 (2 df, two-tailed); HWE (exact p-values): CONTROLS, p=0.70; SZ, p=0.66
| Genotypes | Alleles | |||||
|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||
| CONT | 166 (.79) | 45 (.21) | 0 (0) | 377 (.89) | 45 (.11) | |
| BD | 135 (.81) | 24 (.14) | 8 (.05) | 294 (.88) | 40 (.12) | |
| CONT | 135 (.78) | 37 (.21) | 1 (.006) | 307 (.89) | 39 (.11) | |
| SZ | 143 (.82) | 30 (.17) | 2 (.01) | 316 (.90) | 34 (.10) | |
The marked increase in homozygotes, given the allele frequency, and the relative decrease in heterozygotes resulted in strong deviation from HWE (exact p=0.0004). The genotype distribution from the control group for this population, and both the SZ and control samples from the US were, however, in HWE (table 3).
To exclude genotyping error as a cause of HWE deviation in the bipolar subjects, the homozygotes were reanalyzed; no errors were detected. In addition, the entire data set was analyzed for two other markers that are in strong LD with rs31745; rs6876364 (140,387,178) and rs17119385 (140,402,321) (LD between rs31745 and rs6876364 is D’=1.0, r2=1.0 in HapMap CEPH families, and D’=1.0, r2=0.849 for rs31745 and rs17119385). Similar results were obtained when the entire data set was genotyped (supplementary table 4). Three other SNPs, rs10036519, rs3756337 and rs59479 located ~160–270kb upstream from the enhancer regions were also analyzed; no significant differences were found in allele or genotype distribution and no deviation from HWE was detected (supplementary table 4). We conclude that the association signal detected using rs31745 is not a genotyping artifact and the deviation from HWE appears to be restricted to SNPs in LD with the PCDHα enhancer region.
Another cause for deviation from HWE is copy variation. Several CNVs have been found in the PCDHαβγ region, including the 16.7 kb deletion involving PCDHα exons 8–10 discovered by Noonan et al., (2003), which we previously analyzed in SZ and BD (in press). An extensive polymorphic copy gain variant involving ~670kb, and a relative rare (<0.045% allele frequency) copy gain variant both appear to affect the 3’ enhancer region (variations 9522 and 3578, respectively; see Database of Genomic Variants, http://projects.tcag.ca/variation, based on Wang et al., 2007; Redon et al., 2006). Although copy loss, not gain, would account for the apparent increase in T allele homozygosity, we wanted to determine whether a deletion involving the enhancer region accounted for the increase in the number of apparent homozygotes (i.e, hemizygosity) we detected using rs31745 and neighboring SNPs. No evidence for copy loss was detected in the homozygotes using a variation of the 2−ΔΔCt technique (supplementary table 5).
3.3. EMSA
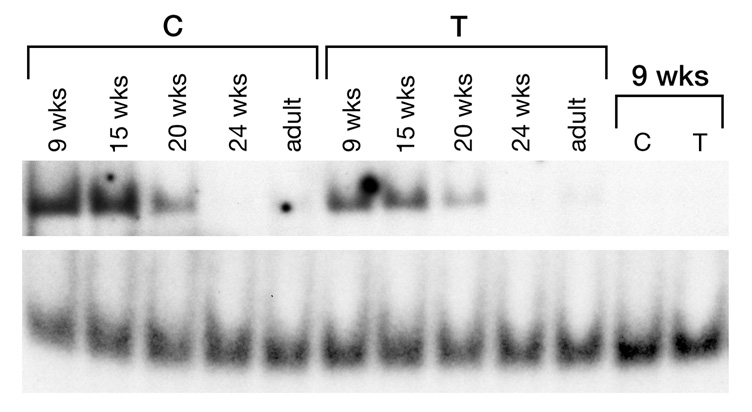
As a first step towards determining the functional significance of the enhancer polymorphism, EMSA was carried out. Crude nuclear protein extract from fetal brain tissue was used. Protein was annealed to labeled double stranded, allele-specific probes, and the samples were resolved by non-denaturing gel electrophoresis, as described in the methods section. As seen in figure 3, band shifts were detected for both alleles. These were specifically bound DNA-protein complexes as seen by the loss of signal when unlabeled competitor oligonucleotide was added during the annealing reaction (last two lanes). The most striking observation is the loss of signal with fetal age. The strongest signal was detected in nine-week old fetuses and the weakest at 20 weeks. Signal was weak in adult brain as well. This is consistent with the higher level of PCDHα expression found in developing brain compared with adults. Second, allele specific differences were also detected. Radiolabeled signals generated from the protein-DNA complexes were scanned and normalized. A 49% decrease in signal intensity was detected for the minor allele-containing probe (T) compared with the wild type allele (C) (p=0.044; paired T-test, supplementary table 6). A decrease in the DNA-protein complex generated with the T-allele probe is supported by the EMSA experiment carried out using different concentrations of unlabeled competitor, which shows a more rapid reduction in signal compared with C-allele probe (supplementary figure 3).
