Abstract
The caryopses of barley (Hordeum vulgare), as of all cereals, are complex sink organs optimized for starch accumulation and embryo development. While their early to late development has been studied in great detail, processes underlying the caryopses’ diurnal adaptation to changes in light, temperature, and the fluctuations in phloem-supplied carbon and nitrogen have remained unknown. In an attempt to identify diurnally affected processes in developing caryopses at the early maturation phase, we monitored global changes of both gene expression and metabolite levels. We applied the 22 K Barley1 GeneChip microarray and identified 2,091 differentially expressed (DE) genes that were assigned to six major diurnal expression clusters. Principal component analysis and other global analyses demonstrated that the variability within the data set relates to genes involved in circadian regulation, storage compound accumulation, embryo development, response to abiotic stress, and photosynthesis. The correlation of amino acid and sugar profiles with expression trajectories led to the identification of several hundred potentially metabolite-regulated DE genes. A comparative analysis of our data set and publicly available microarray data disclosed suborgan-specific expression of almost all diurnal DE genes, with more than 350 genes specifically expressed in the pericarp, endosperm, or embryo tissues. Our data reveal a tight linkage between day/night cycles, changes in light, and the supply of carbon and nitrogen. We present a model that suggests several phases of diurnal gene expression in developing barley caryopses, summarized as starvation and priming, energy collection and carbon fixation, light protection and chaperone activity, storage and growth, and embryo development.
Metabolic processes in plants and most other organisms operate in concert with day/night cycles. This coordination is accomplished by diurnal oscillations in transcriptional and posttranscriptional activities integrated by light, temperature, carbon status, and circadian signaling (Usadel et al., 2008, and refs. therein). Through this arrangement, metabolic pathways in the cell can sense and anticipate environmental cues.
The plant body is generally divided into net carbon-exporting tissues, like mature leaves, and net carbon-importing tissues, such as the endosperm of developing seeds. These source and sink tissues alike can sense day/night cycles. For sink organs, this sensing is partially mediated via diurnal fluctuations in metabolite availability or circadian rhythms in the source. A prime example is Suc export from photosynthetically active tissues to sink organs, which slows down at night when carbon export depends on mobilization of transitory starch (Geigenberger and Stitt, 2000). In addition, since leaves export not only sugars but also amino acids, it follows that sink organs should experience shifts in nitrogen supply as well. This communication between source and sink has a major impact on cellular functions in both types of organs and is further integrated with hormonal, particularly abscisic acid, signaling (Gutierrez et al., 2007; Baguma et al., 2008). In addition, sink organs exhibit circadian or circadian-like rhythms in gene activities independently of the metabolic output from the source (Baguma et al., 2008).
The cereal seed, called caryopsis, is an economically important sink organ. It comprises the filial embryo and endosperm that are surrounded by a maternal seed coat, which consists of the photosynthetically active palea, lemma, and pericarp tissues. Palea and lemma form the outermost layer of caryopses and are involved in the seed's protection from light damage and pathogen attack (Abebe et al., 2004). The pericarp, which is vital until late stages of caryopsis development, contains the vascular tissues and acts as the central tissue for phloem unloading into the caryopsis (Patrick and Offler, 2001). The filial endosperm, which constitutes the largest part of the caryopsis during middle to late stages of development, accumulates storage starch and nourishes the embryo during seed germination. In addition to these compartments, thin layers of specialized cells that connect the different suborgans and/or function in transporting metabolites from vascular tissues into filial tissues add to the complexity of the caryopsis (Patrick and Offler, 2001; Weschke et al., 2003; Abebe et al., 2004; Thiel et al., 2008; Sabelli and Larkins, 2009).
Metabolic and regulatory changes throughout the development of the barley (Hordeum vulgare) caryopsis, from early flowering to late seed maturation and germination processes, have been studied to a great extent (Sreenivasulu et al., 2006, 2008; Radchuk et al., 2009). However, little is known about diurnal adaptations in the cereal caryopses, and several aspects pose intriguing questions regarding the regulation of these processes.
To obtain a comprehensive understanding of the interplay between major metabolic processes in the caryopsis, like phloem unloading, embryo development, storage starch biosynthesis, photosynthate production, and turnover of transient starch in the green tissues, we set out to follow the transcriptional activities during a 24-h light/dark regime in intact barley caryopses. We used the 22 K Barley1 GeneChip array (Close et al., 2004) and monitored levels of transcripts at different time points. A total of 2,091 differentially expressed (DE) genes were clustered into coexpressed and functional groups. To correlate oscillations in gene expression with metabolic activities, we measured metabolite levels for sugars and amino acids and found several hundred genes that are potential targets for metabolite-dependent gene regulation. A comparison of our DE genes and publicly available microarray data (Sreenivasulu et al., 2008) disclosed that the pericarp, endosperm, and embryo contributed equally to the observed day/night diversity. While the majority of diurnal DE genes were expressed in all suborgans tested, about 350 diurnal DE genes were assigned to specific parts of the caryopsis.
RESULTS
Diurnal Expression Profiling of Developing Barley Caryopses
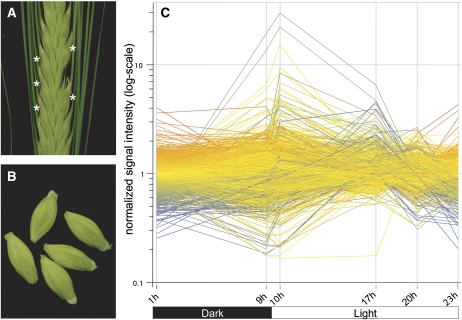
We used barley caryopses at 11 to 12 d post anthesis (d.p.a.) to study diurnal changes in gene expression. This developmental stage, also referred to as the early maturation phase, is characterized by high Suc levels and a rapid accumulation of storage starch in filial tissues of the caryopsis (Borisjuk et al., 2004; Sabelli and Larkins, 2009). Caryopses from the mid region of the spike (Fig. 1, A and B) were sampled at six time points during the day. The first sample was harvested at 11 d.p.a., 1 h after the onset of darkness. The following day (12 d.p.a.), samples were taken 0.5 h before and after the shift from dark to light phase (9 and 10 h), at two time points in the middle of the light phase (17 and 20 h), and 1 h before the end of the light phase (23 h after the onset of darkness). Using pooled plant material, we obtained highly reproducible data for two replicates at each time point. Correlation coefficients for the replicates ranged from 0.968 to 0.989 (data not shown). In total, 17,416 probe sets (hereafter referred to as genes) were scored present and used for further data analysis (Supplemental Table S1). For data verification, quantitative (q)PCR experiments were performed for a subset of genes and revealed similar expression patterns (Supplemental Fig. S1).
Figure 1.
Sampling of plant material and profiles of DE genes. A, Central region of a barley spike. One floret was removed to determine the onset of flowering. At 12 d.p.a., the five barley caryopses closest to the floret used for determination of the onset of anthesis were harvested (white asterisks). B, Prior to pooling and freezing the samples, awns and possibly remaining anthers were removed from the caryopses. C, Expression profile of 2,091 DE genes during the diurnal cycle. Six time points of harvest in dark and light phases are indicated on the x axis as follows: beginning and end of the dark phase (1 and 9 h), beginning of the light phase (10 h), middle of the light phase (17 and 20 h), and end of the light phase (23 h).
Cluster Analysis of Genes Showing Diurnal Expression Patterns
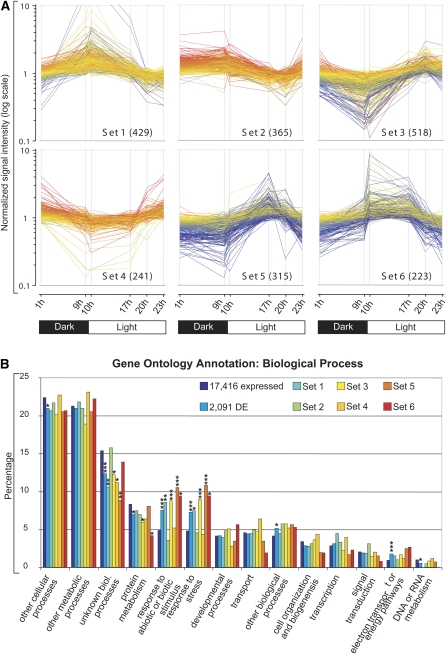
To examine expressional changes in more detail, we calculated the total number of DE genes. On the basis of the 17,416 expressed genes, a total of 2,091 (11%) DE genes could be assigned with significance (Fig. 1C; Supplemental Table S2). Subsequently, the expression trajectories of all 2,091 DE genes were subjected to k-means clustering, and six main clusters (sets 1–6) were chosen, with each of them exhibiting a distinct minimum and maximum of expression (Fig. 2A; Supplemental Table S3). Set 1 (429 genes) can generally be described by an increase of expression during the night and a constant decrease of expression during the light phase. Interestingly, the expression trajectories of cluster set 3 (518 genes) are complementary to set 1, with a steady increase of transcript levels during the light phase and a decrease during the dark. Together, these two inversely correlated clusters represent about 45% of all DE genes. A remarkable feature of both clusters is that the major up- or down-regulation of gene expression occurs in anticipation of the dawn (i.e. before the actual onset of light).
Figure 2.
Expression patterns of k-means clusters of DE genes and their classification into functional categories of GO. A, Gene expression trajectories of the k-means clusters named sets 1 to 6. The respective six time points are indicated on the x axis. Total numbers of assigned probe sets are given in parentheses. Colors of the profiles depict the normalized transcript level of the probe set at 1 h: red, high expression; yellow, moderate expression; blue, low expression. B, Classification of genes according to GO groups for Biological Processes. The number of genes belonging to a particular GO group is shown as a percentage for all present probe sets detected by the microarray (17,416 expressed), all DE genes (2,091 genes), and the subsets of genes belonging to clusters 1 to 6 (sets 1–6). Asterisks indicate a significant overrepresentation or underrepresentation of functional categories compared with the functional categories of all expressed genes with putative orthologs in Arabidopsis (* P ≤ 0.01, ** P ≤ 0.001, *** P ≤ 0.0001).
Genes clustered in set 6 (223 genes) are characterized by a rapid induction upon transition from darkness to light at the onset of day, and their expression is significantly reduced after the middle of the day. In contrast, the genes contained in set 4 (241 genes) exhibit a rapid reduction of gene expression at the shift from darkness to light and a maximum of gene expression at the onset of the night. Genes of set 2 (365 genes) can be described by rather high and stable transcript levels during the night and a constant reduction during the day. It is noteworthy that set 3 transcript levels are restored during the last 3 h of the light phase before the onset of the night. The genes contained in set 5 (315 genes) are characterized by a maximum expression in the middle of the day followed by a decrease in transcript abundance during the later light phase and stable expression during the night.
