Abstract
We present the Spherical Demons algorithm for registering two spherical images. By exploiting spherical vector spline interpolation theory, we show that a large class of regularizors for the modified Demons objective function can be efficiently approximated on the sphere using iterative smoothing. Based on one parameter subgroups of diffeomorphisms, the resulting registration is diffeomorphic and fast. The Spherical Demons algorithm can also be modified to register a given spherical image to a probabilistic atlas. We demonstrate two variants of the algorithm corresponding to warping the atlas or warping the subject. Registration of a cortical surface mesh to an atlas mesh, both with more than 160k nodes requires less than 5 minutes when warping the atlas and less than 3 minutes when warping the subject on a Xeon 3.2GHz single processor machine. This is comparable to the fastest non-diffeomorphic landmark-free surface registration algorithms. Furthermore, the accuracy of our method compares favorably to the popular FreeSurfer registration algorithm. We validate the technique in two different applications that use registration to transfer segmentation labels onto a new image: (1) parcellation of in-vivo cortical surfaces and (2) Brodmann area localization in ex-vivo cortical surfaces.
Index Terms: Surface Registration, Spherical Registration, Cortical Registration, Vector Field Interpolation, Demons, Diffeomorphism
I. Introduction
Motivated by many successful applications of the spherical representation of the cerebral cortex, this paper addresses the problem of registering two spherical images. Cortical folding patterns have been shown to correlate with both cytoarchitectural [25], [68] and functional regions [64], [27]. In group studies of cortical structure and function, determining corresponding folds across subjects is therefore important. There has been much effort focused on registering cortical surfaces in 3D [22], [23], [30], [58]. Since cortical areas – both structure and function – are arranged in a mosaic across the cortical surface, an alternative approach is to model the surface as a 2D closed manifold in 3D and to warp the underlying spherical coordinate system [27], [50], [59], [60], [64], [67]. Warping the spherical coordinate system establishes correspondences across the surfaces without actually deforming the surfaces in 3D.
Deformation Model
There is frequently a need for invertible deformations that preserve the topology of structural or functional regions across subjects. Unfortunately, this causes many spherical warping algorithms to be computationally expensive. Previously demonstrated methods for cortical registration [27], [60], [67] rely on soft regularization constraints to encourage invertibility. These require unfolding the mesh triangles, or limit the size of optimization steps to achieve invertibility [27], [67]. Elegant regularization penalties that guarantee invertibility exist [5], [46] but they explicitly rely on special properties of the Euclidean image space that do not hold for the sphere.
An alternative approach to achieving invertibility is to work in the group of diffeomorphisms [4], [7], [9], [22], [31], [43], [66]. In this case, the underlying theory of flows of vector fields can be extended to manifolds [11], [44], [47]. The Large Deformation Diffeomorphic Metric Mapping (LDDMM) [7], [9], [22], [31], [43] is a popular framework that seeks a time-varying velocity field representation of a diffeomorphism. Because LDDMM optimizes over the entire path of the diffeomorphism, the resulting method is slow and memory intensive. By contrast, Ashburner [4] and Hernandez et al. [33] consider diffeomorphic transformations parameterized by a single stationary velocity field. While restricting the space of solutions reduces the memory needs relative to LDDMM, these algorithms still have to consider the entire trajectory of the deformation induced by the velocity field when computing the gradients of the objective function, leading to a long run time. We note that recent algorithmic advances [34], [43] promise to improve the speed and relieve the memory requirements of both LDDMM and the stationary velocity approach.
In this work, we adopt the approach of the Diffeomorphic Demons algorithm [66], demonstrated in the Euclidean image space, which constructs the deformation space that contains compositions of diffeomorphisms, each of which is parameterized by a stationary velocity field. Unlike the Euclidean Diffeomorphic Demons, the Spherical Demons algorithm utilizes velocity vectors tangent to the sphere and not arbitrary 3D vectors. This constraint need not be taken care of explicitly in our algorithm since we directly work with the tangent spaces. In each iteration, the method greedily seeks the locally optimal diffeomorphism to be composed with the current transformation. As a result, the approach is much faster than LDDMM [7], [9], [22], [31] and its simplifications [4], [33]. A drawback is that the path of deformation is no longer a geodesic in the group of diffeomorphisms.
Image Similarity vs. Regularization Tradeoffs
Another challenge in registration is the tradeoff between the image similarity measure and the regularization in the objective function. Since most types of regularization favor smooth deformations, the gradient computation is complicated by the need to take into account the deformation in neighboring regions. For Euclidean images, the popular Demons algorithm [57] can be interpreted as optimizing an objective function with two regularization terms [14], [66]. The special form of the objective function facilitates a fast two-step optimization where the second step handles the warp regularization via a single convolution with a smoothing filter.
Using spherical vector spline interpolation theory [31] and other differential geometric tools, we show that the two-stage optimization procedure of Demons can be efficiently approximated on the sphere. We note that the problem is not trivial since tangent vectors at different points on the sphere are not directly comparable. We also emphasize that this decoupling of the image similarity and the warp regularization could also be accomplished with a different space of admissible warps, e.g., spherical thin plate splines [72].
Interpolation
Yet another reason why spherical image registration is slow is because of the difficulty in performing interpolation on a spherical grid, unlike a regular Euclidean grid. In this paper, we use existing methods for interpolation, requiring about one second to interpolate data from a spherical mesh of 160k vertices onto another spherical mesh of 160k vertices. Recent work on using different coordinate charts of the sphere [63] promises to further speed up our implementation of the Spherical Demons algorithm.
While most discussion in this paper concentrates on pairwise registration of spherical images, the proposed Spherical Demons algorithm can be modified to incorporate a probabilistic atlas. We derive and implement two variants of the algorithm for registration to an atlas corresponding to whether we warp the atlas or the subject. On a Xeon 3.2GHz single processor machine, registration of a cortical surface mesh to an atlas mesh, both with more than 160k nodes, requires less than 5 minutes when warping the atlas and less than 3 minutes when warping the subject. Note that the registration runtime reported includes registration components dealing with rotation, which takes up one quarter of the total runtime. The total runtime is comparable to other nonlinear landmark-free cortical surface registration algorithms whose runtime ranges from minutes [23], [60] to more than an hour [27], [67]. However, the other fast algorithms suffer from folding spherical triangles [60] and intersecting triangles in 3D [23] since only soft constraints are used. No runtime comparison can be made with spherical registration algorithm of the LDDMM type because to the best of our knowledge, no landmark-free LDDMM spherical registration algorithm that handles cortical surfaces has been developed yet.
Unlike landmark-based methods for surface registration [8], [22], [31], [50], [58], [64], we do not assume the existence of corresponding landmarks. Landmark-free methods have the advantage of allowing for a fully automatic processing and analysis of medical images. Unfortunately, landmark-free registration is also more challenging, because no information about correspondences are provided. The difficulty is exacerbated for the cerebral cortex since different sulci and gyri appear locally similar. Nevertheless, we demonstrate that our algorithm is accurate in both cortical parcellation and cytoarchitectonic localization applications.
The Spherical Demons algorithm for registering cortical surfaces presented here does not take into account the metric properties of the original cortical surface. FreeSurfer [27] uses a regularization that penalizes deformation of the spherical coordinate system based on the distortion computed on the original cortical surface. Thompson et al. [59] suggest the use of Christoffel symbols [39] to correct for the metric distortion of the initial spherical coordinate system during the registration process. However, it is unclear whether correcting for the metric properties of the cortex is important in practice, since we demonstrate that the accuracy of the Spherical Demons algorithm compares favorably to that of FreeSurfer. A possible reason is that we initialize the registration with a spherical parametrization that minimizes metric distortion between the spherical representation and the original cortical surface [27].
This paper is organized as follows. In the next section, we discuss the classical Demons algorithm [57] and its diffeomorphic variant [66]. In Section III, we present the extension of the Diffeomorphic Demons algorithm to the sphere. We conclude with experiments in Section IV and further discussion in Section V. The appendices provide technical and implementation details of the Spherical Demons algorithm and the extension to probabilistic atlases. This paper extends a previously presented conference article [69] and contains detailed derivations and discussions that were left out in the conference version. We note that our Spherical Demons code is freely available1. To summarize,
We demonstrate that the Demons algorithm can be efficiently extended to the sphere.
We demonstrate that the use of a limited class of diffeomorphisms combined with the Demons algorithm yields a speed gain of more than an order of magnitude compared with other landmark-free invertible spherical registration methods, such as [27], [67].
We validate our algorithm by demonstrating an accuracy comparable to that of the popular FreeSurfer algorithm [27] in two different applications.
II. Background - Demons Algorithm
We choose to work with the modified Demons objective function [14], [66]. Let F be the fixed image, M be the moving image and Γ be the desired transformation that deforms the moving image M to match the fixed image F. Throughout this paper, we assume that F and M are scalar images, even though it is easy to extend the results to vector images [70]. We introduce a hidden transformation ϒ and seek
Algorithm 1. Demons Algorithm.
| Data: A fixed image F and moving image M. | ||
| Result: Transformation Γ so that M ∘ Γ is “close” to F. | ||
| Set ϒ0 = identity transformation (or some a-priori transformation, e.g., from a previous registration) | ||
| repeat | ||
| Step 1. Given ϒ(t), | ||
| Minimize the first two terms of Eq. (3) | ||
| ||
| where u is any admissible transformation. Compute Γ(t) = ϒ(t) ∘ u(t). | ||
| Step 2. Given Γ(t), | ||
| Minimize the last two terms of Eq. (3): | ||
| ||
| until convergence; |
| (3) |
In this case, the fixed image F and warped moving image M ∘ Γ are treated as N × 1 vectors. Typically, dist(ϒ, Γ) = ‖ϒ − Γ‖2, encouraging the resulting transformation Γ to be close to the hidden transformation ϒ and Reg(ϒ) = ‖ ∇(ϒ − Id)‖2, i.e., the regularization penalizes the gradient magnitude of the displacement field ϒ − Id of the hidden transformation ϒ. σx and σt provide a tradeoff among the different terms of the objective function. Σ is typically a diagonal matrix that models the variability of a feature at a particular voxel. It can be set manually or estimated during the construction of an atlas.
This formulation facilitates a two-step optimization procedure that alternately optimizes the first two (first and second) and last two (second and third) terms of Eq. (3). Starting from an initial displacement field ϒ0, the Demons algorithm iteratively seeks an update transformation to be composed with the current estimate, as summarized in Algorithm 1.
In the original Demons algorithm [57], the space of admissible warps includes all 3D displacement fields, and the composition operator ∘ corresponds to the addition of displacement fields. The resulting transformation might therefore be not invertible. In the Diffeormorphic Demons algorithm [66], the update u is a diffeormorphism from ℝ3 to ℝ3 parameterized by a stationary velocity field υ⃗. Note that υ⃗ is a function that associates a tangent vector with each point in ℝ3. Under certain mild smoothness conditions, a stationary velocity field υ⃗ is related to a diffeomorphism through the exponential mapping u = exp(υ⃗). In this case, the stationary ODE ∂x(t)/∂t = υ⃗(x(t)) with the initial condition x(0) ∈ ℝ3 yields exp(υ⃗) as a solution at time 1, i.e., x(1) = exp(υ⃗)(x(0)) ∈ ℝ3. In this case, exp(υ⃗)(x(0)) maps point x(0) to point x(1).
