Abstract
Interactions between sulfur (S) nutritional status and sulfate transporter expression in field-grown wheat (Triticum aestivum) were investigated using Broadbalk +S and −S treatments (S fertilizer withheld) at Rothamsted, United Kingdom. In 2008, S, sulfate, selenium (Se), and molybdenum (Mo) concentrations and sulfate transporter gene expression were analyzed throughout development. Total S concentrations were lower in all tissues of −S plants, principally as a result of decreased sulfate pools. S, Se, and Mo concentrations increased in vegetative tissues until anthesis, and thereafter, with the exception of Mo, decreased until maturity. At maturity, most of the S and Se were localized in the grain, indicating efficient remobilization from vegetative tissues, whereas less Mo was remobilized. At maturity, Se and Mo were enhanced 7- and 3.7-fold, respectively, in −S compared with +S grain, while grain total S was not significantly reduced. Enhanced expression of sulfate transporters, for example Sultr1;1 and Sultr4;1, in −S plants explains the much increased accumulation of Se and Mo (7- and 3.7-fold compared with +S in grain, respectively). Sultr5;2 (mot1), thought to be involved in Mo accumulation in Arabidopsis (Arabidopsis thaliana), did not fully explain patterns of Mo distribution; it was expressed in all tissues, decreasing in leaf and increasing in roots under −S conditions, and was expressed in florets at anthesis but not in grain at any other time. In conclusion, S fertilizer application has a marked impact on Mo and Se distribution and accumulation, which is at least partially a result of altered gene expression of the sulfate transporter family.
Crops produced in many regions in the world, including western Europe, north Africa, and some parts of China, are low in selenium (Se) due to low availability of Se in soil (Hawkesford and Zhao, 2007; Zhu et al., 2009). Between 0.5 and 1 billion people worldwide are estimated to have insufficient intake of Se (Combs, 2001). Biofortification of crops through Se fertilization is a feasible strategy to enhance human Se intake, as has been practiced in Finland since the mid 1980s (Broadley et al., 2006). However, wheat (Triticum aestivum) crops recover only 20% to 35% of the Se fertilizer applied (Broadley et al., 2010; Stroud et al., 2010), indicating a low utilization efficiency, with the underlying reasons still unknown.
Se may be taken up by plants in the form of selenate, selenite, or organic Se. Selenate uptake occurs through sulfate transporters in the plasma membrane of plant roots. The assimilation of selenate follows the sulfate pathway (Sors et al., 2005a, 2005b; Kopsell and Kopsell, 2007), as the selenate molecule has a similar size and charge to sulfate. It is widely known that the uptake and assimilation of sulfate is regulated by the nutrient status of the plant (Smith et al., 1995, 1997; Buchner et al., 2004a). Laboratory studies have shown that a decrease in sulfate availability results in a several-fold enhanced expression of sulfate transporter genes, which enhances the capacity for sulfate uptake (Hawkesford, 2000; Hawkesford et al., 2003a, 2003b). Arabidopsis (Arabidopsis thaliana) mutants containing a lesion in the SULTR1;2 gene that encodes a high-affinity sulfate transporter are selenate resistant, indicating that selenate uptake occurs by this sulfate transporter (Shibagaki et al., 2002; Kassis et al., 2007). However, there are few studies concerned with other sulfate transporters or Se transport to grain in crop species such as wheat. Less is known about selenite uptake, which may be related to the phosphate transport pathway in the plasma membrane (Hopper and Parker, 1999; Li et al., 2008). Selenate and selenite uptake are enhanced in sulfur (S)-starved and phosphorus-starved plants, respectively (Li et al., 2008). The antagonistic impact of S on Se fertilization of crops has been shown in many studies (Terry et al., 2000), but not all (Stroud et al., 2010).
The sulfate transporter gene family in plants was described by Hawkesford (2003), and in Arabidopsis the gene family consists of 14 isoforms that may be subdivided into at least five groups. Genome analysis and systematic cloning studies indicate that wheat, Brassica oleracea, and rice (Oryza sativa) have similar sulfate transporter gene groups, and it is probable that close homologs have similar functions (Buchner et al., 2004a, 2004b, 2004c, 2010). The phylogenetic relationships of Arabidopsis and wheat sulfate transporters are shown in Figure 1. Group 1 comprises high-affinity transporters located in the plasma membrane, group 2 contains low-affinity transporters in the plasma membrane, group 3 is of unknown function but may participate in heterodimer associations (Kataoka et al., 2004a), group 4 catalyzes the efflux of sulfate from the vacuole into the cytoplasm across the tonoplast (Kataoka et al., 2004b), and a member of group 5 (Sultr5;2) is involved in molybdenum (Mo) metabolism in Arabidopsis, probably as an intracellular transporter, and has been termed mot1 (Tomatsu et al., 2007; Baxter et al., 2008). As high-affinity sulfate transporters are able to transport molybdate (Fitzpatrick et al., 2008), Mo uptake, as for selenate, is likely to be via the sulfate uptake pathway.
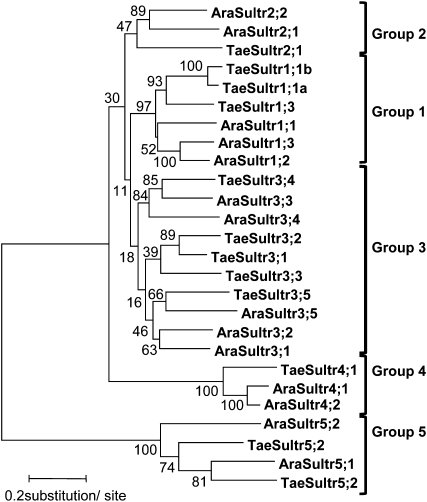
Figure 1.
