Abstract
The pRNA (packaging RNA) of bacteriophage phi29 DNA packaging motor has been reported to have novel applications in nanotechnology and nanomedicine. The unique ability of pRNA to form dimers, trimers, hexamers and patterned superstructures via the interaction of two reengineered interlocking loops makes it a promising polyvalent vehicle to load siRNA and other therapeutic molecules and be applied as a therapeutic nanoparticle in tumor therapy. In this study, several tumor cell lines were used to evaluate the previously reported pRNA nanotechnology for specific siRNA delivery and for the silencing of targeted genes. It was found that MCF-7 and HeLa cells, out of twenty-five tested tumor cell lines, expressed high levels of folate receptors and exhibited specific binding of the FITC-folate-pRNA nanoparticles, while the others expressed low levels and thus, for these, delivery was not feasible using folate as a targeting agent. Folate receptor positive tumor cells were then incubated with the chimeric pRNA dimer harboring both the folate-pRNA and the chimeric pRNA/siRNA (survivin). Knock down effects of survivin expression in these tumor cells were detected at the mRNA level by real time-PCR and at the protein level by western blot. Apoptosis was detected by flow cytometry analysis with dual staining of annexinV-FITC and PI. The data suggest that the chimeric pRNA nanoparticles containing folate-pRNA and pRNA/siRNA (survivin) could be specifically taken up by tumor cells through folate receptor-mediated endocytosis, resulting in significant inhibition of both transcription and expression of survivin in tumor cells and triggering cell apoptosis. Using such protein-free nanoparticles as therapeutic reagents would not only allow specific gene delivery and extend the in vivo retaining time but also allow long-term administration of therapeutic particles, therefore avoiding the induction of antibodies caused by repeated treatment for chronic diseases.
Introduction
The self-assembly of nanoparticles from RNA or RNA/chemical conjugates is a prominent bottom-up approach (for reviews see ref. 1 and 2) to obtain nanostructure complexes for a variety of RNA nanotechnology and nanomedicine applications.3–7 A combination of chemical and biological techniques can be successfully integrated into nanotechnology. Such approaches for RNA nanotechnology rely upon cooperative intra- or inter-RNA interactions, which result in spontaneous folding or assembly into larger two- or three-dimensional complexes with the appropriate structure and stoichiometry. The bacteriophage phi29 DNA packaging RNA (pRNA) molecule8 has the unique ability to serve as a building block to build nanoparticles via bottom-up assembly. This pRNA can form dimers, trimers, hexamers, and patterned superstructures via the interaction of two interlocking loops.9–12
Over the past decade, siRNA (small interfering RNA)-mediated degradation of the complementary homologous mRNA has shown significant potential as a novel molecular approach to down-regulate specific gene expression.13–15 Studies have demonstrated that siRNA can be successfully used to knock down related gene expression of respiratory syncytial virus (RSV),16 human immunodeficiency virus (HIV)17,18 and tumor development.7,19–22 However, for successful application in the treatment of diseases, siRNA must be delivered to targeted cells safely, efficiently, specifically, and in a non-toxic manner. It has been demonstrated that synthetic materials, such as liposomes and lipids, cationic polymers and siRNA conjugates, can be successfully used for nucleic acid delivery in vivo.23–25 To address the current challenge of specific delivery of siRNA to targeted cells or tissues, we developed the bacteriophage phi29 pRNA dimer as a difunctional gene delivery vehicle.3–6 The bacteriophage phi29 pRNA contains two domains, an intermolecular interacting domain and a double-stranded helical domain located at the 5′/3′-paired ends.26–28 The 5′/3′ proximate double-stranded helical region of pRNA can be re-designed to carry additional sequences without altering its secondary structure or intermolecular interactions on the condition that the 5′ and 3′ ends are paired. Hence, the 5′/3′ double-stranded helical domain of pRNA can be utilized to carry foreign sequences.1,3–6 Moreover, connection of the CD4-binding RNA aptamer, folate, ribozyme, or other chemical components to the pRNA 5′/3′ helical region does not interfere with the folding of the pRNA.
In this study, we further evaluated the feasibility of the previously reported pRNA nanotechnology in siRNA delivery using 26 cell lines. We fabricated a chimeric pRNA dimer in which one pRNA monomer harbored folate for cell recognition, while the other monomer harbored a siRNA sequence targeting specific genes such as green fluorescent protein (GFP), luciferase, or survivin. As such, a pRNA dimer includes a targeting function and gene silencing effect within one self-assembled molecular structure; it can achieve specific gene knock-down of target cells or tissues. Furthermore, because the pRNA dimer gene delivery vehicle is protein-free, it might not induce the production of antibody. Since pRNA is also biodegradable, cytotoxicity is reduced. These unique advantages will enable pRNA to be used as a drug delivery carrier in future tumor therapeutic applications.