The Demons algorithm and its variants are fast because for certain forms of dist(ϒ, Γ) and Reg(ϒ), Step 1 reduces to a non-linear least-squares problem that can be efficiently minimized via Gauss-Newton optimization and Step 2 can be solved by a single convolution of the displacement field Γ with a smoothing kernel. The proof of the reduction of Step 2 to a smoothing operation is illuminating and holds for dist(ϒ, Γ) = ‖ϒ − Γ‖2 and any Sobolev norm Reg(ϒ) = Σi σi‖ ∇i(ϒ − Id)‖2 [14], [45]. In practice, a Gaussian filter is used without consideration of the actual induced norm [14], [66]. The proof uses Fourier transforms and is therefore specific to the Euclidean domain. Due to differences between the geometry of the sphere and Euclidean spaces, we will see in Section III-D that the reduction of Step 2 to a smoothing operation is only an approximation on the sphere.
III. Spherical Demons
In this section, we demonstrate suitable choices of dist(ϒ, Γ) and Reg(ϒ) that lead to efficient optimization of the modified Demons objective function in Eq. (3) on the unit sphere S2. We construct updates u as diffeomorphisms from S2 to S2 parameterized by a stationary velocity field υ⃗. We emphasize that unlike Diffeomorphic Demons [66], υ⃗ is a tangent vector field on the sphere and not an arbitrary 3D vector field. A glossary of common terms used throughout the paper is found in Table I.
TABLE I.
Glossary of terms used throughout the paper.
| F, M | Fixed image F, moving image M. |
| Σ | Typically a diagonal matrix that models variability of feature values at a particular vertex. |
| σx, σT | Parameters of Demons cost function in Eq. (3). |
| Γ, ϒ | Transformations from S2 to S2. Γ is the transformation we are seeking. ϒ is the smooth hidden transformation close to Γ. |
| Γ⃗ ≜ {Γ⃗n}, ϒ⃗ ≜ {ϒ⃗n} | Discrete tangent vector representation of the deformations (see Fig. 1 and Eq. (5)). For example, given the tangent vector Γ⃗n at xn ∈ S2, one can compute Γ(xn). |
| υ⃗ ≜ {υ⃗n} | We parameterize diffeomorphic transformations from S2 to S2 by a composition of diffeomorphisms, each parameterized by a stationary velocity field υ⃗. υ⃗n is the velocity vector at xn. |
| u(·) ≜ exp(υ⃗)(·) | The diffeomorphism parameterized by the stationary velocity field υ⃗ is the solution of a stationary ODE at time 1. |
| En ≜ [e⃗n1 e⃗n2] | e⃗n1 and e⃗n2 are orthonormal vectors tangent to the sphere at xn |
| Ψn | Coordinate chart defined in Eq.(10): . Ψn is a diffeomorphism between ℝ2 and a hemisphere centered at xn ∈ S2. |
| z⃗n | z⃗n is an arbitrary tangent vector at the origin of ℝ2. At xn, the velocity vector υ⃗n = Enz⃗n via the coordinate chart Ψn (see Eq. (14)). |
A. Choice of dist(ϒ, Γ)
Suppose the transformations Γ and ϒ map a point xn ∈ S2 to two different points Γ(xn) ∈ S2 and ϒ(xn) ∈ S2 respectively. An intuitive notion of distance between Γ(xn) and ϒ(xn) would be the geodesic distance between Γ(xn) and ϒ (xn). Therefore, we could define geodesic(ϒ(xn), Γ (xn)). For reasons that will become clear in Section III-D, we prefer to define dist(ϒ, Γ) in terms of a tangent vector representation of the transformations Γ and ϒ, illustrated in Fig. 1, where the length of the tangent vector encodes the amount of deformation.
Fig. 1.
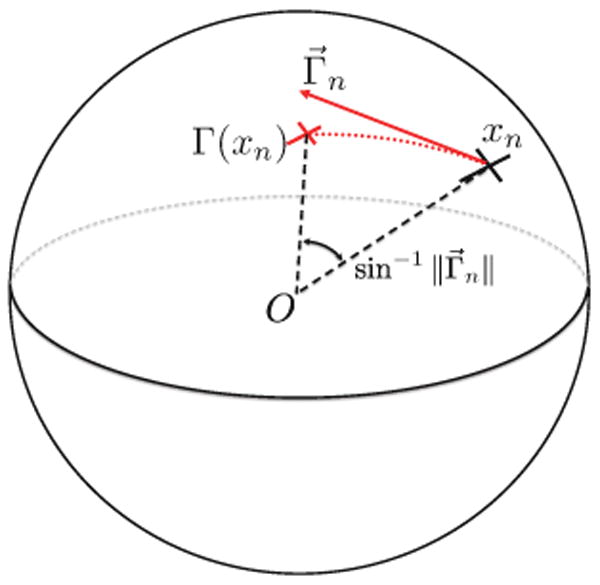
Tangent vector representation of transformation Γ. See text for more details.
Let Txn S2 be the tangent space at xn. We define Γ⃗n ∈ Txn S2 to be the tangent vector at xn pointing along the great circle connecting xn to Γ(xn). In this work, we set the length of Γ⃗n to be equal to the sine of the angle between xn and Γ(xn). With this particular choice of length, there is a one-to-one correspondence between Γ(xn) and Γ⃗n, assuming the angle between xn and Γ(xn) is less than π/2, which is a reasonable assumption even for relatively large deformations. The choice of this length leads to a compact representation of Γ⃗n via vector products. We define Gn to be the 3 × 3 skew-symmetric matrix representing the cross-product of xn with any vector:
| (4) |
where xn(i) is the i-th coordinate of xn. Thus, xn × Γ(xn) = GnΓ(xn). Then on a unit sphere, we obtain
| (5) |
A more intuitive choice for the length of Γ⃗n might be the geodesic distance between xn and Γ(xn). If we restrict Γ⃗n to be at most length π, there is a one-to-one mapping between this choice of the tangent vector Γ⃗n and the resulting transformation Γ(xn). Indeed, such a choice of a tangent vector corresponds to an exponential map of S2 [39]. The resulting expression for is feasible, but more complicated than Eq. (5). In this paper, for simplicity, we follow the definition in Eq. (5).
Given N vertices , the set of transformed points – or equivalently the tangent vectors – together with a choice of an interpolation function define the transformation Γ everywhere on S2. Similarly, we can define the transformation ϒ or the equivalent tangent vector field ϒ⃗ through a set of N tangent vectors . We emphasize that these tangent vector fields are simply a convenient representation of the transformations ϒ and Γ and should not be confused with the stationary velocity field υ⃗ that will be used later on. We now set
| (6) |
which is well-defined since both Γ⃗n and ϒ⃗n belong to Txn S2 for each n = 1,…, N.
B. Spherical Demons Step 1
In this section, we show that the update for Step 1 of the Spherical Demons algorithm can be computed independently for each vertex. With our choice of dist(ϒ, Γ), step 1 of the algorithm becomes a minimization with respect to the velocity field . By substituting u = exp(υ⃗) and into Eq. (1), we obtain
| (7) |
| (8) |
| (9) |
where is the n-th diagonal entry of Σ and ∘ denotes warp composition.
Defining Coordinate Charts on the Sphere
The cost function in Eq. (9) is a mapping from the tangent bundle TS2 to the real numbers ℝ. We can think of each tangent vector υ⃗n as a 3 × 1 vector in ℝ3 tangent to the sphere at xn. Therefore υ⃗n has 2 degrees of freedom and Eq. (9) represents a constrained optimization problem. Instead of dealing with the constraints, we employ coordinate charts that are diffeomorphisms (smooth and invertible mappings) between open sets in R2 and open sets on S2. The differential of the coordinate chart establishes correspondences between the tangent bundles Tℝ2 and TS2 [39], [44], so we can reparameterize the constrained optimization problem into an unconstrained one in terms of Tℝ2 (see Fig. 2).
Fig. 2.
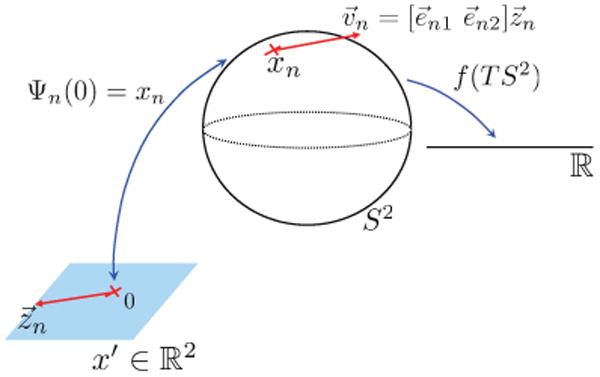
Coordinate chart of the sphere S2. The chart allows a reparameterization of the constrained optimization problem f in step 1 of the Spherical Demons algorithm into an unconstrained one.
It is a well-known fact in differential geometry that covering S2 requires at least two coordinate charts. Since the tools of differential geometry are coordinate-free [39], [44], our results are independent of the choice of the coordinate charts. Let e⃗n1, e⃗n2 be any two orthonormal 3 × 1 vectors tangent to the sphere at xn, where orthonormality is defined via the usual Euclidean inner product in 3D. In this work, for each mesh vertex xn, we define a local coordinate chart Ψn: ℝ2 ↦ S2,
| (10) |
As illustrated in Fig. 2, Ψn(0) = xn. Let z⃗n be a 2 × 1 tangent vector at the origin of ℝ2. With the choice of the coordinate chart above, the corresponding tangent vector at xn is given by the differential of the mapping DΨn(·) evaluated at x′ = 0:
| (11) |
| (12) |
| (13) |
| (14) |
The above equation defines the mapping of a tangent vector z⃗n at the origin of ℝ2 to the tangent vector υ⃗n at xn via the differential of the coordinate chart DΨn at x′ = 0. We note that for a tangent vector at an arbitrary point in ℝ2, the expression for the corresponding tangent vector on the sphere is more complicated. This motivates our definition of a separate chart for each mesh vertex, to simplify the derivations.
Gauss-Newton Step of Spherical Demons
From Eq. (14), we obtain exp(υ⃗) = exp({υ⃗n}) = exp({Enz⃗n}) and rewrite Eq. (9) as an unconstrained optimization problem:
| (15) |
| (16) |
This non-linear least-squares form can be optimized efficiently with the Gauss-Newton method, which requires finding the gradient of both terms with respect to {z⃗n} at {z⃗n = 0} and solving a linearized least-squares problem.
We let be the 1 × 3 spatial gradient of the warped moving image M ∘ ϒ(t)(·) at xn and note that is tangent to the sphere at xn. The computation of is discussed in Appendix A-A. Defining un ≜ exp({Enz⃗n})(xn), we differentiate the first term of the cost function fn(z⃗) in Eq. (15) using the chain rule, resulting in the 1 × 2 vector:
| (17) |
| (18) |
| (19) |
| (20) |
where δ(k, n) = 1 if k = n and 0 otherwise. Eq. (20) uses the fact that the differential of exp(υ⃗) at υ⃗ = 0 is the identity [47], i.e, [D exp(0)] υ⃗ = υ⃗. In other words, a change in velocity υ⃗k. at vertex xk does not affect exp(υ⃗)(xn) for n ≠ k up to the first order derivatives.
Similarly, we define to be the 3 × 3 Jacobian of ϒ(t)(·) at xn. The computation of is discussed in Appendix A-B. Differentiating the second term of the cost function gn(z⃗) in Eq. (15) using the chain rule, we get the 3 × 2 matrix:
| (21) |
where Gn is the skew-symmetric matrix defined in Eq. (4).