4. Discussion
SZ is viewed as a neurodevelopmental disorder in which fetal brain development is adversely affected by genetic factors, possibly in combination with an environmental component, such as neonatal hypoxia, maternal infection, malnutrition, and micronutrient deficiency (reviewed by Jarskog et al., 2007). Although prenatal complications have generally not been implicated in BD (Scott et al., 2006), a neurodevelopmental problem may underlie disease pathogenesis in a subgroup of such patients as well, as suggested by the involvement of both SZ and BD by DISC1, which codes for various protein isoforms that influence neuronal migration and neurite outgrowth (Kamiya et al., 2005; Maier et al., 2006; Sawamura and Sawa, 2006; Mackie et al., 2007; Roberts, 2007).
Based on the neurodevelopmental model, we and others are beginning to analyze cadherin and protocadherin-encoding genes in SZ and BD. Analysis of nonsynonymous SNPs in PCDH11X/Y, a brain expressed, hominoid-specific protocadherin member that maps to the Xq21.3 pseudoautosomal region revealed no significant differences in patients with SZ and controls, although the data sets may have been underpowered (Ross et al., 2003; Giouzeli et al., 2004; Durand et al., 2006). Analysis of a nonsynonymous SNP at PCDH8 codon 7 showed a trend towards significance for the minor allele in a large case control comparison, but not in a family based association sample (Bray et al., 2002). Another rare nonsynonymous SNP was found in one patient and an affected sibling, but no other patients, so its significance could be not determined. Recently published studies suggest that genetic variation in the cadherin member FAT is involved in BD (Blair et al., 2006), and haplotypes within 22q11-linked ARVCF, a member of the catenin family that maps immediately 3’ to COMT, are associated with SZ (Sanders et al., 2005); catenins interact with cadherins to modulate synaptic function (reviewed by Kwiatkowski et al., 2007).
Molecular analysis of protocadherins in SZ and BD has been limited so far. Dean et al., (2007), found an increase in PCDH17 mRNA in BA 46 in patients with SZ, but only in subjects with a short history of disease. No changes were found in other brain areas. In our previous study of a polymorphic copy deletion variant involving PCDHα exons 8–10, the frequency was similar in patients with SZ and BD, as well as controls, although the sample was underpowered to detect a small effect (in press). Similarly, no association was detected in SZ in the analysis of rs31745 in this study, which maps to a putative PCDHα enhancer (although there were 2 patients and only 1 control who were homozygous for the minor allele). An underpowered sample size could have resulted in the absence of statistical significance for minor allele homozygosity. Nevertheless, a much more thorough investigation of the PCDHα and γ loci is justified in SZ, considering the allele-specific pattern of expression found for PCDH α and γ variable exons; allele-specific expression could explain the MZ concordance rate of ~50% found in SZ and the similar risk of SZ found in the children of discordant and concordant MZ twins (Gottesman and Bertelsen, 1989; Onstad et al., 1991; Tsuang, 2000; Petronis, 2003; Kato et al., 2005; Procopio, 2005).
In contrast to the negative findings in SZ, however, a strong association was detected in the bipolar cohort from the Czech Republic we analyzed. One caveat to the finding is the significant deviation from HWE in the patient sample. We ruled out several experimental artifacts and genetic phenomena that could account for the finding including genotyping error and copy variation. In addition, the clinical records and demographic profiles for each of the rs31745 homozygotes were examined to determine if population stratification could have artificially caused the deviation from HWE. However, these subjects did not appear to differ from other bipolar subjects analyzed in this study. Nevertheless, more subtle forms of population stratification could exist which would require much more extensive genotyping. Another possibility, of course, is that homozygosity for the minor allele in the enhancer SNP is a risk genotype for BD, and selection for this genotype in the bipolar cohort could have resulted in HWE deviation.