Results
Expression of folate-receptor in twenty-five cell lines
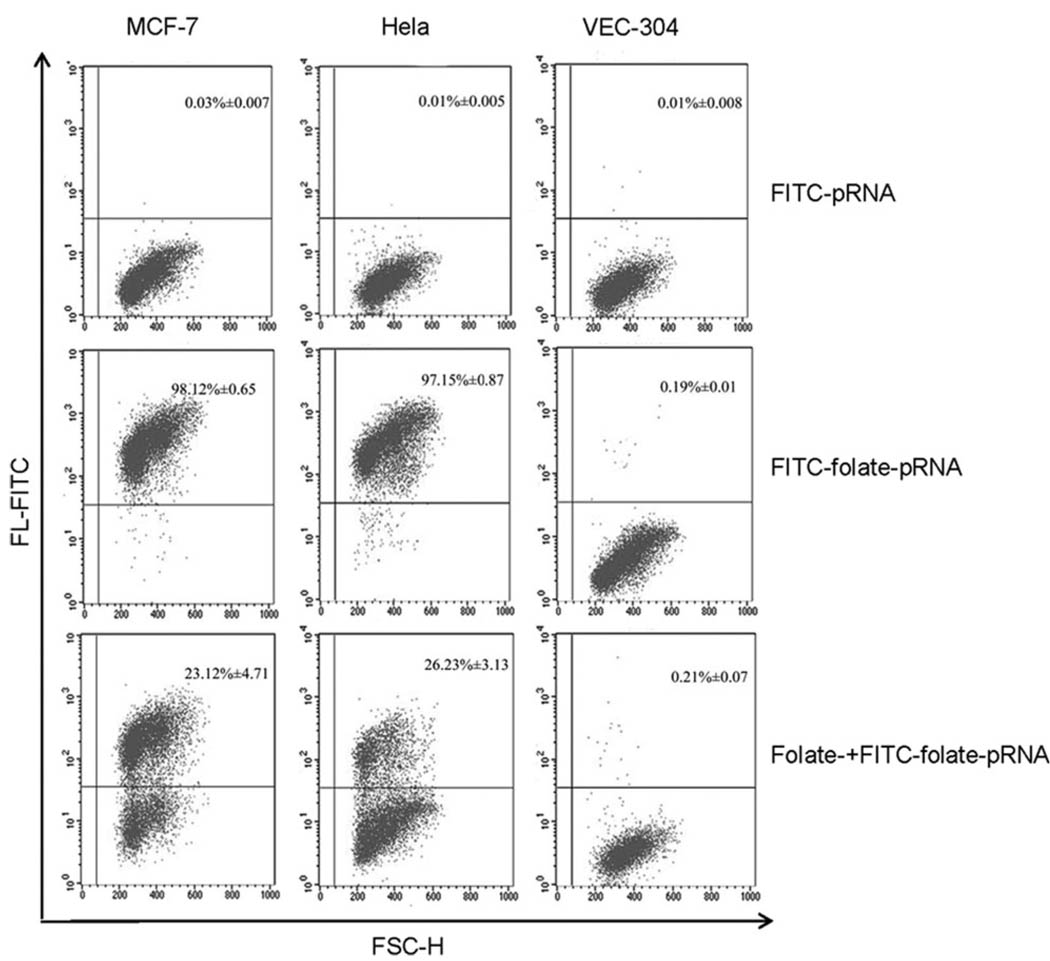
Specific cell binding activity of FITC-folate-pRNA (FITC = fluorescein isothiocyanate) was investigated in twenty-five tumor cell lines by flow cytometry. VEC-304 cells were used as the negative control. Of those investigated, MCF-7, HeLa, KB, A2780, JAR, SKOV3, and A375 cells exhibited high affinity for FITC-folate-pRNA. Adding free folate as a competitive blocking reagent significantly decreased the amount of FITC-folate-pRNA binding cells. However, folate-free FITC-pRNA did not exhibit cell binding (Table 1). These results indicated that MCF-7, HeLa, KB, A2780, JAR, SKOV3 and A375 cells highly expressed folate receptors. Binding behavior of FITC-folate-pRNA to cells was specific and folate-receptor dependent.
Table 1.
The percentage of FITC-positive cells binding FITC-folate-pRNA by flow cytometry assay
| Cell lines | FITC-folate-pRNA | FITC-folate-pRNA + folate | FITC-pRNA |
|---|---|---|---|
| VEC-304 | 0.19% ± 0.01 | 0.21% ± 0.07 | 0.01% ± 0.008 |
| MCF-7 | 98.12% ± 0.65 | 23.12% ± 4.71 | 0.03% ± 0.007 |
| HeLa | 97.15% ± 0.87 | 26.23% ± 3.13 | 0.01% ± 0.005 |
| A549 | 0.21% ± 0.05 | 0.34% ± 0.04 | 0.01% ± 0.007 |
| HepG2 | 1.56% ± 0.56 | 0.12% ± 0.07 | 0.03% ± 0.009 |
| K562 | 0.16% ± 0.008 | 0.34% ± 0.06 | 0.05% ± 0.008 |
| KB | 68.13% ± 6.71 | 11.34% ± 4.41 | 0.02% ± 0.005 |
| SK-BR-3 | 8.05% ± 0.65 | 0.06% ± 0.006 | 0.03% ± 0.004 |
| MG-63 | 0.43% ± 0.05 | 0.06% ± 0.005 | 0.06% ± 0.007 |
| HL60 | 0.67% ± 0.07 | 0.11% ± 0.05 | 0.02% ± 0.008 |
| A2780 | 56.64% ± 8.68 | 11.03% ± 4.91 | 0.04% ± 0.005 |
| PC3M | 0.06% ± 0.007 | 0.05% ± 0.004 | 0.06% ± 0.005 |
| MKN45 | 0.17% ± 0.003 | 0.07% ± 0.009 | 0.07% ± 0.003 |
| GRC-1 | 1.02% ± 0.26 | 0.04% ± 0.005 | 0.05% ± 0.006 |
| MOLT-4 | 0.08% ± 0.005 | 0.06% ± 0.009 | 0.01% ± 0.005 |
| JAR | 67.91% ± 7.58 | 21.23% ± 5.06 | 0.04% ± 0.009 |
| Raji | 0.06% ± 0.005 | 0.07% ± 0.001 | 0.05% ± 0.005 |
| Daudi | 0.67% ± 0.05 | 0.01% ± 0.003 | 0.05% ± 0.004 |
| LM-8 | 0.47% ± 0.07 | 0.05% ± 0.007 | 0.08% ± 0.003 |
| SLC-89 | 0.03% ± 0.009 | 0.07% ± 0.009 | 0.07% ± 0.002 |
| LO2 | 0.78% ± 0.07 | 0.44% ± 0.05 | 0.05% ± 0.004 |
| SKOV3 | 76.22% ± 8.64 | 18.84% ± 5.24 | 0.06% ± 0.002 |
| CHO-K1 | 0.43% ± 0.01 | 0.06% ± 0.007 | 0.02% ± 0.009 |
| MDA-MB-231 | 1.74% ± 0.68 | 0.06% ± 0.008 | 0.01% ± 0.007 |
| A375 | 52.12% ± 6.92 | 19.04% ± 4.37 | 0.01% ± 0.006 |
Of the twenty-five tumor cell lines tested, MCF-7 and HeLa cell lines exhibited the highest expression of folate receptors and high affinity for FITC-folate-pRNA. When MCF-7 and HeLa cells were incubated with FITC-folate-pRNA, 98.12% ± 0.65 of MCF-7 cells and 97.15% ± 0.87 of HeLa cells were FITC positive. When free folate was added, the amount of FITC-positive cells reduced to 23.12% ± 4.71 and 26.23% ± 3.13, respectively (Fig. 1). As a result, these two cell lines were chosen to carry out the following experiments.