Phylogenetic analysis. Neighbor-joining tree (MEGA4; Tamura et al., 2007) from the multiple alignment (ClustalX version 1.81; Thompson et al., 1997) of the coding cDNAs of the wheat (AJ512821, AJ512820, BT009249, TC366953/ TC291347, FN432835, TC272130/ TC259376, TC318325/ TC314180, AM747385, BT009340, FN601348, FN601349) and Arabidopsis (AB018695, AB042322, AB049624, AB003591, D85416, D89631, AB004060, AB023423, AB054645, AB061739, AB008782, AB052775, AC018848, AC006053) sulfate transporter gene family. The bootstrap values, expressed as percentages, were obtained from 1,000 replicate trees. Ara, Arabidopsis; Tae, wheat.
Under controlled laboratory conditions utilizing agar plates, hydroponic culture systems, or greenhouse pot experiments, the expression of group 1, group 2, and group 4 sulfate transporter genes is induced by S starvation in plant species including barley (Hordeum vulgare; Smith et al., 1997), wheat (Buchner et al., 2004b), Stylosanthes hamata (Smith et al., 1995), Brassica (Buchner et al., 2004a), and Arabidopsis (Takahashi et al., 1997, 2000; Yoshimoto et al., 2003). Analysis of sulfate transporter expression in relation to the S nutritional status in field-grown crops has not previously been investigated and is reported here, to our knowledge, for the first time.
Wheat was sampled from the Broadbalk winter wheat field experiment at Rothamsted Research (Harpenden, United Kingdom), the world's oldest continually running agronomic experiment, which has investigated wheat crop nutrition for 165 years (Poulton, 1995). S, Se, and Mo concentrations and sulfate transporter gene expression were determined in plant tissues, from seedling to grain-filling stage, from plots in which S fertilization has been withheld since 2000 and compared with +S plots, with the aim of relating mineral accumulation patterns to expression of the transport systems.
RESULTS
Total S and Sulfate Ion Concentrations
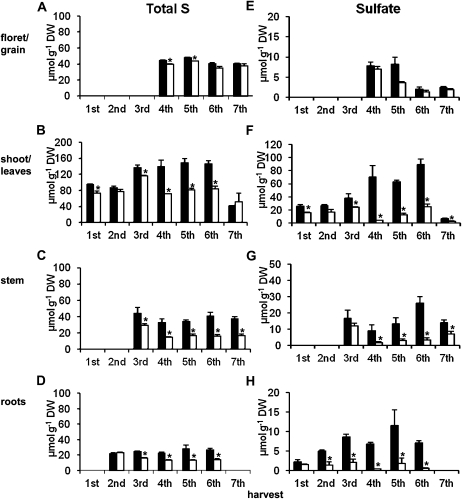
Figure 2 shows total S and sulfate ion concentrations in defined wheat tissues from field plots with or without S fertilization. In developing (fourth to seventh harvest) florets and grains, there was little difference in S concentration between S-fertilized (+S) and non-S-fertilized (−S) plants (Fig. 2A). In leaves of +S plants, the total S concentration increased at flag leaf emergence (third harvest) and remained at a high concentration until 4 weeks post anthesis (wpa; sixth harvest) but was substantially reduced at the final harvest (seventh harvest). In the −S plants, the total S concentration was slightly lower than the +S until third harvest and thereafter did not increase after anthesis, resulting in a substantial difference between the treatments (Fig. 2B). In stems (Fig. 2C), total S concentration was lower in −S plants compared with the +S plants at all time points. In roots (Fig. 2D), total S concentration was similar in +S and −S treatments at the second harvest, but after the third harvest it increased slightly in the +S plants and decreased in −S plants.
Figure 2.
Total S (A–D) and sulfate ion (E–H) concentrations in selected tissues of wheat from S-fertilized field plots (black bars) and non-S-fertilized plots (white bars). First to seventh harvests were as follows: first, seedling growth; second, stem elongation; third, starting flag leaf; fourth, anthesis; fifth, 2 wpa; sixth, 4 wpa; seventh, 60 d post anthesis. The total S concentrations of the roots at first and last harvest were not analyzed; significant stem tissue was not present before stem extension (first and second harvest); no floret/grain tissues could be obtained before emergence (first to third harvest). All samples were collected in 2008. Results are mean values of plants (10 for first and second harvest and five to seven for later harvests) obtained from three replicate areas of each plot. Error bars indicate se. The ±S comparisons were submitted to t test variance analysis with two-tailed distribution and two-sample equal variance, and asterisks represent significance at P < 0.05. DW, Dry weight.
In grain (florets at fourth harvest), sulfate concentrations decreased after anthesis and the absence of S fertilization had little influence on the sulfate concentrations (Fig. 2E). In leaves (Fig. 2F), an increase of the sulfate ion concentration was found at successive sampling points in the +S plants and the sulfate concentration in −S plants was smaller at all sample times, particularly after anthesis, when the sulfate ion concentration of +S leaves continued to increase. The observed difference in total S concentration between +S and −S plants (Fig. 2B) could be approximately attributed to the difference in sulfate concentrations (Fig. 2F). In stems (Fig. 2G), the patterns of changes of concentrations were similar to leaves. In roots (Fig. 2H), sulfate concentration increased in +S plants until the fifth harvest and decreased slightly thereafter. In the −S plants, sulfate concentrations in the roots remained low.