Once the derivatives are known, we can compute the corresponding gradients based on our choice of the metric of vector fields on S2. In this work, we assume an l2 inner product, so that the inner product of vector fields is equal to the sum of the inner products of the individual vectors. The inner product of individual vectors is in turn specified by the choice of the Riemannian metric on S2. Assuming the canonical metric, so that the inner product of two tangent vectors is the usual inner product in the Euclidean space [39], the gradients are equal to the transpose of the derivatives Eqs. (20), (21) (see Appendix A-C). We can then rewrite Eq. (15) as a linearized least-squares objective function:
| (22) |
| (23) |
| (24) |
Because of the delta function δ(k, n) in the derivatives in Eqs. (20), (21), z⃗n only appears in the n-th term of the cost function Eq. (24). The solution of Eq. (24) can therefore be computed independently for each z⃗n. Solving this linear least-squares equation yields an update rule for z⃗n:
| (25) |
For each vertex, we only need to perform matrix-vector multiplication of up to 3 × 3 matrices and matrix inversion of 2 × 2 matrices. This implies the update rule for v⃗n:
| (26) |
| (27) |
In practice, we use the Levenberg-Marquardt modification of Gauss-Newton optimization [49] to ensure matrix invertibility:
| (28) |
where ε is a regularization constant. We note that in the classical Euclidean Demons [57], [14], the term turns out to be the identity, so it can also be seen as utilizing Levenberg-Marquardt optimization. Once again, we emphasize that a different choice of the coordinate charts will lead to the same update.
Given , we use “scaling and squaring” to compute exp(υ⃗(t)) [3], which is then composed with the current transformation estimate ϒ(t) to form Γ(t) = ϒ(t) ∘ exp(υ⃗(t)). Appendix D discusses implementation details of extending the “scaling and squaring” procedure in Euclidean spaces to S2.
C. Choice of Reg(ϒ)
We now define the Reg(ϒ) term using the corresponding tangent vector field representation ϒ⃗. Following the work of [31], [61], we let H be the Hilbert space of square integrable vector fields on the sphere defined by the inner product:
| (29) |
where u⃗1, u⃗2 ∈ H and 〈·,·〉R refers to the canonical metric. Because vector fields from H are not necessarily smooth, we restrict the deformation ϒ⃗ to belong to the Hilbert space V ⊂ H of vector fields obtained by the closure of the space of smooth vector fields on S2 via a choice of the so-called energetic inner product denoted by
| (30) |
where L could for example be the Laplacian operator on smooth vector fields on S2 [31], [61].
We define Reg(ϒ) ≜ ‖ϒ⃗‖V. With a proper choice of the energetic inner product (e.g., Laplacian), a smaller value of ‖ϒ⃗‖V corresponds to a smoother vector field and thus smoother transformation ϒ. As we will see later in this section, the exact choice of the inner product is unimportant in our implementation.
D. Optimizing Step 2 of Spherical Demons
With our choice of dist(ϒ, Γ) in Section III-A and Reg(ϒ) in Section III-C, the optimization in Step 2 of the Spherical Demons algorithm
| (31) |
seeks a smooth vector field ϒ⃗ ∈ V that approximates the tangent vectors . This problem corresponds to the inexact vector spline interpolation problem solved in [31], motivating our use of tangent vectors in the definition of dist(ϒ, Γ) in Section III-A, instead of the more intuitive choice of geodesic distance.
We can express the tangent vectors Γ⃗n and ϒ⃗n as EnΓn and Enϒn respectively. Essentially, this represents Γ⃗n and ϒ⃗n in terms of the tangent space basis En at xn, where Γn and ϒn are the components of the tangent vectors with respect to this basis. Γ̂ and ϒ̂ be 2N × 1 vectors corresponding to stacking Γn and ϒn respectively. The particular optimization formulated in Eq. (31) has a unique optimum [31], given by
| (32) |
where K is a 2N × 2N matrix consisting of N × N blocks of 2 × 2 matrices: the (i,j) block corresponds to k(xi,xj)Txi,xj. The 2 × 2 linear transformation Txi,xj(·) parallel transports a tangent vector along the great circle from TxiS2 to TxjS2. k(xi,xj) is a non-negative scalar function uniquely determined by the choice of the energetic norm. Typically, k(xi,xj) monotonically decreases as a function of the distance between xi and xj. The proof of the uniqueness of the global optimum and the form of solution in Eq. (32) follow from the fact that the Hilbert space V is a reproducing kernel hilbert space (RKHS), allowing the exploitation of the Riesz representation theorem [31]. This offers a wide range of choices of regularization depending on the choice of the energetic norm and the corresponding RKHS.
In [31], the spherical vector spline interpolation problem was applied to landmark matching on S2, resulting in a reasonable sized linear system of equations. Solving the matrix inversion shown in Eq. (32) is unfortunately prohibitive for cortical surfaces with more than 100,000 vertices. If one chooses a relatively wide kernel k(xi,xj), the system is not even sparse.
Inspired by the convolution method of optimizing Step 2 in the Demons algorithm [14], [57], [66] and the convolution-based fast fluid registration in the Euclidean space [12], we propose an iterative smoothing approximation to the solution of the spherical vector spline interpolation problem.
In each smoothing iteration, for each vertex xi, tangent vectors of neighboring vertices xj are parallel transported to xi and linearly combined with the tangent vector at xi. The weights for the linear combination are set to and for i ≠ j, where |Ni| is the number of neighboring vertices of xi. Therefore, larger number of iterations m and values of γ results in greater amount of smoothing.
We note that the iterative smoothing approximation to spline interpolation is not exact because parallel transport is not transitive on S2 due to the non-flat curvature of S2 (unlike in Euclidean space), i.e., parallel transporting a tangent vector from point a to b to c results in a vector different from the result of parallel transporting a tangent vector from a to c. Furthermore, the approximation accuracy degrades as the distribution of points becomes less uniform. In Appendix B, we provide a theoretical bound on the approximation error and demonstrate empirically that iterative smoothing provides a good approximation of spherical vector spline interpolation for a relatively uniform distribution of points corresponding to those of the subdivided icosahedron meshes used in this work.
E. Remarks
The Spherical Demons algorithm is summarized in Algorithm 2.
We run the Spherical Demons algorithm in a multi-scale fashion on a subdivided icosahedral mesh. We begin from a subdivided icosahedral mesh (ic4) that contains 2,562 vertices and work up to a subdivided icosahedral mesh (ic7) that contains 163,842 vertices, which is roughly equal to the number of vertices in the cortical meshes we work with. We perform 15 iterations of Step 1 and Step 2 at each level. Because of the fast convergence rate of the Gauss-Newton method, we find that 15 iterations are more than sufficient for our purposes. We also perform a rotational registration at the beginning of each multi-scale level via a sectioned search of the three Euler angles.
Empirically, we find the computation time of the Spherical Demons algorithm is roughly divided equally among the four components: registration by rotation, computing the Gauss-Newton update, performing “scaling and squaring” and smoothing the vector field.
Algorithm 2. Spherical Demons Algorithm.
| Data: A fixed spherical image F and moving spherical image M. |
| Result: Diffeomorphism Γ so that M ∘ Γ is “close” to F. |
| Set ϒ0 = identity transformation (or some a-priori transformation, e.g., from a previous registration) |
| repeat |
| Step 1. Given ϒ(t), |
| foreach vertex n do |
| Compute using Eq. (28). |
| end |
| Compute Γ(t) = exp(v̄) using “scaling and squaring”. |
| Step 2. Given Γ(t), |
| foreach vertex n do |
| Compute using Eq. (48) implemented via iterative smoothing. |
| end |
| until convergence; |
In practice, we work with spheres that are normalized to be of radius 100, because we find that at ic7, the average edge length of 1mm corresponds to that of the original cortical surface meshes. This allows for meaningful interpretation of distances on the sphere. This requires slight modification of the equations presented previously to keep track of the radius of the sphere.
The Spherical Demons algorithm presented here registers pairs of spherical images. To incorporate a probabilistic atlas defined by a mean image and a standard deviation image, we modify the Demons objective function in Eq. (3), as explained in Appendix C. This requires a choice of warping the subject or warping the atlas. We find that interpolating the atlas gives slightly better results, compared with interpolating the subject. However, interpolating the subject results in a runtime of under 3 minutes, while the runtime for interpolating the atlas is less than 5 minutes. In the next section, we report results for interpolating the atlas.
IV. Experiments
We use two sets of experiments to evaluate the performance of the Spherical Demons algorithm by comparing it to the widely used and freely available FreeSurfer [27] software. The FreeSurfer registration algorithm uses the same similarity measure as Demons, but explicitly penalizes for metric and areal distortion. As we will show, even though the Spherical Demons algorithm does not specifically take into account the original metric properties of the cortical surface, we still achieve comparable if not better registration accuracy than FreeSurfer. Furthermore, FreeSurfer runtime is more than an hour while Spherical Demons runtime is less than 5 minutes.
There are four parameters in the algorithm. and ε appear in Eq. (28). Larger values of and ε decrease the size of the update taken in Step 1 of the Spherical Demons algorithm. In the experiments that follow, we set and set their values such that the largest vector of the update velocity field is roughly two times the edge lengths of the mesh. The number of iterations m and the weight determine the degree of smoothing. We set γ = 1 and explore a range of smoothing iterations m in the following experiments.
A. Parcellation of In-Vivo Cortical Surfaces
We validate Spherical Demons in the context of automatic cortical parcellation. Automatic labeling of cortical brain surfaces is important for identifying regions of interests for clinical, functional and structural studies [20], [52]. Recent efforts have ranged from the identification of sulcal/gyral ridge lines [56], [62] to the segmentation of sulcal/gyral basins [20], [28], [38], [41], [42], [51], [52], [67]. Similar to these prior studies, we are interested in parcellation of the entire cortical surface meshes, where each vertex is assigned a label.
We consider a set of 39 left and right cortical surface models extracted from in-vivo MRI [19]. Each surface is spherically parameterized and represented as a spherical image with geometric features at each vertex: mean curvature of the cortical surfaces, mean curvature of the inflated cortical surfaces and average convexity of the cortical surfaces, which roughly corresponds to sulcal depth [26]. These features are intrinsic and thus independent of the parameterization of the surface. The tools used for segmentation [19] and spherical parameterization [26] are freely available [29]. Both hemispheres of each subject were manually parcellated by a neuroanatomist into 35 labels, corresponding to the main sulci and gyri, enumerated in Table II.
TABLE II.
List of Parcellation Structures
| 1. Sylvian Fissure / Unknown | 2. Bank of the Superior Temporal Sulcus | 3. Caudal Anterior Cingulate |
| 4. Caudal Middle Frontal Gyrus | 5. Corpus Callosum | 6. Cuneus |
| 7. Entorhinal | 8. Fusiform Gyrus | 9. Inferior Parietal Complex |
| 10. Inferior Temporal Gyrus | 11. Isthmus Cingulate | 12. Lateral Occipital |
| 13. Lateral Orbito Frontal | 14. Lingual | 15. Medial Orbito Frontal |
| 16. Middle Temporal Gyrus | 17. Parahippocampal | 18. Paracentral |
| 19. Parsopercularis | 20. Parsorbitalis | 21. Parstriangularis |
| 22. Peri-calcarine | 23. Post-central Gyrus | 24. Posterior Cingulate |
| 25. Pre-central Gyrus | 26. Pre-cuneus | 27. Rostral Anterior Cingulate |
| 28. Rostral Middle Frontal | 29. Superior Frontal Gyrus | 30. Superior Parietal Complex |
| 31. Superior Temporal Gyrus | 32. Supramarginal | 33. Frontal Pole |
| 34. Temporal Pole | 35. Transverse Temporal |
We co-register all 39 spherical images of cortical geometry with Spherical Demons by iteratively building an atlas and registering the surfaces to the atlas. The atlas consists of the mean and variance of cortical geometry represented by the surface features described above. We then perform 4-fold cross-validation of the parcellation of the co-registered cortical surfaces. In each iteration of cross-validation, we leave out ten subjects and use the remainder of the subjects to train a classifier [20], [28] that predicts the labels based on location and geometric features. We then apply the classifier to the hold-out set of ten subjects. We perform each iteration with a different hold-out set, i.e., subjects 1-10, 11-20, 21-30 and 31-39.
