Abstract
Serine/arginine-rich (SR) proteins are essential nuclear-localized splicing factors. We have investigated the dynamic subcellular distribution of the Arabidopsis (Arabidopsis thaliana) RSZp22 protein, a homolog of the human 9G8 SR factor. Little is known about the determinants underlying the control of plant SR protein dynamics, and so far most studies relied on ectopic transient overexpression. Here, we provide a detailed analysis of the RSZp22 expression profile and describe its nucleocytoplasmic shuttling properties in specific cell types. Comparison of transient ectopic- and stable tissue-specific expression highlights the advantages of both approaches for nuclear protein dynamic studies. By site-directed mutagenesis of RSZp22 RNA-binding sequences, we show that functional RNA recognition motif RNP1 and zinc-knuckle are dispensable for the exclusive protein nuclear localization and speckle-like distribution. Fluorescence resonance energy transfer imaging also revealed that these motifs are implicated in RSZp22 molecular interactions. Furthermore, the RNA-binding motif mutants are defective for their export through the CRM1/XPO1/Exportin-1 receptor pathway but retain nucleocytoplasmic mobility. Moreover, our data suggest that CRM1 is a putative export receptor for mRNPs in plants.
In eukaryotes, the protein-encoding genes are transcribed as long precursor molecules (pre-mRNAs) containing intervening sequences or introns. These introns are excised by a macromolecular complex, the spliceosome, consisting of five small nuclear ribonucleoproteins (U1, U2, U4/U6, and U5 snRNPs) and a large number of non-snRNP-associated proteins (Wahl et al., 2009). The Ser/Arg-rich (SR) proteins are described as a phylogenetically highly conserved family of non-snRNP essential splicing factors that mediate numerous events in pre-mRNA splicing (Reddy, 2007; Long and Caceres, 2009). In human, seven canonical SR proteins have been described with sizes ranging from 20 to 75 kD: SRp20, 9G8, ASF/SF2 (SRp30a), SC35 (SRp30b), SRp40, SRp55, and SRp75 (Bourgeois et al., 2004; Long and Caceres, 2009). SR proteins dynamically participate in spliceosome assembly through both protein-protein and protein-RNA interactions and have been recently reported to associate on nascent pre-mRNAs for cotranscriptional splicing (Das et al., 2007; Wahl et al., 2009). They promote the recruitment of the heterodimeric splicing factor U2AF and the U1 snRNP to the 3′ and 5′ splice sites, respectively. They also regulate alternative splicing by determining splice site selection, and this activity is antagonized mainly by heterogeneous nucleoproteins of the A and B groups (Graveley, 2000; Matlin et al., 2005). Although SR proteins appear to have redundant functions in vitro, individual proteins have specific activities in (pre-)mRNA metabolism (e.g. alternative splicing, mRNA export, translation) during the development of metazoans (Long and Caceres, 2009).
SR proteins have a modular structure consisting of one or two N-terminal RNA recognition motifs (RRMs) and a C-terminal RS domain rich in Arg and Ser residues with extensive repetition of SR or RS dipeptides. Some SR proteins, such as 9G8, contain an RNA-binding CCHC zinc (Zn)-knuckle motif located between the RRM and RS domains. The RRM is thought to determine RNA-binding specificity by recognizing exonic splicing enhancers, whereas the RS domain is involved in protein-protein and protein-RNA interactions (Shen et al., 2004). The role of the Zn-knuckle is less well understood, and the functional relationship between RRM and Zn-knuckle domains has still to be explored.
At steady state, SR proteins accumulate in nuclear speckles that may act as storage, assembly, and/or modification sites from which splicing factors are recruited to regulate splicing at the active transcription sites (Misteli et al., 1997). Moreover, a number of SR proteins, including ASF/SF2, 9G8, and SRp20, shuttle continuously between the nucleus and the cytoplasm, and this dynamic behavior has been tied to their postsplicing activities such as mRNA export or control of translation efficiency (Huang and Steitz, 2001; Huang et al., 2003; Lai and Tarn, 2004; Zhang and Krainer, 2004; Swartz et al., 2007; Michlewski et al., 2008). SR proteins have also been involved in genome stability, RNA stability, and nonsense-mediated decay (NMD; Zhang and Krainer, 2004). NMD is an mRNA degradation pathway that detects aberrant transcripts containing premature termination codons and triggers their degradation (Kim et al., 2009).
The nucleocytoplasmic transport of macromolecules occurs through the nuclear pore complex, which is a specialized edifice embedded in the nuclear envelope. Several nucleocytoplasmic trafficking pathways have been described that require so-called karyopherins for trafficking of molecules larger than approximately 40 to 90 kD. These specific transport receptors bind to cargo molecules that carry either nuclear localization signals for nuclear import or nuclear export signals (NES) for nuclear export (Wang and Brattain, 2007; Xu and Meier, 2008). The best known import pathway is mediated by the importin-α /importin-β karyopherins that bind to nuclear localization signals. One of the best characterized mammalian export pathways is mediated by CRM1/XPO1/Exportin-1, which recognizes short Leu-rich-type NES on proteins and is involved in the export of U snRNAs and ribosomal subunits (Hutten and Kehlenbach, 2007; Sleeman, 2007; Carmody and Wente, 2009). Identification of CRM1-specific cargoes has been facilitated by the use of leptomycin B (LMB), a CRM1-specific inhibitor (Hutten and Kehlenbach, 2007). While the bulk of mRNA is exported via the nonkaryopherin Nxf1(TAP)/Nxt1(p15) heterodimer, CRM1 was also shown to mediate the nuclear export of a subset of endogenous transcripts and of unspliced (or partially spliced) viral mRNAs (Gallouzi and Steitz, 2001; Carmody and Wente, 2009). SR protein-specific nuclear import receptors, the transportin-SR, interact with the phosphorylated RS domain (Lai et al., 2000), which is also necessary but not sufficient for the cytoplasmic export of shuttling SR proteins (Caceres et al., 1997). The human 9G8-shuttling SR protein associates with mRNA, providing an adapter function in recruiting the export receptor Nxf1 (Huang et al., 2003).
The Arabidopsis (Arabidopsis thaliana) genome contains 19 genes encoding SR-related proteins, some of which are homologous to the metazoan prototypes ASF/SF2, SC35, and 9G8, whereas others are plant specific (Kalyna and Barta, 2004). Many studies have focused on the cellular distribution of plant SR splicing factors; like their animal counterparts, they localize into nuclear irregular dynamic domains similar to speckles (Ali et al., 2008). Interestingly, a recent report demonstrated that in tobacco (Nicotiana tabacum) protoplasts, SR proteins may localize into distinct populations of speckles, with only partial or no colocalization (Lorkovic et al., 2008). These recent findings add further complexity to our understanding of SR splicing factor localization and emphasize the importance of investigating the determinants that regulate their dynamic distribution in vivo.