Fig. 1.
Flow cytometry analyses of the binding of FITC-labeled folate-pRNA. Upper: binding was tested using folate-free pRNA labeled with FITC as a negative control. The percentages of FITC-positive cells were shown in the upper right quadrants. Middle: MCF-7, HeLa and VEC-304 cells were incubated with folate-pRNA labeled with FITC. Lower: cells were pre-incubated with free folate, which served as a blocking agent to compete with FITC-folate-pRNA for binding to the receptor.
Gene silencing effects of chimeric pRNA monomers
1 pRNA/siRNA (GFP) monomer
To determine the silencing activity of pRNA/siRNA (GFP) monomer, GFP-expressing plasmid was co-transfected with various pRNAs into MCF-7 and HeLa cells, respectively. Fluorescence microscopy demonstrated that chimeric pRNA/siRNA (GFP) specifically suppressed the expression of GFP compared to other control pRNAs. The flow cytometry (FCM) data also revealed that pRNA/siRNA (GFP) effectively inhibited GFP expression in MCF-7 and HeLa cells. In contrast, significant inhibition was not observed in cells treated without pRNA, with normal pRNA, with chimeric pRNA/siRNA (luciferase) or with 18s RNA. In both assays, pRNA/siRNA (GFP) monomer exhibited superior and specific inhibition of GFP expression compared with controls (Fig. 2A).
Fig. 2.
Silencing effect of pRNA/siRNA (GFP) monomer by transfection and folate-pRNA-pRNA/siRNA (GFP) dimer by incubation in MCF-7 and HeLa cells. (A) By transfection: both fluorescence microscopy and the flow cytometry assay proved the inhibitory effect of pRNA/siRNA (GFP) monomer on GFP expression. (B) By incubation: chimeric pRNA dimer complex containing folate-pRNA and chimeric pRNA/siRNA (GFP) was incubated with cells for 3 h to allow the binding and entry of RNA. Data are shown as means ± SEs for five experiments.
2 pRNA/siRNA (luciferase) monomer
Chimeric pRNA/siRNA (luciferase) monomer targeting firefly luciferase was introduced into MCF-7 and HeLa cells by transfection and the expression levels of both firefly and Renilla luciferase were measured simultaneously by a dual reporter assay system. When targeted luciferase was determined, the non-targeted one served as the internal control. As shown in Fig. 3A, the chimeric pRNA/siRNA (luciferase) monomer was found to efficiently and specifically suppress the firefly luciferase gene.
Fig. 3.
Silencing effect of chimeric pRNA/siRNA monomer targeting firefly luciferase by transfection and folate-pRNA-pRNA/siRNA (firefly) dimer by incubation with MCF-7 and HeLa cells (A, B). Dual reporter luciferase assays showed the specific knock down of firefly luciferase expression by pRNA/siRNA (firefly) monomer and folate-pRNA-pRNA/siRNA (firefly) dimer. Data are shown as means ± SEs for five experiments.