We next performed an unbiased approach to estimate the contribution of the DE genes to the total variability observed within the experiment and subjected the data set to principal component analysis. We assumed that the genes contained in the six sets derived by the k-means clustering should make a distinct contribution to the entire variability of the data set, which in turn would constitute an independent validation of the number of inferred gene clusters. Indeed, most DE genes overlapped with principal components PC1 to PC3 and contributed to more than 90% of the expression variability observed in our data set (Supplemental Table S4). In addition, the six clusters mark three pairs of antagonistic sets to be either highly correlated or anticorrelated to PC1, PC2, and PC3, which is supportive of our clustering procedure. Supplemental Table S4 lists those genes that exhibit high positive or negative correlation coefficients for the antagonistic pairs sets 3 and 1 (for PC1), sets 6 and 4 (for PC2), and sets 5 and 2 (for PC3).
Functional Annotations of the Expression Clusters Using Gene Ontology
To determine whether the expression profiles in the six clusters correlated with functional activities, they were classified into functional groups using the Gene Ontology (GO) terms. We caution that GO annotations could only be made for those probe sets on the microarray that had been assigned in putative orthology to Arabidopsis (Arabidopsis thaliana) based on their HarvEST annotation (Close et al., 2007). This was the case for about 70% of all probe sets (Supplemental Table S1). In addition, we did not compute the error from different probe sets that share the identical Arabidopsis gene as a best match in the HarvEST annotation. Hence, these genes contribute only once to the GO annotation and may thus add a bias to the analysis.
Ontologies for Biological Process, Cellular Component, and Molecular Function are shown in Figure 2B and Supplemental Figure S3. For the individual six clusters, all exhibit a characteristic distribution of the GO categories. For example, genes related to nucleic acid binding (P ≤ 10−5) and transcription factor activities (P ≤ 0.001) are overrepresented in set 1, whereas they are underrepresented in the antagonistic set 3 (P ≤ 0.01; Fig. 2A). Particularly, the presence of transcription-related genes in set 1 is in line with the idea of the plants’ strategy to anticipate the dawn. Furthermore, we note that there is an overrepresentation of DE genes in the GO groups for stress responses (P ≤ 10−8). A closer look at the genes grouped in the stress-related categories revealed that a common stimulus for most of these genes was temperature stress. This was particularly true for genes in sets 3 and 5 (P ≤ 10−4), which depict maximum transcript levels during the light phase. Another finding was that the GO category Extracellular is dominated by set 4 genes (P ≤ 0.01) and may thus indicate that biosynthesis processes in the cell wall or transport predominantly occur during the later light and early dark phases. Notably, genes with Nucleotide Binding (P ≤ 0.001) capabilities are significantly depleted in set 6, which translates into a lower necessity for ATP-dependent processes, like energized transport over membranes.
MapMan Analysis of Present Genes
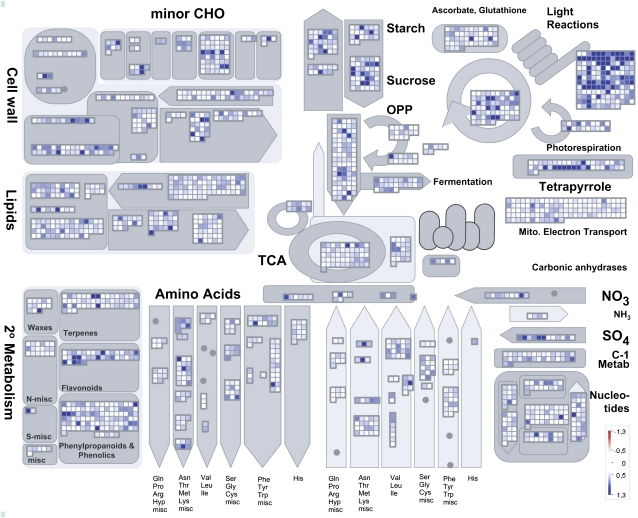
To aid visualization, the data set was processed with the MapMan software (Thimm et al., 2004) that has recently been adapted to the Barley1 microarray (Sreenivasulu et al., 2008). Out of the 17,416 genes scored as present, there are 833 genes (5%) with a diurnal amplitude exceeding log2 = 1 (Supplemental Table S1). As indicated in the MapMan screenshot shown in Figure 3, many genes involved in photosynthesis, carbohydrate metabolism, and secondary metabolism undergo strong diurnal fluctuations. A Wilcoxon rank test (P ≤ 0.05) was performed to list those functional categories, so-called BINs, which showed an overrepresentation of highly fluctuating genes. As a result, BINs for genes involved in the response to abiotic stress, particular heat stress, and BINs related to photosynthesis were among the most fluctuating ones (Supplemental Table S5), thus being consistent with the overrepresentation of GO categories for abiotic stress and plastid localization among the DE genes (Fig. 2B; Supplemental Fig. S4). A strong adaptation of metabolic processes in caryopses to the availability of incoming carbon and nitrogen sources is indicated by significant fluctuations of BINs for metabolism of proteins, amino acids, and major carbohydrates. Significant fluctuations in BINs for biosynthesis of abscisic acid and BINs summarizing abscisic acid-responding genes, such as late embryo abundant (LEA) and hordein genes, demonstrate the diurnal importance of abscisic acid-regulated processes.
Figure 3.
MapMan display showing diurnal amplitudes of 17,416 present genes in a metabolism overview map. Genes on the Barley1 microarray are represented by individual squares. Genes with increasingly large amplitudes are shown as an increasingly intense blue saturating at an amplitude of 1.3 (log2 value). Genes with no significant change in the amplitude are shown in white. CHO, Carbohydrate; OPP, oxidative pentose phosphate; TCA, tricarboxylic acid.
Diurnal Fluctuations of Sugar and Amino Acid Levels in Developing Barley Caryopses
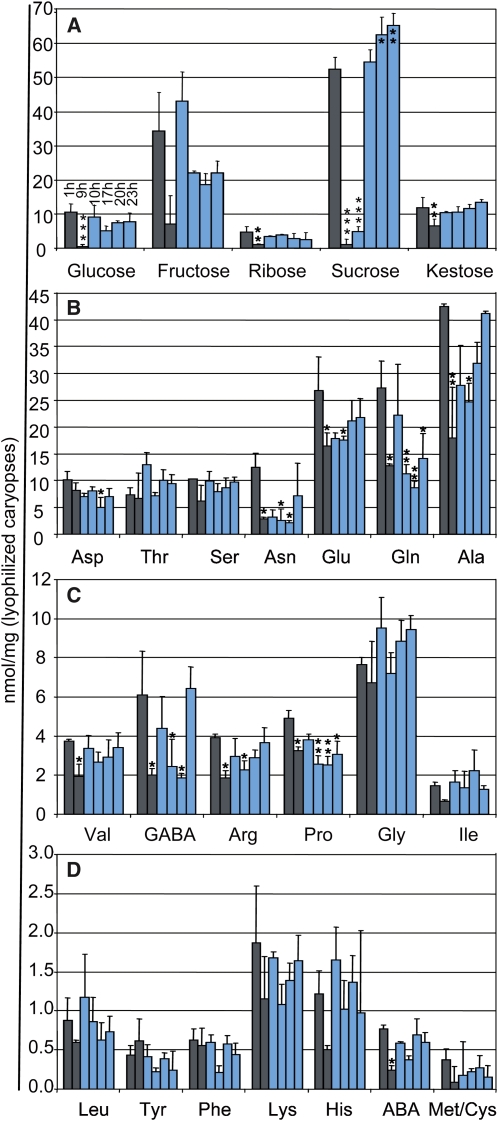
In leaf tissues, transitory starch is accumulated during the light phase and degraded during the night for respiration and in order to support cell viability of both source and sink tissues. Complex regulatory and metabolic networks exist that enable the accurate partitioning of the carbon sources in the plant during the day (Lu et al., 2005; Smith et al., 2005; Zeeman et al., 2007). Nevertheless, the daily shift from carbon assimilation during the light phase to starch degradation at night results in substantial variations in leaf sugar levels during the day (Smith and Stitt, 2007). We wondered whether similar fluctuations of sugar contents occur in the cereal caryopsis or whether the supply from the breakdown of transitory starch in source tissues would result in the maintenance of steady-state sugar levels in the caryopsis. As displayed in Figure 4A and Supplemental Table S6, barley caryopses underwent significant fluctuations in sugar contents during the light and dark phases, with Suc showing the biggest diurnal shifts in amplitude. The sugar levels were generally rather constant during the middle and end of the day and then declined during the night. The pools for Suc, Glc, and Rib were almost depleted at the end of the dark phase. Just 0.5 h into the light phase, the amounts of Rib, Glc, Fru, and kestose were restored to levels similar to those at the end of the photosynthetic active light phase. This indicates that as little as 30 min of light is enough to refill the supply of these sugars in the caryopsis. In contrast, refilling of the Suc pool advanced considerably more slowly. After 30 min of light induction, the Suc levels were still roughly 10 times lower than at the end of the previous day. The cycling pattern for Suc suggests a rapid turnover in the caryopsis during the early phase of the light period (Fig. 4; Supplemental Table S6), which can be explained by Suc being the main carbon supply for the endosperm. However, in the middle of the day, Suc levels are restored and Suc accumulates until the end of the day. This, in turn, may reflect the saturation of Suc-demanding pathways and a consistent supply of photosynthate during the later light phase. Levels of kestose, the primary product of fructan biosynthesis, showed a slight but steady increase during the day, which is consistent with the function of fructans in the photosynthetic partitioning process (Nagaraj et al., 2004).
Figure 4.
Levels of sugars and amino acids undergoing diurnal fluctuations in developing caryopses of barley. A, The amount of sugars was determined at six times during the day. B to D, The amount of indicated amino acids was determined at six times during the day. Sampling times were as indicated for the Glc samples in A. Gray and light blue bars depict times in darkness and light, respectively. The data are displayed as means + sd. Significant differences compared with the first sample (1 h) are indicated by asterisks (* P ≤ 0.01, ** P ≤ 0.001, *** P ≤ 0.0001).
Compared with the investigated sugars, changes in amino acid contents were less pronounced (Fig. 4, B–D; Supplemental Table S6). A number of amino acids had constant levels during the night or showed minimum levels in the middle or end of the light phase. However, Gln and Glu are known to constitute the dominating transport forms of amino acids throughout the light phase of the day (Lam et al., 1995); accordingly, their contents exhibited significant fluctuations. Moreover, we noted a generally high variability even between the pooled individuals for Gln that was especially pronounced in the early morning samples.
In agreement with previous reports, we found elevated amounts of Asn, which serves as an important nitrogen storage and transport compound for the allocation of nitrogen resources between source and sink organs, at the beginning of the night (Lam et al., 1995, 2003).