RSZp22, a prototypic member of the 9G8 subgroup of the Arabidopsis SR protein family, exhibits an unusual dynamic distribution in speckles and also within the nucleolus. Phosphorylation of the RSZp22 RS domain might influence its subcellular distribution, including its nucleolar localization (Tillemans et al., 2005, 2006). This dynamic distribution of a plant SR factor was unexpected, although human and plant nucleolar proteome analysis identified many pre-mRNA-binding proteins, including SR proteins (Andersen et al., 2002; Scherl et al., 2002; Pendle et al., 2005). We previously suggested that the nucleocytoplasmic shuttling of RSZp22 is at least partly controlled by the CRM1-dependent export pathway (Tillemans et al., 2006). Regulatory mechanisms underlying the nucleocytoplasmic transport of plant SR factors remain poorly understood, and knowledge about the functions of their RNA-binding motifs in transport processes is limited. Most previous studies on the dynamics of plant mRNP-binding proteins were carried out using ectopic overexpression of GFP-tagged proteins, which may have changed their dynamic properties (Tillemans et al., 2005, 2006; Ali et al., 2008; Lorkovic et al., 2008). Here, we investigate the dynamics of RSZp22 in stable transformants enabling tissue-specific expression of the transgene. To determine the role of RNA-binding domains in its dynamics, we performed site-directed mutagenesis on functional residues within these domains and assessed the dynamics of the mutant proteins. Our findings further extend previous results on the dynamic fate of RSZp22 and demonstrate that RSZp22 is a bona fide nucleocytoplasmic shuttling SR protein. Our approach constitutes a very sensitive assay to gain insights into the relationships between protein domains and dynamics and suggests that each RSZp22 structural RNA-binding domain has a functional role in protein mobility in vivo.
RESULTS
RSZp22 Nuclear Efflux and Influx Using a Photoactivatable Reporter Protein
We previously developed a fluorescence loss in photobleaching (FLIP)-shuttling assay to investigate the dynamics of SR proteins in vivo after transient expression. This imaging approach consists of measuring the fluorescence intensity of GFP-tagged proteins within the nucleus during a continuous photobleaching of a large adjacent cytoplasmic area. The rate of fluorescence loss in the nucleus is dependent on the nucleocytoplasmic mobility of the protein. Using this approach, we suggested that RSZp22 is shuttled between nucleus and cytoplasm (Tillemans et al., 2006). To further support this finding, we used the monomeric Dendra2 protein, which can be irreversibly photoactivated from a green to a red fluorescence upon UV and/or blue light irradiation (Gurskaya et al., 2006). Therefore, a RSZp22-Dendra2 fusion protein was transiently expressed in tobacco leaf cells (Fig. 1). Before photoactivation, green fluorescence of RSZp22-Dendra2 was only detectable within the nucleus and no red fluorescence was observed above background level. Upon continuous photoactivation of a cytoplasmic area using a 405-nm laser line, the green fluorescence intensity gradually decreased whereas the red fluorescence proportionally increased in the nucleus (Fig. 1). The use of photoactivatable fluorescent protein thus enabled the direct monitoring of both nuclear efflux and influx of RSZp22 very efficiently. It validates the FLIP-shuttling assay in transiently transformed cells by demonstrating that RSZp22 is a nucleocytoplasmic shuttling protein. However, in our hands, Dendra2 photoactivation by the harmless 488-nm blue light was never achieved at the end of the time-lapse experiment, even upon very intense blue irradiation, and required repetitive intense UV light irradiation that may be harmful to the cells. In addition, we failed to obtain Arabidopsis transformants stably expressing Dendra2 fusion proteins. Because of these drawbacks, we performed all subsequent dynamic experiments using the FLIP-shuttling assay on GFP-tagged RSZp22, which proved to be particularly suitable for monitoring protein dynamics in vivo.
Figure 1.
Nuclear efflux and influx of RSZp22-Dendra2 in living plant cells. A, Time course of Dendra2 green-to-red UV photoactivation in tobacco leaf cells transiently expressing P35S:RSZp22-Dendra2. The UV-irradiated cytoplasmic area surrounding the nucleus is boxed (top right panel). A nucleus that shows a progressive loss of green fluorescence and concomitant increase of red fluorescence is indicated by the arrows. Bar = 2 μm. B, The kinetics of green and red fluorescences were fitted with nonlinear regression curves.
Expression Analysis of RSZp22 in Arabidopsis
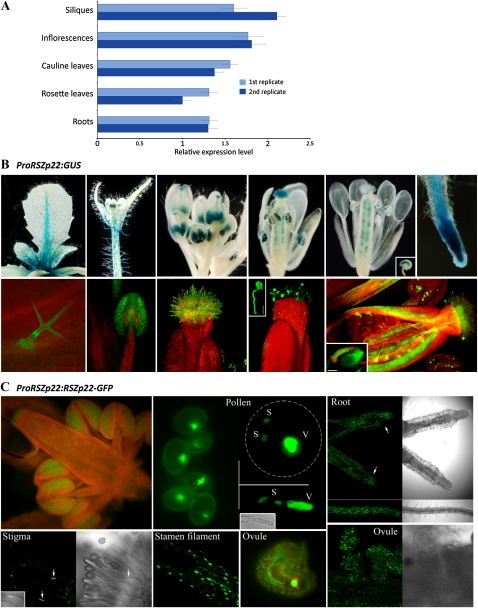
Although our data validate transient ectopic expression to study SR protein localization, the use of a nonnative and strong promoter may alter the dynamic distribution of SR proteins. There is thus a need to report on the dynamics of RSZp22 expressed under the control of its endogenous promoter in stable transgenic plants where native dosage and tissue specificity of expression would be better reflected. To date, information on the RSZp22 gene expression profile is scarce (Lopato et al., 2002), and no data are available on RSZp22 protein expression during Arabidopsis development. Therefore, we initially investigated in detail the expression pattern of RSZp22. We first measured RSZp22 transcript levels in different tissues by quantitative reverse transcription (RT)-PCR. The gene was expressed at relatively low and similar levels in all Arabidopsis vegetative and floral organs examined (Fig. 2A). We also generated promoter-GUS reporter lines (PRSZp22:GUS). Using both classical blue staining and fluorescence GUS detection, the PRSZp22:GUS expression profile was identical in several independent homozygous lines, with the exception of a single line that displayed expression restricted to developing pollen. During vegetative growth, GUS activity was observed in various tissues, including primary and lateral roots (root tips and stele), stems, petioles, abaxial and adaxial epidermis cells particularly along the leaf midrib, and in some trichomes. During floral development, PRSZp22:GUS expression was observed in unopened flowers (stages 7 and 8; Smyth et al., 1990) in anther filaments, in anthers, in stigma, and in pollen tube-transmitting tissue. The expression in immature anthers was initially stronger in tapetal cells, and as stamen matured, expression was seen in developing pollen. At later floral stages, GUS activity was observed in the upper part of filaments, in stigmatic papillae, in pollen and germinating pollen, in ovule funiculi and integuments, and in the embryo sac (Fig. 2B). After pollination and fertilization, GUS activity was barely detectable in maternal tissues and was not visible in the embryo. These findings confirm and significantly extend earlier RSZp22 expression studies (Lopato et al., 1999, 2002).