3 pRNA/siRNA (survivin) monomer
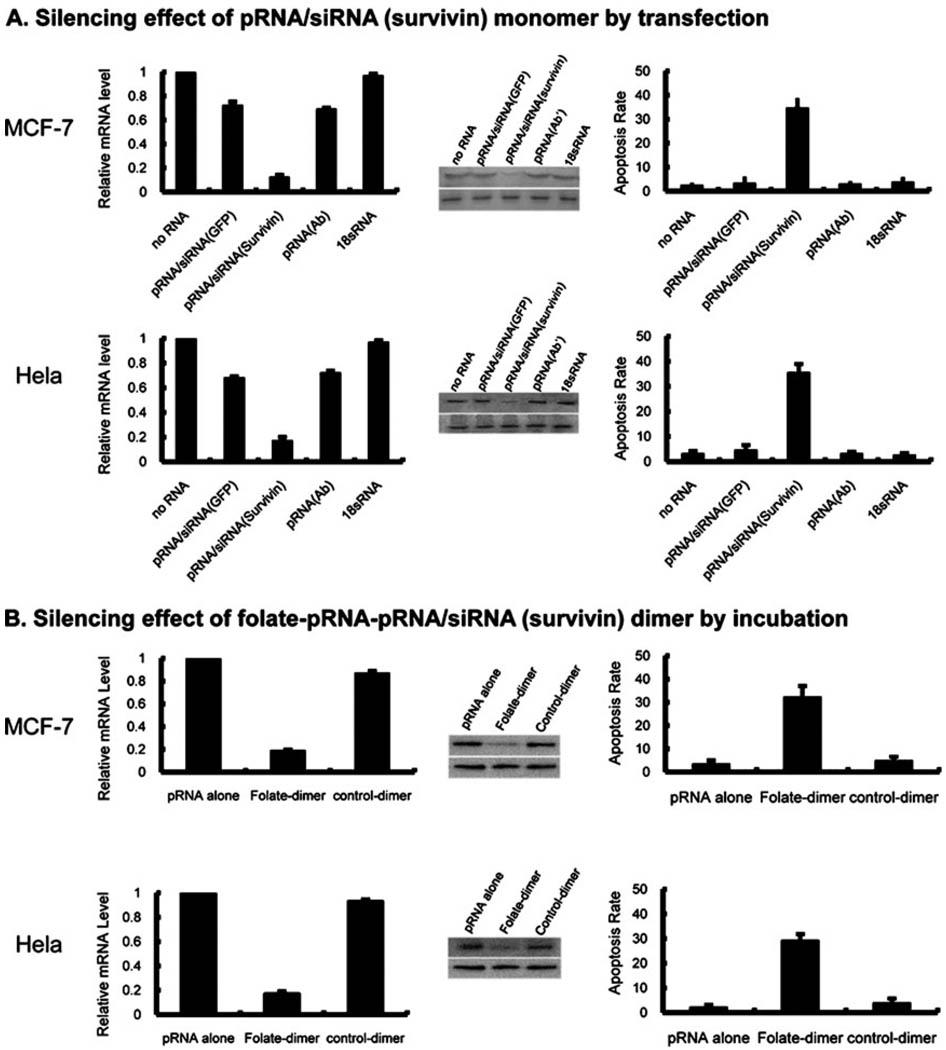
Since chimeric pRNA/siRNA targeting reporter genes have demonstrated possession of specific silencing effects, pRNA/siRNA (survivin) was further designed to perform the desired down-regulation or silencing of the survivin gene, which is highly expressed in most tumor cells. pRNA/siRNA (survivin) monomer was introduced by lipofectamine transfection into MCF-7 and HeLa cells in which survivin was abundantly expressed. Both real-time PCR and western blot analysis revealed that the mRNA and protein expression of survivin were significantly inhibited 48 h after transfection. In contrast, the non-specific pRNA/siRNA control, normal pRNA and 18sRNA treated and un-treated cells did not show a significant decrease in survivin expression.
To determine whether pRNA/siRNA (survivin) monomer could silence the anti-apoptosis factor survivin to induce tumor apoptosis, annexin V–propidium iodide (PI) double-staining was performed, followed by flow cytometry analysis, on both MCF-7 and HeLa cells. As shown in Fig. 4A, 34.52% ± 4.21 of MCF-7 cells and 35.42% ± 3.51 of HeLa cells underwent apoptosis after pRNA/siRNA (survivin) treatment. On the other hand, MCF-7 cells treated with non-specific pRNA/siRNA (GFP) showed 3.14% ± 2.91 apoptotic cells, and cells treated with normal pRNA and 18sRNA demonstrated values of 2.87% ± 1.32 and 3.59% ± 2.35, respectively. HeLa cells treated with non-specific pRNA/siRNA (GFP) showed 4.49% ± 2.09 apoptotic cells, compared to 3.07% ± 0.83 and 2.44% ± 0.94 of cells treated with normal pRNA and 18sRNA. These results indicated that pRNA/siRNA (survivin) monomer was functional specific to survivin (Fig. 4A).
Fig. 4.