The levels of Ala, which serves as a precursor for several other amino acids, appeared to be tightly coupled to Suc levels (correlation coefficient of +0.68) and thus to carbohydrate metabolism (Fig. 4, A and B). The correlation between Suc and Ala levels is probably explained by their connection through glycolysis with the α-keto acid pyruvate, which is converted to Ala by a transamination (Pratelli and Pilot, 2007). Similar accumulation patterns were displayed for amino acids produced from the same or intersecting pathways. This was particularly evident for Ala, Val, and Ser as one group and, to a lesser extent, His, Phe, and Tyr as another. The amounts of Gly were notably lower than expected, which might be explained by the generally lower levels of photorespiration in Poaceae (Engel et al., 2007; Perez-Lopez et al., 2009).
Correlation of Gene Expression and Metabolite Levels
To address the question of whether metabolite levels are related to gene expression, we calculated the correlation coefficients of metabolite profiles and the trajectories of all expressed genes (Figs. 5 and 6; Supplemental Table S7). Using a minimum correlation coefficient of 0.8 as a threshold to correlate the levels of amino acids and sugars to gene expression profiles, we found 877 positively and 878 negatively correlated gene expression trajectories (Supplemental Table S7). The total number of genes in the cereal caryopsis that responded to sugars is likely underestimated because the correlation study of gene expression and metabolite levels was conducted on the DE genes only.
Figure 5.
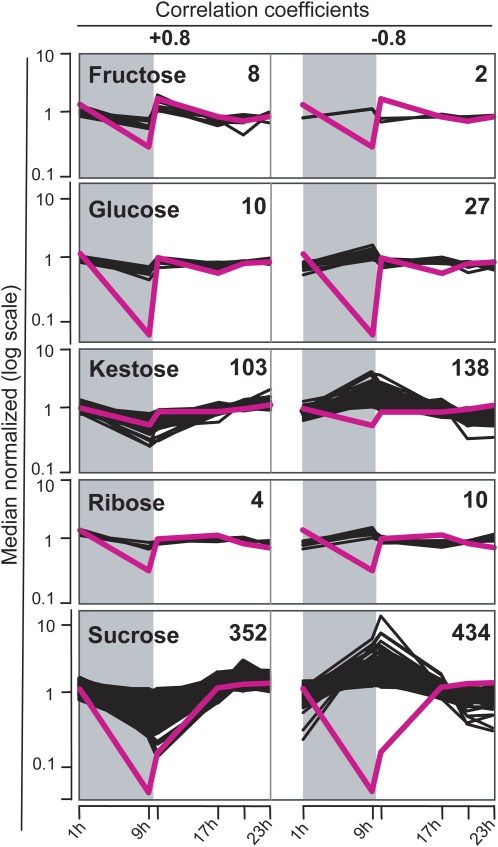
Correlation of sugar contents and expression profiles. Profiles of the indicated sugars and their correlating and anticorrelating gene expression profiles are shown as pink and black lines, respectively. Time points are indicated on the x axis. Numbers of correlating genes are displayed for each diagram. Sugar contents and signal intensities are normalized to the mean and plotted in log scale.
Figure 6.
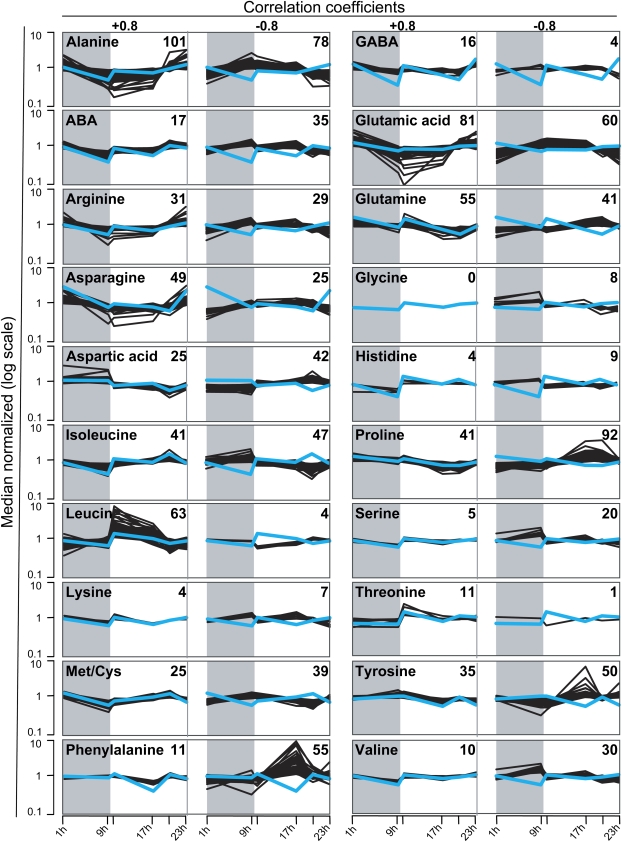
Correlation of amino acid contents and expression profiles. Profiles of the indicated amino acids and their correlating and anticorrelating gene expression profiles are shown as blue and black lines, respectively. Numbers of correlating genes are displayed for each diagram, and time points are indicated at the bottom. The amino acid contents and signal intensities are normalized to the mean and plotted in log scale. ABA, α-Aminobutyric acid; GABA, γ-aminobutyric acid.
The amino acids Gln, Glu, Asn, and Asp were linked with large numbers of correlating and anticorrelating genes. As these amino acids are known as the major transport forms of nitrogen in the phloem (Lam et al., 1995), this suggests an impact of incoming metabolites on gene expression in the caryopsis. As expected by their connected pathways, a large overlap of 47 correlated and 54 anticorrelated genes existed for Suc and Ala. The high number of genes showing a positive correlation to Pro, which acts as a compatible solute, further indicates a connection between diurnal gene regulation and adaptation to changes in osmotic conditions (Ashraf and Foolad, 2006).
Among the investigated sugars, Suc had by far the highest linkage number of correlated and anticorrelated transcript profiles (352 and 434, respectively). Among the positively correlating genes, we found those that have been shown to be Suc responsive in other contexts or species. These were, for example, the genes for granule-bound starch synthase1b (HvGBSS1b; Contig12208_at) and for the Suc:fructan 6-fructosyltransferase (Hv6-SFT; Contig3392_at; Dian et al., 2003; Nagaraj et al., 2004). Also, expression of a trehalose-6-phosphate phosphatase-like gene (TPP-like; HS16E24u_s_at) appeared to follow the Suc levels, suggesting its potential role in sugar signaling (Gomez et al., 2005). Several genes putatively involved in the biosynthesis of the cell wall, such as a callose synthase-like gene (Contig19065_at) and two arabinofuranosidase-like genes (Contig5239_at; S0001000058H07F1_s_at), were also found among the positively correlated genes (Supplemental Table S7). A dozen genes related to photosynthesis were found to be negatively correlated to Suc, as expected by the inhibitory effect of Suc on photosynthetic processes (Koch, 1996; Paul and Pellny, 2003). The negative correlation for the β-amylase gene HvBAM5 (Contig11522_at) is indicative of starch degradation occurring in times of Suc starvation (Smith et al., 2005). For a more global investigation, a GO classification of genes with an expression profile that correlated to Suc levels was performed (Supplemental Fig. S4). As a result, genes of the GO categories Developmental Processes and Cell Organization and Biogenesis had a greater tendency to positively correlate to Suc levels in the caryopsis, whereas genes negatively correlated to Suc levels were more often related to the GO terms Transcription and Transport.
Expression Profiles for Genes Involved in Suc and Starch Metabolism
As we expected Suc and starch metabolism to be affected by diurnal changes, we screened the Barley1 GeneChip for unambiguous probe sets that represent genes involved in these processes. In order to assign genes to a given probe set, the Barley1 GeneChip was annotated using the HarvEST software version 1.73 (Close et al., 2007). Moreover, probe sets representing previously described isoforms of genes involved in carbohydrate metabolism in barley were annotated manually to reduce the risk of false annotations. Out of 70 genes with a documented or potential role in Suc, starch, and fructan metabolism and regulation, 58 genes were represented on the microarray and 23 genes were DE in our experiment. Table I provides their common gene names, corresponding probe sets, and diurnal log2 amplitudes as well as the expression cluster group for DE genes. The precise expression trajectories of the genes are shown in Figure 7.
Table I. Genes investigated in this study with known or putative involvement in sugar signaling and Suc-to-starch conversion.