As mentioned previously, increasing the number of iterations of smoothing results in smoother warps. As discussed in [67], the choice of the tradeoff between the similarity measure and regularization is important for segmentation accuracy. Estimating the optimal registration regularization tradeoff is an active area of research [1], [18], [48], [65], [67], [68] that we do not deal with in this paper. Here, we simply repeat the above experiments using {6, 8, 10, 12, 14} iterations of smoothing. For brevity, we will focus the discussion on using 10 iterations of smoothing and comment on results obtained with the other levels of smoothing.
We repeat the above procedure of performing co-registration and cross-validation with the FreeSurfer registration algorithm [27] using the default FreeSurfer settings. Once again, we use the same features and parcellation algorithm [20], [28]. As before, the atlas consists of the mean and variance of cortical geometry.
To compare the cortical parcellation results, we compute the average Dice measure, defined as the ratio of cortical surface area with correct labels to the total surface area averaged over the test set. Because the average Dice can be misleading by suppressing small structures, we also compute the Dice measure for each structure.
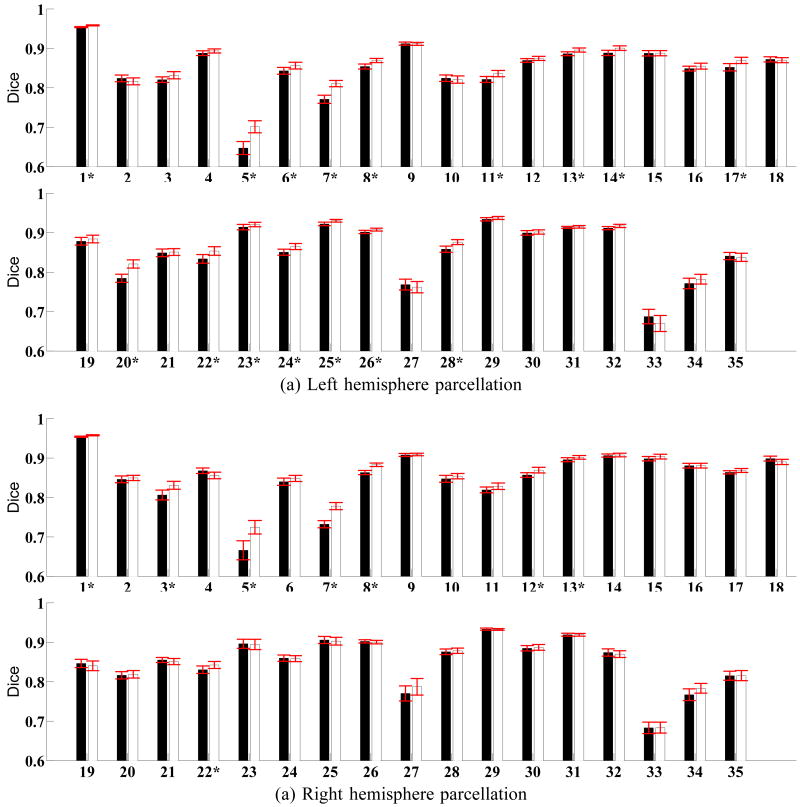
On the left hemisphere, FreeSurfer achieves an average Dice of 88.9, while Spherical Demons achieves an average Dice of 89.6 with 10 iterations of smoothing. While the improvement is not big, the difference is statistically significant for a onesided t-test with the Dice measure of each subject treated as an independent sample (p = 2 × 10−6). Furthermore, the overall Dice is statistically significantly better than FreeSurfer for all levels of smoothing we considered, with the best overal dice achieved with 12 iterations of smoothing.
On the right hemisphere, FreeSurfer obtains a Dice of 88.8 and Spherical Demons achieves 89.1 with 10 iterations of smoothing. Here, the improvement is smaller, but still statistically significant (p = 0.01). Furthermore, the overall dice is statistically significantly better than FreeSurfer for all levels of smoothing we considered, except when 6 iterations of smoothing is used (p = 0.06). All results we report in the remainder of this section use 10 iterations of smoothing.
We analyze the segmentation accuracy separately for each structure. To compare Spherical Demons with FreeSurfer, we perform a one-sided paired-sampled t-test treating each subject as an independent sample and correct for multiple comparisons using a False Discovery Rate (FDR) of 0.05 [10]. On the left (right) hemisphere, the segmentations of 16 (8) structures are statistically significantly improved by Spherical Demons with respect to FreeSurfer, while no structure is significantly worse.
Fig. 3 shows the percentage improvement of individual structures over FreeSurfer. Fig. 4 displays the average Dice per structure for FreeSurfer and Spherical Demons (10 iterations of smoothing) for the left and right hemispheres. Standard errors of the mean are displayed as red bars. The numbering of the structures correspond to Table II. Parcellation improvements suggest that our registration is at least as accurate as FreeSurfer.
Fig. 3.
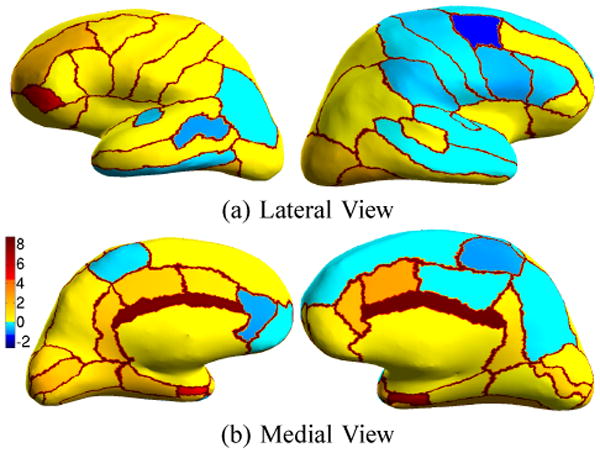
Percentage Improvement over FreeSurfer. Yellow regions indicate structures scoring better than FreeSurfer. Blue regions correspond to decrease in accuracy. Note that none of these blue regions are statistically significant. The boundaries between parcellation regions are set to reddish-brown to improve visualization of the regions.
Fig. 4.
(a) Dice measure for each structure in the left hemisphere. (b) Dice measure for each structure in the right hemisphere. Black columns correspond to FreeSurfer. White columns correspond to Spherical Demons. * indicates structures where Spherical Demons shows statistically significant improvement over FreeSurfer (FDR = 0.05). No structure exhibit significant decrease in accuracy.
The structures with the worst Dice are the frontal pole and entorhinal cortex. These structures are small and relatively poorly defined by the underlying cortical geometry. For example, the entorhinal cortex is partially defined by the rhinal sulcus, a tiny sulcus that is only visible on the pial surface. The frontal pole is defined by the surrounding structures, rather than by the underlying cortical geometry.
B. Brodmann Area Localization on Ex-vivo Cortical Surfaces
Brodmann areas are cyto-architectonically defined parcellations of the cerebral cortex [13]. They can be observed through histology and more recently, through ex-vivo high resolution MRI [6]. Unfortunately, much of the cytoarchitectonics cannot be observed with current in-vivo imaging. Nevertheless, most studies today report their functional findings with respect to Brodmann areas, usually estimated by visual comparison of cortical folds with Brodmann's original drawings without quantitative analysis of local accuracy. By combining histology and MRI, recent methods for creating probabilistic Brodmann area maps in the Talairach and Colin27 normalized space promise a more principled approach [2], [24], [54], [55], [71].
In this experiment, we consider a data set that contains Brodmann labels mapped to the corresponding MRI volume. Specifically, we work with postmortem histological images of ten brains created using the techniques described in [54], [71]. The histological sections were aligned to postmortem MR with nonlinear warps to build a 3D histological volume. These volumes were segmented to separate white matter from other tissue classes, and the segmentation was used to generate topologically correct and geometrically accurate surface representations of the cerebral cortex using FreeSurfer [19]. The eight manually labeled Brodmann area maps (areas 2, 4a, 4p, 6, 44, 45, 17 and 18) were sampled onto the surface representations of each hemisphere, and errors in this sampling were manually corrected (e.g., when a label was erroneously assigned to both banks of a sulcus). A morphological close was then performed on each label to remove small holes. We note that Brodmann areas 4a, 4p and 6 were mapped in only eight of the ten subjects. Fig. 5 shows these eight Brodmann areas on the resulting cortical representations for two subjects. Finally, we map the folding patterns and the Brodmann area labels onto a spherical coordinate system [27].
Fig. 5.
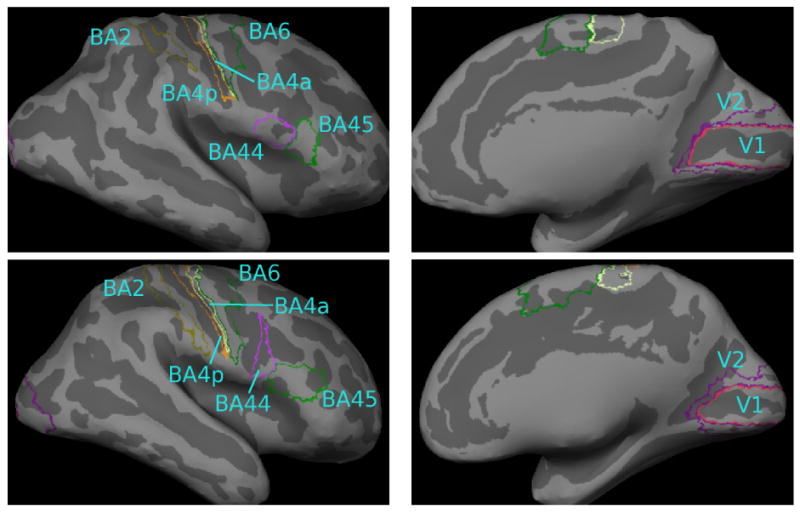
Brodmann areas 17 (V1), 18 (V2), 2, 4a, 4p, 6, 44 and 45 shown on inflated cortical surfaces of two subjects. Notice the variability of BA44 and BA45 with respect to the underlying folding pattern.
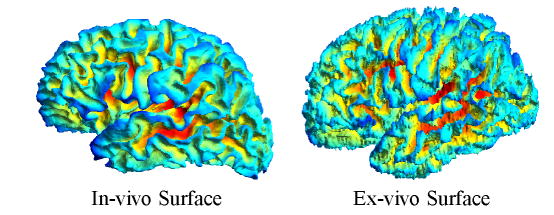
It has been shown that nonlinear surface registration of cortical folds can significantly improve Brodmann area overlap across different subjects [25], [68] compared with volumetric registration. Registering the ex-vivo surfaces is more difficult than in-vivo surfaces because the reconstructed volumes are extremely noisy due to the distortions introduced by the histology, resulting in noisy geometric features, as shown in Fig. 6.
Fig. 6.
Left: example in-vivo surface used in the parcellation study. Right: example ex-vivo surface used in the Brodmann area study.
We consider two strategies for aligning Brodmann areas. For both strategies, we will use 10 iterations of smoothing for Spherical Demons as it proved reasonable in the previous set of experiments. The first strategy involves co-registering the 10 ex-vivo surfaces using cortical geometry by repeatedly building an atlas and registering the surfaces to the atlas, similar to the previous experiment on cortical parcellation. We use either Spherical Demons or FreeSurfer for registration. We refer to the co-registration using Spherical Demons and FreeSurfer as SD10 and FS10 respectively (10 refers to the number of subjects in the study, not the number of smoothing iterations).