Real-time PCR (left), western blotting (middle), and flow cytometry (right) assays to test the silencing effect of pRNA/siRNA (survivin) monomer and folate-pRNA/siRNA (survivin) dimer on MCF-7 and HeLa cells. Gene expression levels were compared to the level of gene expression found in non-transfected samples, arbitrarily assigned the value 1. Bars represent the fold number in gene expression over the expression level in the non-transfected samples. (A) By transfection. Left: for samples transfected with pRNA/siRNA (survivin), the mRNA level decreased to 12.45% ± 2.19 and 17.14% ± 3.13 compared with that of the non-transfected cells in MCF-7 and HeLa cells. Middle: survivin protein levels were compared by western blot with survivin antibody (upper). β-Actin was used as internal control (lower). Right: 34.52% ± 4.21 of MCF-7 cells and 35.42% ± 3.51 of HeLa cells transfected with pRNA/siRNA (survivin) monomer showed apoptosis. (B) By incubation. Left: mRNA level of MCF-7 and Hela cells treated with folate-pRNA dimer significantly reduced to 18.73% ± 1.21 and 16.74% ± 2.03, respectively, compared to that of cells treated by pRNA alone and control dimer. Middle: survivin protein levels of MCF-7 and Hela cells, treated by folate-dimer, pRNA alone and control dimer, were compared by western blot with survivin antibody. β-actin was used as internal control. Right: apoptosis rate of MCF-7 and Hela cells induced by folate containing chimeric pRNA dimer was 32.12% ± 4.91 and 29.12% ± 2.74 respectively. Data were shown as means ± SEs for five experiments.
Specific, targeted gene silencing of chimeric pRNA dimers
1 Dimers harboring both folate and siRNA (GFP)
The strategy of pRNA dimer-mediated gene delivery is that the receptor-binding moiety mediates cell recognition and subsequent internalization, and the siRNA is then released to down-regulate a specific gene. To determine whether the folate moiety on the chimeric pRNA dimer could mediate the entry of the complex into MCF-7 and HeLa cells, a chimeric RNA dimer containing folate-pRNA and pRNA/siRNA(GFP) was fabricated in vitro in the presence of 5 mM Mg2+ and incubated, rather than transfected, with MCF-7 and HeLa cells. The dimer formed by folate-pRNA and pRNA/siRNA (GFP) showed inhibitory efficacy as revealed by fluorescence microscopy imaging and flow cytometry analysis. The GFP fluorescence in MCF-7 and HeLa cells, which had been co-incubated with chimeric RNA dimer containing folate-pRNA and pRNA/siRNA(GFP), was remarkably weaker than that of cells co-incubated with pRNA/siRNA alone or with the control dimer (no folate-labeling). The fluorescence density did not decrease when folate-free pRNA/siRNA dimer or pRNA monomer alone were used. Flow cytometry assays demonstrated that the expression level of GFP in MCF-7 and HeLa cells treated with RNA dimer containing folate-pRNA and pRNA/siRNA(GFP) reduced to 8.43% ± 1.13 and 10.43% ± 3.01, respectively. In contrast, MCF-7 and HeLa cells treated with a control folate-free pRNA dimer retained 51.78% ± 5.58 and 49.78% ± 4.78, respectively, of GFP expression. These results indicate that specific knock down of GFP expression was achieved by folate receptor-mediated internalization of chimeric pRNA dimer harboring siRNA (GFP) in the absence of transfection reagents (Fig. 2B).
2 Dimers harboring both folate and siRNA (firefly luciferase)
The specific binding and gene-silencing effects of folate-mediated siRNA targeting were further investigated using firefly luciferase as a marker in MCF-7 and HeLa cells. A dual luciferase reporter system assay revealed a significant decrease in firefly luciferase expression after incubation with the dimer containing both the folate-pRNA and pRNA/siRNA (firefly luciferase). The control pRNA dimer without folate did not inhibit the expression of the luciferase gene (Fig. 3B).
3 Dimer harboring both folate and siRNA (survivin)
Dimeric pRNA containing folate-pRNA and pRNA/siRNA (survivin) was incubated with MCF-7 and HeLa cells, and both cells responded strongly. Analysis by real-time PCR revealed that the expression of the survivin gene in both MCF-7 and HeLa cells reduced to 18.73% ± 1.21 and 16.74% ± 2.03, respectively, compared to that of cells treated with normal pRNA. Western blot data exhibited that the survivin protein was strongly inhibited by the folate-pRNA dimer in both MCF-7 and HeLa cells. The induced apoptosis rate of MCF-7 and HeLa cells was 32.12% ± 4.91 and 29.12% ± 2.74 respectively. These results suggest that the folate-mediated entry of pRNA/siRNA (survivin) into MCF-7 and HeLa cells induced apoptosis, and this effect occurred only in cells that overexpressed the folate receptor (Fig. 4B). Therefore, apoptosis was specifically induced by pRNA/siRNA (survivin).
Discussion
RNA interference (RNAi) is currently one of the most promising therapeutic approaches for gene silencing. Various synthetic siRNAs have been reported to have gene silencing effects on diverse diseases, such as liver cirrhosis,29 hepatitis B virus,23 ovarian cancer30 and hypercholesterolaemia.31 The delivery of siRNA by virus vectors,32,33 synthetic lipids34,35 and polymers24,36–38 have been attempted. Still, in vivo delivery of siRNA to specific cells using safe vehicles remains challenging.