The function of the enhancer SNP has not yet been determined. However, a significant decrease in binding to an unknown brain protein was detected by EMSA, which is suggestive, but by no means proof of an in vivo effect. The SNP is of interest because the major allele appears to be unique to humans and the surrounding 30–35 bases are only conserved in higher animals. The effect of this SNP on enhancer function and the nature of the DNA binding protein detected in the EMSA experiments are currently under investigation. A TRANSFAC search for potential binding sites for transcription factors shows that rs31745 is within a motif for the POU transcription factor Brn2 (POU3F2), which regulates neural development (motif is CATNSRWAATNMR; Core match 0.987, matrix match 0.842 for C allele; Core match 0.987, matrix match 0.790 for T allele). Two NGAAN motifs suggestive of HSF1 (heat shock factor 1) binding sites are also found for the C allele (opposite orientation), while the T allele has only one. However, the random frequency of this motif is very high, so the detection does not have much statistical significance. Also, two motifs are usually insufficient for HSF binding.
The 3’ enhancer in mice referred to as HS5–1, which corresponds to peak 4 in our ChIP-chip experiments, has been shown to affect the expression of all PCDHα variable exons, as well as PCDHαC1, but not PCDHαC2 or the adjacent PCDHβ locus (Ribich et al., 2006). Whether the same pattern of regulation exists in humans is not known. An effect on an enhancer regulating the expression of each PCDHα variable exon and PCDHαC1, to the exclusion of an affect on PCDHαC2, could influence synaptogenesis in brain development, and in the post-development brain as well, by reducing expression of PCDHα variable exons and changing the dynamics of constant region splicing.
Overall, our findings suggest that the PCDHαβγ is an interesting gene family to consider in SZ and BD susceptibility, and that rs31745 is a candidate allelic variant.
Supplementary Material
Colored peaks correspond to increase in log2 ratio signal intensity (test immunoprecipitate/control) above threshold. The color code corresponds to the false discovery rate (FDR) based on randomizing the log2 ratio data 20 times, as described in methods. The numbers correspond to the peaks or peak clusters described in manuscript table 1. PCDHα extends from ~140,140,000–140,350,000; PCDHβ from ~140,420,000–140,140,620,000 and from ~140,700,000–140,850,000. Horizontal lines under peaks correspond to the primary transcripts generated from each member of the PCDHαβγ family. Direction of transcription for PCDHαβγ is left to right (sense strand).
Same as legend to supplementary figure 1
EMSA was carried out as shown in figure 1. Protein extract was incubated with equal amounts of labeled probe along with a 10 to 100-fold excess of unlabled oligonucleotide probe (“cold” competitor). The signal shown in the first two lanes contains no competitor. As seen in the figure, the signal generated with the “T” allele was diminished in each experimental setting, as shown in figure 1. In addition, at this exposure, a very weak signal is seen with the “T” allele when a 25-fold molar excess of cold competitor was added, and no signal is seen with a 50-fold excess, while the “C” allele signal is plainly visible.
Protocol for immunoprecipitation adapted from Oberley et al (2004) and Xu et al (2007).
A. Primer sets 1 + 2, and 3 + 4 were annealed to form double-stranded probes for EMSA. G and C overhangs are generated which are filled in with 32PdGTP and 32PdCTP. Probes 1 and 2 differ from 3 and 4 by a single nucleotide shown in bold and underlined to generate allele-specific probes.
B. H6PD (hexose-6-phosphate dehydrogenase – the autosomal or H-form of glucose 6-phosphate dehydrogenase [G6PD]) was used as a control gene for copy number determination, as described by Weksberg et al, 2005; Lachman et al, 2007). Amplicon is 220bp. The test primer was the TaqMan® probe for rs17119385
method for calculating DNA copy number from Weksberg et al, 2005 and Lachman et al, 2007.
HWE for additional markers (see methods for map position). Bold indicates deviation from HWE for bipolar subjects at rs6876364 and rs17119385. Genotype and allele frequency distributions were not significant except for BD vs controls for rs6876364 and rs17119385 which showed significant differences in genotype distribution: χ2 test for independence, Pearson χ2 statistic=11.28, exact p=0.0025; and 11.27, p=0.0024, respectively. Ten Czech control samples could not be genotyped for rs17119385 because of poor performance after two attempts or lack of availability.
Determination of copy number: Two copy controls were identified as described in the text, which served as reference samples. Two copy controls should have ΔKCt = 0.00 (actual value in 6 experiments, 0.07 +/− 0.34, upper panel). Three copy controls should have ΔKCt values ~+0.50 (actual value in 3 experiments, 0.65 +/− 0.19). Analysis of individual alleles in two heterozygotes was used as single copy controls. An analysis of six PCR reactions resulted in a mean ΔKCt −0.98, very close to the expected value of −1.00 (C1-C6, C-allele; T1–T6, T-allele). Test samples, homozygotes for the minor allele at rs17119385, are shown at bottom of table: no evidence for copy loss was detected. Two samples, BD8 (a Czech bipolar patient) and SZ2 (a patient with SZ) did not perform well in the assay and their copy number could not be determined (N/A).