| Protein Name | Genea | Barley1 Probe Set | DEb | Organc | Foldd | Reference |
| ADP-glucose pyrophosphorylase large subunit | HvAPL1 | Contig5267_at | No | U | 1.37 | Villand et al. (1992) |
| ADP-glucose pyrophosphorylase large subunit | HvAPL2 | Contig3390_at | (1) | U | 1.83 | Eimert et al. (1997) |
| ADP-glucose pyrophosphorylase small subunit | HvAPS1a/b | Contig2267_s_at | No | U | 1.35 | Thorbjornsen et al. (1996) |
| ADP-glucose pyrophosphorylase small subunit | HvAPS2 | No probe set | U | Johnson et al. (2003) | ||
| ADP-glucose transporter | HvNST1 | Contig7529_at | (6) | U | 2.39 | Patron et al. (2004) |
| α-Amylase 1 | HvAMY1 | Contig7088_at | n.d. | U | Radchuk et al. (2009) | |
| α-Amylase 2 | HvAMY2 | Contig3952_at | n.d | n.d. | Radchuk et al. (2009) | |
| α-Amylase 3 | HvAMY3 | Contig3953_s_at | No | n.d. | 1.12 | Radchuk et al. (2009) |
| α-Amylase 4 | HvAMY4 | EBma03_SQ002_M07_at | No | P | 1.35 | Radchuk et al. (2009) |
| α-Glucan phosphorylase | HvPHO1 | No probe set | U | Radchuk et al. (2009) | ||
| α-Glucan phosphorylase, cytosolic | HvPHO2 | Contig6517_at | U | 0.50 | Radchuk et al. (2009) | |
| β-Amylase 1 | HvBAM1 | Contig1406_at | No | U | 1.41 | Radchuk et al. (2009) |
| β-Amylase 2 | HvBAM2 | Contig1411_s_at | No | U | 1.34 | Radchuk et al. (2009) |
| β-Amylase 3 | HvBAM3 | Contig8246_at | n.d. | U | Radchuk et al. (2009) | |
| β-Amylase 4 | HvBAM4 | No probe set | U | Radchuk et al. (2009) | ||
| β-Amylase 5 | HvBAM5 | Contig11522_at | (1) | U | 5.21 | Radchuk et al. (2009) |
| β-Amylase 6 | HvBAM6 | No probe set | U | Radchuk et al. (2009) | ||
| β-Amylase 7 | HvBAM7 | No probe set | U | Radchuk et al. (2009) | ||
| β-Amylase 8 | HvBAM8 | Contig10917_at | No | U | 1.38 | Edner et al. (2007) |
| Cell wall invertase 1 | HvCWINV1 | Contig11241_at | (3) | U | 2.28 | Weschke et al. (2003) |
| Cell wall invertase 2 | HvCWINV2 | Contig6787_at | No | U | 1.48 | Weschke et al. (2003) |
| Disproportionating enzyme 1 | HvDPE1 | Contig26451_at | No | U | 1.49 | Radchuk et al. (2009) |
| Disproportionating enzyme 2 | HvDPE2 | Contig6654_at | No | U | 1.48 | Radchuk et al. (2009) |
| Ferredoxin-NADP reductase, putative | FNR1-like | baak21d06_s_at | (4) | U | 1.96 | Balmer et al. (2006) |
| Ferredoxin-thioredoxin reductase, putative | FTR-like | Contig14916_at | No | P | 1.16 | Balmer et al. (2006) |
| Fructan 1-exohydrolase | Hv1-FEH | Contig7811_s_at | (3) | U | 2.21 | Nagaraj et al. (2004) |
| Glucan, water dikinase 1 | HvGWD1 | Contig18313_at | (4) | U | 2.08 | Edner et al. (2007) |
| Glucan, water dikinase 2 | HvGWD2 | Contig14761_at | n.d. | U | Edner et al. (2007) | |
| Granule-bound starch synthase 1a | HvGBSS1a | Contig694_at | No | U | 1.32 | Patron et al. (2002) |
| Granule-bound starch synthase 1b | HvGBSS1b | Contig12208_at | (4) | U | 1.80 | Patron et al. (2002) |
| Hexokinase 1 | HvHXK1 | Contig12296_at | (2) | P | 1.72 | E. Mangelsen (unpublished data) |
| Hexokinase 2 | HvHXK2 | Contig17401_at | No | P | 1.42 | E. Mangelsen (unpublished data) |
| Hexokinase 3 | HvHXK3 | Contig14657_at | No | U | 1.38 | E. Mangelsen (unpublished data) |
| Hexokinase 5 | HvHXK5 | Contig4153_at | No | U | 1.20 | E. Mangelsen (unpublished data) |
| Isoamylase 1 | HvISA1 | Contig7560_at | No | P | 1.11 | Sun et al. (1999) |
| Isoamylase 2 | HvISA2 | Contig13547_at | No | E | 1.24 | Radchuk et al. (2009) |
| Isoamylase 3 | HvISA3 | Contig14990_at | (6) | U | 1.38 | Radchuk et al. (2009) |
| Limit dextrinase (pullulanase) | HvLD | Contig11648_at | (3) | U | 1.86 | Burton et al. (1999) |
| NADPH-dependent thioredoxin reductase 2 | HvNTR2 | HB22M22r_s_at | No | U | 1.50 | Shahpiri et al. (2008) |
| Phosphoglucan, water dikinase | HvPWD | No probe set | U | Kotting et al. (2005) | ||
| Plant glycogenin-like starch initiation protein 1 | HvPGSIP1 | rbags11k20_s_at | No | U | 1.28 | Chatterjee et al. (2004) |
| Plant glycogenin-like starch initiation protein 2 | HvPGSIP2 | Contig3977_at | (3) | U | 4.01 | Chatterjee et al. (2004) |
| SNF1-related protein kinase a | HvSnRK1a | Contig3529_at | No | E | 1.33 | Hannappel et al. (1995) |
| SNF1-related protein kinase b | HvSnRK1b | HVSMEc0011O22r2_at | No | U | 1.54 | Hannappel et al. (1995) |
| Starch-branching enzyme I | HvSBEI | No probe set | U | Sun et al. (1997) | ||
| Starch-branching enzyme IIa | HvSBEIIa | Contig3761_at | No | U | 1.42 | Sun et al. (1998) |
| Starch-branching enzyme IIb | HvSBEIIb | Contig3762_s_at | (5) | E | Sun et al. (1998) | |
| Starch synthase I | HvSSI | Contig1808_at | No | U | 1.59 | Li et al. (2003) |
| Starch synthase IIa | HvSSIIa | Contig20469_at | No | U | 1.85 | Li et al. (2003) |
| Starch synthase IIb | HvSSIIb | No probe set | U | Radchuk et al. (2009) | ||
| Starch synthase IIIa | HvSSIIIa | No probe set | U | Radchuk et al. (2009) | ||
| Starch synthase IIIb | HvSSIIIb | Contig10722_at | (6) | U | 2.96 | Radchuk et al. (2009) |
| Starch synthase IV | HvSSIV | Contig13138_at | No | U | 1.26 | Radchuk et al. (2009) |
| Sucrose synthase 1 | HvSuSy1 | Contig361_s_at | No | U | 2.05 | Guerin and Carbonero (1997) |
| Sucrose synthase 2 | HvSuSy2 | Contig823_at | No | U | 1.40 | Guerin and Carbonero (1997) |
| Sucrose transporter 1 | HvSUT1 | Contig4612_at | No | U | 1.44 | Weschke et al. (2000) |
| Sucrose transporter 2 | HvSUT2 | rbah43d09_s_at | (3) | U | 1.66 | Weschke et al. (2000) |
| Sucrose:fructan-6-fructosyltransferase | Hv6-SFT | Contig3392_at | (4) | U | 2.64 | Nagaraj et al. (2004) |
| Sucrose:sucrose-1-fructosyltransferase | Hv1-SST | Contig4521_s_at | No | U | 1.41 | Nagaraj et al. (2004) |
| Sugar transporter 1 | HvSTP1 | Contig3479_at | (5) | U | 1.91 | Weschke et al. (2003) |
| Sugar transporter 2 | HvSTP2 | No probe set | U | Weschke et al. (2003) | ||
| Sugar transporter 3 | HvSTP3 | Contig6135_s_at | No | U | 1.26 | Weschke et al. (2003) |
| Thioredoxin f | Trx f-like | Contig4022_at | (6) | U | 4.06 | |
| Thioredoxin h1 | HvTrx h1 | Contig171_at | No | P | 1.38 | Maeda et al. (2003) |
| Thioredoxin h2 | HvTrx h2 | Contig2980_s_at | No | P | 1.09 | Maeda et al. (2003) |
| Thioredoxin h, putative | Trx h-like | HV06G09u_s_at | (6) | P | 1.63 | Maeda et al. (2003) |
| Thioredoxin m, putative | Trx m-like | Contig26351_at | (3) | E | 2.09 | Balmer et al. (2006) |
| Thioredoxin-like 1 | Trx-like 1 | Contig5236_at | (6) | U | 2.15 | Balmer et al. (2006) |
| Trehalose-6-phosphate phosphatase, putative | TPP-like | HS16E24u_s_at | (3) | U | 2.81 | Ramon et al. (2009) |
| Trehalose-6-phosphate synthase, putative | TPS-like | Contig3373_s_at | (6) | U | 2.14 | Ramon et al. (2009) |
| WRKY transcription factor 34 | HvWRKY34 | Contig10471_at | No | U | 1.61 | Mangelsen et al. (2008) |
| WRKY transcription factor 41 | HvWRKY41 | Contig12033_at | (3) | U | 1.78 | Mangelsen et al. (2008) |
| WRKY transcription factor SUSIBA2 | HvWRKY46 | Contig7243_at | No | U | 1.10 | Sun et al. (2003) |
Previously annotated barley genes carry the prefix “Hv”; gene annotations based on best BLAST matches in Arabidopsis or rice (Oryza sativa) carry the suffix “-like.”
DE according to the calculation of 2,091 DE probe sets. Number of the set is given in parentheses. n.d., No signal detected.
Presence of transcript according to Sreenivasulu et al. (2008). E, Endosperm; n.d., no signal detected; P, pericarp; U, unspecific.
Amplitude of the diurnal fluctuation of gene expression given as the ratio of the median normalized maximum signal intensity to the median normalized minimum signal intensity.
Figure 7.
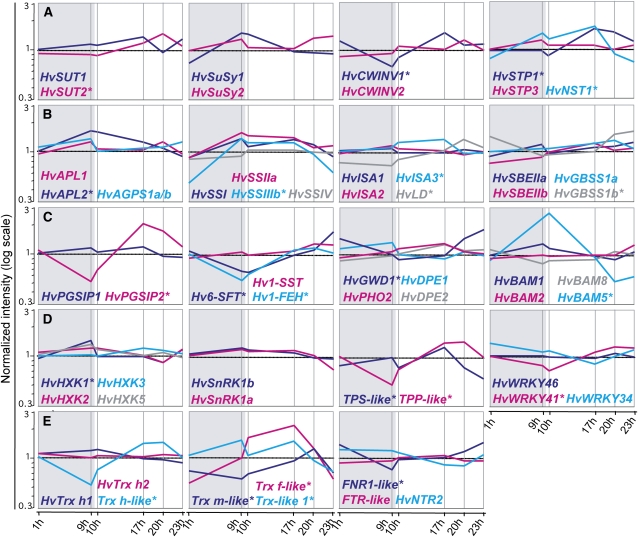
Gene expression profiles of genes involved in sugar metabolism and signaling. Signal intensities at indicated time points during the day are normalized to the mean expression values and plotted in log scale. Genes are grouped based on their involvement in different pathways as follows: initial sugar transport and processing (A), starch and fructan synthesis and degradation (B and C), potential sugar signaling (D), and ferredoxin/thioredoxin system (E). DE genes are indicated by asterisks. For complete gene names, see Table I.
Contained within the 2,091 DE genes, we identified several of the key genes involved in Suc transport and metabolism, such as the cell wall invertase gene 1 (HvCWINV1) and genes for a Suc transporter (HvSUT2), hexose transporter (HvHSTP1), and Suc:fructan 6-fructosyltransferase (Hv6-SFT). For genes related to starch metabolism, those encoding the large subunit of AGPase (HvAPL2), granule-bound starch synthase 1b (HvGBSS1b), starch-branching enzyme 2b (HvSBEIIb), limit dextrinase (HvLD), glucan, water dikinase 1 (HvGWD1), isoamylase 3 (HvISA3) and β-amylase 5 (HvBAM5) were among the DE genes. In addition, the fructan 1-exohydrolase gene (Hv1-FEH), probably involved in fructan biosynthesis and degradation (Van Den Ende et al., 2003; Lothier et al., 2007), underwent strong diurnal fluctuation in expression. However, there were no pronounced diurnal variations in transcript levels for many of the genes responsible for metabolic or regulatory steps in starch biosynthesis, such as genes for Suc synthases 1 and 2 (HvSuSy1 and HvSuSy2), isoamylases 1 and 2 (HvISA1 and HvISA2), starch synthases 1 and 2 (HvSs1 and HvSs2), and SUSIBA2 (HvWRKY46).
Next to genes with a known function in the conversion from Suc to starch, we found several candidate genes potentially involved in Suc signaling to be DE, such as genes for a hexokinase (HvHXK1), two trehalose-phosphate metabolic enzymes (TPS-like and TPP-like), and the WRKY transcription factor gene HvWRKY41 (Rolland et al., 2006).