Figure 2.
RSZp22 expression profile and subcellular localization. A, Quantitative RT-PCR analyses of RSZp22 gene expression in Arabidopsis vegetative and floral organs. Data (means ± se) are normalized transcript levels relative to four reference genes (see “Materials and Methods”). B, Detection of GUS activity (blue staining, top; green fluorescence, bottom) directed by the RSZp22 promoter in root, leaf, and floral tissues. The top row shows histochemical blue staining, and the bottom row shows GUS Imagene Green fluorescence and chlorophyll red autofluorescence. The insets show a germinating pollen (middle; bar = 20 μm), an ovule showing GUS staining in the embryo sac (top right), and an ovule with its funiculus (bottom right; bar = 100 μm). C, Expression pattern and subcellular localization of RSZp22-GFP fusion proteins in root and reproductive tissues of PRSZp22:RSZp22-GFP transgenic plants. The pollen panel shows pollen cells (left), trinucleate pollen cell (top right), and germinating pollen (bottom right). S, Sperm; V, vegetative. In the root panel, the arrows indicate the root tip. In the stigma panel, the arrows point to the nuclei of stigmatic papillae.
Next, we generated transgenic lines expressing RSZp22-GFP under the control of the RSZp22 promoter (PRSZp22:RSZp22-GFP). Heterozygous and homozygous PRSZp22:RSZp22-GFP lines were indistinguishable from wild-type plants, indicating that the expression of the transgene did not alter plant development. In agreement with the GUS staining results, the RSZp22 promoter-driven RSZp22-GFP expression was observed in the nuclei of distinct cell types (Fig. 2C). Altogether, these data strongly suggest that the expression of RSZp22 is restricted to specific cell types at distinct developmental stages.
RSZp22 Localization and Dynamics in Arabidopsis
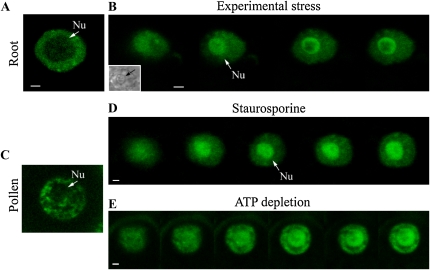
Having determined the expression pattern of RSZp22 promoter-driven RSZp22-GFP in transgenic lines, we next investigated the dynamic localization of RSZp22 in specific tissues. We first assessed whether the RSZp22-GFP can relocate into the nucleolus. Indeed, we previously showed using our overexpression transient assay that RSZp22-GFP concentrates in the nucleolus upon phosphorylation inhibition. This nucleolar localization was specific to RSZp22 compared with the other Arabidopsis SR proteins studied (Tillemans et al., 2006). In root and pollen cells, we consistently observed a dynamic relocalization and accumulation of RSZp22-GFP within nucleoli upon staurosporine treatment, ATP depletion, and experimental stress (Fig. 3; see “Materials and Methods”). However, the strand-like concentration of RSZp22 within the nucleoli was not observed, suggesting that this extreme RSZp22 nucleolar retention may be due to protein overexpression in the transient assay.
Figure 3.
Nucleolar relocalization of RSZp22 upon experimental stress and phosphorylation inhibition. Selection of confocal series images of nuclei in root (A and B) and pollen (C–E) cells of Arabidopsis stably expressing PRSZp22:RSZp22-GFP. RSZp22 nuclear localization is shown in the absence of treatment (A and C) and upon experimental stress (B), staurosporine (D), and ATP-depletion (E) treatments. The inset in B shows a transmission image of the nucleus. Arrows position the nucleolus (Nu). Bars = 2 μm. [See online article for color version of this figure.]
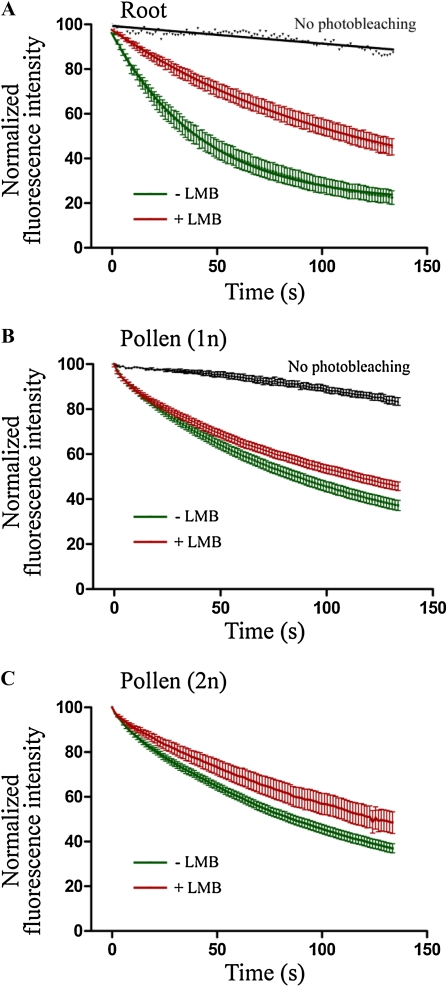
We then asked whether RSZp22 export from the nucleus to the cytoplasm is CRM1 dependent in distinct cell types. We thus performed FLIP-shuttling experiments in root and pollen cells at different developmental stages and upon LMB treatment (Fig. 4). RSZp22-GFP nucleocytoplasmic shuttling was evident from the curves of fluorescence exponential decay in the nucleus, confirming constant exchange between nucleus and cytoplasm. The fluorescence of unbleached control cells was only very slightly decreased by photobleaching of the GFP during the time-lapse experiment (Fig. 4, A and B). LMB treatment had a more intense inhibitory effect on RSZp22 export in root cells than in pollen cells, maybe due to the more impermeable cell wall of pollen (Fig. 4). The fitting of the FLIP curves revealed faster shuttling kinetics in root cells than in pollen, with a half-life of approximately 30 and approximately 60 s, respectively. In roots, nucleocytoplasmic exchange of half of the RSZp22-GFP mobile fraction thus required only approximately 30 s, and approximately 80% of RSZp22-GFP fluorescence in the nucleus was lost in about 130 s. Upon LMB inhibition, the half-life of fluorescence was significantly slower in both cell types (Fig. 4). These results demonstrate that (1) RSZp22 is a bona fide nucleocytoplasmic shuttling protein, with a rapid exchange between nucleus and cytoplasm, in PRSZp22:RSZp22-GFP plants, and (2) its shuttling is at least partly controlled by the CRM1 receptor.
Figure 4.