Changes of Se and Mo Concentrations
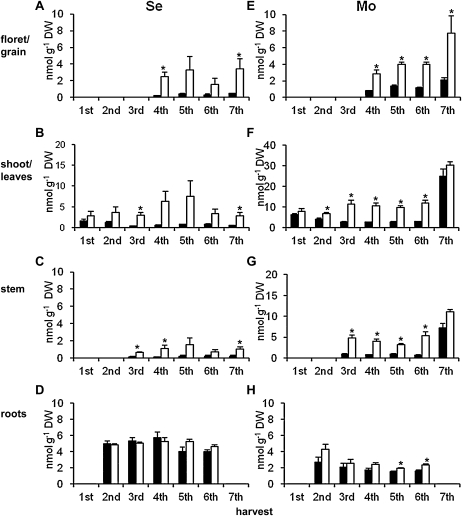
Se concentrations in −S plants in floret and grain tissues (Fig. 3A) were much higher than in +S plants. Similarly, in leaves (Fig. 3B), high concentrations of Se were observed in −S plants. In the final (sixth and seventh) harvests, the Se concentration decreased in the leaves. In stems (Fig. 3C), the Se concentration of −S plants was higher than +S plants and showed similar changes to leaves. There were no significant differences in Se concentration in root tissues between +S and −S plants (Fig. 3D).
Figure 3.
Se (A–D) and Mo (E–H) concentrations in selected tissues of wheat from S-fertilized field plots (black bars) and non-S-fertilized plots (white bars). Sample details are as in legend for Figure 2. Error bars indicate se. The data were submitted to t test variance analysis with two-tailed distribution and two-sample equal variance, and asterisks represent significance at P < 0.05. DW, Dry weight.
The Mo concentration of floret and grain tissues (Fig. 3E) was higher in −S plants compared with +S plants. In leaves (Fig. 3F) at the first harvest, the Mo concentration was slightly higher in −S plants, and this difference increased until the sixth harvest. By the final (seventh) harvest, the Mo concentrations were high in both +S and −S plants. Observed changes of Mo concentrations in leaves were different from those observed for Se in that they did not decrease at the later harvests. In stems (Fig. 3G), a similar pattern to that seen in leaves was observed, although absolute concentrations were lower. There were no clear differences in the Mo concentrations in roots between +S and −S plants (Fig. 3H).
Tissue Distributions of S, Se, and Mo after Anthesis
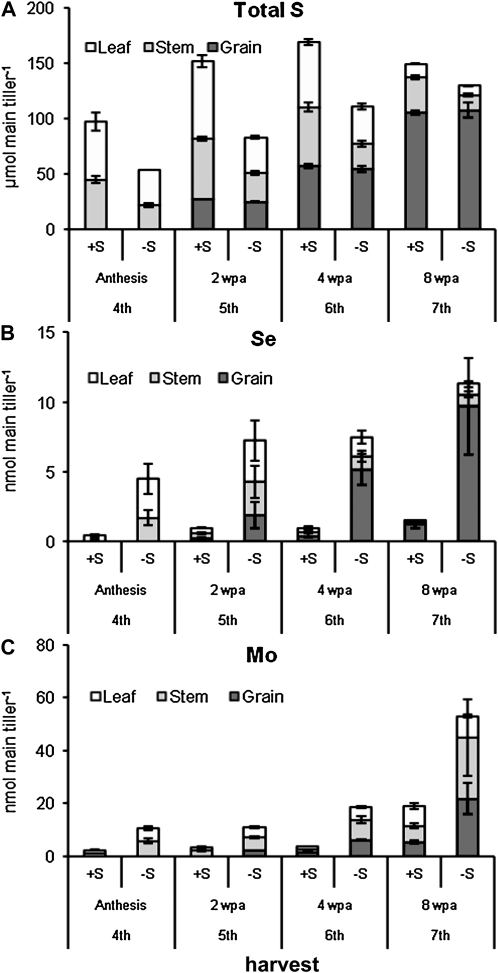
Distributions of the S, Se, and Mo contents of grain, leaf, and stem fractions were determined on a per (main) tiller basis after anthesis, as calculated from the concentrations and mean fraction weight (Fig. 4). The +S plants had a greater quantity of S than the non-S-fertilized plants (Fig. 4A), and this S was evenly distributed between leaves and stems at anthesis. At 2 and 4 wpa, the S content of the S-fertilized plants increased in leaves, stems, and grains. In −S plants, only the grain total S content increased, and no additional accumulation was observed in the stem or leaf fractions compared with anthesis. At the final harvest, grain S contents were similar in +S and −S plants but higher in the stem fraction of the +S plants.
Figure 4.
Distribution of total S (A), Se (B), and Mo (C) between plant parts in the main tiller. Plant parts are leaf (white bars), expanded leaves, stem (without sheaths; gray bars), and grain (except fourth harvest = florets; black bars). Total tissue content was calculated using the concentrations (Figs. 2 and 3) and mass data for individual plant parts from corresponding samples collected in 2005 (mean of 10 plants; data not shown). Error bars indicate se.
The Se contents were much higher in the −S plants compared with the +S plants (Fig. 4B). At 2 wpa, Se was evenly distributed between leaves, stems, and grains, although the distribution switched to being predominantly in the grain from 4 wpa. The total quantity of Se in the tiller increased during maturation of the grain in the weeks after anthesis in both +S and −S plants.
Mo distribution (Fig. 4C) was different from S and Se. Similar to Se, Mo contents were much higher in the −S plants compared with the +S plants at all time points after anthesis. The total tiller Mo content continued to increase after anthesis, although by the final harvest, much of the Mo still remained in the nongrain fraction (leaves and stems).