Survivin is a structurally unique member of the inhibitors of the apoptosis protein (IAP) family, which is involved in the control of cell division and the inhibition of apoptosis.39–41 As a result, it has been widely considered as a target in tumor therapy. Inclusion of ligands in the delivery particles to target appropriate cell surface receptors could improve the therapeutic efficacy and reduce the accompanying drug toxicity and side effects. Folate receptors42–45 have been demonstrated to be excessively expressed in a broad spectrum of tumor cells, while normal cells express no or low levels of folate receptors. Therefore, folate receptors represent an attractive target for selective drug delivery.46–49
In this work, we tested the expression of folate receptors in 25 tumor cell lines. The results showed that MCF-7 and HeLa cells expressed the highest level of folate receptors. Concurrent experiments by transfection demonstrated the gene silencing efficiency of three pRNA/siRNA monomers on GFP, luciferase and survivin, respectively. Incubation of MCF-7 and HeLa cells with pRNA dimers containing folate-pRNA and specific pRNA/siRNA targeting GFP, luciferase or survivin, respectively, showed remarkable inhibition. Thus, the advantage of using folate-conjugated pRNA in the dimer is the specific delivery of siRNA to target cells. Folate-mediated gene delivery is triggered by endocytosis of the folate receptor. It should be noted that folate-mediated delivery is less effective than the lipofectamine-based route. Our results showed the folate-mediated knock down effect results in target gene activity of 20–25%, which coincides with the efficiency of other siRNA delivery vehicles.50,51
In this work, the specific delivery of siRNA mediated by folate was demonstrated by two individual experiments. Firstly, FCM screening showed that FITC-folate-pRNA binding to MCF-7 and HeLa cells can be significantly blocked by adding free folate. Secondly, the control dimer containing the folate-free pRNA and pRNA/siRNA did not show a gene silencing effect. These suggest that the gene silencing by the folate-containing dimer was specifically triggered by the folate receptors.
Experimental
Preparation of RNA
The preparation of the RNAs was described previously.52 The characterizations of the dimer were shown in previous publications.9 Briefly, the RNAs were prepared by in vitro transcription using T7 RNA polymerase. Magnesium (5 mM) was included in all buffers to maintain the folding of pRNA and the formation of the dimer.53,54 The nomenclature of pRNA and the resulting chimeric pRNA subunits for the construction of deliverable RNA nanoparticles have been reported. To label the 5′ end of RNA with folate, both 4 mM folate-AMP and 0.25 mM ATP were included in a transcription reaction, together with 1 mM UTP, CTP and GTP.5 FITC-folate-pRNA was formed by annealing 5′ labeled FITC oligo with the 3′ end of folate-pRNA.
Cell lines
Cell lines were obtained from the Department of Immunology, Tongji Medical College, Huazhong University of Science and Technology.
Flow cytometry analysis of folate receptor expression screening
Cells were seeded into a six-well plate and grown for 24 h. After two rinses with PBS, the cells were incubated with 100 nM folate-FITC for 20 min at room temperature, with or without the presence of free folate as blocking reagent. Cells were then washed and analyzed by flow cytometry.
Functional assay of monomeric pRNA/siRNA subunits by transfection
Various chimeric pRNA/siRNA (0.2 µg) monomers and plasmid EGFP-N1 were co-transfected using Lipofectamine2000 (1.5 µl) in a 24-well plate. After 24 h, the expression of GFP was detected by fluorescence microscopy and flow cytometry.
For the luciferase assay of the pRNA/siRNA monomer, various chimeric pRNA/siRNAs (0.2 µg) were co-transfected into cells with plasmid DNA pGL3 encoding firefly luciferase (0.7 µg) and pRL-TK (0.1 µg) encoding Renilla luciferase. Luciferase expressions were measured by a dual reporter assay system (Promega) 1 day after transfection. Cells were washed once with PBS and lysed with passive lysis buffer. A volume of 20 µl of the lysate was added to 100 µl of luciferase assay reagent (LAR II) in a luminometer tube, and firefly luciferase activity was measured. Upon addition of twenty microlitres of Stop & Glo reagent, control measurements of Renilla luciferase activity were obtained. The previously obtained data were then normalized to the Renilla activity by determining the average ratio of firefly to Renilla activity over several trials.
For the survivin knock down assay, cells were transfected with various pRNA/siRNA (0.5 µg) in 24-well plates. Expression of survivin was detected by real-time PCR and western blot. Apoptosis of tumor cells was assayed by flow cytometry.
Real-time PCR
Cells were seeded into 24-well plates at a density of 105 cells/well, and transfected with 0.5 µg pRNA. Cells were harvested 48 h after transfection and total RNA was extracted with Trizol (Invitrogen, USA) according to the manufacturer’s protocol. Reverse transcription was carried out on 1 µg of RNA with Revert Aid™. Equal amounts of cDNA were submitted to PCR, in the presence of SYBR Green dye with the QuantiTect SYBR Green RT-PCR Kit and the ABI PRISM 6700 Real-time PCR detection machine. Primers for survivin were 5′-AAA GAG CCA AGA ACAAAA TTG C-3′ and 5′-GAG AGAGAA GCA GCC ACT GTT AC-3′. PCR was performed by 40 cycles of 0.5 s at 95 °C, 10 s at 60 °C and 10 s at 72 °C. PCR without template was used as the negative control. The β-actin endogenous housekeeping gene was used as the internal control. Both β-actin and the negative control were amplified on the same plate as the experimental gene of interest. Each sample was normalized by using the difference in critical thresholds (CT) between survivin and β-actin. The following equation was used to describe the result: ΔΔCT survivin = ΔCT survivin – ΔCT β-actin, where ΔCT survivin and ΔCT β-actin represent the difference in CT between survivin and the negative control and the difference between β-actin and the negative control, respectively. The mRNA levels of each sample were then compared using the expression 2−ΔΔCT survivin. The results of each group were averaged. The expression level for the non-transfected sample was arbitrarily assigned a value of 1 and the final results were expressed as a fold number compared to the non-transfected sample.