EMSA autoradiograms were scanned and normalized ratios obtained for C vs T containing probes, as described in methods. Differences in signal between the two different probes for the different samples of tissue shown in figure 1 were compared using paired t-test (column 1 vs column 2).
Figure 1.
EMSA. Double-stranded oligonucleotide probes (32-mers) containing either the C or T alleles for rs31745 were annealed to nuclear brain protein extracts from fetal (different ages shown) and adult tissue. Top panel, DNA-protein complex; bottom panel, unused probe. Last two lanes show loss of signal when 100-fold molar excess of unlabeled probe was added to annealing step (protein from 9 week old fetus used in cold competition).
Acknowledgments
We would like to thank Hana Fridrichova for help in recruiting patients and administrative support for the Czech bipolar cohort, and Petr Turek, M.D., Ph.D. for providing Czech blood bank controls. The authors would also like to thank the Albert Einstein College of Medicine Human Genetics Program and the Department of Molecular Genetics for their assistance, and Dr. Joseph Locker of the Department of Pathology, for discussions related to proteins that could potentially bind to the enhancer region. PS is supported by a research grant from IGA MZ CR NR8564; HML is supported by the NIMH (R01MH073164) and by the Juvenile Bipolar Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blair IP, Chetcuti AF, Badenhop RF, Scimone A, Moses MJ, Adams LJ, Craddock N, Green E, Kirov G, Owen MJ, Kwok JB, Donald JA, Mitchell PB, Schofield PR. Positional cloning, association analysis and expression studies provide convergent evidence that the cadherin gene FAT contains a bipolar disorder susceptibility allele. Mol Psychiatry. 2006;11(4):372–383. doi: 10.1038/sj.mp.4001784. [DOI] [PubMed] [Google Scholar]
- Bonn S, Seeburg PH, Schwarz MK. Combinatorial Expression of α- and γ-Protocadherins Alters Their Presenilin-Dependent Processing. Mol Cell Biol. 2007;27(11):4121–4132. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NJ, Kirov G, Owen RJ, Jacobsen NJ, Georgieva L, Williams HJ, Norton N, Spurlock G, Jones S, Zammit S, O'Donovan MC, Owen MJ. Screening the human protocadherin 8 (PCDH8) gene in schizophrenia. Genes Brain Behav. 2002;1(3):187–191. doi: 10.1034/j.1601-183x.2002.10307.x. [DOI] [PubMed] [Google Scholar]
- Dean B, Keriakous D, Scarr E, Thomas EA. Gene expression profiling in Brodmann's area 46 from subjects with schizophrenia. Aust NZ J Psych. 2007;41(4):308–320. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K, Relja M, Byerley W, Sherrington R. Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet. 2002;114(5):497–508. doi: 10.1002/ajmg.10538. [DOI] [PubMed] [Google Scholar]
- Devlin B, Bacanu SA, Roeder K, Reimherr F, Wender P, Galke B, Novasad D, Chu A, Tcuenco K, Tiobek S, Otto C, Byerley W. Genome-wide multipoint linkage analyses of multiplex schizophrenia pedigrees from the oceanic nation of Palau. Mol Psychiatry. 2002;7(7):689–694. doi: 10.1038/sj.mp.4001056. [DOI] [PubMed] [Google Scholar]
- Durand CM, Kappeler C, Betancur C, Delorme R, Quach H, Goubran-Botros H, Melke J, Nygren G, Chabane N, Bellivier F, Szoke A, Schurhoff F, Rastam M, Anckarsater H, Eichler EE. Widening the spectrum of human genetic variation. Nat Genet. 2006;38(1):9–11. doi: 10.1038/ng0106-9. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35(7):837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37(2):171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Chen X, Wang X, Amdur RL, O'Neill FA, Walsh D, Kendler KS Related Articles. Association between the 5q31.1 gene neurogenin1 and schizophrenia. Am J Med Genet B (Neuropsychiatr Genet) 2007;144(2):207–214. doi: 10.1002/ajmg.b.30423. [DOI] [PubMed] [Google Scholar]
- Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14(5):557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- Fujii K, Matsubara Y, Akanuma J, Takahashi K, Kure S, Suzuki Y, Imaizumi M, Iinuma K, Sakatsume O, Rinaldo P, Narisawa K. Mutation detection by TaqMan-allele specific amplification: application to molecular diagnosis of glycogen storage disease type Ia and medium-chain acyl-CoA dehydrogenase deficiency. Human Mutation. 2002;15(2):189–196. doi: 10.1002/(SICI)1098-1004(200002)15:2<189::AID-HUMU8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Gillberg C, Leboyer M, Bourgeron T. Expression and genetic variability of PCDH11Y, a gene specific to Homo sapiens and candidate for susceptibility to psychiatric disorders. Am J Med Genet B (Neuropsychiatr Genet) 2006;141(1):67–70. doi: 10.1002/ajmg.b.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giouzeli M, Williams NA, Lonie LJ, DeLisi LE, Crow TJ. ProtocadherinX/Y, a candidate gene-pair for schizophrenia and schizoaffective disorder: a DHPLC investigation of genomic sequence. Am J Med Genet B (Neuropsychiatr Genet) 2004;129(1):1–9. doi: 10.1002/ajmg.b.30036. [DOI] [PubMed] [Google Scholar]