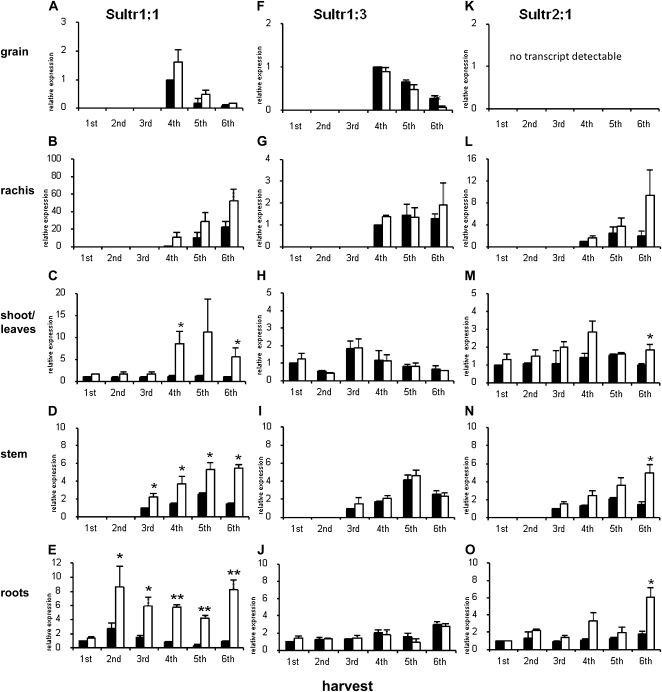
Expression of Sultr1;1, Sultr1;3, and Sultr2;1
Sultr1;1, Sultr1;3, and Sultr2;1 are sulfate ion transporters located in the plasma membrane. Sultr1;1, a high-affinity-type transporter, had higher expression (as measured by corresponding mRNA abundance) in all tissues examined in the samples isolated from the −S field plots (Fig. 5, A–E). Sultr1;1 expression decreased in the grain tissues following anthesis, whereas expression increased in the rachis in successive samples after anthesis. In leaf and stem fractions, Sultr1;1 expression increased after anthesis in non-S-fertilized leaves. There was a high expression of Sultr1;1 in the roots from the second harvest onward in the −S roots.
Figure 5.
Expression analysis of Sultr1;1, Sultr1;3, and Sultr2;1 by semiquantitative RT-PCR. Relative expression of Sultr1;1, Sultr1;3, and Sultr2;1 in relation to the first S-fertilized time point set to 1 is shown in A to E, F to J, and K to O, respectively. S-fertilized plants are shown by black bars, and non-S-fertilized plants are shown by white bars. Sample details are as in legend for Figure 2. Results are mean values of three replicates from independent areas within the plots. Error bars indicate se. The ±S comparisons were submitted to t test variance analysis with two-tailed distribution and two-sample equal variance, and asterisks represent significance at P < 0.05.
Sultr1;3, a high-affinity sulfate ion transporter expressed in the phloem of Arabidopsis (Yoshimoto et al., 2003), was expressed in all tissues and was unaffected by the S nutritional status of the plants (Fig. 5, F–J). In the grain, Sultr1;3 expression deceased at successive sample times after anthesis.
Sultr2;1, a low-affinity sulfate ion transporter expressed in the vascular tissue, the central cylinder, and the root cap of Arabidopsis (Takahashi et al., 2000), was also expressed in all tissues examined except the grain (Fig. 5, K–O). Up-regulation of expression was detectable in the roots, stems, and rachis of the −S plants. In shoots and leaves, Sultr2;1 expression was not influenced by S fertilization.
Expression of Sultr1;1 and Sultr1;3, but not Sultr2;1, was detected in grain tissues after anthesis (fourth harvest). Expression decrease with time and was higher under S-limiting conditions for Sultr1;1.
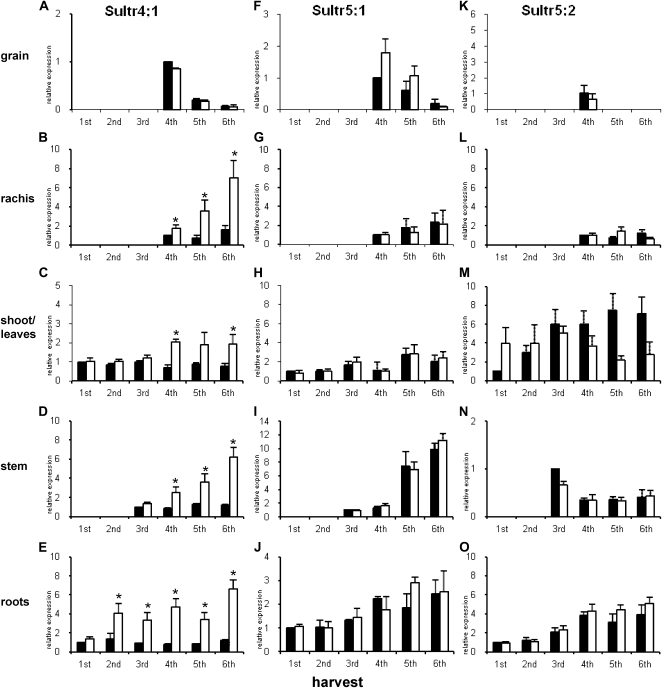
Expression of Sultr4;1, Sultr5;1, and Sultr5;2
Sultr4;1 is a sulfate ion transporter that, at least in Arabidopsis, is located in the tonoplast of vacuoles and facilitates the efflux of sulfate from the vacuole into the cytoplasm, influencing the capacity for vacuole storage of sulfate (Kataoka et al., 2004b). In all tissues except grain, Sultr4;1 expression increased relative to the control (+S) in the −S plants from the second harvest in the root tissues and following anthesis in the leaf, rachis, and stem (Fig. 6, A–E). Sultr4;1 expression decreased in the grain after anthesis.
Figure 6.
Expression analysis of Sult4;1, Sultr5;1, and Sultr5;2 by semiquantitative RT-PCR. Relative expression of Sultr4;1, Sultr5;1, and Sultr5;2 expression in relation to the first S-fertilized time point set to 1 is shown in A to E, F to J, and K to O, respectively. S-fertilized plants are shown by black bars, and non-S-fertilized plants are shown by white bars. Sample details are as in legend for Figure 2. Results are mean values obtained from three replicate sets of plants from independent areas within the plots. Error bars indicate se. The ±S comparisons were submitted to t test variance analysis with two-tailed distribution and two-sample equal variance, and asterisks represent significance at P < 0.05.