We also noted that several genes of the ferredoxin/thioredoxin system underwent diurnal regulation. These were a ferredoxin-NADP reductase gene and four putative thioredoxin (Trx) genes, among them the amyloplast-located Trx m. Several of the 43 identified Trx m targets in wheat (Triticum aestivum) amyloplasts (Balmer et al., 2006) were found to be DE in our study, among them the ADP-Glc transporter gene HvNST1 and the gene for the large subunit of the ADP-Glc phosphorylase HvAPL1.
Combined Microarray Analysis of Spatial Gene Expression Patterns
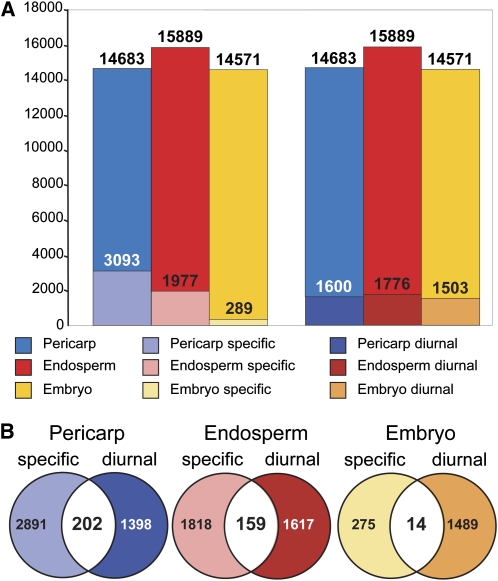
To gain a more global overview of the spatial distribution of diurnal changes in gene expression, we combined our data set with previously published microarray data that investigate the changes in gene expression in endosperm, embryo, and pericarp tissues during caryopsis development (Sreenivasulu et al., 2008). Rather than looking at the developmental expression profiles of the three suborgans separately, we intended to identify those genes that were either commonly expressed in all suborgans or specific to a single suborgan. As shown in Figure 8A and Supplemental Table S8, we found 3,093, 1,977, and 289 genes to be specifically expressed in pericarp, endosperm, and embryo tissues, respectively. Hence, about one-third of all present genes in the study of Sreenivasulu et al. (2008) had a suborgan-specific spatial expression pattern. In addition, all three suborgans investigated contributed equally to the set of diurnally expressed genes identified in our study, with 11% of pericarp and endosperm genes (1,600 and 1,776 genes, respectively) and 10% of the embryo genes (1,503 genes) exhibiting significant day/night fluctuations (Fig. 8A; Supplemental Table S8). Nevertheless, the majority of DE genes were not tissue specific but expressed in two or three of the suborgans tested. The overlap of tissue-specific and diurnally regulated genes was relatively low, indicating that only 7% (202 genes) of pericarp-specific, 8% (159 genes) of endosperm-specific, and 5% (14 genes) of embryo-specific genes underwent diurnal fluctuations (Fig. 8B; Supplemental Table S8). Only 224 diurnal DE genes were not detected in the data set of Sreenivasulu et al. (2008), suggesting either that they were expressed below detection levels in the sampled developmental stages or that they belong to a group of palea/lemma-specific genes that have not been probed. In summary, the combined analysis suggests that the majority of diurnally regulated genes have a function in several of the caryopsis tissues. On the other hand, many tissue-specific genes were identified among those that were diurnally regulated, prompting speculation about tissue-specific pathways for diurnal regulation.
Figure 8.
Diurnal gene expression in suborgan tissues of the caryopses. A, The total number of genes that were expressed in the suborgans pericarp, endosperm, and embryo are shown in blue, red, and yellow, respectively. Subsets of genes that were exclusively expressed in one suborgan are shown in light-shaded colors. Diurnally expressed genes are depicted in dark-shaded colors. B, Venn diagrams depicting the exclusive and intersecting suborgan-specific and diurnally regulated genes.
Quantification of Spatial Gene Expression
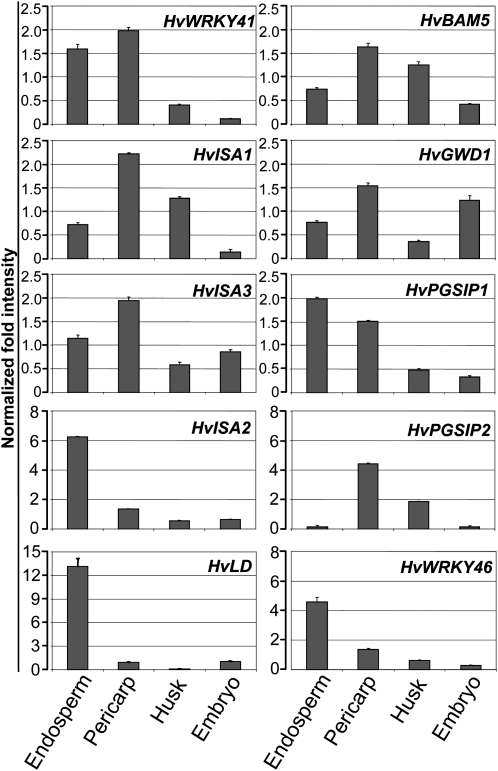
In order to verify the suborgan presence predicted by the comparative microarray analysis described above, we applied qPCR to investigate the spatial expression of a number of gene candidates. As shown in Figure 9, the selected genes showed distinct spatial expression patterns in endosperm, pericarp, husk, and embryo tissues. In accordance with their role in mobilization of transitory starch (Smith et al., 2005), HvBAM5, HvGWD1, and HvISA3 were predominantly expressed in the photosynthetically active pericarp tissues. A potential role of plant glycogenin-like starch initiation proteins (PGSIP) in starch biosynthesis has previously been suggested by Chatterjee et al. (2004). An orthologous gene, HvPGSIP2, which was found to be DE during the day (Table I), was mainly expressed in green tissues of the caryopsis. In contrast, its paralog HvPGSIP1, which showed an expression profile negatively correlated to Suc levels of the caryopsis (Supplemental Table S7), was mainly expressed in endosperm tissue.
Figure 9.
Presence of transcripts in four different tissues of the developing barley caryopsis. Normalized transcript abundance of selected genes was determined by qPCR. Expression levels in endosperm, pericarp, husk, and embryo tissues are shown. Results are displayed for genes coding for the following proteins: WRKY transcription factor 41 (HvWRKY41), β-amylase 5 (HvBAM5), isoamylase 1 (HvISA1), glucan, water dikinase 1 (HvGWD1), isoamylase 3 (HvISA3), plant glycogenin-like starch initiation protein 1 (HvPGSIP1), isoamylase 2 (HvISA2), plant glycogenin-like starch initiation protein 2 (HvPGSIP2), limit dextrinase (HvLD), and WRKY transcription factor SUSIBA2 (HvWRKY46).
High expression levels in endosperm tissues were also found for the genes encoding SUSIBA2 (HvWRKY46) and limit dextrinase (HvLD), confirming earlier findings (Burton et al., 1999; Sun et al., 2003). The diurnally fluctuating transcripts for HvWRKY41 (Mangelsen et al., 2008) could mainly be detected in endosperm and pericarp tissues. As was previously observed for the isoamylase 1 gene (HvISA1; Sun et al., 1999), the isoamylase 2 gene in barley (HvISA2) was preferentially expressed in the endosperm, whereas HvISA3 transcripts were located in the pericarp.
DISCUSSION
Photocycles, thermocycles, and the circadian clock together phase cellular activities in plants to generate diurnal oscillations in gene expression, metabolism, and physiology (Wijnen and Young, 2006; Michael et al., 2008). We expected that relatively few DE genes in a sink organ such as the cereal caryopsis should be responsive to light signals, as only genes in the pericarp and the husk would be directly influenced by photocycles. Thus, we assumed that in the barley caryopsis, a major portion in diurnal oscillations of gene expression is conveyed through fluctuations in metabolite availability.
To gain an insight into these presumed relationships, we analyzed day/night cycles in gene expression and monitored the metabolite accumulation alongside, which subsequently allowed the correlation of metabolite availability and its putative effects on gene expression in barley caryopses. However, we are aware that observed transcript levels do not necessarily reflect protein levels or protein activity; thus, our conclusions on gene function and interactions must be considered as tentative.
Diurnal Regulation of Starch Metabolism and Sugar Signaling
Nearly all steps in the conversion from Suc to starch or fructan have the potential to be diurnally regulated (Table I; Fig. 7). The majority of genes involved in these processes were expressed in all suborgans of the caryopsis and, hence, are of general importance in various cell types (Supplemental Table S8). The combined analysis with the data set of Sreenivasulu et al. (2008) revealed only a few genes that were unambiguously located to the pericarp and endosperm (Table I). However, the comparison with recent published studies on the spatial distribution of barley transcripts (Radchuk et al., 2009) and suborgan localization using qPCR (Fig. 8) allowed us to identify the predominant locations of several of these genes and to shed light on several diurnal regulatory aspects occurring in the developing caryopsis.
Of the genes encoding enzymes involved in starch elongation, branching, and trimming (Smith, 1999, 2001; Smith et al., 2005; Hannah and James, 2008), HvLD, HvISA3, HvSBEIIb, and the gene for starch synthase IIIb (HvSSIIIb) displayed diurnal regulation (Table I; Fig. 7). As reported earlier (Sun et al., 1998; Burton et al., 1999; Radchuk et al., 2009), the transcripts of HvSBEIIb and HvLD were mainly found in the endosperm and are thus likely to be involved in the biosynthesis of storage starch (Table I; Fig. 8). The expression of these genes peaked during the middle to late light phase, when sugar supply was at its maximum. The HvSSIIIb gene (Radchuk et al., 2009), which is preferentially expressed in the endosperm, was strongly induced in anticipation of the light rather than being correlated to high sugar levels (Figs. 2 and 7). Hence, its transcript levels peaked prior to those of HvLD and HvSBEIIb (Fig. 7).
Furthermore, our data suggest an involvement of HvISA3 in the production of transient starch, especially in the pericarp, during the light phase (Figs. 7, 8, and 10). A similar function was recently demonstrated for isoamylase paralogs in Arabidopsis (Streb et al., 2008). The specific roles of HvISA1 and HvISA2 remain unclear (Burton et al., 2002), and neither seems to be under diurnal control (Table I). However, the high expression levels of HvISA2 in the endosperm suggest a prominent role during storage starch synthesis (Fig. 8).
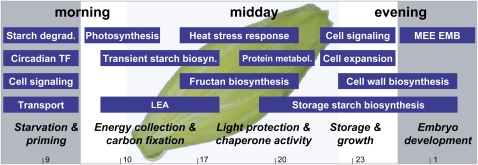
Figure 10.
Schematic overview of the diurnal expressional trends in developing caryopses of barley. Functional groups of DE genes with high transcript levels at the investigated times of the day, summarized as morning, midday, and evening, are depicted as blue boxes. The length of the blue boxes is indicative for the duration of high expression levels over just one or several investigated time points that are indicated at the bottom of the model. Five phases of diurnal adaptation, named starvation and priming, energy collection and carbon fixation, light protection and chaperone activity, storage and growth, and embryo development, were defined and are indicated in italics. White and gray areas depict light and dark phases of the day, respectively. LEA, Late embryo abundant proteins; TF, transcription factors.