- Goidts V, Cooper DN, Armengol L, Schempp W, Conroy J, Estivill X, Nowak N, Hameister H, Kehrer-Sawatzki H. Complex patterns of copy number variation at sites of segmental duplications: an important category of structural variation in the human genome. Hum Genet. 2006;120(2):270–284. doi: 10.1007/s00439-006-0217-y. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Bertelsen A. Confirming unexpressed genotypes for schizophrenia. Risks in the offspring of Fischer's Danish identical and fraternal discordant twins. Arch Gen Psychiatry. 1989;46(10):867–872. doi: 10.1001/archpsyc.1989.01810100009002. [DOI] [PubMed] [Google Scholar]
- Gould TD, Dow ER, O'Donnell KC, Chen G, Manji HK. Targeting signal transduction pathways in the treatment of mood disorders: recent insights into the relevance of the Wnt pathway. CNS Neurol Disord Drug Targets. 2007;6(3):193–204. doi: 10.2174/187152707780619308. [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68(3):661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39(2):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herzberg I, Jasinska A, Garcia J, Jawaheer D, Service S, Kremeyer B, et al. Convergent linkage evidence from two Latin-American population isolates supports the presence of a susceptibility locus for bipolar disorder in 5q31-34. Hum Mol Genet. 2006;15(21):3146–3153. doi: 10.1093/hmg/ddl254. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Yagi T. The role and expression of the protocadherin-alpha clusters in the CNS. Curr Opin Neurobiol. 2006;16(3):336–342. doi: 10.1016/j.conb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Hong KS, McInnes LA, Service SK, Song T, Lucas J, Silva S, Fournier E, Leon P, Molina J, Reus VI, Sandkuijl LA, Freimer NB. Genetic mapping using haplotype and model-free linkage analysis supports previous evidence for a locus predisposing to severe bipolar disorder at 5q31-33. Am J Med Genet B (Neuropsychiatr Genet) 2004;125(1):83–86. doi: 10.1002/ajmg.b.20091. [DOI] [PubMed] [Google Scholar]
- Hope BT, Kelz MB, Duman RS, Nestler EJ. Chronic electroconvulsive seizure (ECS) treatment results in expression of a long-lasting AP-1 complex in brain with altered composition and characteristics. J Neurosci. 1994;14:4318–4328. doi: 10.1523/JNEUROSCI.14-07-04318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Iwata C, Suzuki T, Kitajima T, Yamanouchi Y, Kinoshita Y, Inada T, Ujike H, Ozaki N. Association analysis of chromosome 5 GABAA receptor cluster in Japanese schizophrenia patients. Biol Psychiatry. 2005;58(6):440–445. doi: 10.1016/j.biopsych.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Jamra RA, Becker T, Klopp N, Dahdouh F, Schulze TG, Gross M, Deschner M, Schmal C, Illig T, Rietschel M, Propping P, Cichon S, Nothen MM, Schumacher J. No evidence for an association between variants at the gamma-amino-n-butyric acid type A receptor beta2 locus and schizophrenia. Psychiatr Genet. 2007;17(1):43–45. doi: 10.1097/YPG.0b013e32801118cd. [DOI] [PubMed] [Google Scholar]
- Jarskog LF, Miyamoto S, Lieberman JA. Schizophrenia: new pathological insights and therapies. Annu Rev Med. 2007;58:49–61. doi: 10.1146/annurev.med.58.060904.084114. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7(12):1067–1078. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. 2006;281(41):30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- Kato T, Iwamoto K, Kakiuchi C, Kuratomi G, Okazaki Y. Genetic or epigenetic difference causing discordance between monozygotic twins as a clue to molecular basis of mental disorders. Mol Psychiatry. 2005;10(7):622–630. doi: 10.1038/sj.mp.4001662. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Toyama T, Kaneko R, Hirayama T, Kawamura Y, Yagi T. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. 2008 Jan 18; doi: 10.1074/jbc.M709648200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers JM, O'Neill FA, Martin R, Murphy B, MacLean CJ, Walsh D, Straub RE. Clinical features of schizophrenia and linkage to chromosomes 5q, 6p, 8p, and 10p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry. 2000;157(3):402–408. doi: 10.1176/appi.ajp.157.3.402. [DOI] [PubMed] [Google Scholar]