Sultr5;1, which belongs to the SulP gene family (Hawkesford, 2003) but has not been shown, to date, to have any specific transporter function, was expressed in all tissues but was not affected by S fertilization except in the floret at anthesis (fourth harvest) and grain (fifth harvest), where expression was higher in the −S plants (Fig. 6, F–J). Expression in the grain decreased during development, while there was a marked developmental increase in expression in rachis, stem, and root but not in leaf tissues.
Sultr5;2 (mot1), which has been implicated in the ability of Arabidopsis to accumulate Mo (Tomatsu et al., 2007; Baxter et al., 2008), was expressed in all tissues, with decreasing expression at successive sampling points under S-limiting conditions in the leaf fraction, paralleling increasing expression in the roots, which was higher in −S plants. Expression of Sultr5;2 was observed in florets at anthesis (fourth harvest) and not at any other time in grain tissues (caryopses).
DISCUSSION
S is essential to wheat crops, for both optimal growth and breadmaking quality, with crops requiring 2 to 3 kg S per tonne grain produced (Zhao et al., 1999). No S has been applied to one specific Broadbalk plot since 2000, which has resulted in a small but consistent effect on yield compared with the +S plot in this experiment (http://www.rothamsted.bbsrc.ac.uk/eRA/), but with a clear reduction in grain S concentration at harvest concomitant with an enrichment of Se and Mo. The decrease in grain S causes a higher N-S ratio (Godfrey et al., 2010) and will likely impact protein quality (Zhao et al., 1999).
There is good evidence that both selenate and molybdate are transported via sulfate transporters and that these anions will interact with the transporters competitively with one another (Leggett and Epstein, 1956; Fitzpatrick et al., 2008). The high-affinity sulfate transporter, SHST1, from S. hamata, which is primarily responsible for the uptake of sulfate from the soil solution (Smith et al., 1995), has definitively been shown to transport molybdate (Fitzpatrick et al., 2008). The enhancement of Se and Mo uptake under conditions of reduced S inputs (no S fertilizer) would be expected if low sulfate concentrations in the soil favored uptake of the other anions. In addition, it is well documented that expression of many sulfate transporters, including those involved in uptake, is enhanced by S limitation coincident with reduced tissue sulfate pools (Smith et al., 1997; Buchner et al., 2004a). This effect is also demonstrated in field-grown wheat in this study. It is probable that competition between ions for the transporters and the observed up-regulation of expression (Fig. 5) contributed to the observed increased uptake of both Se and Mo in the −S plants relative to the S-fertilized plants. Prior to the S fertilizer application, soil sulfate levels were similar in the two plots (data not shown) and no differential uptake would be expected, as was observed at the earliest sampling time points. In S-fertilized plots, following application, soil sulfate would be increased and would effectively compete against the much lower Se and Mo levels in the soil.
Induction of expression of Sultr1;1 in the roots occurred by the second harvest, particularly in the −S plants, coincident with an acceleration of plant growth (stem elongation stage). At this time point, there was a noticeable difference in the sulfate pools of all tissues between control and −S plants. The almost complete absence of sulfate pools in tissues from the nonfertilized plots indicated that sulfate taken up was directly assimilated and utilized for biosynthesis (Fig. 2, E–H).
The depletion of tissue sulfate content resulted in the induction of the inducible sulfate transporters in the respective tissues. As selenate and molybdate are either nonessential or only required in small amounts, the active sulfate transporters would transport these anions around the plant and into subcellular storage compartments such as the vacuole in place of free sulfate ions. Interestingly, the root tissues showed little enrichment of Se and Mo, indicating that the two elements were effectively translocated to the aboveground part of the plant. This would be facilitated by enhanced expression of the key sulfate transporters. The high expression of Sultr4;1, particularly in the roots, and thought to be responsible for vacuole unloading (Kataoka et al., 2004b), might mitigate against high accumulations of vacuolar anions under these conditions in the roots. During this period, any sulfate ions, as well as selenate and molybdate, would be directed toward the aboveground tissues and ultimately to the grain, and selenate followed this pattern.
In contrast, a large proportion of the Mo remained in the vegetative tissues, indicating that either the transport pathways were not effective for molybdate or that the Mo was in other storage pools not influenced by the remobilization processes. A member of the sulfate transporter gene family, Sultr5;2 (also named mot1), has been reported to be involved either in Mo uptake/translocation (Tomatsu et al., 2007) or specifically in Mo transport and shoot accumulation (Baxter et al., 2008). MOT1 has been localized to either the plasma membrane/vesicular fraction (Tomatsu et al., 2007) or the mitochondrial fraction (Baxter et al., 2008), and although apparently essential for Mo accumulation in Arabidopsis, a precise role remains unclear. Expression of a homolog was detected in wheat to our knowledge for the first time, and expression was both developmentally and nutritionally regulated with tissue-specific patterns of expression. The developmental increase in expression in roots was coincident with a decreasing Mo concentration (compare Figs. 3H and 6O). The highest concentrations in the leaf fraction were coincident with lower levels of expression (Figs. 3F and 6M). High expression appears to coincide with lower Mo concentration. However, the apparent restricted expression of Sultr5;2 in the grain had no correlation with the observed grain accumulation of Mo (Figs. 3E and 6K). Taken together, this indicates that while Sultr5;2 may be involved in leaf Mo accumulation, it is unlikely to be solely responsible for uptake and distribution in the maturing wheat plant. However, it is highly probable that Mo uptake will be via the high-affinity transporters (Fitzpatrick et al., 2008).
While S from aerial sources and from mineralization of soil organic matter provides the crop with marginally sufficient S to minimize yield losses in the nonfertilized plots, there is a major impact on the accumulation of Se and Mo. Efficient capture of S by the crop is in part facilitated by the up-regulation of the transporter systems. However, the absence of an excess of sulfate in the soil and enhanced expression of sulfate transporters result in a remarkable accumulation of Se and Mo as a result of their uptake in the form of the analogous anions, selenate and molybdate.