Not only biosynthesis but also the degradation of starch follows a diurnal pattern. We found the expression of HvGWD1 and HvBAM5, both with a predicted role in starch degradation (Smith et al., 2005; Smith and Stitt, 2007; Zeeman et al., 2007), to be diurnally regulated. Expression of HvGWD1 peaks at the onset of night and, thus, prior to HvBAM5 (Fig. 7), which is in line with the idea that phosphorylation by GWD1 prepares insoluble glucans for degradation by β-amylases (Hejazi et al., 2008). The HvBAM5 gene, encoding a presumed plastidial enzyme, exhibited high expression levels in the pericarp with a profile that was negatively correlated to Suc content (Fig. 9; Supplemental Table S7). This observation lends support to a role of HvBAM5 in the degradation of transitory starch during the night with granular starch and/or linear maltodextrins as substrate (Fulton et al., 2008; Radchuk et al., 2009). Moreover, activation of HvBAM5 by Suc starvation at the end of the night contrasted to the activity of genes involved in the initial metabolism of Suc, such as HvCWINV1, HvSUT2, and HvSTP1 (Fig. 7).
Little is known about HXK function in barley despite its enrollment in sugar signaling and targeting of carbon into downstream metabolic pathways (Rolland et al., 2006). Although at least seven HvHXK genes are encoded in the barley genome (E. Mangelsen, unpublished data), only HvHXK1 followed a distinct diurnal expression profile with high transcript levels at night (set 2; Fig. 7). Interestingly, HvHXK1 and HvHXK2 transcripts showed exclusive expression in pericarp tissues (Table I; Supplemental Table S8).
Sugar Signaling Networks May Cause Diurnal Regulation of a Large Subset of Genes in the Barley Caryopsis
A total number of 882 DE gene expression profiles exhibited positive or negative correlation to sugar levels (Fig. 5; Supplemental Table S7). Hence, a significant fraction of those genes might act downstream of, or participate in, a sugar-dependent signaling cascade. In Arabidopsis seedlings, more than 4,000 diurnally fluctuating genes are affected by changing Suc levels and many of the rapidly Suc-induced or -repressed genes correlate either positively or negatively to diurnal Suc levels (Blasing et al., 2005; Osuna et al., 2007).
As expected, positive correlation to Suc was found for genes encoding enzymes in the starch biosynthetic pathway (Supplemental Table S7) and for genes involved in Suc transport. The Suc transporter HvSUT1 is thought to play a central role in the determination of endospermal Suc levels, while the role of its paralog HvSUT2, which is expressed in all parts of the caryopsis, is yet unknown (Weschke et al., 2000; Endler et al., 2006; Thiel et al., 2008). Expression of HvSUT2 peaked when Suc levels were at a maximum and expression levels of HvSUT1 were at a transient minimum. Thus, we speculate that in times of excess Suc, HvSUT2 facilitates a redirection of the Suc flux into suborgans other than the endosperm (Fig. 7). Coexpression of HvSUT2 and the fructan biosynthesis genes Hv6-SFT and Hv1-FEH suggests a concerted role for an intermediate carbon storage in vacuoles to provide sink strength of the caryopsis (Vijn and Smeekens, 1999). The positive correlation of Hv6-SFT and Hv1-FEH and Suc further indicates that these genes are Suc inducible also in sink organs, as described previously for leaves (Nagaraj et al., 2005). Finally, more than 240 genes were found to correlate to kestose levels, and several of these genes are likely to be involved in fructan metabolism.
We notice that several genes with negative correlation to Suc encode photosynthesis-related proteins, which highlights the inhibitory effect of Suc on photosynthesis (Pego et al., 2000). Other negatively correlated genes are glycosyl hydrolases with β-glucosidase activities and glycosyl hydrolases that presumably act on Man and Xyl (Supplemental Table S7). This implies hemicellulose degradation at low Suc levels and supports the idea of hemicellulose and cellulose mobilization to supply energy under Suc-limiting conditions (Contento et al., 2004; Hoch, 2007; Lee et al., 2007). Anticorrelation of profiles for several transcription factors and kinase genes with Suc levels supports the idea that the absence of Suc as the main energy-providing carbon molecule, rather than its presence, has an impact on cellular signaling (Blasing et al., 2005).
Nitrogen Signaling May Regulate Gene Expression in Developing Caryopses of Barley
In accordance with the pronounced diurnal fluctuations of the major transport forms of amino acids, Gln, Glu, Asn, and Asp, we identified several genes related to their metabolism to undergo strong diurnal fluctuations (Supplemental Tables S2 and S5). Whether this hints at a dual function of these amino acids, as both substrates for protein biosynthesis and signaling molecules, is unclear and needs to be addressed by further investigation as, generally, most of the predicted nitrogen and carbon/nitrogen signaling matrices in plants are still far from being resolved (Lam et al., 1995; Coruzzi and Zhou, 2001; Zheng, 2009). Nevertheless, we found five genes for amino acid permeases (AAPs) to be diurnally regulated, out of which three showed a strong correlation to Gln and Glu levels (Supplemental Tables S2 and S7). The diurnal and spatial expression profiles of AAP genes are diverse, and further studies are required to unravel their potential roles in the control of amino acid levels in the developing seeds, as recently described for orthologous AAPs in Arabidopsis and pea (Pisum sativum; Weigelt et al., 2008; Sanders et al., 2009). In addition to amino acids, phloem-derived peptides contribute significantly to the nitrogen status and regulate development in developing seeds (Miranda et al., 2003; Li et al., 2009). The diurnal expression of 11 peptide transporter genes points at a tight diurnal control of the nitrogen status in seeds via peptide import (Supplemental Table S3). Finally, the observed positive correlation of a potential ortholog to TIMING OF CAB EXPRESSION1 (Contig11872_at) and Glu (Supplemental Table S7) raises the question of whether there is a link between nitrogen and circadian signaling in barley caryopses, as postulated previously for Arabidopsis leaves (Gutierrez et al., 2008).
Embryo Development Genes Undergo Diurnal Regulation
A noteworthy feature of the list of DE genes (Supplemental Table S1) is the overrepresentation of genes potentially crucial to embryo development, such as putative orthologs to so-called embryo-defective (EMB) and maternal effect embryo arrest (MEE) genes from Arabidopsis (Tzafrir et al., 2004). Indeed, a minimum of 21 genes, which could be linked to embryo development via their annotations, were found to exhibit diurnal expression patterns. While all MEE-like DE genes found have unknown functions, the protein products of several EMB-like DE genes have enzymatic activities in nucleotide and amino acid biosynthesis. Others are described as constituents of the nuclear pore and the proteasome complex, underlining their pivotal cellular roles. Interestingly, the majority of embryo developmental genes fall into sets 3 and 2, with eight and four genes, respectively (Supplemental Table S3). This suggests that processes vital for embryo development are mainly activated during the late light period or at night (expression peaks of sets 3 and 2, respectively). In line with this notion, only two embryo development-related genes are found in set 6 and none in set 5, clusters characterized by low expression levels at late day and night times.
Additionally, a minimum of 11 LEA genes were diurnally expressed. Strikingly, seven LEA genes cluster into set 6, whereas the other clusters contain only one or even not a single LEA gene (Supplemental Table S3). The trajectories of set 6 genes suggest that LEA gene expression and, hence, the accumulation of storage proteins preferably occur at times of maximum photosynthate supply, starting immediately after the onset of light. In addition, one of the long-term functions of LEA proteins is the induced desiccation tolerance of seeds (Battaglia et al., 2008); therefore, it is tempting to speculate that LEA proteins are enrolled in a diurnal, thus short-term, protection against potential temperature or drought stresses in developing seeds.
Global Gene Expression in Barley Caryopses Suggests Different Phases of Diurnal Adaptation to Environmental Input Factors
Finally, we tried to summarize our findings on the predominant functional processes and corresponding genes in Figure 10, which provides a better overview of the network of several interconnected phases of diurnal gene expression.
At the end of the dark phase, depleted pools of sugars and many amino acids as well as low expression of genes involved in phloem unloading, such as HvCWINV1 and HvSTP1(Weschke et al., 2003), are characteristic for a phase of starvation. This is supported by the induced expression of HvBAM5 as a potential central player in starch degradation. In contrast, during the late light and early dark phases, metabolite pools are saturated and expression of genes related to protein metabolism, storage starch biosynthesis, cell wall expansion, and embryo development are indicative of extensive cell growth and storage compound accumulation (sets 2 and 3). Interestingly, we see indications for the existence of priming in barley caryopses, as altered gene expression of genes impaired in cellular signaling in sets 1 and 4 occurred in anticipation of dawn and dusk rather than in response to changes in light conditions (Davis and Millar, 2001). An involvement of the circadian clock in the regulation of gene expression in barley caryopses is further suggested by the expression of a putative ortholog of the potential central circadian regulator gene LATE ELONGATED HYPOCOTYL (Contig3875_s_at) in set 1, which was negatively correlated to the evening genes GIGANTEA (Contig5247_at) and PHYTOCLOCK1/LUX ARRHYTHMO (Contig13480_at; Yakir et al., 2007) contained in set 4.
During the early light phase and around midday, the induced expression of genes involved in photosynthesis and transient starch and fructan biosynthesis prompts the assumption that incoming light signals and metabolite availability regulate gene expression (sets 5 and 6). Indeed, the biosynthesis of fructans plays an important role in carbon partitioning in leaves by buffering the excess Suc that is produced during times of maximum photosynthesis (Pollock et al., 2003), and a similar function might exist in caryopses. Expression of heat stress-related genes is typical during the middle of the day and highlights their possible dual roles: on the one hand, they are involved in the heat stress response; on the other hand, they play roles in general cellular processes as chaperones in higher order complexes (Bosl et al., 2006; Rajan and D'Silva, 2009). A similar dual functionality for genes related to other stresses has previously been suggested (Kreps et al., 2002).
CONCLUSION
To the best of our knowledge, this study provides the first global monitoring of diurnal fluctuations in gene expression in a developing cereal seed. On the basis of the experimental work and meta-data analysis, we conclude that all suborgans are subject to diurnal regulation. This regulation includes processes such as phloem unloading, embryo development, and biosynthesis of storage products, which are independent of the photosynthetically active tissues. Thus, in addition to available developmental and spatial gene expression data published by Sreenivasulu et al.(2008) and Radchuk et al.(2009), the diurnal data set presented here adds a third dimension of complexity to the regulation of gene expression in the barley caryopsis. The combined use of the three available data sets aids the understanding of the function and interplay of thousands of genes expressed in a barley seed, which provides a model for other cereal crops.