The second strategy involves registering the 10 ex-vivo surfaces to the in-vivo “Buckner40” atlas, constructed from 40 in-vivo subjects, that is distributed with the FreeSurfer software. Once again, we use either Spherical Demons or FreeSurfer for the registration. We refer to the co-registration using Spherical Demons and FreeSurfer as SD40 and FS40 respectively.
To measure the quality of alignment of the Brodmann areas after cortical registration, we use an adaptation of the modified Hausdorff distance [21]. For each pair of registered subjects, we project each Brodmann area from the first subject onto the second subject and compute the mean distance between the boundaries, measured on the original cortical surface of the second subject. We obtain a second measurement by projecting each Brodmann area from the second subject onto the first subject. Since we have 10 surfaces, we get 90 ordered pairs and 90 alignment errors for each Brodmann area.
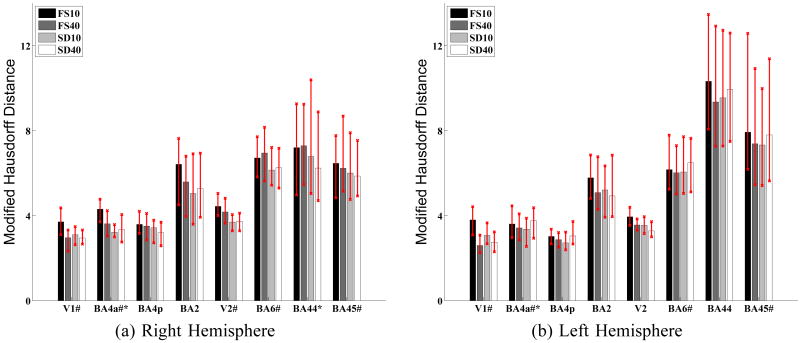
Table III reports the mean alignment errors for each Brodmann area and for each method. The lowest errors for each Brodmann area are shown in bold. We see that for almost all Brodmann areas, the best alignment come from SD10 or SD40. Similarly, Fig. 7 shows the median alignment error for each Brodmann area. The error bars indicate the lower and upper quartile alignment errors.
TABLE III.
Mean alignment errors of Brodmann areas in mm for the four registration methods. Lowest errors are shown in bold. SD refers to Spherical Demons; FS refers to FreeSurfer.
| Right Hemisphere |
Left Hemisphere |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | BA4a | BA4p | BA2 | V2 | BA6 | BA44 | BA45 | V1 | BA4a | BA4p | BA2 | V2 | BA6 | BA44 | BA45 | |
| FS10 | 3.8 | 4.4 | 3.8 | 6.3 | 4.6 | 7.0 | 7.4 | 6.8 | 3.8 | 3.8 | 3.1 | 5.9 | 4.0 | 6.5 | 11.5 | 9.9 |
| FS40 | 2.9 | 3.8 | 3.6 | 5.6 | 4.2 | 7.1 | 7.6 | 6.9 | 2.7 | 3.6 | 2.9 | 5.7 | 3.6 | 6.3 | 10.5 | 9.2 |
| SD10 | 3.1 | 3.3 | 3.3 | 5.4 | 3.7 | 6.4 | 7.7 | 6.4 | 3.2 | 3.4 | 2.8 | 5.5 | 3.5 | 6.4 | 10.4 | 8.6 |
| SD40 | 3.0 | 3.4 | 3.2 | 5.5 | 3.8 | 6.4 | 6.8 | 6.3 | 2.8 | 3.8 | 3.7 | 5.6 | 3.4 | 6.6 | 10.7 | 9.0 |
Fig. 7.
Median alignment errors of Brodmann areas in mm for the four registration methods. The error bars indicate the upper and lower quartile alignment errors. “#” indicates that the median errors of SD10 are statistically lower than those of FS10 (FDR = 0.05). “*” indicates SD40 outperforms FS40. For no Brodmann area does FreeSurfer outperform Spherical Demons.
We use permutation testing to evaluate statistical significance of the results. We cannot use the t-test because the 90 alignment errors are correlated - since the subjects are co-registered together, good alignment between subjects 1 and 2 and between subjects 2 and 3 necessarily implies a higher likelihood of good alignment between subjects 1 and 3.
The tests show that SD10 is better than FS10 and SD40 is slightly better than FS40. SD10 and SD40 are comparable. Compared with FS10, SD10 improves the median alignment errors of 5 (4) Brodmann areas on the right (left) hemisphere (FDR = 0.05) and no structure gets worse. Compared with FS40, SD40 statistically improves the alignment of 2 (1) Brodmann areas on the right (left) hemisphere (FDR = 0.05) with no structure getting worse. Permutation tests on the mean alignment errors show similar results, except that FS40 performs better than SD40 for BA4p on the left hemisphere when using the mean statistic. These results suggest that the Spherical Demons algorithm is at least as accurate as FreeSurfer in aligning cortical folds and Brodmann areas.
V. Discussion
The Demons algorithms [57], [66] discussed in Section II and the Spherical Demons algorithm proposed in this paper use a regularization term that modulates the final deformation. Motivated by [12], [14], the Diffeomorphic Demons algorithm [66] admits a fluid prior on the velocity fields. This involves smoothing the velocity field updates before computing the exponential map to obtain the displacement field updates to be composed with the current transformation. The resulting algorithm is very similar to the fast implementation [12] of Christensen's well-known fluid registration algorithm [16], except that Christensen's algorithm does not employ a higher-order update method like Gauss-Newon. The Spherical Demons algorithm can similarly incorporate a fluid prior by smoothing the velocity field υ⃗(t) in Eq. (28) before computing the exponential map to obtain the displacement updates exp(υ⃗(t)).
An alternative interpretation of the smoothing implementation of Christensen's algorithm comes from choosing a different metric for computing the gradient from the derivatives [9]. The choice of the metric also arises in our problem when computing the gradient as discussed in Appendix A-C. This suggests that the Spherical Demons algorithm can incorporate a fluid prior by modifying the Gauss-Newton update step Eq. (28). Unfortunately, this process introduces coupling among the vertices resulting in the loss of the speed-up advantage of Spherical Demons (see for example the derivations of [34]). The exploration of the performance of the different fluid prior implementations is outside the scope of this paper.
Because the tools of differential geometry are general, the Spherical Demons algorithm can be in principle extended to arbitrary manifolds, besides the sphere. One challenge is that the definition of coordinate charts for an arbitrary manifold is more difficult than that for the sphere. Approaches of utilizing the embedding space [15] or the intrinsic properties of manifolds [40] are promising avenues of future work.
VI. Conclusion
In this paper, we presented the fast Spherical Demons algorithm for registering spherical images. We showed that the two-step optimization of the Demons algorithm can also be applied on the sphere. By utilizing the one parameter subgroups of diffeomorphims, the resulting deformation is invertible. We tested the algorithm extensively in two different applications and showed that the accuracy of the algorithm compares favorably with the widely used FreeSurfer registration algorithm [27] while offering more than one order of magnitude speedup. Both matlab and ITK versions of the Spherical Demons algorithm are publicly available2.
A clear future challenge is to take into account the original metric properties of the cortical surface in the registration process, as demonstrated in previously proposed registration methods [27], [59].
We note that while fast algorithms are useful for deploying the developed tool on large datasets, they can further allow for complex applications that were previously computationally intractable. For example, we have incorporated the ideas behind Spherical Demons into a meta-registration framework that learns registration cost functions which are optimal for specific applications [68].
Acknowledgments
We thank Hartmut Mohlberg, Katrin Amunts and Karl Zilles for providing the histological dataset. We also like to thank Xavier Pennec, Wanmei Ou and Serdar Balci for helpful discussions and Tamar Riklin Raviv for reading the conference draft of this paper. We also thank Luis Ibanez and Michel Audette for reading the journal draft of this paper and for their help in implementing the ITK version of the Spherical Demons algorithm. Finally, we thank the reviewers for their many helpful suggestions on improving and clarifying the paper.
Support for this research is provided in part by the NAMIC (NIH NIBIB NAMIC U54-EB005149), the NAC (NIH NCRR NAC P41-RR13218), the mBIRN (NIH NCRR mBIRN U24-RR021382), the NIH NINDS R01-NS051826 grant, the NSF CAREER 0642971 grant, the National Institute on Aging (AG02238), NCRR (P41-RR14075, R01 RR16594-01A1), the NIBIB (R01 EB001550, R01EB006758), the NINDS (R01 NS052585-01) and the MIND Institute. Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation. B.T. Thomas Yeo is funded by the A*STAR, Singapore. Short visits of B.T. Thomas Yeo and Nicholas Ayache between MIT and INRIA were partially funded by the CompuTumor associated teams funding.
Appendix A: Step 1 Gradient Derivation
In this appendix, we provide details on the computation of the spatial gradient of the warped moving image M ∘ ϒ(t) and the Jacobian of the deformation ϒ(t). We also compute the gradients of the demons cost function using the derivatives computed in Eq. (20) and Eq. (21), assuming the l2 inner product space for vector fields and the canonical metric.
A. Computing Spatial Gradient of M ∘ ϒ(t)
In this appendix, we discuss the computation of , the spatial gradient of the warped moving image M ∘ ϒ(t) at the point xn. We can think of M ∘ ϒ(·) as an image Ms ≜ M ∘ ϒ defined on the mesh vertices {xn}. This image is made continuous by the choice of an interpolation method. In this work, we assume that we are working with a triangular mesh. To evaluate Ms at a point x ∈ S2, we first find the triangle that contains the intersection between the vector representing the point x (i.e., the vector between the center and the point x of the sphere) and the mesh. The image value at x is then given by the barycentric interpolation of the image values at the intersection point. Mathematically, we can write
| (33) |
where p(x) is the intersection point and I(·) is the barycentric interpolation. Let p1, p2, p3 denote the vertices of the triangle containing p(x) and n⃗ denote the 3 × 1 normal vector to the triangle. Since p(x) = αx for some α and 〈p(x) − p1, n⃗〉 = 0, we can write
| (34) |
and
| (35) |
where A1(p), A2(p) and A3(p) are the areas of the triangles △pp2p3, △pp1p3 and △pp1p2 respectively. Note that A = A1(p) + A2(p) + A3(p). Ms(p1), Ms(p2) and Ms(p3) are the image values at the mesh vertices p1, p2 and p3 respectively.
Computing the derivative of the image value at x follows easily from the chain rule:
| (36) |
| (37) |
where ∇pAi(p) is the derivative of the triangle area Ai. For example, ∇pA1(p) is a 1 × 3 vector in the plane of the triangle pp2p3, perpendicular and pointing to the edge p2p3, with magnitude half the length of p2p3. Combining Eq. (36) and Eq. (37) gives the spatial gradient of the warped moving image.
A complication arises when x corresponds to one of the mesh vertices, since the spatial gradient is not defined in this case. The same problem arises in Euclidean space with linear interpolation and the spatial gradient is typically defined via finite central difference. It is unclear what the equivalent definition on a mesh is. Here, for a mesh vertex x, we compute the spatial gradient for each of the surrounding triangles and linearly combine the spatial gradients using weights corresponding to the areas of the triangles.
B. Computing the Jacobian of Deformation ϒ(t)
In this appendix, we discuss the computation of , the Jacobian of the deformation ϒ(t) at xn. We can think of ϒ(t) as a vector function on S2 that maps each mesh vertex {xn} to a new point on the sphere. This vector image is made continuous by the choice of an interpolation method. We use the same interpolation as in Appendix A-A, except we need to normalize the barycentric interpolation so that the interpolated point is constrained to be on the sphere:
| (38) |
where p(x) is the same as in the previous section and
| (39) |
The Jacobian is computed via chain rule, just like in the previous section.