MATERIALS AND METHODS
Plant Materials
Winter wheat (Triticum aestivum var Hereward [RAGT Seeds]) was sampled from the Broadbalk continuous wheat experiment in 2008 from plots 9 (+S) and 14 (−S, after 2001) section 1 (last fallowed in 1965). The Broadbalk wheat experiment is located at Rothamsted, Hertfordshire, United Kingdom. Winter wheat was sown 400 seeds per m2 on October 15, 2007.
Nitrogen was applied to both plots in a single dressing in mid April at 192 kg ha−1 (the typical rate used in UK winter wheat). Plot 14 received KCl at a rate of 90 kg ha−1, and plot 9 received K2SO4 at the same amount. Plot 9 received 12 kg of magnesium as kieserite, and plot 14 received none since 1999; neither plot is limited in soil magnesium.
Harvesting was carried out at seven time points (Table I). Five to 10 plants were taken from each of three areas (pseudoreplicates) with a trowel to remove the whole plant, including roots, from soil. Roots were rinsed twice in deionized water to remove remaining soil. Roots as well as other separated plant parts, shoot and root (first and second harvest), leaf, stem, and root (third harvest), floret, rachis, leaf, stem, and root (fourth harvest), grain, rachis, leaf, stem, and root (fifth and sixth harvest), were frozen in liquid nitrogen in the field. Frozen plant tissues were ground to a fine powder with a mortar and pestle with liquid nitrogen. Ground plant tissues were stored at −80°C and used for RNA extraction and chemical analysis.
Table I. Plant harvesting.
Fertilizer was applied on April 4, 2008, as sulfate of potash and Kieserite.
| Harvest | Growth Stage (Code) | Date |
| First | Seedling growth, five leaves unfold (15) | April 9, 2008 |
| Second | Stem elongation, second node detectable (32) | April 28, 2008 |
| Third | Booting, flag leaf sheath opening (47) | May 28, 2008 |
| Fourth | Anthesis, anthesis halfway (64) | June 17, 2008 |
| Fifth | Milk development, early milk (73) | July 1, 2008 (2 wpa) |
| Sixth | Dough development, early dough (83) | July 15, 2008 (4 wpa) |
| Seventh | Ripening, caryopsis hard (92) | August 16, 2008 (60 d post anthesis) |
The seventh harvest comprised five main tillers from each area. The separated plant parts, grain, leaf, and stem, were dried at 80°C for 2 d and weighed. Dried plant materials were ground with a mill and used for chemical analysis.
Sulfate Determination
Ground plant tissue samples were freeze dried and used for sulfate extraction. One milliliter of deionized water was added to 0.02 g of dried tissue and incubated at 80ºC for 2 to 4 h with shaking. After incubation, the extracts were centrifuged and the supernatant was filtered into a 1.5-mL tube using a 0.2-μm membrane syringe filter. Extracted solutions were diluted further (15-fold) prior to analysis by ion chromatography (Dionex DX 500 with G50 gradient pump and ED 40 conductivity detector). The eluent was 1.8 mm Na2CO3 and 1.7 mm NaHCO3, which was pumped isocratically over an AG9SC guard column coupled to an AS9SC separation column.
Analysis of Elements
Ground plant tissue samples were transferred into weighed digestion test tubes and dried at 70°C for 2 d. Dried plant tissues (0.2–0.5 g) were digested with 5 mL of nitric acid:perchloric acid (87:13, v/v; 70% concentration, trace analysis grade; Fisher Scientific; Zhao et al., 1994). The digest solution samples were analyzed for Se and Mo by inductively coupled plasma mass spectrometry (ICP-MS) and for S by inductively coupled plasma atomic emission spectrometry (ICP-AES) analysis. Repeat samples were carried out every 10 samples; blanks and standard reference material (NIST 1567, a wheat flour) were used for quality control.
Inductively coupled plasma analysis was carried out using a 7500ce Octopole Reaction System ICP-MS apparatus (Agilent Technologies) to determine concentrations of Se and Mo. The sample introduction system consisted of a micromist glass concentric nebulizer, quartz Scott-type double-pass spray chamber at 2°C, and nickel sample (1 mm) and skimmer (0.4-mm cones). Operating parameters were optimized daily using a tune solution containing 1 μg L−1 cerium, lithium, tellurium, and yttrium. Other instrument conditions were radiofrequency forward power of 1,550, sample depth of 8.0 mm, carrier gas flow rate of 0.89 L min−1, reaction gas flow rate (H2) of 4 mL min−1 or (helium) of 4.5 mL min−1. An internal standard (500 μg L−1 germanium) was used to correct for signal drift. The analytical procedures gave satisfactory values for the standard reference materials. Concentrations of S were determined by an Accuris ICP-AES system (ARL).
RNA Extraction
RNA extraction was conducted by a modified hot phenol method (Verwoerd et al., 1989). One milliliter of hot (80°C) phenol:extraction buffer (1:1, v/v) was added to approximately 1 mL of frozen ground plant tissue and mixed. After further addition of 0.5 mL of chloroform:isoamyl alcohol (IAA; 24:1, v/v), mixing, and centrifugation for 5 min at 4°C, the aqueous phases were transferred to new tubes. After an additional chloroform:IAA extraction, the total RNA was precipitated by 2 m LiCl overnight at 4°C. After collection of the total RNA by centrifugation and a 70% ethanol wash, putative DNA contamination was removed by DNase treatment. After further purification by phenol:chloroform:IAA and chloroform:IAA extraction, the final total RNA was collected by ethanol precipitation and dissolved in diethylpyrocarbonate-treated water.