MATERIALS AND METHODS
Plant Growth, Identification of Developmental State, Harvest, and Pooling Strategies
Barley plants (Hordeum vulgare ‘Golden Promise’) were grown in a greenhouse with automated shading in soil in pots of 10 cm diameter with additional lighting from xenon light bulbs (15 h of light [6:00 am–9:00 pm] and 9 h of dark [9:00 pm–6:00 am], 22°C–24°C, and 40% relative humidity). The age of the caryopses was determined as described by Weschke et al. (2000). Spikes were labeled at the onset of anthesis and harvested 12 d.p.a. Five adjacent caryopses to the spikelet used for the determination of age were used for the studies. At each time point, a total of 30 caryopses of spikes from six different plants (five each) were harvested. Awns (i.e. the projection of the tip of the lemma) and possibly still adhesive anthers were removed from the caryopsis before freezing it in liquid nitrogen. To allow for rapid harvesting (30 s per spike) and to avoid wounding-induced gene expression, the remaining parts of the husk were not removed from the caryopsis. Caryopses of individual plants were homogenized in liquid nitrogen. Tissue powder of homogenized caryopses of three plants was pooled and used for RNA extraction.
Microarray Hybridization
RNA extraction of caryopses was performed as described by Singh et al. (2003). Briefly, 100 mg of pooled homogenized plant material was mixed with 900 μL of sarcosyl buffer (1% sodium lauryl sarcosinate, 0.1 m Tris-HCl, pH 7.5, 0.1 m NaCl, 20 mm EDTA, and 50 mm β-mercaptoethanol), and purification of nucleic acids was done by three steps of phenol:chloroform:isoamyl alcohol (25:24:1) extractions. Precipitation of nucleic acids was performed with sodium acetate (pH 5.2) and 1 volume of isopropanol. After centrifugation, sediment was washed with ethanol and purified RNA was dissolved in diethyl pyrocarbonate-treated water. Incubation at 65°C for 5 min, followed by centrifugation at 4°C, was performed to remove residual high-Mr polysaccharides. The obtained RNA solution was purified following the RNeasy (Qiagen) cleanup protocol. RNA quantity and integrity were assessed using the RNA 6000 Nano LabChip Kit and BioAnalyzer software (Agilent).
Two independent samples of 1 μg of total RNA were used to synthesize amplified complementary RNA (cRNA; MessageAmp II aRNA Amplification Kit; Ambion). After fragmentation of cRNA using the RNA Fragmentation Reagents (Ambion), the cRNA was hybridized to Affymetrix Barley1 GeneChips. Hybridization and scanning of the GeneChips were performed according to the manufacturer's instructions.
Data Processing
The Affymetrix CEL files were imported into GeneSpring software version 7 (Agilent Technologies). The data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (Edgar et al., 2002) and are accessible through Gene Expression Omnibus series accession number GSE20034 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20034). Per-chip normalization to the median was applied to obtain comparability. The arrays were adjusted for background optical noise using sequence-dependent robust multiarray averaging (GC-RMA) software and normalized using quantile normalization (Wu et al., 2004).
Identification of DE genes was performed with LIMMA (P ≤ 0.05) as part of the R (http://cran.r-project.org/) software package: The GC-RMA summary file, a design matrix, and a contrast matrix were used as input for LIMMA all-against-all time point comparisons. After fitting, a decide test was performed. Multiple testing for control of false discovery rate was conducted (Benjamini and Hochberg, 1995). Due to a lack of significant present calls (less than 34%), one replicate was removed from the analysis (6:00 am, replicate A).
A k-means clustering of the DE genes for six major clusters was conducted, where clusters converged after 28 iterations. To independently validate the number of k-means clusters, the figure of merit was computed using MeV (Saeed et al., 2003) with varying settings and both Euclidean distance and Pearson correlation.
The principal component analysis was performed with mean centering on DE genes as input. Principal component analysis, hierachical clustering, and k-means clustering were conducted with GeneSpring software version 7 (Agilent Technologies).
Annotations of the Barley1 GeneChip were updated from the HarvEST database (version 1.73) with a stringency set to a minimum of 11 matching probes (Close et al., 2007).
A functional categorization using GO terms was performed on The Arabidopsis Information Resource Web site using the best BLAST matches for Arabidopsis (Arabidopsis thaliana) retrieved from the HarvEST annotation tool. Percentages of all functional categories in the different gene lists were calculated in Excel. For the significant overrepresentation or underrepresentation of a particular GO category, the P values were calculated using the hypergeometric distribution in regular spreadsheet analysis programs. The GO categories of the universe (all present genes with orthologs to Arabidopsis) were compared with the sampling distribution of the respective gene lists.
The MapMan software version 2.2.0 was used to annotate functional BINs of barley genes. The mapping file for the Barley1 GeneChip was downloaded from the MapMan Web site (http://gabi.rzpd.de/projects/MapMan/). Default settings were used to perform an uncorrected Wilcoxon rank test.
Comparison with Data Sets of Barley Maturating Grains (Meta-Analysis)
The Affymetrix CEL file GSE9365 (Sreenivasulu et al., 2008) was retrieved from the Gene Expression Omnibus database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) and imported into GeneSpring software version 7. The arrays were adjusted for background optical noise using GC-RMA software and normalized using quantile normalization. Genes are considered to be expressed in the caryopsis in cases where normalized signal intensities were 50 or higher in at least one of the time points and one of the tissues.
Evaluation of gene expression in the pericarp, endosperm, or embryo was conducted for the 4- and 8-d.p.a. data sets only. To identify genes that are exclusively expressed in the pericarp, the endosperm, or the embryo, we performed a subtractive approach. First, the signal intensities were normalized to the mean expression. Second, all DE genes required at least a 3-fold difference from the mean expression in at least one of the time points for each of the tissues independently. Third, the DE genes from all tissues were subsequently subtracted from the expressed genes of the tissue under investigation.
Independent from the above procedures, significantly regulated genes in one of the caryopsis tissues were identified with LIMMA (P ≤ 0.05) as part of the R (http://cran.r-project.org/) software package. Multiple testing for control of false discovery rate was conducted (Benjamini and Hochberg, 1995).
Data sets were derived for each of the different tissues (DE genes and tissue-specific genes) and subsequently compared with our DE genes (Supplemental Table S8).
Metabolite Extraction
For HPLC sample preparation, pooled plant material of three caryopses from two plants was lyophilized. Powdered samples (35 mg) were taken up in 400 μL of methanol solution (80%), incubated at 60°C for 10 min, and centrifuged at 13,000g for 15 min at 4°C. The resulting supernatant was transferred to a new vial. This extraction procedure was repeated, resulting in a final volume of 1,200 μL. The extract was split into 2 volumes of 600 μL and vacuum dried (samples A and B). Sample A was used for sugar analysis. The dried pellet was dissolved in 250 μL of Milli-Q water and centrifuged (1 min, 13,000g, 4°C). The resulting supernatants were filtered through a 0.22-μm Millex-GV filter. Tenfold dilutions were used for the measurement of Suc, Fru, Rib, and kestose. One hundred-fold dilutions were used for the measurement of Glc. Sample B was used for amino acid analysis. The dried pellet was resuspended in 138 μL of lithium diluent (Pickering Laboratories). As an internal standard, 12 μL of a 2.5 mm norleucine solution was used. Samples were centrifuged (1 min, 13,000g, 4°C), and the resulting supernatants were filtered through 0.22-μm Millex-GV filter.
All samples (A + B) were stored at –20°C. Quantification of amino acids and sugars by HPLC was carried out as described by Pilot et al. (2004) and Horak et al. (2008).
Metabolite contents of three independent samples were normalized to the median, and the sd was calculated. Identification of significant fluctuations in metabolite content was performed with LIMMA (P ≤ 0.05, P ≤ 0.01, P ≤ 0.001) as part of the R (http://cran.r-project.org/) software package, with raw data of the metabolite levels as input. Next, metabolite data were converted into trajectories over time. A Pearson correlation to the normalized expression profiles was performed to identify genes with a correlation coefficient of greater than 0.8.
qPCR
For detection of transcripts at suborgan resolution, caryopses were harvested at the middle of the day and samples of endosperm, pericarp, husk, and embryo were hand dissected. Caryopses at 8 d.p.a. were used to harvest pericarp samples to enable separation from aleurone layers. All remaining suborgans were harvested at 12 d.p.a. Material of 15 caryopses was pooled prior to RNA extraction. RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA quantity and integrity were assessed as described for microarray hybridization.
For each suborgan, three independent samples of 1 μg of RNA were used for reverse transcription with an oligo(dT) primer for first-strand synthesis and SuperScript III RNase Reverse Transcriptase (Invitrogen). cDNA equivalent to a starting material of 10 ng of total RNA was amplified in 20-μL reaction volumes using the Dynamo Flash SYBR Green qPCR kit (Finnzymes). All qPCRs were carried out in an iQ5 Real-Time PCR Detection System (Bio-Rad) following the manufacturer's instructions. Reaction conditions were as follows: initial denaturation at 95°C for 7 min, followed by 40 cycles of 95°C for 10 s and annealing/extension at 60°C for 30 s, followed by final extension at 60°C for 1 min. A melting curve was obtained by gradually increasing the temperature from 60°C to 98°C in steps of 0.5°C and 10 s of ramp time. Forward and reverse primers are listed in Supplemental Table S9. A cDNA pool of all samples was analyzed for a standard dilution series to monitor the qPCR efficiency for each particular primer pair. Based on a geNorm analysis (Vandesompele et al., 2002), tubulin, HSP70, and cyclophilin were chosen as multiple reference genes for further quantification of gene expression. Mathematical operations on qPCR data were performed with regular spreadsheet analysis using Microsoft Excel.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Verification of microarray data by qPCR.
Supplemental Figure S2. Principal component analysis of the data set.
Supplemental Figure S3. GO classification of genes belonging to six k-means clusters for Cellular Component and Molecular Function.
Supplemental Figure S4. GO classification of Suc-correlated genes.
Supplemental Table S1. Normalized signal intensities of all 17,416 present probe sets.
Supplemental Table S2. List of 2,091 DE genes.
Supplemental Table S3. List of genes sorted to sets 1 to 6.
Supplemental Table S4. Overlap of principal components and k-means clusters.
Supplemental Table S5. Results of an uncorrected Wilcoxon rank test performed in MapMan.
Supplemental Table S6. Contents of sugars and amino acids in caryopses of barley at 11 to 12 d.p.a.
Supplemental Table S7. List of genes that were positively or negatively correlated to sugars and amino acids.
Supplemental Table S8. Presence of transcripts in the different suborgan tissues of the caryopsis.
Supplemental Table S9. List of primers used for qPCR experiments.
Supplementary Material
Acknowledgments
We thank Markus Schmid for support with the microarray hybridizations and Bettina Stadelhofer for excellent help with HPLC analysis. We also thank Friederike Ladwig for scientific discussions and critical comments on the manuscript and Tom Martin for proofreading.