Nucleocytoplasmic shuttling of RSZp22 in stable PRSZp22:RSZp22-GFP Arabidopsis transgenics. FLIP-shuttling was monitored in the absence (−LMB) and upon LMB (+LMB) treatment in root cells (A), uninucleate pollen (1n; B), and binucleate pollen (2n; C). The curves show a significant inhibitory effect of LMB on shuttling (Student's t test, P < 0.0001). One hundred percent fluorescence indicates prebleach fluorescence intensity. As a control, cells were repeatedly scanned under no-photobleaching conditions and fluorescence was quantified. Half-time of fluorescence decay for –LMB and +LMB curves were as follows (in seconds): 30.12 and 81.65 (A), 60.33 and 65.48 (B), and 64.75 and 90.04 (C), respectively. Values are means ± se for at least 10 nuclei for roots and 15 nuclei for pollen.
Differential Dynamics of RSZp22 RNA-Binding Domain Mutants
Comparing the FLIP-shuttling kinetics obtained from transient assays and stable transgenics, we established that the transient assay does accurately assess whether a specific SR protein shuttles between nucleus and cytoplasm. A major advantage of this assay is that it can provide reproducible and comparable FLIP decay curves from cell to cell due to rather constant leaf cell morphology and therefore constant volume of photobleached cytoplasmic area. Herein, we specifically mutagenized amino acid residues mediating RNA binding in the CCHC Zn-knuckle and RNP1 motifs to assess whether these RSZp22 structural domains are required for the functional dynamics of the protein (Fig. 5). It has been shown that the Zn-knuckle provides RNA binding specificity to RSZp22 (Lopato et al., 1999) and its human homolog 9G8 (Cavaloc et al., 1999). A NMR spectroscopy study of the 9G8 RRM-RNA structure revealed that the aromatic residues of the RRM β-sheet are involved in direct RNA contact (Hargous et al., 2006). Zn-knuckle alterations consisted of point mutations of (1) each single conserved Cys into Ser (C101S, C104S, and C114S), generating SCHC, CSHC, and CCHS mutants, (2) all Cys into Ser (SSHS), and (3) all Cys into Ser and His (H109G) into Gly (SSGS). The rnp1 mutant consisted of two point mutations of the aromatic Tyr and Phe residues (Y40A and F42A; Fig. 5A). It has been previously shown for other RNA-binding proteins that these mutations impair the RNA-binding capacity of either the RRM or the Zn-knuckle but do not induce the total destructuring of the overall protein (Caceres and Krainer, 1993; Varani and Nagai, 1998; Gama-Carvalho et al., 2003; Shomron et al., 2004). Hence, point mutations of RNP1 aromatic residues of the ASF/SF2 first RRM domain induce a decrease of cross-linking efficiency to pre-mRNA (Caceres and Krainer, 1993). Similar mutations in the U2AF65 RRM1 impaired its binding to RNA (Gama-Carvalho et al., 2001).
Figure 5.
Differential dynamics of RSZp22 wild-type and mutant proteins in tobacco leaf cells. A, Diagram depicting the structure of wild-type RSZp22 and generated mutant derivatives. RSZp22 contains an N-terminal RRM domain (dark blue) followed by a CCHC-type Zn-knuckle motif (ZnK; light blue) embedded in an RGG domain (orange). Two highly conserved motifs of six (RNP2; not shown in the diagram) and eight (RNP1) amino acid residues are present within the approximately 90-amino acid RRM domain. The RS domain is boxed in red. The mutagenized residues within the RNP1 and Zn-knuckle motifs are indicated in boldface and underlined letters. B, Selected images of nuclear fluorescence distribution of GFP-tagged RSZp22 (left) and SSGS mutant (right) proteins. C, Intranuclear mobility of RSZp22 (left) and SSGS mutant (right) proteins. FRAP was monitored within speckles in the absence of treatment (top) and upon staurosporine (middle) and ATP depletion (bottom) treatments. D, Nucleocytoplasmic shuttling of RSZp22 (left) and SSGS mutant (right) proteins. FLIP-shuttling was monitored in the absence (−LMB) and upon LMB (+LMB) treatment. In C and D, insets show the overlay of wild-type and mutant curves. Values are means ± se for at least 20 nuclei.
The mutant proteins retained their exclusive nuclear localization and showed a speckle-like intranuclear distribution similar to wild-type RSZp22 (Fig. 5B; Supplemental Fig. S1A). We then analyzed the dynamics of wild-type and mutant RSZp22 proteins using fluorescence recovery after photobleaching (FRAP) experiments. Speckles were selectively photobleached and the fluorescence recovered rapidly after photobleaching with nearly identical recovery curves between RSZp22 and mutant proteins (Fig. 5C; Supplemental Fig. S1B). In contrast to our previous data obtained with domain-deleted mutants (Tillemans et al., 2006), our observations here using point mutations of essential residues were highly reproducible and constant between experiments and did not vary from cell to cell. Upon staurosporine treatment and ATP depletion, we observed the absence (RSZp22 and SCHC) and a very weak (CSHC, CCHS, and rnp1) fluorescence recovery for bleached proteins. Interestingly, FRAP analysis of the SSHS and SSGS mutants indicated that these two mutated proteins remain highly dynamic upon inhibition of phosphorylation using staurosporine and ATP-depletion treatments (Fig. 5C; Supplemental Fig. S1B). Altogether, these data suggest that functional RNP1 and Zn-knuckle motifs are dispensable for the exclusive nuclear localization and speckle-like distribution of RSZp22. Assuming that speckles are assembly sites of splicing factors, our data further suggest that these mutations weaken but do not completely abolish the ability of RSZp22 to interact with other splicing partners (see also fluorescence resonance energy transfer [FRET] experiments below).
To assess further the mobility and nuclear export of wild-type and mutated RSZp22 proteins, we then performed FLIP-shuttling studies. Quantitative analysis showed nearly identical and rapid rates of nuclear export of wild-type and all mutant RSZp22 proteins (Fig. 5D; Supplemental Fig. S1C), indicating that functional RNP1 and Zn-knuckle motifs are dispensable for RSZp22 nucleocytoplasmic shuttling. LMB treatment blocked the nucleocytoplasmic export of wild-type RSZp22 and SCHC mutant proteins, which both show identical shuttling kinetics. In agreement with the FRAP experiment described herein, this suggests that the mutation of the first Cys does not impair the integrity of the Zn-knuckle. On the contrary, all other mutant forms of RSZp22 preserved their ability to shuttle upon LMB treatment, albeit at a slower rate than without the inhibitor (Fig. 5D; Supplemental Fig. S1C). It is worth mentioning that identical results were obtained with the SSGS mutant expressed in Arabidopsis leaf cells after transient transformation (Supplemental Fig. S2).