Semiquantitative Reverse Transcription-PCR
Sulfate transporter gene expression was analyzed by two-step semiquantitative reverse transcription (RT)-PCR. First-strand cDNA synthesis was performed with 2-μg aliquots of total RNA and a dT-adapter primer using Invitrogen SuperScript III according to the standard protocol except for a 2-h synthesis time. Subsequently, semiquantitative PCR was performed as a 15-μL reaction using 1 μL of each first-strand cDNA solution, specific primer combinations for the respective sulfate transporters (Table II), and Red Taq mix (Sigma-Aldrich) containing loading dye. The PCR amplification for expression analysis was stopped during the linear amplification phase of the expressed analyzed sulfate transporter. In all tissues apart from the grain, linear amplification of Sultr1;1, Sultr1;3, Sultr2;1, and Sultr4;1 was in the range of 20 to 30 cycles; for grain tissues, this was in the range of 32 to 38 cycles. The linear amplification of Sultr5;1 was between 30 and 36 cycles in all tissues, and the linear amplification of Sultr5;2 could be determined in the range of 30 to 36 cycles in roots and between 36 to 40 cycles in nonroot tissues. As a constitutive control, semiquantitative RT-PCR was performed using wheat actin2-specific oligonucleotide sense and antisense primers in nongrain tissues. For grain tissues, oligonucleotide sense and antisense primers corresponding to the proteasome subunit family protein (GenBank accession no. BQ806121) gene were used as a constitutive control (Wan et al., 2008). For both actin2 and the proteasome subunit family protein, linear amplification occurred in the range of 22 to 28 cycles in all tissues analyzed.
Table II. Sequences of primers used in this work.
| Gene | Accession/Gene Index No. | Primer Sequence (5′→3′) | Strand | Product Size |
| bp | ||||
| TaSultr1;1 | AJ512821 | ACGTATCCATCTGCACATAGG | Forward | 508 |
| GACCGATGGCTATATCCCTGG | Reverse | |||
| TaSultr1;3 | FN432835 | GGATTGACCATCGCAAGTCTCT | Forward | 505 |
| CCAGGAAAGATACGCCAATCAC | Reverse | |||
| TaSultr2;1 | TC366953/TC291347 | CCGGATCTCTATCCTCGTGCTA | Forward | 507 |
| GATGAAAGTCGCGTTGATGAAGC | Reverse | |||
| TaSultr4;1 | BT009340 | GCTGTCACTGGCCTGGTAGATT | Forward | 498 |
| CGCTATAGCAATCTGGATGTCG | Reverse | |||
| TaSultr5;1 | FN601348 | GGCCTCTCCTTCGCCTTCAC | Forward | 504 |
| TTCATGAGCCCCACGCTGAC | Reverse | |||
| TaSultr5;2 | FN601349 | AGGACCTTGGCAAGGGCAAG | Forward | 485 |
| ATGGTCACCGACACCGACGTC | Reverse | |||
| Ta actin | TC234027 | CCTTCAATGTTCCAGCCATGTA | Forward | 538 |
| ATAGTTGAGCCACCACTGAGCA | Reverse | |||
| Ta proteasome subunit | BQ806121 | CTTCTCCACGCGAATCAGCTA | Forward | 452 |
| CTGCTCGCAGCCGATAACTAC | Reverse |
PCR products (11 μL) were applied to a 1.2% (w/v) agarose gel of Tris-acetate EDTA buffer containing 0.3 μg mL−1 ethidium bromide. After electrophoresis, the gel was analyzed by a digital image system using Gene Snap software (Syngene, Synoptics Ltd.). The fluorescence values of products were analyzed by Gene Tools software (Syngene, Synoptics Ltd.). Gene expression values were determined as peak volumes. Specific gene expression values were normalized to the actin2 or proteasome subunit family protein expression value, and relative values (with respect to the −S at the first time point measured for any specific tissue) are presented. A comparison of absolute expression levels for the different transporters in different wheat tissues is presented by Buchner et al. (2010).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaSultr1;1a (AJ512821), TaSultr1;1b (AJ512820), TaSultr1;3 (BT009249), TaSultr2;1 (TC366953/TC291347), TaSultr3;1 (FN432835), TaSultr3;2 (FN599528), TaSultr3;3 (TC272130/TC259376), TaSultr3;4 (TC318325/TC314180), TaSultr3;5 (AM747385), TaSultr4;1 (BT009340), TaSultr5;1 (FN601348), TaSultr5;2 (FN601349), AtSultr1;1 (AB018695), AtSultr1;2 (AB042322), AtSultr1;3 (AB049624), AtSultr2;1 (AB003591), AtSultr2;2 (D85416), AtSultr3;1 (D89631), AtSultr3;2 (AB004060), AtSultr3;3 (AB023423), AtSultr3;4 (AB054645), AtSultr3;5 (AB061739), AtSultr4;1 (AB008782), AtSultr4;2 (AB052775), AtSultr5;1 (NM_106680), AtSultr5;2 or mot1 (NM_128127), Ta Actin (TC234027), and Ta Proteasome subunit (BQ806121).
Acknowledgments
We thank Caroline Shepherd for assistance with field harvesting.