References
- Abebe T, Skadsen RW, Kaeppler HF. (2004) Cloning and identification of highly expressed genes in barley lemma and palea. Crop Sci 44: 942–950 [Google Scholar]
- Ashraf M, Foolad MR. (2006) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59: 206–216 [Google Scholar]
- Baguma Y, Sun C, Boren M, Olsson H, Rosenqvist S, Mutisya J, Rubaihayo PR, Jansson C. (2008) Sugar-mediated semidian oscillation of gene expression in the cassava storage root regulates starch synthesis. Plant Signal Behav 3: 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, Buchanan BB. (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H, Radchuk R, Weschke W, Wobus U, Weber H. (2004) Seed development and differentiation: a role for metabolic regulation. Plant Biol 6: 375–386 [DOI] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S. (2006) The molecular chaperone Hsp104: a molecular machine for protein disaggregation. J Struct Biol 156: 139–148 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, van Wegen S, et al. (2002) Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J 31: 97–112 [DOI] [PubMed] [Google Scholar]
- Burton RA, Zhang XQ, Hrmova M, Fincher GB. (1999) A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley. Plant Physiol 119: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Berbezy P, Vyas D, Coates S, Barsby T. (2004) Reduced expression of a protein homologous to glycogenin leads to reduction of starch content in Arabidopsis leaves. Plant Sci 168: 501–509 [Google Scholar]
- Close TJ, Wanamaker S, Roose ML, Lyon M. (2007) HarvEST: an EST database and viewing software. Methods Mol Biol 406: 161–178 [DOI] [PubMed] [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP. (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC. (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Millar AJ. (2001) Watching the hands of the Arabidopsis biological clock. Genome Biol 2: reviews10081–reviews1008.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Chen Q, Liu F, Wu P. (2003) Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 218: 261–268 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, et al. (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial β-amylases. Plant Physiol 145: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert K, Luo C, Dejardin A, Villand P, Thorbjornsen T, Kleczkowski LA. (1997) Molecular cloning and expression of the large subunit of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. Gene 189: 79–82 [DOI] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Parnik T, Keerberg O, Bauwe H. (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. (2000) Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J 23: 795–806 [DOI] [PubMed] [Google Scholar]
- Gomez LD, Baud S, Graham IA. (2005) Metabolite sensing in plants: a role for trehalose metabolism in seed development and embryo development. FEBS J 272: 460 [Google Scholar]
- Guerin J, Carbonero P. (1997) The spatial distribution of sucrose synthase isozymes in barley. Plant Physiol 114: 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC, James M. (2008) The complexities of starch biosynthesis in cereal endosperms. Curr Opin Biotechnol 19: 160–165 [DOI] [PubMed] [Google Scholar]
- Hannappel U, Vicente-Carbajosa J, Barker JH, Shewry PR, Halford NG. (1995) Differential expression of two barley SNF1-related protein kinase genes. Plant Mol Biol 27: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G. (2008) Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J 55: 323–334 [DOI] [PubMed] [Google Scholar]
- Hoch G. (2007) Cell wall hemicelluloses as mobile carbon stores in non-reproductive plant tissues. Funct Ecol 21: 823–834 [Google Scholar]
- Horak J, Grefen C, Berendzen KW, Hahn A, Stierhof YD, Stadelhofer B, Stahl M, Koncz C, Harter K. (2008) The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PE, Patron NJ, Bottrill AR, Dinges JR, Fahy BF, Parker ML, Waite DN, Denyer K. (2003) A low-starch barley mutant, riso 16, lacking the cytosolic small subunit of ADP-glucose pyrophosphorylase, reveals the importance of the cytosolic isoform and the identity of the plastidial small subunit. Plant Physiol 131: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi G. (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Wong P, Chan HK, Yam KM, Chen L, Chow CM, Coruzzi GM. (2003) Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol 132: 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Matsumura Y, Soga K, Hoson T, Koizumi N. (2007) Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol 48: 405–413 [DOI] [PubMed] [Google Scholar]
- Li S, Qian Q, Fu Z, Zeng D, Meng X, Kyozuka J, Maekawa M, Zhu X, Zhang J, Li J, et al. (2009) Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58: 592–605 [DOI] [PubMed] [Google Scholar]
- Li Z, Sun F, Xu S, Chu X, Mukai Y, Yamamoto M, Ali S, Rampling L, Kosar-Hashemi B, Rahman S, et al. (2003) The structural organisation of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthases in plants. Funct Integr Genomics 3: 76–85 [DOI] [PubMed] [Google Scholar]
- Lothier J, Lasseur B, Le Roy K, Van Laere A, Prud'homme MP, Barre P, Van den Ende W, Morvan-Bertrand A. (2007) Cloning, gene mapping, and functional analysis of a fructan 1-exohydrolase (1-FEH) from Lolium perenne implicated in fructan synthesis rather than in fructan mobilization. J Exp Bot 58: 1969–1983 [DOI] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138: 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Finnie C, Ostergaard O, Svensson B. (2003) Identification, cloning and characterization of two thioredoxin H isoforms, HvTrxh1 and HvTrxh2, from the barley seed proteome. Eur J Biochem 270: 2633–2643 [DOI] [PubMed] [Google Scholar]
- Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, Wanke D. (2008) Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus U. (2003) Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol 132: 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj VJ, Altenbach D, Galati V, Lüscher M, Meyer AD, Boller T, Wiemken A. (2004) Distinct regulation of sucrose:sucrose-1-fructosyltransferase (1-SST) and sucrose:fructan-6-fructosyltransferase (6-SFT), the key enzymes of fructan synthesis in barley leaves: 1-SST as the pacemaker. New Phytol 161: 735–748 [DOI] [PubMed] [Google Scholar]
- Nagaraj VJ, Galati V, Lüscher M, Boller T, Wiemken A. (2005) Cloning and functional characterization of a cDNA encoding barley soluble acid invertase (HvINV1). Plant Sci 168: 249–258 [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Blasing OE, Hohne M, Gunter M, Kamlage B, Trethewey R, Scheible WR, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Patrick JW, Offler CE. (2001) Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52: 551–564 [PubMed] [Google Scholar]
- Patron NJ, Greber B, Fahy BF, Laurie DA, Parker ML, Denyer K. (2004) The lys5 mutations of barley reveal the nature and importance of plastidial ADP-Glc transporters for starch synthesis in cereal endosperm. Plant Physiol 135: 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K. (2002) The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5′-non-coding region. Plant Physiol 130: 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK. (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Pego JV, Kortstee AJ, Huijser C, Smeekens SC. (2000) Photosynthesis, sugars and the regulation of gene expression. J Exp Bot (Spec No) 51: 407–416 [DOI] [PubMed] [Google Scholar]
- Perez-Lopez U, Robredo A, Lacuesta M, Sgherri C, Munoz-Rueda A, Navari-Izzo F, Mena-Petite A. (2009) The oxidative stress caused by salinity in two barley cultivars is mitigated by elevated CO2. Physiol Plant 135: 29–42 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB. (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Farrar J, Tomos D, Gallagher J, Lu CG, Koroleva O. (2003) Balancing supply and demand: the spatial regulation of carbon metabolism in grass and cereal leaves. J Exp Bot 54: 489–494 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Pilot G. (2007) Altered amino acid metabolism in glutamine dumper1 plants. Plant Signal Behav 2: 182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk VV, Borisjuk L, Sreenivasulu N, Merx K, Mock HP, Rolletschek H, Wobus U, Weschke W. (2009) Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol 150: 190–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan VB, D'Silva P. (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9: 433–446 [DOI] [PubMed] [Google Scholar]
- Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, Rolland F, Beeckman T, Thevelein JM. (2009) Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ 32: 1015–1032 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M. (2009) AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Shahpiri A, Svensson B, Finnie C. (2008) The NADPH-dependent thioredoxin reductase/thioredoxin system in germinating barley seeds: gene expression, protein profiles, and interactions between isoforms of thioredoxin h and thioredoxin reductase. Plant Physiol 146: 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kumar S, Singh P. (2003) A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Mol Biol Rep 21: 93a–93f [Google Scholar]
- Smith AM. (1999) Making starch. Curr Opin Plant Biol 2: 223–229 [DOI] [PubMed] [Google Scholar]
- Smith AM. (2001) The biosynthesis of starch granules. Biomacromolecules 2: 335–341 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM. (2005) Starch degradation. Annu Rev Plant Biol 56: 73–98 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA- regulated maturation in developing barley seeds. Plant J 47: 310–327 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M.et al (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC. (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C. (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Sathish P, Ahlandsberg S, Deiber A, Jansson C. (1997) Identification of four starch-branching enzymes in barley endosperm: partial purification of forms I, IIa and IIb. New Phytol 137: 215–222 [DOI] [PubMed] [Google Scholar]
- Sun CX, Sathish P, Ahlandsberg S, Jansson C. (1998) The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiol 118: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Sathish P, Ahlandsberg S, Jansson C. (1999) Analyses of isoamylase gene activity in wild-type barley indicate its involvement in starch synthesis. Plant Mol Biol 40: 431–443 [DOI] [PubMed] [Google Scholar]
- Thiel J, Weier D, Sreenivasulu N, Strickert M, Weichert N, Melzer M, Czauderna T, Wobus U, Weber H, Weschke W. (2008) Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: a microdissection-based transcriptome study of young barley grains. Plant Physiol 148: 1436–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thorbjornsen T, Villand P, Kleczkowski LA, Olsen OA. (1996) A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase small subunit from barley (Hordeum vulgare). Biochem J 313: 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Ende W, Clerens S, Vergauwen R, Van Riet L, Van Laere A, Yoshida M, Kawakami A. (2003) Fructan 1-exohydrolases: β-(2,1)-trimmers during graminan biosynthesis in stems of wheat? Purification, characterization, mass mapping, and cloning of two fructan 1-exohydrolase isoforms. Plant Physiol 131: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1–research0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijn I, Smeekens S. (1999) Fructan: more than a reserve carbohydrate? Plant Physiol 120: 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand P, Olsen OA, Kilian A, Kleczkowski LA. (1992) ADP-glucose pyrophosphorylase large subunit cDNA from barley endosperm. Plant Physiol 100: 1617–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Kuster H, Radchuk R, Muller M, Weichert H, Fait A, Fernie AR, Saalbach I, Weber H. (2008) Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J 55: 909–926 [DOI] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U. (2003) The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. Plant J 33: 395–411 [DOI] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U. (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21: 455–467 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW. (2006) Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F. (2004) A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Yakir E, Hilman D, Harir Y, Green RM. (2007) Regulation of output from the plant circadian clock. FEBS J 274: 335–345 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. (2007) The diurnal metabolism of leaf starch. Biochem J 401: 13–28 [DOI] [PubMed] [Google Scholar]
- Zheng ZL. (2009) Carbon and nitrogen nutrient balance signaling in plants. Plant Signal Behav 4: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.