Importantly, our results demonstrate that the nuclear retention of RSZp22 upon LMB treatment is not due to an indirect effect of the inhibition of the CRM1-dependent nuclear export. Furthermore, the remaining shuttling of RSZp22 mutant proteins upon LMB treatment demonstrates that these point mutants can be exported through a CRM1-independent export pathway. This raises the question whether wild-type RSZp22 uses alternative nuclear export pathways simultaneously. If this is true, differential loss of kinetics should be observed under LMB treatment between RSZp22 and a cargo molecule that is strictly dependent on the CRM1 receptor. To address this question, we performed FLIP-shuttling experiments on the GFP-fused tomato (Solanum lycopersicum) heat stress transcription factor HsfA1, bearing a C-terminal NES sequence and described as strictly CRM1 dependent for shuttling (Heerklotz et al., 2001; Kotak et al., 2004; Tillemans et al., 2006). The effect of a short-term LMB treatment was analyzed and the differential inhibition of nuclear export of RSZp22 compared with HsfA1 is evident from the fluorescence decay curves. LMB treatment induced a rapid and almost complete nuclear retention of GFP-HsfA1 but did not slow the RSZp22 shuttling rate to the same extent (Supplemental Fig. S3). Interestingly, these observations suggest that at least two pathways are responsible for RSZp22 nuclear export.
Although the extreme RSZp22 nucleolar accumulation upon LMB treatment may be due to protein overexpression, we decided to exploit this feature to determine the protein determinants required for such localization. RNA-binding mutants, therefore, were examined for their dynamic distribution upon LMB inhibition (Supplemental Fig. S4; Supplemental Table S1). Our data first confirmed the nucleolar concentration of wild-type RSZp22 in a high number of cells (approximately 82%). Mutation within either the RRM or Zn-knuckle reduced the nucleolar translocation of all mutant proteins and abolished the formation of strand-like structures (except for the SCHC mutation). Altogether, our data suggest that the nucleolar retention of RSZp22 is determined by its ability to bind RNA.
RSZp22 RNA-Binding Domains and Molecular Interactions
The FRAP and FLIP-shuttling kinetics suggest that site-directed mutagenesis of the RNA-binding domains partly impairs the ability of mutant RSZp22 proteins to assemble into molecular complexes. Therefore, we investigated the interactions of wild-type and mutant RSZp22 proteins using FRET, which monitors molecular interactions in vivo by evaluating the transfer of energy from an excited donor (cyan fluorescent protein [CFP]) to an acceptor (yellow fluorescent protein [YFP]). Since RSZp22 and RSZ33 have been previously shown to interact using yeast two-hybrid assay (Lopato et al., 2002), we studied heterodimer formation of RSZp22-CFP, RSZp22/SSGS-CFP, and RSZp22/rnp1-CFP with RSZ33-YFP in living cells with FRET-sensitized emission imaging. As positive controls, we monitored FRET efficiency between RSZ33-YFP and either SCL30-CFP or SCL30a-CFP, two SC35-like plant SR proteins, for which strong interactions have been demonstrated by yeast two-hybrid and pull-down assays (Lopato et al., 2002). Significant FRET efficiencies were observed for RSZp22-CFP and RSZ33-YFP in both nucleoplasm and speckles as well as for RSZ33-YFP and either SCL30-CFP or SCL30a-CFP, whereas no FRET was observed in cells coexpressing CFP and YFP (Fig. 6). The overall FRET signals for RSZ33-YFP and either RSZp22/SSGS-CFP or RSZp22/rnp1-CFP were about half of those measured with wild-type RSZp22, confirming an important role for both the Zn-knuckle and RNP1 motifs in molecular interactions between splicing factors and/or (pre-)mRNAs.
Figure 6.
Molecular interactions of RSZp22 wild-type and mutant proteins. FRET-sensitized emission was measured for the interaction between RSZ33-YFP and RSZp22-CFP, RSZp22/SSGS-CFP, RSZp22/rnp1-CFP, SCL30-CFP, and SCL30a-CFP, respectively. The YFP/CFP pair served as a negative control. Data in the histogram and the table are means ± se of FRET efficiency values in nucleoplasm (n > 30) and speckles (n > 16). Different letters denote significant differences between means (Student's t test, P < 0.05). [See online article for color version of this figure.]
DISCUSSION
Splicing is regulated by trans-acting factors that recognize and bind to cis-acting elements along pre-mRNAs (Chen and Manley, 2009). The dynamic properties of SR splicing factors have been extensively studied, especially in mammalian cells. In addition to splicing, the human nucleocytoplasmic shuttling SR proteins also function as regulators of mRNA export, stability, and translation (Zhong et al., 2009). Our knowledge is more limited on the dynamic distribution of plant SR proteins, which appear to play important roles in constitutive and alternative splicing (Reddy, 2007). In particular, the sequence determinants underlying the control of plant SR protein dynamics and nucleocytoplasmic transport are poorly defined. Here, we have extended our understanding of the dynamic distribution of RSZp22, an Arabidopsis homolog of the human 9G8 SR protein, using a range of functional imaging approaches in transient expression assays and stable Arabidopsis transgenic plants. We demonstrate the functional role of RSZp22 RNA-binding domains in intranuclear mobility, nucleocytoplasmic shuttling, and molecular interactions.
SR Protein Dynamics: Contribution of the FLIP-Shuttling Assay
In mammals, classical assays have used heterokaryons to analyze nucleocytoplasmic shuttling of splicing factors, including SR proteins (Gama-Carvalho et al., 2001; Cazalla et al., 2002; Sapra et al., 2009). To monitor the mobility of Arabidopsis SR proteins, transient expression-based methods have been developed (Ali et al., 2008; Lorkovic et al., 2008), and cytoplasmic FLIP was adapted to identify RSZp22 as a shuttling protein (Tillemans et al., 2006). Tissue-specific expression of distinct Arabidopsis SR proteins has already been successful in uncovering their dynamic organization (Fang et al., 2004). However, most studies were carried out by ectopically overexpressing SR proteins in Arabidopsis cell-based assay or in heterologous systems. With such an experimental approach, it cannot be fully excluded that ectopic overexpression may alter the dynamics of SR proteins, including shuttling kinetics. Here, we have obtained very similar results concerning RSZp22 mobility using both transient and stable tissue-specific expression. The only exception may concern the extreme strand-like RSZp22 nucleolar concentration under LMB treatment after transient overexpression.
Overall, we find that transient ectopic- and tissue-specific expression are complementary with distinct advantages, and both approaches present important and unique features that should drive experimental design. The use of endogenous promoter-driven reporter protein allows experimentation in more native conditions. Furthermore, specific interactions of SR proteins with other molecules, as well as their posttranslational modifications, may occur in specific cell types, and this could consequently influence their dynamic behavior. On the other hand, its simplicity and sensitivity make transient expression a method of choice to investigate the dynamics of numerous GFP-tagged wild-type and/or mutant SR proteins in a relatively short period of time, as long as precise kinetic parameters are not considered. FLIP-shuttling transient assays may be useful to study the dynamics of many proteins or macromolecular edifices, and both Arabidopsis and tobacco are equally suited for such studies. Although transient ectopic overexpression may not fully reflect the exact dynamics of SR proteins, this approach delivers important insights on their dynamic behavior and allows us to understand how the nuclear machinery functions in vivo.
RSZp22 Nucleolar Localization
Consistent with our previous data obtained in transient expression assays (Tillemans et al., 2006), we showed here in stable transgenics that RSZp22 displays a specific nucleolar concentration upon phosphorylation inhibition and experimental stress. RNA binding is likely to play a role in nucleolar accumulation of RSZp22, as a reduced relocalization was observed for the RNP1 and Zn-knuckle mutants upon LMB treatment.