- Kerner B, Brugman DL, Freimer NB. Evidence of linkage to psychosis on chromosome 5q33-34 in pedigrees ascertained for bipolar disorder. Am J Med Genet B (Neuropsychiatr Genet) 2007;144(1):74–78. doi: 10.1002/ajmg.b.30402. [DOI] [PubMed] [Google Scholar]
- Kirov G, Georgieva L, Williams N, Nikolov I, Norton N, Toncheva D, O'Donovan M, Owen MJ. Variation in the protocadherin gamma A gene cluster. Genomics. 2003;82(4):433–440. doi: 10.1016/s0888-7543(03)00167-8. [DOI] [PubMed] [Google Scholar]
- Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20(6):1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Weis WI, Nelson WJ. Catenins: playing both sides of the synapse. Curr Opin Cell Biol. 2007 doi: 10.1016/j.ceb.2007.08.005. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52(1):103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta_analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20(6):1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Liu J, Shi Y, Tang W, Guo T, Li D, Yang Y, Zhao X, Wang H, Li X, Feng G, Gu N, Zhu S, Liu H, Guo Y, Shi J, Sang H, Yan L, He L. Positive association of the human GABA-A-receptor beta 2 subunit gene haplotype with schizophrenia in the Chinese Han population. Biochem Biophys Res Commun. 2005;334(3):817–823. doi: 10.1016/j.bbrc.2005.06.167. [DOI] [PubMed] [Google Scholar]
- Lo WS, Harano M, Gawlik M, Yu Z, Chen J, Pun FW, Tong KL, Zhao C, Ng SK, Tsang SY, Uchimura N, Stober G, Xue H. GABRB2 association with schizophrenia: commonalities and differences between ethnic groups and clinical subtypes. Biol Psychiatry. 2007;61(5):653–660. doi: 10.1016/j.biopsych.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Mackie S, Millar JK, Porteous DJ. Role of DISC1 in neural development and schizophrenia. Curr Opin Neurobiol. 2007 doi: 10.1016/j.conb.2007.01.007. [Epub ahead of print] PMID:17258902. [DOI] [PubMed] [Google Scholar]
- Maier W, Zobel A, Wagner M. Schizophrenia and bipolar disorder: differences and overlaps. Curr Opin Psychiatry. 2006;19(2):165–170. doi: 10.1097/01.yco.0000214342.52249.82. [DOI] [PubMed] [Google Scholar]
- Meijerink J, Mandigers C, van de Locht L, Tonnissen E, Goodsaid F, Raemaekers J. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real time PCR. J Mol Diagnostics. 2001;3(2):55–61. doi: 10.1016/S1525-1578(10)60652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon B, Johnson JN, Ross RS, Singh M, Singh K. Glycogen synthase kinase-3beta plays a pro-apoptotic role in beta-adrenergic receptor-stimulated apoptosis in adult rat ventricular myocytes: Role of beta1 integrins. J Mol Cell Cardiol. 2007;42(3):653–661. doi: 10.1016/j.yjmcc.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki R, Hattori K, Taguchi Y, Tada MN, Isosaka T, Hidaka Y, Hirabayashi T, Hashimoto R, Fukuzako H, Yagi T. Identification and characterization of coding single nucleotide polymorphisms within human protocadherin alpha and -beta gene clusters geneclusters. Gene. 2005;349:1–14. doi: 10.1016/j.gene.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, Akutsu H, Yamaguchi T, Yagi T, Ikegami T. Structure of the cadherin related neuronal receptor/protocadherin alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem. 2006;281(44):33650–33663. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, Bhangoo AP, Ten S, Weigel J, Pfäffle RW, Rhodes SJ. Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol. 2007:265–266. 190–195. doi: 10.1016/j.mce.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Li J, Nguyen L, Caoile C, Dickson M, Grimwood J, Schmutz J, Feldman MW, Myers RM. Extensive linkage disequilibrium, a common 16.7_kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am J Hum Genet. 2003;72(3):621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onstad S, Skre I, Torgersen S, Kringlen E. Twin concordance for DSMIIIR schizophrenia. Acta Psychiatr Scand. 1991;83(5):395–401. doi: 10.1111/j.1600-0447.1991.tb05563.x. [DOI] [PubMed] [Google Scholar]
- Paunio T, Ekelund J, Varilo T, Parker A, Hovatta I, Turunen JA, Rinard K, Foti A, Terwilliger JD, Juvonen H, Suvisaari J, Arajarvi R, Suokas J, Partonen T, Lonnqvist J, Meyer J, Peltonen L. Genome_wide scan in a nationwide study sample of schizophrenia families in Finland reveals susceptibility loci on chromosomes 2q and 5q. Hum Mol Genet. 2001;10(26):3037–3048. doi: 10.1093/hmg/10.26.3037. [DOI] [PubMed] [Google Scholar]