C. Computing the Gradients from the Derivatives
In this appendix, we seek to compute the gradients of fn(z⃗) ≜ F(xn) − M ∘ {ϒ(t) ∘ exp({Enz⃗n})}(xn) and , assuming a l2 inner product for vector fields and the canonical metric R for S2. These assumptions imply that the inner product of two vector fields and are given by
| (40) |
| (41) |
| (42) |
where
Eq. (40) follows from the equivalence of the tangent bundles Tℝ2 and TS2 induced by the coordinate charts{Ψn}.
Eq. (41) is the result of the l2 assumption that the inner product of vector fields is given by the sum of the inner products of individual vectors.
Because we assume the canonical metric, each term in the inner product in Eq. (41) is simply the usual inner product between 3 × 1 vectors and . Since the columns of Ek are orthonormal with respect to the usual inner product and using linearity of the inner product, Eq. (41) implies Eq. (42), i.e., the inner product 〈z⃗1, z⃗2〉l2 can be computed by the sum of the usual inner product between 2 × 1 tangent vectors and .
Let dfn(z⃗) be the directional derivative of fn for any z⃗ = {z⃗k}. The directional derivative is independent of the choice of metric. Since the derivative of fn(z⃗) with respect to z⃗k is a 1 × 2 vector (Eq. (20)), we get
| (43) |
Recall that the gradient ∇l2fn of fn(z⃗) is defined to be a tangent vector field such that dfn(z⃗) = 〈∇l2fn, z⃗〉l2 for any z⃗ = {z⃗k}. The gradient is therefore dependent on the choice of the inner product. From Eq. (42) and Eq. (43), we can write
| (44) |
| (45) |
| (46) |
Therefore, the gradient ∇l2fn can be written as a 2N × 1 vector consisting of N blocks of 2 × 1 vectors, where all the blocks are zeros, except the n-th block is equal to .
Similarly, we denote the gradient of gn(z⃗) as ∇l2gn(j) for j = 1, 2, 3 corresponding to the 3 output components of gn(z⃗). The derivative of ∇gn with respect to z⃗k is a 3 × 2 matrix (Eq. (21)), where is a 1 × 2 vector corresponding to the derivative of the j-th component of gn with respect to z⃗k. Using the same derivation as before, we can show that ∇l2gn(j) can be written as a 2N × 1 vector consisting of N blocks of 2 × 1 vectors, where all the blocks are zeros, except the n-th block is equal to a⃗jn.
Appendix B: Approximating Spline Interpolation with Iterative Smoothing
In this appendix, we demonstrate empirically that iterative smoothing provides a good approximation of spherical vector spline interpolation for a relatively uniform distribution of points corresponding to those of the subdivided icosahedron meshes used in this work. Once again, we work with spheres that are normalized to be of radius 100.
Recall that we seek {ϒ⃗n} = {Enϒn}, which is a smooth approximation of the input vector field {Γ⃗n} = {EnΓn}. The solution of the spherical vector spline interpolation problem is given in Eq. (32) as
| (47) |
where K is a 2N × 2N matrix consisting of N × N blocks of 2 × 2 matrices: the (i, j) block corresponds to k(xi, xj)Txi,xj. Txi,xj is the parallel transport operator from xi to xj. k(xi, xj) is a non-negative scalar function uniquely determined by the choice of the energetic norm that monotonically decreases as a function of the distance between xi and xj.
In constrast, the iterative smoothing approximation we propose can be formalized as follows:
| (48) |
where m is a positive integer and K′ is a 2N × 2N matrix consisting of N × N blocks of 2 × 2 matrices: the (i, j) block corresponds to λ(xi, xj)Txi,xj if xi and xj are neighboring vertices and is a zero matrix otherwise. and for i ≠ j, where |Ni| is the number of neighboring vertices of xi.
A. Reverse Engineering the Kernel
We now demonstrate empirically that for a range of values of γ, iterations m and the relatively uniform distribution of points corresponding to those of the subdivided icosahedron mesh, there exist kernels k(xi, xj) that are well approximated by iterative smoothing. Technically, the resulting k(xi, xj) might not correspond to a true choice of the energetic norm. However, in practice, this does not appear to be a problem.
More specifically, given a configuration of mesh points, iterations m and value of γ, we seek k̃(xi, xj), such that is “close” to (K′)m. We propose a two-stage estimation of k̃(xi, xj):
-
In the first stage, we seek k*(xi, xj) that is not constrained to be a function of the distance between xi and xj, such that
(49) Rearranging the terms, we get
(50) To make the “≈” concrete, we optimize for
(51) where ‖ · ‖F is the Frobenius norm.
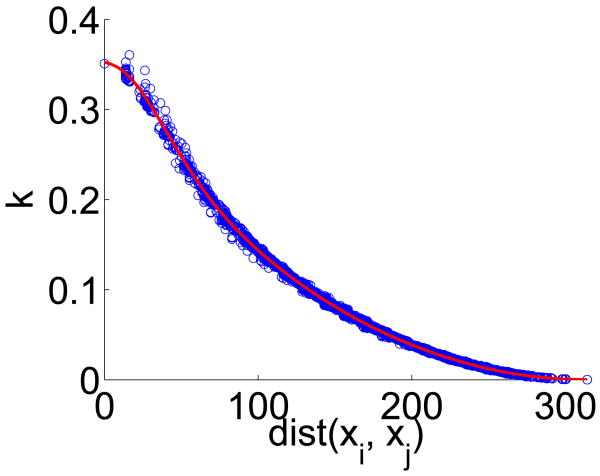
The cost function Eq. (51) can be optimized componentwise, i.e., we can solve for k*(xi, xj) for each pair xi, xj. For γ = 1, m = 10 and a subdivided icosahedron mesh with 642 vertices, we plot the resulting k*(xi, xj) as a function of the geodesic distance between xi and xj in Fig. 8.
In the second stage, we perform a least-squares fit of a b-spline function to the estimated k*(xi, xj) to obtain the final estimate of k̃(xi, xj). Fig. 8 illustrates an example kernel k̃(xi, xj) we obtain (c.f., the kernel illustrated in [31]). We note that an alternative to b-spline interpolation is to fit the coefficients of the general kernel function suggested in Appendix A of [31]. This will guarantee that the estimated kernel corresponds to an energetic norm. We leave exploring this direction to future work.
Fig. 8.
Approximating the kernel function k(xi, xj). The scattered points corresponds to the estimation of k*(xi, xj) via Eq. (51). The red curve corresponds to fitting the scattered points so that k̃(xi, xj) is strictly a function of the geodesic distance between xi and xj.
B. Evaluating Approximation
We now investigate the quality of the estimate k̃(xi, xj) by computing:
| (52) |
where ‖ · ‖2 is the l2 matrix operator norm. The difference metric Eq. (52) measures the maximum l2 difference between smoothed vector fields obtained from iterative smoothing and spherical vector spline interpolation for any possible input vector field {Γ⃗n} of unit l2 norm, i.e., Σn ‖Γ⃗n‖2 = 1. We note that k̃(xi, xj) can be in principle estimated by minimizing Eq. (52) instead of the proposed 2-stage process. However, the optimization is difficult since evaluating the cost function itself requires finding the largest singular value of a large, non-sparse matrix.
Fig. 9 displays the difference metric we obtained with different values of γ and iterations m for meshes ic2, ic3, ic4 and ic5. Each of the meshes is obtained from recursively subdividing a lower resolution mesh: ic2 indicates that the mesh was obtained from subdividing an icosahedron mesh twice. The number of vertices quadruples with each subdivision, so that ic5 corresponds to 10,242 vertices.
Fig. 9.
Difference metric as a function of the number of iterations m and value of γ.
We conclude from the figure that the differences between the two smoothing methods are relatively small and increase with mesh resolution. As discussed in the next section, we run Spherical Demons on different mesh resolutions, including ic7. Unfortunately, because of the large non-sparse matrices we are dealing with, we were only able to compute the differences up to ic5. Computing the difference metric for ic5 took an entire week on a machine with 128GB of RAM. However, the plots in Fig. 9 indicate that the differences appear to have converged by ic5.
To better understand the incurred differences, Fig. 10 illustrates the outputs and differences of the two smoothing methods for different inputs on ic4. The first row illustrates an input vector field which is zero everywhere except for a single tangent vector of unit norm. The results of spline interpolation and iterative smoothing correspond to our intuition that smoothing a single tangent vector propagates tangent vectors of smaller magnitudes to the surronding areas. The two methods also produce almost identical results as shown by the clean difference image in the fourth column.
Fig. 10.

Comparison of spline interpolation and iterative smoothing (m = 10, γ = 1). (a) Input vector field (b) Spline Interpolation Output (c) Iterative Smoothing Output (d) Difference between the second and third columns (e) l2 norm of the difference. The first row uses an input vector field which is zero everywhere except for a single tangent vector of unit norm. Second row illustrates the worst unit norm input as measured by the difference metric Eq. (52). This worst unit norm input vector field corresponds to the largest eigenvector in Eq. (52). The third row uses a vector field from an actual warped image from the experimental section. Note that the input vector field in the first two rows are scaled down for the purpose of display. The vector fields in the entire third row are of the same scale, but are scaled down relative to the first two rows, since the vector field from the warped image is substantially larger in magnitude than the unit norm inputs of the first two rows. This explains the substantially larger difference metric on the third row.
The second row of Fig. 10 demonstrates the worst unit norm input vector field as measured by the difference metric Eq. (52). This worst unit norm input vector field corresponds to the largest eigenvector in Eq. (52). The pattern of large differences correspond to the original 12 vertices of the uniform icosahedron mesh. These original 12 vertices are the only vertices in the subdivided icosahedron meshes with five, instead of six neighbors, as shown by the pentagon pattern. The fact that these 12 vertices are local maxima of differences suggest that these vertices are treated differently by the two smoothing techniques.
The last row of Fig. 10 demonstrates an input vector field that represents the deformation of an actual registration performed in Section IV. The norm of the input vector field is 700 times that in the first two rows, but the discrepancies between spline interpolation and iterative smoothing are less than expected. The differences of 90% of the vectors are less than 0.2mm, with larger differences in the neighborhoods of the 12 vertices identified previously. Since we conclude previously that the difference metric appears to have converged after ic4, the discrepancies are likely to be acceptable at ic7, whose mesh resultion is 1mm.
We should emphasize that the discrepancies between spline interpolation and iterative smoothing do not necessarily imply registration errors. The differences only indicate the deviations of the deformations from true local optima of the Demons registration cost function Eq. (3) assuming the estimated kernel. Approximating smoothing kernels by iterative smoothing is an active area of research in medical imaging [17], [32]. Future work would involve understanding the interaction between the number of smoothing iterations m and the choice of the weights on the quality of the spherical registration.
Appendix C: Atlas-Based Spherical Demons
In this section, we demonstrate how an atlas consisting of a mean image and a standard deviation image can be incorporated into the Spherical Demons algorithm. The standard deviation image replaces Σ in Eq. (3). We first discuss a probabilistic interpretation of the Demons objective function and its relationship to atlases. We then discuss the optimization of the resulting probabilistic objective function.
A. Probabilistic Demons Objective Function
The Demons objective function reviewed in Section II is defined for the pairwise registration of images. To incorporate a probabilistic atlas, we now reformulate the objective function. Consider the following Maximum-A-Posteriori objective function:
| (53) |
| (54) |
| (55) |
Assuming a Gaussian noise model, we define
| (56) |
| (57) |
| (58) |
where Reg(ϒ) is defined via the energetic norm as discussed in Section III-C and for reasons that will soon be clear, we are being purposefully agnostic about the form of σ(Γ, xn). The objective function in Eq. (55) becomes
| (59) |
which is the instantiation of the Demons objective function Eq. (3), except for the extra term . Note that we have omitted the partition functions and because σx and σT are constant with respect to the deformations Γ and (ϒ). In this probabilistic interpretation, the two regularization terms p(Γ∣(ϒ)) and p(ϒ) act as a hierarchical prior on Γ, with the hidden transformation (ϒ) as a hyperparameter.