The relocation of RSZp22 to the nucleolus raises the question of its potential roles in RNA metabolism in this nuclear compartment. Indeed, recent data suggest that the nucleolus may play a role in postsplicing events such as mRNA export and NMD: (1) human and Arabidopsis nucleolar proteomics identified proteins involved in mRNA metabolism regulation, including SR 9G8-like splicing factors (Andersen et al., 2002; Scherl et al., 2002; Pendle et al., 2005); (2) components of the Arabidopsis EJC, which is involved in mRNA splicing, export, stability, and NMD, were shown to localize to the nucleolus under hypoxia conditions (Koroleva et al., 2009), and Arabidopsis AtPym, which interacts with both Mago and Y14 EJC core proteins, concentrates in the nucleolus upon LMB treatment (Park and Muench, 2007); (3) both aberrant mRNA transcripts and NMD factors UPF2 and UPF3 have recently been identified in nucleolar fractions in Arabidopsis (Kim et al., 2009). The fact that SR proteins could colocalize with EJC factors within the nucleolus (or speckles) emphasizes a putative functional association of these proteins. Taking this into account, we may hypothesize that RSZp22 dynamics within the nucleolus underlies either its passive binding to partially spliced or unspliced transcripts or its direct function in RNA stability and NMD. Indeed, it was shown that overexpression of several human SR proteins enhances NMD, the presence of an intact RS domain being required for this activity (Zhang and Krainer, 2004).
Role of RSZp22 Domains in Nucleocytoplasmic Shuttling in Vivo
The roles of SR protein domains in subnuclear localization and dynamic distribution in vivo have been extensively studied in mammals. In particular, the RS domain of shuttling SR proteins has been shown to be necessary for nucleocytoplasmic transport (Caceres et al., 1998), a stretch of 10 consecutive RS dipeptides being sufficient for shuttling (Cazalla et al., 2002). The RS domain of the nonshuttling human SC35 protein contains a dominant nuclear retention signal, which is sufficient to confer nuclear retention to the otherwise shuttling ASF/SF2 (Cazalla et al., 2002). Point mutations in the RNP1 motif of the first RRM domain of ASF/SF2 impair its ability to shuttle (Caceres et al., 1998). It was also shown that point mutations of the hSLU7 Zn-knuckle disrupt the zinc atom binding at this motif and modify its nucleocytoplasmic shuttling balance (Shomron et al., 2004). The 9G8, ASF/SF2, and SRp20 shuttling SR proteins directly bind to Nxf1(TAP) by a short Arg-rich peptide adjacent to RRMs (Hargous et al., 2006; Tintaru et al., 2007). These proteins can function as mRNA export factors, exhibiting a higher affinity to mRNAs when hypophosphorylated (Huang and Steitz, 2001; Huang et al., 2003). In mammals, the Nxf1/Nxt1 receptor promotes the export of bulk mRNA, whereas CRM1, through binding via adapter proteins, can mediate nuclear export of a subset of endogenous transcripts and viral mRNAs (Gallouzi and Steitz, 2001; Kimura et al., 2004; Carmody and Wente, 2009). In plants, an Nxf1 homolog has yet to be identified (Hernandez-Pinzon et al., 2007).
In this report, we demonstrate that RSZp22-GFP is a bona fide nucleocytoplasmic SR protein. Cell-specific expression allowed us to reveal precise shuttling kinetic parameters. Fluorescence loss curves indicate a very rapid nuclear export and import of RSZp22 in metabolically active tissues such as Arabidopsis root cells. Interestingly, we provide evidence that continuous RSZp22 nuclear export is mediated by the CRM1-dependent pathway, even if RSZp22 has no obvious NES known to bind to CRM1. The presence of functional determinants underlying nucleocytoplasmic shuttling remained to be clarified. We previously attempted to identify such determinants by deleting entire RSZp22 domains. RSZp22 lacking the RS domain localizes both in the nucleus and the cytoplasm, revealing that this domain is crucial for its nuclear import (Tillemans et al., 2005). In FLIP experiments, dynamics of RSZp22 mutants either lacking the entire RRM domain or the RGG and Zn-knuckle motifs was highly variable from cell to cell, making it impossible to accurately evaluate their roles in protein shuttling (Tillemans et al., 2006). Here, this limitation has been circumvented by introducing point mutations at essential amino acid residues either in RRM or Zn-knuckle. Our data thus emphasize the benefits of using targeted mutagenesis rather than domain deletion to accurately measure the mobility parameters of mutant proteins. We showed that neither RNP1 nor Zn-knuckle motifs are key to nuclear localization and speckle-like organization. We further established that molecular interactions between splicing factors were strongly destabilized, although not entirely inhibited, with Zn-knuckle and RNP1 mutants. This suggests that both RNA-binding domains might either be needed for direct protein-protein interactions or for mRNA-mediated interactions independent of splicing factor preassembly. We also provide evidence that both RSZp22 RNA-binding domains play important roles in specifying shuttling properties and nuclear export through the CRM1-dependent pathway. Interestingly, in contrast to ASF/SF2 (Caceres et al., 1998), mutations of each individual RNA-binding domain do not inhibit the nuclear export of RSZp22 mutants even upon inhibition of CRM1, suggesting the existence of a yet unknown alternative export pathway. This could also reflect simple passive diffusion of RSZp22-GFP. In plants, mRNA export might occur through different pathways, and the use of one of these could depend on the cellular state or mRNA nature.
Assuming that RSZp22 plays a role in splicing in vivo (Lopato et al., 1999, 2002; Lorkovic et al., 2008), it is very likely that this protein is shuttled tethered to (pre-)mRNAs. Our results strongly suggest that either RSZp22 is directly involved in mRNA nuclear export or remains tethered to (aberrantly processed) mRNA transcripts during transport. Two Arabidopsis CRM1 receptors, termed XPO1a and XPO1b, have been identified (Merkle, 2003), and whether and how they regulate the nuclear export of specific subpopulations of mRNAs remain unknown. Nevertheless, it has been shown that the export of uncapped mRNAs is blocked by LMB in tobacco cells (Stuger and Forreiter, 2004).
CRM1 might have a more extended role in the active nuclear export of SR splicing factors and pre-mRNAs, since shuttling of the Arabidopsis ASF/SF2-like SRp34 protein is also significantly inhibited by LMB in transient as well as in stable expression assays (N. Stankovic and P. Motte, unpublished data). Interestingly, SRp34 colocalizes with AtMago EJC protein in speckles (Park and Muench, 2007). Altogether, our findings reveal CRM1 as a putative transport receptor for mRNPs in plants. Further work should aim to identify the receptor(s) involved in the alternative export pathway and investigate the spatial associations and interconnections between plant export receptors, mRNAs, EJC proteins, and SR splicing factors. In the future, the study of the nuclear compartmentalization linked to nucleocytoplasmic export will shed light on the regulation of gene expression in plants.