- Petronis A. Epigenetics and bipolar disorder: new opportunities and challenges. Am J Med Genet C (Semin Med Genet) 2003;123(1):65–75. doi: 10.1002/ajmg.c.20015. [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Middleton FA, Tahl AR, Rockwell GN, Purcell S, Aldinger KA. Genetic investigation of chromosome 5q GABAA receptor subunit genes in schizophrenia. Mol Psychiatry. 2005;10(12):1074–1088. doi: 10.1038/sj.mp.4001739. [DOI] [PubMed] [Google Scholar]
- Pimm J, McQuillin A, Thirumalai S, Lawrence J, Quested D, Bass N, Lamb G, Moorey H, Datta SR, Kalsi G, Badacsonyi A, Kelly K, Morgan J, Punukollu B, Curtis D, Gurling H. The Epsin 4 gene on chromosome 5q, which encodes the clathrin-associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am J Hum Genet. 2005;76(5):902–907. doi: 10.1086/430095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ, Robinson PA, Inglehearn CF, Isaacs JD, Markham AF. Real_time PCR based on SYBR_Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procopio M. Does god play dice with schizophrenia? A probabilistic model for the understanding of causation in mental illness. Med Hypotheses. 2005;64(4):872–877. doi: 10.1016/j.mehy.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-{alpha} gene cluster. Proc Natl Acad Sci. 2006;103(52):19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RC. Schizophrenia in translation: disrupted in schizophrenia (DISC1): integrating clinical and basic findings. Schizophr Bull. 2007;33(1):11–15. doi: 10.1093/schbul/sbl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17(1):74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19(5):542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross NL, Mavrogiannis LA, Sargent CA, Knight SJ, Wadekar R, DeLisi LE, Crow TJ. Quantitation of X-Y homologous genes in patients with schizophrenia by multiplex polymerase chain reaction. Psychiatr Genet. 2003;13(2):115–119. doi: 10.1097/01.ypg.0000056683.89558.1c. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Rusu I, Duan J, Vander Molen JE, Hou C, Schwab SG, Wildenauer DB, Martinez M, Gejman PV. Haplotypic association spanning the 22q11.21 genes COMT and ARVCF with schizophrenia. Mol Psychiatry. 2005;10(4):353–365. doi: 10.1038/sj.mp.4001586. [DOI] [PubMed] [Google Scholar]
- Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, Taketani S, Suzuki S. Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12(6):2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura N, Sawa A. Disrupted-in-schizophrenia 1 (DISC1): a key susceptibility factor for major mental illnesses. Ann N Y Acad Sci. 2006;1086:126–133. doi: 10.1196/annals.1377.018. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Eckstein GN, Hallmayer J, Lerer B, Albus M, Borrmann M, Lichtermann D, Ertl MA, Maier W, Wildenauer DB. Evidence suggestive of a locus on chromosome 5q31 contributing to susceptibility for schizophrenia in German and Israeli families by multipoint affected sib-pair linkage analysis. Mol Psychiatry. 1997;2(2):156–160. doi: 10.1038/sj.mp.4000263. [DOI] [PubMed] [Google Scholar]
- Scott J, McNeill Y, Cavanagh J, Cannon M, Murray R. Exposure to obstetric complications and subsequent development of bipolar disorder: Systematic review. Br J Psychiatry. 2006;189:3–11. doi: 10.1192/bjp.bp.105.010579. [DOI] [PubMed] [Google Scholar]
- Sklar P, Pato MT, Kirby A, Petryshen TL, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Ferreira CP, Lei M, Verner A, Hudson TJ, Morley CP, Kennedy JL, Azevedo MH, Lander E, Daly MJ, Pato CN. Genome-wide scan in Portuguese Island families identifies 5q31-5q35 as a susceptibility locus for schizophrenia and psychosis. Mol Psychiatry. 2004;9(2):213–218. doi: 10.1038/sj.mp.4001418. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Endicott J, Robins E. Research diagnostic criteria: Rationale and reliability. Arch Gen Psychiatry. 1978;35:773–782. doi: 10.1001/archpsyc.1978.01770300115013. [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7(6):542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O'Neill FA, Walsh D, Kendler KS. Support for a possible schizophrenia vulnerability locus in region 5q22-31 in Irish families. Mol Psychiatry. 1997;2(2):148–155. doi: 10.1038/sj.mp.4000258. [DOI] [PubMed] [Google Scholar]
- Tang RQ, Zhao XZ, Shi YY, Tang W, Gu NF, Feng GY, Xing YL, Zhu SM, Sang H, Liang PJ, He L. Family-based association study of Epsin 4 and Schizophrenia. Mol Psychiatry. 2006;11(4):395–399. doi: 10.1038/sj.mp.4001780. [DOI] [PubMed] [Google Scholar]
- Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10(1):21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry. 2000;47(3):210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes and Dev. 2002;16(15):1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M, Penn CNV. An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007 doi: 10.1101/gr.6861907. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, Squire JA. A method for accurate detection of genomic microdeletions using real_time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17(11):1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14(10):1169–1180. [PubMed] [Google Scholar]
- Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144(2):579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colored peaks correspond to increase in log2 ratio signal intensity (test immunoprecipitate/control) above threshold. The color code corresponds to the false discovery rate (FDR) based on randomizing the log2 ratio data 20 times, as described in methods. The numbers correspond to the peaks or peak clusters described in manuscript table 1. PCDHα extends from ~140,140,000–140,350,000; PCDHβ from ~140,420,000–140,140,620,000 and from ~140,700,000–140,850,000. Horizontal lines under peaks correspond to the primary transcripts generated from each member of the PCDHαβγ family. Direction of transcription for PCDHαβγ is left to right (sense strand).
Same as legend to supplementary figure 1
EMSA was carried out as shown in figure 1. Protein extract was incubated with equal amounts of labeled probe along with a 10 to 100-fold excess of unlabled oligonucleotide probe (“cold” competitor). The signal shown in the first two lanes contains no competitor. As seen in the figure, the signal generated with the “T” allele was diminished in each experimental setting, as shown in figure 1. In addition, at this exposure, a very weak signal is seen with the “T” allele when a 25-fold molar excess of cold competitor was added, and no signal is seen with a 50-fold excess, while the “C” allele signal is plainly visible.
Protocol for immunoprecipitation adapted from Oberley et al (2004) and Xu et al (2007).
A. Primer sets 1 + 2, and 3 + 4 were annealed to form double-stranded probes for EMSA. G and C overhangs are generated which are filled in with 32PdGTP and 32PdCTP. Probes 1 and 2 differ from 3 and 4 by a single nucleotide shown in bold and underlined to generate allele-specific probes.
B. H6PD (hexose-6-phosphate dehydrogenase – the autosomal or H-form of glucose 6-phosphate dehydrogenase [G6PD]) was used as a control gene for copy number determination, as described by Weksberg et al, 2005; Lachman et al, 2007). Amplicon is 220bp. The test primer was the TaqMan® probe for rs17119385
method for calculating DNA copy number from Weksberg et al, 2005 and Lachman et al, 2007.
HWE for additional markers (see methods for map position). Bold indicates deviation from HWE for bipolar subjects at rs6876364 and rs17119385. Genotype and allele frequency distributions were not significant except for BD vs controls for rs6876364 and rs17119385 which showed significant differences in genotype distribution: χ2 test for independence, Pearson χ2 statistic=11.28, exact p=0.0025; and 11.27, p=0.0024, respectively. Ten Czech control samples could not be genotyped for rs17119385 because of poor performance after two attempts or lack of availability.
Determination of copy number: Two copy controls were identified as described in the text, which served as reference samples. Two copy controls should have ΔKCt = 0.00 (actual value in 6 experiments, 0.07 +/− 0.34, upper panel). Three copy controls should have ΔKCt values ~+0.50 (actual value in 3 experiments, 0.65 +/− 0.19). Analysis of individual alleles in two heterozygotes was used as single copy controls. An analysis of six PCR reactions resulted in a mean ΔKCt −0.98, very close to the expected value of −1.00 (C1-C6, C-allele; T1–T6, T-allele). Test samples, homozygotes for the minor allele at rs17119385, are shown at bottom of table: no evidence for copy loss was detected. Two samples, BD8 (a Czech bipolar patient) and SZ2 (a patient with SZ) did not perform well in the assay and their copy number could not be determined (N/A).
EMSA autoradiograms were scanned and normalized ratios obtained for C vs T containing probes, as described in methods. Differences in signal between the two different probes for the different samples of tissue shown in figure 1 were compared using paired t-test (column 1 vs column 2).