Western blot analysis
Cells were rinsed and harvested in lysis buffer 48 h after transfection. Protein concentrations were determined and equal amounts of protein were loaded onto a 12% polyacrylamide gel. Membranes were blocked, incubated with primary antibody for survivin and β-actin (Santa Cruz), and conjugated to a secondary antibody according to the manufacturer’s instructions (Pierce ECL kit). Membranes were exposed to film.
Apoptosis analysis
The apoptosis assay for pRNA/siRNA (survivin) was performed using annexinV–PI double-staining in flow cytometry. Cells in 24-well plates were rinsed with PBS and transfected with various pRNA. Cells were stained with annexin V and PI followed by flow cytometry assay.
Assay of gene knock down targeted by folate receptor
Chimeric pRNA/siRNA dimers were prepared by mixing folate-pRNA Ba’ and chimeric pRNA/siRNA Ab’ in the presence of 5 mM Mg2+. Cells were seeded in a 96-well plate in folate-free cell culture medium. After washing with PBS containing MgCl2, the premixed dimer (1.75 µM) was then added to the cells and incubated with the cells for 3 h at 37 °C. RNase inhibitor (SUPERaseIN, 1 unit l−1; Ambion, Austin, TX) was added to the binding buffer. After incubation, EGFP-N1, PGL3 and PRL-TK plasmids were introduced into the cells by Lipofectamine2000. Cells were further incubated for 24 or 48 h. GFP expression, firefly luciferase activity, and survivin knock down were measured, respectively.
Conclusion
In summary, we report systematically that the phi29 pRNA dimer containing folate-pRNA and pRNA/siRNA (survivin) could be specifically delivered into MCF-7 and HeLa cells that exhibited high expression of folate receptors. It produced significant inhibition of transcription and expression of survivin in tumor cells as well as promotion of tumor cell apoptosis. As we know, phi29 pRNAs have a tendency to form dimers, as a result of the interaction of the interlocking loops of each pRNA. This paper has demonstrated the production of the dimer to deliver therapeutic siRNA to specific cells. In the future, chimeric pRNA multimers could also be assembled via hand-in-hand interaction. Such fabricated RNA nanodevices could hold different RNA building blocks with controllable stoichiometries ranging from one, two, three or six copies up to thousands of copies.4,55 The features of multiplicity and assortment make such RNA nanodevices capable of carrying polyvalent therapeutic molecules to enhance therapeutic efficacy.1,3,4,55,56
Acknowledgements
This work was supported by NIH grant EB00370 (Nanosciences and Nanotechnology in Biology and Medicine) and NIH grant GM59944 to Peixuan Guo, the Hi-Tech Research and Development Program of China (No. 2006AA02Z158) to Guanxin Shen, the State Scholar Fund by China Scholarship Council (CSC) to Jing Liu. We thank Huifen Zhu and Zhihui Liang for the flow cytometry assay.
Contributor Information
Peixuan Guo, Email: guop@purdue.edu.
Guanxin Shen, Email: guanxin_shen@yahoo.com.cn.
References
- 1.Guo P. J. Nanosci. Nanotechnol. 2005;5(12):1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeger L, Chworos A. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Khaled A, Guo S, Li F, Guo P. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo S, Tschammer N, Mohammed S, Guo P. Hum. Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo S, Huang F, Guo P. Gene Ther. 2006;13:814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Guo S, Roll R, Li J, Diao Z, Shao N, Riley MR, Cole AM, Robinson JP, Snead NM, Shen G, Guo P. Cancer Biol. Ther. 2007;6:697–704. doi: 10.4161/cbt.6.5.3962. [DOI] [PubMed] [Google Scholar]
- 7.Hoeprich S, ZHou Q, Guo S, Qi G, Wang Y, Guo P. Gene Ther. 2003;10:1258–1267. doi: 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- 8.Guo P, Erickson S, Anderson D. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 9.Guo P, Zhang C, Chen C, Trottier M, Garver K. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, Lemieux S, Wu X, St.-Arnaud S, McMurray CT, Major F, Anderson D. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Trottier M, Guo P. Nucleic Acids Symp. Ser. 1997;36:190–193. [Google Scholar]
- 12.Chen C, Zhang C, Guo P. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton AJ, Baulcombe DC. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 17.Akkina R, Banerjea A, Bai J, Anderson J, Li MJ, Rossi J. Anticancer Res. 2003;23:1997–2005. [PubMed] [Google Scholar]
- 18.Sano M, Li H, Nakanishi M, Rossi JJ. Mol. Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- 19.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 20.Atkins KM, Thomas L, Youker RT, Harriff MJ, Pissani F, You HH, Thomas G. J. Biol. Chem. 2008;283:11772–11784. doi: 10.1074/jbc.M707572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang SF, Chang CA, Lee DY, Leet PL, Yeh YM, Yeh CR, Cheng CK, Chien S, Chiu JJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3927–3932. doi: 10.1073/pnas.0712353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsung AJ, Kargiotis O, Chetty C, Lakka SS, Gujrati M, Spomar DG, Dinh DH, Rao JS. Int. J. Oncol. 2008;32:557–564. [PMC free article] [PubMed] [Google Scholar]