References
- Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, et al. (2008) Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet 4: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, Alcock A, Alford J, Cartwright P, Foot I, Fairweather-Tait SJ, Hart DJ, Hurst R, Knott P, McGrath SP, et al. (2010) Selenium biofortification of high-yielding winter wheat (Triticum aestivum L.) by liquid or granular Se fertilisation. Plant Soil (in press) [Google Scholar]
- Broadley MR, White PJ, Bryson RJ, Meacham MC, Bowen HC, Johnson SE, Hawkesford MJ, McGrath SP, Zhao FJ, Breward N, et al. (2006) Biofortification of UK crops with selenium. Proc Nutr Soc 65: 169–181 [DOI] [PubMed] [Google Scholar]
- Buchner P, Elisabeth C, Stuiver E, Westerman S, Wirtz M, Hell R, Hawkesford MJ, De Kok LJ. (2004a) Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol 136: 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P, Parmar S, Kriegel A, Carpentier M, Hawkesford MJ. (2010) The sulfate transporter family in wheat: tissue-specific gene expression in relation to nutrition. Mol Plant (in press) [DOI] [PubMed] [Google Scholar]
- Buchner P, Prosser IM, Hawkesford MJ. (2004b) Phylogeny and expression of paralogous and orthologous sulphate transporter genes in diploid and hexaploid wheats. Genome 47: 526–534 [DOI] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. (2004c) Plant sulphate transporters: co-ordination of uptake, intracellular and long -distance transport. J Exp Bot 55: 1765–1773 [DOI] [PubMed] [Google Scholar]
- Combs GF. (2001) Selenium in global food systems. Br J Nutr 85: 517–547 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick KL, Tyerman SD, Kaise BN. (2008) Molybdate transport through the plant sulfate transporter SHST1. FEBS Lett 582: 1508–1513 [DOI] [PubMed] [Google Scholar]
- Godfrey D, Hawkesford MJ, Powers S, Millar S, Shewry PR. (2010) Nutritional effects on wheat grain composition and end use quality. J Agric Food Chem 58: 3012–3021 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ. (2000) Plant responses to sulfur deficiency and the genetic manipulation of sulfate transporters to improve S-utilization efficiency. J Exp Bot 51: 131–138 [PubMed] [Google Scholar]
- Hawkesford MJ. (2003) Transporter gene families in plants: the sulphate transporter gene family—redundancy or specialization? Physiol Plant 117: 155–165 [Google Scholar]
- Hawkesford MJ, Buchner P, Hopkins L, Howarth JR. (2003a) The plant sulfate transporter family: specialized functions and integration with whole plant nutrition. Davidian JC, Grill D, DeKok LJ, Stulen I, Hawkesford MJ, Schnug E, Rennenberg H, , Sulfur Transport and Assimilation in Plants: Regulation, Interaction and Signalling. Backhuys Publishers, Leiden, The Netherlands, pp 1–10 [Google Scholar]
- Hawkesford MJ, Buchner P, Hopkins L, Howarth JR. (2003b) Sulphate uptake and transport. Abrol YP, Ahmad A, , Sulphur in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 71–86 [Google Scholar]
- Hawkesford MJ, Zhao FJ. (2007) Strategies for increasing the selenium content of wheat. J Cereal Sci 46: 282–292 [Google Scholar]
- Hopper JL, Parker DR. (1999) Plant availability of selenite and selenate as influenced by the competing ions phosphate and sulfate. Plant Soil 210: 199–207 [Google Scholar]
- Kassis EE, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC. (2007) Characterization of a selenate-resistant Arabidopsis mutant. Plant Physiol 143: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. (2004a) Root-to-shoot transport of sulfate in Arabidopsis: evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H. (2004b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopsell DA, Kopsell DE. (2007) Selenium. Barker AE, Pilbeam DJ, , Handbook of Plant Nutrition. CRC Press, New York, pp 515–549 [Google Scholar]
- Leggett JE, Epstein E. (1956) Kinetics of sulfate absorption by barley roots. Plant Physiol 31: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, McGrath SP, Zhao FJ. (2008) Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytol 178: 92–102 [DOI] [PubMed] [Google Scholar]
- Poulton PR. (1995) The importance of long-term trials in understanding sustainable farming systems: Rothamsted experience. Aust J Exp Agric 35: 825–834 [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP. (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, van den Berg PJ, Belcher AR, Warrilow AGS. (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Na GN, Lahner B, Lee S, Leustek T, Pickering IJ, Salt DE. (2005a) Analysis of sulfur and selenium assimilation in Astragalus plants with varying capacities to accumulate selenium. Plant J 42: 785–797 [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Salt DE. (2005b) Selenium uptake, translocation, assimilation and metabolic fate in plants. Photosynth Res 86: 373–389 [DOI] [PubMed] [Google Scholar]
- Stroud JL, Li HF, Lopez-Bellido FJ, Broadley MR, Foot I, Fairweather-Tait SJ, Hart DJ, Hurst R, Knott P, Mowat H, et al. (2010) Impact of sulphur fertilisation on crop response to selenium fertilisation. Plant Soil (in press) [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sakurai N, Watanabe A, Leustek T, Engler JDA, Engler G, Montagu MV, Saito K. (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Terry N, Zayed AM, de Souza MP, Tarun AS. (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51: 401–432 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatsu H, Takano J, Takahashi H, Watanabe-Takahashi A, Shibagaki N, Fujiwara T. (2007) An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc Natl Acad Sci USA 104: 18807–18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Poole RL, Huttly AK, Toscano-Underwood C, Feeney K, Welham S, Gooding MJ, Mills C, Edwards KJ, Shewry PR, et al. (2008) Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H. (2003) Phloem-localizing sulfate transporter, sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131: 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, McGrath SP, Crosland AR. (1994) Comparison of 3 wet digestion methods for the determination of plant sulfur by inductively-coupled plasma-atomic emission-spectroscopy (ICPAES). Commun Soil Sci Plant Anal 25: 407–418 [Google Scholar]
- Zhao FJ, Hawkesford MJ, McGrath SP. (1999) Sulphur assimilation and effects on yield and quality of wheat. J Cereal Sci 30: 1–17 [Google Scholar]
- Zhu YG, Pilon-Smits EAH, Zhao FJ, Williams PN, Meharg AA. (2009) Selenium in higher plants: understanding mechanisms for biofortification and phytoremediation. Trends Plant Sci 14: 436–442 [DOI] [PubMed] [Google Scholar]