As before, σ(Γ, xn) is the standard deviation of the intensity at vertex n. Given a set of co-registered images, we can create an atlas by computing the mean intensity and standard deviation at each vertex. To incorporate the atlas, we need to make the choice of treating the atlas as the fixed or moving image. If we treat the atlas as the fixed image, then we set F to be the mean image and σ to be the standard deviation. In this case, we do not need to interpolate the mean or standard deviation image. Consequently, σ(Γ, xn) = σ(xn) and log σ(Γ, xn) can be omitted from the optimization. The registration becomes identical to the Spherical Demons algorithm for two images.
However, recent work [1], [53] suggests that treating the atlas as a moving image might be more correct theoretically. This involves setting the moving image M to be the mean image. In this case, σ(Γ, xn) = σ(Γ(xn)) is a function of Γ and we must include log(σ(Γ(xn))) in the optimization. We performed experiments for both choices and found the results from interpolating the atlas, i.e., treating it as a moving image, to be only slightly better than interpolating the subject. However, interpolating the subject results in a faster algorithm, whose computational time is less than 3 minutes. We report the results of interpolating the atlas in the experimental section.
B. Optimization of Atlas-Based Spherical Demons
We now discuss the optimization in Eq. (59). Note that the introduction of the new term only affects Step 1 of the Spherical Demons algorithm. By parameterizing Γ(t) = (ϒ)(t) ∘ exp({Enz⃗n}), we get
| (60) |
| (61) |
The second term is the same as before, while the first term has become more complicated. Using the product rule and the techniques described in Appendix A, we can find the first derivatives of the first and second terms and estimate their second derivatives using the Gauss-Newton method. The difficulty lies in the third term, which is not quadratic and is even strictly concave, so we have to make further approximations.
Consider the problem of optimizing a one-dimensional function f(x). Let the current estimate of x be x0. Newton's optimization [49] involves the following update:
| (62) |
where f′(x0) and f″(x0) are the gradient and the Hessian of f evaluated at x0 respectively. When f″ is negative (positive), the update △x increases (decreases) the objective function, regardless of whether one is attempting to increase or decrease the objective function! The Gauss-Newton approximation of the Hessian for minimizing non-linear quadratic functions actually stabilizes the Newton's method by ensuring the estimated Hessian is positive.
To optimize Eq. (61) with Newton's method, we need to compute the gradient and the Hessian. Because we are using the l2 inner product and the canonical metric (see Appendix A-C), the gradient and the Hessian correspond to the first and second derivatives. The first derivative or gradient corresponds to
| (63) |
and the second derivative corresponds to
| (64) |
| (65) |
where the last approximation was made using the Gauss-Newton method. Not surprisingly, the third term corresponding to log is negative, which can introduce instability in the Gauss-Newton update. Consequently, we drop the last term, resulting in:
| (66) |
Note that the resulting update Eq. (62) is always in the direction of descent since the estimated second derivative is always positive. Theoretically, it is necessary to do a line search along the Gauss-Newton update direction to ensure convergence. In practice, we find that the objective function decreases reliably for each full Newton's step.
Appendix D: Numerics of Diffeomorphism
While υ and Φυ(x) = exp(υ)(x) are technically defined on the entire continuous image domain, in practice, υ and u are represented by vector fields defined on a discrete set of points in the image, such as at each pixel [57], [66] or control points [4], [9] or in our case, vertices of a spherical mesh. From the theories of ODEs [11], we know that the integral curves or trajectories u(t) = Φtυ(·) of a velocity field υ(x, t) exist and are unique if υ(x, t) is Lipschitz continuous in x and continuous in t. This is true in both Euclidean spaces and on manifolds. Uniqueness means that the trajectories do not cross, implying that the deformation is invertible. Furthermore, we know from the theories of ODEs that a Cr continuous velocity field υ produces a Cr continuous deformation field. Therefore, a sufficiently smooth velocity field results in a diffeomorphic transformation.
Since the velocity field υ is stationary in the case of the one parameter subgroup of diffeomorphisms, υ is clearly continuous (and in fact C∞) in t. A smooth interpolation of υ is continuous in the spatial domain and is Lipschitz continuous if we consider a compact domain, which holds since we only consider images that are closed and bounded.
To compute the final deformation of an image, we have to estimate exp(υ) at least at the set of image grid points. We can compute exp(υ) by numerically integrating the smoothly interpolated velocity field υ with Euler integration. In this case, the estimate becomes arbitrarily close to the true exp(υ) as the number of integration time steps increases. With a sufficiently large number of integration steps, we expect the estimate to be invertible and the resulting transformation to be diffeomorphic.
The parameterization of diffeomorphisms by a stationary velocity field is popular due to the “scaling and squaring” approach [3] for computing exp(υ). Instead of Euler integration, the “scaling and squaring” approach iteratively composes displacement fields. Because we are working on the sphere S2, the “scaling and squaring” procedure discussed in [3] has to be slightly modified:
| (67) |
| (68) |
where Ψn is the local coordinate chart defined in Eq. (10), such that Ψn(0) = xn. Eq. (67) differs from “scaling and squaring” in Euclidean space. is the velocity vector at the origin of ℝ2 corresponding to the velocity vector at xn. For large enough K, we can approximate a particle at the origin to move to position via the flow of . Finally, the coordinate chart Ψn maps the point back to the sphere. The correctness of this process follows from the fact that the solution trajectories of the ODEs of a vector field can be consistently transformed via the coordinate charts.
While “scaling and squaring” converges to the true answer as K approaches ∞ in the continuous case, in the discrete case, composition of the displacement fields requires interpolation of displacement fields, introducing errors in the process. In particular, suppose Φt0υ (x) and Φ2t0υ (x) are the true trajectories found by performing an accurate Euler integration up to time t0 and 2t0 respectively. Then, there does not exist a trivial interpolation scheme, so that Φ2t0υ(x) = Φt0υ(Φt0υ(x)). In practice however, it is widely reported that in ℝ2 and ℝ3, “scaling and squaring” tends to preserve invertibility even with rather large deformations [4], [66].
As discussed in Appendix A-B, we employ barycentric interpolation, followed by normalization to ensure the warp stays on the unit sphere. In practice, we find that the resulting transformation is indeed diffeomorphic. Technically speaking, since we use linear interpolation for the displacement field, the transformation is only homeomorphic rather than diffeomorphic. This is because the transformation is continuous, but not differentiable across mesh edges. However, we follow the convention of [3], [4], [66] who call their transformation diffeomorphic even though they are homeomorphic.
Footnotes
There are two versions of the code (matlab and ITK) available at http://sites.google.com/site/yeoyeo02/software/sphericaldemonsrelease. The matlab code is used in the experiments of this paper. The preliminary ITK code [35], [36], [37] can also be found at http://hdl.handle.net/10380/3117.
The matlab code was used for this paper. The ITK code is still preliminary. Please check website http://sites.google.com/site/yeoyeo02/software/sphericaldemonsrelease for updates.
Contributor Information
B.T. Thomas Yeo, Email: ythomas@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, USA.
Mert R. Sabuncu, Email: msabuncu@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, USA.
Tom Vercauteren, Email: tom.vercauteren@maunakeatech.com, Mauna Kea Technologies, Paris, France.
Nicholas Ayache, Email: nicholas.ayache@sophia.inria.fr, Asclepios Group, INRIA, Sophia Antipolis, France.
Bruce Fischl, Email: fischl@nmr.mgh.harvard.edu, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, USA; Department of Radiology, Harvard Medical School, Charlestown, USA and the Divison of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, USA.
Polina Golland, Email: polina@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, USA.
References
- 1.Allassonniere S, Amit Y, Trouvé A. Toward a Coherent Statistical Framework for Dense Deformable Template Estimation. Journal of the Royal Statistical Society, Series B. 2007;69(1):3–29. [Google Scholar]
- 2.Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann's Areas 17 and 18 Brought into Stereotaxic Space - Where and How Variable? NeuroImage. 2000;11:66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- 3.Arsigny V, Commowick O, Pennec X, Ayache N. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 4190. LNCS; 2006. A Log-Euclidean Framework for Statistics on Diffeomorphisms; pp. 924–931. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner J. A Fast Diffeomorphic Image Registration Algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner J, Andersson J, Friston K. High-Dimensional Image Registration using Symmetric Priors. NeuroImage. 1999;9:619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- 6.Augustinack J, Van der Kouwe A, Blackwell M, Salat D, Wiggins C, Frosch M, Wiggins G, Potthast A, Wald L, Fischl B. Detection of Entorhinal Layer ii Using 7 Tesla Magnetic Resonance Imaging. Annals of Neurology. 2005;57(4):489–494. doi: 10.1002/ana.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avants B, Gee J. Geodesic Estimation for Large Deformation Anatomical Shape Averaging and Interpolation. NeuroImage. 2004;23:139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Bakircioglu M, Joshi S, Miller M. Landmark Matching on Brain Surfaces via Large Deformation Diffeomorphisms on the Sphere. Proc SPIE Medical Imaging. 1999;3661:710–715. [Google Scholar]
- 9.Beg M, Miller M, Trouvé A, Younes L. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. International Journal on Computer Vision. 2005;61(2):139–157. [Google Scholar]
- 10.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society B. 1995;57(1):256–300. [Google Scholar]
- 11.Birkhoff G, Rota G. Ordinary Differential Equations. John Wiley and Sons Inc; 1978. [Google Scholar]
- 12.Bro-Nielsen M, Gramkow C. Fast Fluid Registration of Medical Images. Proceedigs Visualization in Biomedical Computing. 1996;1131:267–276. [Google Scholar]
- 13.Brodmann K. Vergleichende Lokalisationslehre der Grohirnrinde in Ihren Prinzipien Dargestellt auf Grund des Zellenbaues. 1909. [Google Scholar]
- 14.Cachier P, Bardinet E, Dormont D, Pennec X, Ayache N. Iconic Feature Based Non-Rigid Registration: The PASHA algorithm. Compute Vision and Image Understanding. 2003;89(2-3):272–298. [Google Scholar]
- 15.Chefd'hotel C. Scale Space and Variational Methods in Computer Vision. Vol. 4485. LNCS; 2007. A Method for the Transport and Registration of Images on Implicit Surfaces; pp. 860–870. [Google Scholar]
- 16.Christensen G, Rabbit R, Miller M. Deformable Templates Using Large Deformation Kinematics. IEEE Transactions on Image Processing. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 17.Chung M, Robbins S, Dalton K, Davidson R, Alexander A, Evans A. Cortical Thickness Analysis in Autism with Heat Kernel Smoothing. NeuroImage. 2005;25(4):1256–1265. doi: 10.1016/j.neuroimage.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 18.Commowick O, Stefanescu R, Fillard P, Arsigny V, Ayache N, Pennec X, Malandain G. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 3750. LNCS; 2005. Incorporating Statistical Measures of Anatomical Variability in Atlas-To-Subject Registration for Conformal Brain Radiotherapy; pp. 927–934. [DOI] [PubMed] [Google Scholar]
- 19.Dale A, Fischl B, Sereno M. Cortical Surface-Based Analysis I: Segmentation and Surface Reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 20.Desikan R, Segonne F, Fischl B, Quinn B, Dickerson B, Blacker D, Buckner R, Dale A, Maguire R, Hyman B, Albert M, Killiany R. An Automated Labeling System for Subdividing the Human Cerebral Cortex on MRI Scans into Gyral Based Regions of Interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Dubuisson M, Jain A. A Modified Hausdorff Distance for Object Matching. Proceedings of the 12th IAPR International Conference on Pattern Recognition. 1994;1:566–568. [Google Scholar]
- 22.Durrleman S, Pennec X, Trouvé A, P, Ayache N. Inferring brain variability from diffeomorphic deformations of currents: an integrative approach. Medical Image Analysis. 2008;12(5):626–637. doi: 10.1016/j.media.2008.06.010. PMID: 18658005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckstein I, Joshi A, Jay Kuo C, Leahy R, Desbrun M. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 4791. LNCS; 2007. Generalized Surface Flows for Deformable Registration and Cortical Matching; pp. 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eickhoff S, et al. A new SPM Toolbox for Combining Probabilistic Cytoarchitectonic Maps and Functional Imaging Data. NeuroImage. 2005;25(4):1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BTT, Mohlberg H, Amunts K, Zilles K. Cortical Folding Patterns and Predicting Cytoarchictecture. Cerebral Cortex. 2008;18(8):1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischl B, Sereno M, Dale A. Surface-Based Analysis II: Inflation, Flattening, and a Surface-Based Coordinate System. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 27.Fischl B, Sereno M, Tootell R, Dale A. High-resolution Inter-subject Averaging and a Coordinate System for the Cortical Surface. Human Brain Mapping. 1999;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat D, Busa E, Seidman L, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale A. Automatically Parcellating the Human Cerebral Cortex. Cerebral Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 29.http://surfer.nmr.mgh.harvard.edu/fswiki.