- 23.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, Maclachlan I, Polisky B. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 24.Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 26.Zhang CL, Trottier M, Guo PX. Virology. 1995;207:442–451. doi: 10.1006/viro.1995.1103. [DOI] [PubMed] [Google Scholar]
- 27.Reid RJD, Bodley JW, Anderson D. J. Biol. Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 28.Reid RJD, Zhang F, Benson S, Anderson D. J. Biol. Chem. 1994;269:18656–18661. [PubMed] [Google Scholar]
- 29.Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y, Takimoto R, Takada K, Miyanishi K, Matsunaga T, Takayama T, Niitsu Y. Nat. Biotechnol. 2008;26:431–442. doi: 10.1038/nbt1396. [DOI] [PubMed] [Google Scholar]
- 30.Halder J, Kamat AA, Landen CN, Jr, Han LY, Lutgendorf SK, Lin YG, Merritt WM, Jennings NB, Chavez-Reyes A, Coleman RL, Gershenson DM, Schmandt R, Cole SW, Lopez-Berestein G, Sood AK. Clin. Cancer Res. 2006;12:4916–4924. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash KN, Maier M, Nechev L, Rajeev KG, Read T, Rohl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, de Fougerolles A, Vornlocher HP, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Proc. Natl. Acad.Sci. U. S. A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barton GM, Medzhitov R. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang LL, Kopinja J, Zhang MD, McManus MT, Gertler FB, Scott ML, van Parijs L. Nat. Genet. 2007;39:803. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 34.Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 35.Kumar P, Lee SK, Shankar P, Manjunath N. PLoS Med. 2006;3:505–514. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zintchenko A, Philipp A, Dehshahri A, Wagner E. Bioconjugate Chem. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 37.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 38.Bartlett DW, Davis ME. Biotechnol. Bioeng. 2008;99:975–985. doi: 10.1002/bit.21668. [DOI] [PubMed] [Google Scholar]
- 39.Arora V, Cheung HH, Plenchette S, Micali OC, Liston P, Korneluk RG. J. Biol. Chem. 2007;282:26202–26209. doi: 10.1074/jbc.M700776200. [DOI] [PubMed] [Google Scholar]
- 40.Al-Joudi FS, Iskandar ZA, Imran AK. Med. J. Malaysia. 2007;62:6–8. [PubMed] [Google Scholar]
- 41.Capalbo G, Rodel C, Stauber RH, Knauer SK, Bache M, Kappler M, Rodel F. Strahlenther. Onkol. 2007;183:593–599. doi: 10.1007/s00066-007-1800-4. [DOI] [PubMed] [Google Scholar]
- 42.Toffoli G, Cernigoi C, Russo A, Gallo A, Bagnoli M, Boiocchi M. Int. J. Cancer. 1997;74:193–198. doi: 10.1002/(sici)1097-0215(19970422)74:2<193::aid-ijc10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 43.Holm J, Hansen SI, Hoier-Madsen M, Christensen TB, Nichols CW. Biosci. Rep. 1999;19:571–580. doi: 10.1023/a:1020219029206. [DOI] [PubMed] [Google Scholar]
- 44.Ross JF, Wang H, Behm FG, Mathew P, Wu M, Booth R, Ratnam M. Cancer. 1999;85:348–357. doi: 10.1002/(sici)1097-0142(19990115)85:2<348::aid-cncr12>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Bueno R, Appasani K, Mercer H, Lester S, Sugarbaker D. J. Thorac. Cardiovasc. Surg. 2001;121:225–233. doi: 10.1067/mtc.2001.111176. [DOI] [PubMed] [Google Scholar]
- 46.Theti DS, Bavetsias V, Skelton LA, Titley J, Gibbs D, Jansen G, Jackman AL. Cancer Res. 2003;63:3612–3618. [PubMed] [Google Scholar]
- 47.Zhang K, Wang Q, Xie Y, Mor G, Sega E, Low PS, Huang Y. RNA. 2008;14:577–583. doi: 10.1261/rna.739308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Low PS. Adv. Drug Delivery Rev. 2002;54:675–693. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 49.Aronov O, Horowitz AT, Gabizon A, Gibson D. Bioconjugate Chem. 2003;14:563–574. doi: 10.1021/bc025642l. [DOI] [PubMed] [Google Scholar]
- 50.Olie RA, Simoes-Wust AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U. Cancer Res. 2000;60:2805–2809. [PubMed] [Google Scholar]
- 51.Paduano F, Villa R, Pennati M, Folini M, Binda M, Daidone MG, Zaffaroni N. Mol. Cancer Ther. 2006;5:179–186. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- 52.Zhang CL, Lee C-S, Guo P. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 53.Chen C, Sheng S, Shao Z, Guo P. J. Biol. Chem. 2000;275(23):17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 54.Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z, Guo P. J. Biol. Chem. 2001;276:32575–32584. doi: 10.1074/jbc.M100045200. [DOI] [PubMed] [Google Scholar]
- 55.Shu D, Moll D, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717–1724. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo P, Coban O, Snead N, Trebley J, Guo S. Adv. Drug Deliv. Rev. doi: 10.1016/j.addr.2010.03.008. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]