MATERIALS AND METHODS
Binary Vector Construction
The construction of P35S:RSZp22-GFP and P35S:GFP-LpHsfA1 in a pBI121 backbone was described in previous reports (Tillemans et al., 2005, 2006). The promoter region of RSZp22 (956 bp upstream of the ATG, corresponding to the entire intergenic region between RSZp22 and the upstream gene At4g31570) was amplified from Arabidopsis (Arabidopsis thaliana) genomic DNA (ecotype Columbia [Col-0]). The PRSZp22 amplicon was ligated at the HindIII/BamHI sites of pBI121 to replace the 35S promoter and create PRSZp22:GUS. To obtain the PRSZp22:RSZp22-GFP construct, the GUS coding sequence was digested out of PRSZp22:GUS and replaced by the RSZp22-GFP cassette using BamHI and SacI restriction sites.
The mutated RSZp22 versions were obtained by PCR-based site-directed mutagenesis on the RSZp22 coding sequence cloned in the pGEM-T Easy vector using overlapping primers containing the desired mutation. The mutated coding sequences were digested out of the pGEM-T vector using BamHI and KpnI and cloned at the corresponding sites of P35S:RSZp22-GFP to replace the wild-type coding sequence.
To generate a Dendra2-tagged RSZp22, the Dendra2 coding sequence was amplified from the Dendra-At-N-vector (Evrogen). The amplicon was then ligated in P35S:RSZp22-GFP at the KpnI/SacI sites to replace the GFP coding sequence. The CFP and YFP coding sequences were PCR amplified from pECFP and pEYFP (Clontech). The P35S:CFP and P35S:YFP constructs were obtained by cloning CFP and YFP into the BamHI/SacI sites of the pBI121:P35S plasmid. The P35S:RSZp22-CFP, P35S:RSZ33-YFP, P35S:RSZp22/SSGS-CFP, and P35S:RSZp22/rnp1-CFP constructs were generated by ligating the CFP or YFP fragment at KpnI/SacI sites to replace GFP in P35S:RSZp22-GFP, P35S:RSZ33-GFP (Tillemans et al., 2005), P35:SSGS-GFP, and P35S:rnp1-GFP, respectively. The SCL30-CFP and SCL30a-CFP fusions were generated as follows: the SCL30 and SCL30a coding sequences were amplified from an Arabidopsis cDNA library (ecotype Col-0) and ligated to the CFP coding sequence using KpnI. The fused sequences were then cloned into the BamHI/SacI sites of pBI121:P35S.
All PCRs were carried out using Pfu polymerase (Promega). A list of primers used in this study is provided in Supplemental Tables S2 and S3. Independent clones were sequenced in order to detect any mutation. All final plasmids were electroporated into the Agrobacterium tumefaciens strain GV3101 (pMP90) and subsequently used for plant transformation.
Plant Growth and Plant Transformation
Tobacco (Nicotiana tabacum ‘Petit Havana’) and Arabidopsis transient transformations by Agrobacterium infiltration were performed as described (Docquier et al., 2004).
Arabidopsis plants were stably transformed by floral dipping. T1 transformants were selected on plates containing sterile solidified half-strength Murashige and Skoog medium (Duchefa) containing kanamycin (50 μg mL−1). Transgenics were potted directly on soil at the two-leaf stage to obtain T2 and then T3 homozygous lines. Observations and imaging were realized on five PRSZp22:GUS and four PRSZp22:RSZp22-GFP independent lines, respectively.
For expression profiling, Arabidopsis plants (ecotype Col-0) were hydroponically grown from seeds in Hoagland medium as described (Talke et al., 2006). After 6 weeks of growth in a climate-controlled chamber at 21°C with a photoperiod of 16 h at a light intensity of 100 μmol m−2 s−1, root, rosette leaf, cauline leaf, inflorescence, and silique tissues were harvested separately from 12 plants. The tissues from the individual plants were pooled, frozen in liquid nitrogen, and stored at −80°C until further processing.
For in vitro pollen germination, drops of solid pollen germination medium were dabbed with anthers and then incubated for the night at 21°C to 22°C as described (Johnson-Brousseau and McCormick, 2004).
RNA Extraction, cDNA Synthesis, and Quantitative RT-PCR
Total DNase-treated RNAs were extracted using the RNeasy Plant Mini Kit and RNase-free DNase set (Qiagen). Quality and quantity of RNAs were checked visually by gel electrophoresis and by spectrophotometric analysis. cDNAs were synthesized from 1.5 μg of total RNAs using oligo(dT) and the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas).
Quantitative RT-PCR was performed on 384-well plates with an ABI Prism 7900HT system (Applied Biosystems) using Maxima SYBR Green qPCR Master Mix (Fermentas) as described (Talke et al., 2006) on material from two independent biological experiments, and a total of six technical repeats were run for each combination of cDNA and primer pair. The quality of the PCRs was checked visually through analysis of dissociation and amplification curves, and reaction efficiencies were determined for each PCR using the LinRegPCR software (Ramakers et al., 2003). Mean reaction efficiencies were then determined for each primer pair from all reactions (60 reactions; Supplemental Table S4) and used to calculate relative gene expression levels by normalization using multiple reference genes with the qBase software (Hellemans et al., 2007). Four reference genes (UBQ10, EF1α, At1g58050, and At1g62930) were selected from the literature (Czechowski et al., 2005). Their adequacy to normalize gene expression in our experimental conditions was verified using the geNorm software (average gene expression stability = 0.56; Vandesompele et al., 2002). Quantitative RT-PCR data evaluation was carried out according to recently published guidelines (Gutierrez et al., 2008; Udvardi et al., 2008).
Analysis of GUS Reporter Lines
Histochemical GUS staining was carried out as described (Jefferson et al., 1987) on Arabidopsis seedlings grown on half-strength Murashige and Skoog medium and on tissues of mature plants grown hydroponically. Harvested tissues were incubated in staining solution for 1 night to 2 d, then ethanol extracted and fixed before observation. Samples were observed with a Nikon SMZ1500 stereomicroscope and equipped with a Nikon Digital Sight DS-U1 camera.
For fluorescent observation of the promoter:GUS expression pattern, samples were incubated in a 50 μm Imagene Green (Molecular Probes), 0.1% Silwet L-77 solution for 2 h at room temperature. Samples were briefly washed in 0.1% Silwet L-77. Analysis was performed by confocal microscopy using GFP settings (see below).
Confocal Microscopy, Image Analysis, Photobleaching Experiments, and Inhibitor Treatments
Leica TCS SP2 and SP5 inverted confocal laser microscopes were used for live cell imaging. The FRAP and FLIP-shuttling experiments were carried out as described previously (Tillemans et al., 2006).
FRET analyses were performed on transiently transformed leaf cells using the Leica FRET-sensitized emission module. Images were collected in a 512- × 512-pixel resolution. Measurements were realized by detection of the fluorescent signals of the CFP donor (excitation at 458 nm and emission at 465–505 nm), the FRET (excitation at 458 nm and emission at 525–600 nm), and the YFP acceptor (excitation at 514 nm and emission at 525–600 nm) in a line-by-line sequential scan acquisition. Once appropriate image sets were obtained, the Leica confocal software generated a FRET efficiency for each plotted region of interest corresponding to nucleoplasm or speckles.