- 30.Geng X, Kumar D, Christensen G. Proceedings of the International Conference on Information Processing in Medical Imaging. Vol. 3564. LNCS; 2005. Transitive Inverse-Consistent Manifold Registration; pp. 468–479. [DOI] [PubMed] [Google Scholar]
- 31.Glaunès J, Vaillant M, Miller M. Landmark Matching via Large Deformation Diffeomorphisms on the Sphere. Journal of Mathematical Imaging and Vision. 2004;20:179–200. [Google Scholar]
- 32.Hagler D, Saygin A, Sereno M. Smoothing and Cluster Thresholding for Cortical Surface-Based Group Analysis for fMRI Data. NeuroImage. 2006;33(4):1093–1103. doi: 10.1016/j.neuroimage.2006.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez M, Bossa M, Olmos S. Registration of Anatomical Images Using Geodesic Paths of Diffeomorphisms Parameterized with Stationary Velocity Fields. Proceedings of the Workshop on Mathematical Methods in Biomedical Image Analysis, International Conference on Computer Vision. 2007:1–9. [Google Scholar]
- 34.Hernandez M, Olmos S. Gauss-Newton Optimization in Diffeomorphic Registration. Proceedings of the International Symposium on Biomedical Imaging: From Nano to Macro. 2008:1083–1086. [Google Scholar]
- 35.Ibanez L, Audette M, Yeo BTT, Golland P. Rotational Registration of Spherical Surfaces Represented as QuadEdge Meshes. Insight Journal. 2009. Jan-Jun. http://hdl.handle.net/10380/3063.
- 36.Ibanez L, Audette M, Yeo BTT, Golland P. Spherical Demons Registration of Spherical Surfaces. Insight Journal. 2009. Jul-Dec. http://hdl.handle.net/10380/3117.
- 37.Ibanez L, Yeo BTT, Golland P. Iterative Smoothing of Field Data in Spherical Meshes. Insight Journal. 2009. Jul-Dec. http://hdl.handle.net/10380/3091.
- 38.Klein A, Hirsch J. Mindboggle: A Scatterbrained Approach to Automate Brain Labeling. NeuroImage. 2005;24:261–280. doi: 10.1016/j.neuroimage.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Kühnel W. Differential Geometry: Curves-Surfaces-Manifolds. second. American Mathematical Society; 2006. [Google Scholar]
- 40.Lefèvre J, Baillet S. Optical Flow and Advection on 2-Riemannian Manifolds: A Common Framework. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2008;30(6):1081–1092. doi: 10.1109/TPAMI.2008.51. [DOI] [PubMed] [Google Scholar]
- 41.Lohmann G, Von Cramon D. Automatic Labelling of the Human Cortical Surface using Sulcal Basins. Medical Image Analysis. 2000;4:179–188. doi: 10.1016/s1361-8415(00)00024-4. [DOI] [PubMed] [Google Scholar]
- 42.Mangin JF, Frouin V, Bloch I, Régis J, Lopez-Krahe J. Automatic Construction of an Attributed Relational Graph Representing the Cortex Topography using Homotopic Transformations. Proceedings of SPIE. 1994;2299:110–121. [Google Scholar]
- 43.Marsland S, McLachlan R. Proceedings of the International Conference on Information Processing in Medical Imaging. Vol. 4548. LNCS; 2007. A Hamiltonian Particle Method for Diffeomorphic Image Registration; pp. 396–407. [DOI] [PubMed] [Google Scholar]
- 44.Munkres J. Analysis on Manifold. Westview Press; 1991. [Google Scholar]
- 45.Nielsen M, Florack L, Deriche R. Regularization, Scale-Space, and Edge Detection Filters. Journal of Mathematical Imaging and Vision. 1997;7:291–307. [Google Scholar]
- 46.Nielsen M, Johansen P, Jackson AD, Lautrup B. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 2489. LNCS; 2002. Brownian Warps: A Least Committed Prior for Non-rigid Registration; pp. 557–264. [Google Scholar]
- 47.Olver P. Applications of Lie Groups to Differential Equations. 2nd. Springer-Verlag; 1993. [Google Scholar]
- 48.Pennec X, Stefanescu R, Arsigny V, Fillard P, Ayache N. Riemannian Elasticity: A Statistical Regularization Framework for Nonlinear Registration. doi: 10.1007/11566489_116. [DOI] [PubMed] [Google Scholar]
- 49.Press W, Teukolsky S, Vetterling W, Flannery B. Numerical Recipes in C: The Art of Scientific Computing. second. Cambridge University Press; 1992. [Google Scholar]
- 50.Qiu A, Miller M. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 4791. LNCS; 2007. Cortical Hemisphere Registration via Large Deformation Diffeomorphic Metric Curve Mapping; pp. 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rettmann M, Han X, Xu C, Prince J. Automated Sulcal Segmentation Using Watersheds on the Cortical Surface. NeuroImage. 2002;15:329–344. doi: 10.1006/nimg.2001.0975. [DOI] [PubMed] [Google Scholar]
- 52.Riviere D, Mangin JF, Papadopoulos-Orfanos D, Martinez J, Frouin V, Regis J. Automatic Recognition of Cortical Sulci of the Human Brain Using a Congregation of Neural Networks. Medical Image Analysis. 2002;6:77–92. doi: 10.1016/s1361-8415(02)00052-x. [DOI] [PubMed] [Google Scholar]
- 53.Sabuncu M, Yeo BTT, Vercauteren T, Van Leemput K, Golland P. Assymetric Image-Template Registration. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2009;5761:565–573. doi: 10.1007/978-3-642-04268-3_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schleicher A, Amunts K, Geyer S, Morosan P, Zilles K. Observer-Independent Method for Microstructural Parcellation of Cerebral Cortex: A Quantitative Approach to Cytoarchitectonics. NeuroImage. 1999;9:165–177. doi: 10.1006/nimg.1998.0385. [DOI] [PubMed] [Google Scholar]
- 55.Schormann T, Zilles K. Three-Dimensional Linear and Non-linear Transformations: An Integration of Light Microscopical and MRI Data. Human Brain Mapping. 1998;6:339–347. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<339::AID-HBM3>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao X, Prince J, Davatzikos C. Using a Statistical Shape Model to Extract Sulcal Curves on the Outer Cortex of the Human Brain. IEEE Transactions on Medical Imaging. 2002;21:513–524. doi: 10.1109/TMI.2002.1009387. [DOI] [PubMed] [Google Scholar]
- 57.Thirion J. Image Matching as a Diffusion Process: an Analogy with Maxwell's Demons. Medical Image Analysis. 1998;2(3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 58.Thompson P, Toga A. A Surface-Based Technique for Warping 3-Dimensional Images of the Brain. IEEE Transactions on Medical Imaging. 1996;15(4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 59.Thompson P, Woods R, Mega M, Toga A. Mathematical/Computational Challenges in Creating Deformable and Probabilistic Atlases of the Human Brain. Human Brain Mapping. 2000;9(2):81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tosun D, Prince J. Proceedings of the International Conference on Information Processing in Medical Imaging. Vol. 3565. LNCS; 2005. Cortical Surface Alignment Using Geometry Driven Multispectral Optical Flow; pp. 480–492. [DOI] [PubMed] [Google Scholar]
- 61.Trouvé A. Diffeomorphisms, Groups and Pattern Matching in Image Analysis. International Journal on Computer Vision. 1998;28(3):213–221. [Google Scholar]
- 62.Tu Z, Zheng S, Yuille A, Reiss A, Dutton R, Lee A, Galaburda A, Dinov I, Thompson P, Toga A. Automated Extraction of the Cortical Sulci Based on a Supervised Learning Approach. IEEE Transactions on Medical Imaging. 2007;26(4):541–552. doi: 10.1109/TMI.2007.892506. [DOI] [PubMed] [Google Scholar]
- 63.Twining C, Davies R, Taylor C. Proceedings of the International Conference on Information Processing in Medical Imaging. Vol. 4584. LNCS; 2007. Non-Parametric Surface-Based Regularisation for Building Statistical Shape Models; pp. 738–750. [DOI] [PubMed] [Google Scholar]
- 64.Van Essen D, Drury H, Joshi S, Miller M. Functional and Structural Mapping of Human Cerebral Cortex: Solutions are in the Surfaces. Proceedings of the National Academy of Sciences. 1996;95(3):788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Leemput K. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 4190. LNCS; 2006. Probabilistic Brain Atlas Encoding Using Bayesian Inference; pp. 704–711. [DOI] [PubMed] [Google Scholar]
- 66.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage. 2009 Mar;45(1, Supp.1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. PMID: 19041946. [DOI] [PubMed] [Google Scholar]
- 67.Yeo BTT, Sabuncu M, Desikan R, Fischl B, Golland P. Effects of registration regularization and atlas sharpness on segmentation accuracy. Medical Image Analysis. 2008;12(5):603–615. doi: 10.1016/j.media.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeo BTT, Sabuncu M, Golland P, Fischl B. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 5761. LNCS; 2009. Task-Optimal Registration Cost Functions; pp. 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeo BTT, Sabuncu M, Vercauteren T, Ayache N, Fischl B, Golland P. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 5241. LNCS; 2008. Spherical Demons: Fast Surface Registration; pp. 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeo BTT, Vercauteren T, Fillard P, Pennec X, Golland P, Ayache N, Clatz O. DT-REFinD: Diffusion Tensor Registration with Exact Finite-Strain Differential. IEEE Transactions on Medical Imaging. 2009 doi: 10.1109/TMI.2009.2025654. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zilles K, Schleicher A, Palomero-Gallagher N, Amunts K. Quantitative Analysis of Cyto- and Receptor Architecture of the Human Brain. Elsevier; 2002. [Google Scholar]
- 72.Zou G, Hua J, Muzik O. Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) Vol. 4791. LNCS; 2007. Non-rigid Surface Registration using Spherical Thin-Plate Splines; pp. 367–374. [DOI] [PubMed] [Google Scholar]