Green Dendra2 was visualized by scanning with a 488-nm excitation light using a minimal light intensity to avoid photobleaching and 500 to 550 nm for detection. Photoconversion was achieved by irradiating a region of interest using a 405-nm UV laser line. After photoconversion, red Dendra2 fluorescence was visualized using the 543-nm laser line and detected between 550 and 670 nm.
Fragments of transiently transformed leaves were used for phosphorylation inhibitor treatments as described previously (Tillemans et al., 2005). Stably transformed Arabidopsis roots and mature flowers were dissected under binocular microscope, and fragments of roots and stamens were incubated in inhibitor solution. Samples were incubated in 50 μm staurosporine (Sigma-Aldrich) or water as a control for various time periods (1–4 h). For ATP depletion, cells were treated with 3 mm sodium azide (Sigma-Aldrich) and 50 mm 2-deoxyglucose (Sigma-Aldrich) for up to 1 h. For LMB treatments, LMB (Sigma-Aldrich) was used at a final concentration of 10 nm for transiently transformed leaves and stably transformed roots and 100 to 200 nm for the pollen grains of stable transgenics. Plant cells were treated with LMB (or water as a control) for up to 4 h (up to 2 h for pollen cells) and processed for imaging as described above. As defined by Tillemans et al. (2006), an experimental stress results in a long observation period (of minimum 2 h) inducing a continuous decrease of cellular ATP level.
All observations and treatments were performed in at least three independent experiments with stable transgenics and at least after three independent transient transformation events. The total number of analyzed nuclei for each experiment (FRAP, FLIP-shuttling, and FRET) is mentioned in the corresponding figure legends.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Differential dynamics of wild-type and RSZp22 mutant proteins in tobacco leaf cells.
Supplemental Figure S2. Nucleocytoplasmic shuttling of wild-type RSZp22 and SSGS mutant proteins in Arabidopsis leaf cells.
Supplemental Figure S3. Comparison of the nucleocytoplasmic shuttling of RSZp22 and HsfA1.
Supplemental Figure S4. Nucleolar relocalization of RSZp22 wild-type and mutant proteins upon LMB treatments in tobacco leaf cells.
Supplemental Table S1. Nucleolar relocalization of RSZp22 wild-type and mutant proteins upon LMB treatments in tobacco leaf cells.
Supplemental Table S2. List of primers used for vector construction.
Supplemental Table S3. List of primers used for site-directed mutagenesis.
Supplemental Table S4. Sequences and reaction efficiencies of primer pairs used for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Pierre Wernimont and Fabienne Clérisse for assistance in vector construction and Leica Mannheim for helpful advice with Dendra2 photoactivation and FRET assays.
References
- Ali GS, Prasad KV, Hanumappa M, Reddy AS. (2008) Analyses of in vivo interaction and mobility of two spliceosomal proteins using FRAP and BiFC. PLoS One 3: e1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J. (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78: 37–88 [DOI] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR. (1993) Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 138: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. (1998) A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev 12: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. (2009) mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. (1999) The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. (2002) Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol 22: 6871–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. (2007) SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- Docquier S, Tillemans V, Deltour R, Motte P. (2004) Nuclear bodies and compartmentalization of pre-mRNA splicing factors in higher plants. Chromosoma 112: 255–266 [DOI] [PubMed] [Google Scholar]
- Fang Y, Hearn S, Spector DL. (2004) Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol Biol Cell 15: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA. (2001) Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Carvalho MP, Kehlenbach A, Valcarcel J, Carmo-Fonseca M. (2001) Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J Biol Chem 276: 13104–13112 [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Condado I, Carmo-Fonseca M. (2003) Regulation of adenovirus alternative RNA splicing correlates with a reorganization of splicing factors in the nucleus. Exp Cell Res 289: 77–85 [DOI] [PubMed] [Google Scholar]
- Graveley BR. (2000) Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24: 461–465 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O. (2008) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20: 1734–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. (2006) Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Doring P, Bonzelius F, Winkelhaus S, Nover L. (2001) The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Yelina NE, Schwach F, Studholme DJ, Baulcombe D, Dalmay T. (2007) SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J 50: 140–148 [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. (2003) SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell 11: 837–843 [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. (2001) Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell 7: 899–905 [DOI] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH. (2007) CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17: 193–210 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S. (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Kalyna M, Barta A. (2004) A plethora of plant serine/arginine-rich proteins: redundancy or evolution of novel gene functions? Biochem Soc Trans 32: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. (2009) Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Hashimoto I, Nagase T, Fujisawa J. (2004) CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J Cell Sci 117: 2259–2270 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JW, Shaw PJ. (2009) Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P. (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. (2000) A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem 275: 7950–7957 [DOI] [PubMed] [Google Scholar]
- Lai MC, Tarn WY. (2004) Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem 279: 31745–31749 [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorkovic ZJ, Barta A. (2002) Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J Biol Chem 277: 39989–39998 [DOI] [PubMed] [Google Scholar]
- Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A. (1999) A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol Biol 39: 761–773 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Hilscher J, Barta A. (2008) Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp Cell Res 314: 3175–3186 [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2003) Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44: 231–260 [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. (2008) The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 30: 179–189 [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527 [DOI] [PubMed] [Google Scholar]
- Park NI, Muench DG. (2007) Biochemical and cellular characterization of the plant ortholog of PYM, a protein that interacts with the exon junction complex core proteins Mago and Y14. Planta 225: 625–639 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ. (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reddy AS. (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. (2009) SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell 34: 179–190 [DOI] [PubMed] [Google Scholar]
- Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR. (2004) Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell 13: 367–376 [DOI] [PubMed] [Google Scholar]
- Shomron N, Reznik M, Ast G. (2004) Splicing factor hSlu7 contains a unique functional domain required to retain the protein within the nucleus. Mol Biol Cell 15: 3782–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J. (2007) A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 120: 1540–1550 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuger R, Forreiter C. (2004) Uncapped mRNA introduced into tobacco protoplasts can be imported into the nucleus and is trapped by leptomycin B. Plant Cell Rep 23: 99–103 [DOI] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. (2007) The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem 282: 19844–19853 [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Kramer U. (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142: 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillemans V, Dispa L, Remacle C, Collinge M, Motte P. (2005) Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J 41: 567–582 [DOI] [PubMed] [Google Scholar]
- Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. (2006) Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18: 3218–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, Wilson SA. (2007) Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep 8: 756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Nagai K. (1998) RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct 27: 407–445 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang R, Brattain MG. (2007) The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett 581: 3164–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Meier I. (2008) The nuclear pore comes to the fore. Trends Plant Sci 13: 20–27 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. (2004) Involvement of SR proteins in mRNA surveillance. Mol Cell 16: 597–607 [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. (2009) SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.