Abstract
The CLAVATA3/embryo-surrounding region (CLE) peptides control the fine balance between proliferation and differentiation in plant development. We studied the role of CLE peptides during indeterminate nodule development and identified 25 MtCLE peptide genes in the Medicago truncatula genome, of which two genes, MtCLE12 and MtCLE13, had nodulation-related expression patterns that were linked to proliferation and differentiation. MtCLE13 expression was up-regulated early in nodule development. A high-to-low expression gradient radiated from the inner toward the outer cortical cell layers in a region defining the incipient nodule. At later stages, MtCLE12 and MtCLE13 were expressed in differentiating nodules and in the apical part of mature, elongated nodules. Functional analysis revealed a putative role for MtCLE12 and MtCLE13 in autoregulation of nodulation, a mechanism that controls the number of nodules and involves systemic signals mediated by a leucine-rich repeat receptor-like kinase, SUNN, which is active in the shoot. When MtCLE12 and MtCLE13 were ectopically expressed in transgenic roots, nodulation was abolished at the level of the nodulation factor signal transduction, and this inhibition involved long-distance signaling. In addition, composite plants with roots ectopically expressing MtCLE12 or MtCLE13 had elongated petioles. This systemic effect was not observed in transgenic roots ectopically expressing MtCLE12 and MtCLE13 in a sunn-1 mutant background, although nodulation was still strongly reduced. These results suggest multiple roles for CLE signaling in nodulation.
In the symbiotic interaction between legume plants and rhizobia, root nodules develop within which the bacteria fix atmospheric nitrogen. Nodule development requires the spatiotemporal orchestration of developmental programs for infection and organ formation (Jones et al., 2007). In Medicago truncatula, the microsymbiont Sinorhizobium meliloti enters via curled root hairs and transcellular infection threads. While infection is taking place, inner cortical and pericycle cells divide and form the nodule primordium. The infection threads penetrate primordium cells, and bacteria are released into the plant cytoplasm within membrane-enclosed symbiosomes. Inside the symbiosomes, the bacteria differentiate into bacteroids and start the nitrogen fixation process (Jones et al., 2007). Meanwhile, an apical meristem develops and provides new cells for bacterial internalization. The nodules are of the indeterminate type and have a cylindrical shape.
The perception of bacterial signaling molecules, the nodulation factors (NFs), by specific LysM-type receptor-like kinases (RLKs) in the epidermis of the host plant elicits various responses to allow root hair invasion and cell division (Madsen et al., 2003; Radutoiu et al., 2003, 2007; Jones et al., 2007; Oldroyd and Downie, 2008). While the inner cortical cells divide, outer cortical cells arrest in the G2 phase of the cell cycle, resulting in cytoplasmic bridges, the preinfection threads, through which the infection threads grow (Yang et al., 1994; van Spronsen et al., 2001).
Opposite preexisting and NF-induced signal gradients have been proposed to rule the cortical cell responses (Smit et al., 1995; Heidstra et al., 1997; van Spronsen et al., 2001). Uridine and ethylene are diffusive signals originating from the vasculature that have been identified as positive and negative regulators, respectively. In white clover (Trifolium repens), the auxin flow within the root vasculature was transiently inhibited at the site of infection, leading to auxin accumulation in the cortical region where the nodule primordia form (Mathesius et al., 1998). A reduction in auxin flow has been confirmed by radioactive auxin tracer experiments for M. truncatula and vetch (Vicia faba) but not Lotus japonicus (Boot et al., 1999; Pacios-Bras et al., 2003; van Noorden et al., 2006; Wasson et al., 2006). Cytokinins are essential for nodule development because L. japonicus knockout mutants for the cytokinin receptor gene, LHK1, or M. truncatula transgenic plants with suppressed expression of the ortholog, CRE1, were defective in nodule primordia formation (Gonzalez-Rizzo et al., 2006; Murray et al., 2007). Additionally, a L. japonicus gain-of-function mutant for the LHK1 receptor provoked spontaneous nodules, indicating that cytokinin signaling is both necessary and sufficient for nodule formation (Tirichine et al., 2007).
Several components that link NF signaling to the initiation of cortical cell division have been identified. In M. truncatula, NF perception by LysM-type RLKs at the epidermis activates a signaling cascade that is mediated by the leucine-rich repeat (LRR)-RLK, Doesn't Make Infections2 (DMI2), and the nuclear potassium channel, DMI1. This signaling cascade triggers Ca2+ spiking in and around the nucleus. Decoding of this Ca2+ signature by a Ca2+ calmodulin-binding protein (DMI3) results in the activation of the transcription factors Nodulation Signaling Pathway1 (NSP1), NSP2, Ethylene-Responsive Binding Domain Factor Required for Nodulation1 (ERN1), and Nodule Inception (NIN; Schauser et al., 1999; Catoira et al., 2000; Borisov et al., 2003; Oldroyd and Long, 2003; Gleason et al., 2006; Andriankaja et al., 2007; Marsh et al., 2007; Middleton et al., 2007; Oldroyd and Downie, 2008). NSP2, ERN1, and NIN also play a role downstream of the cytokinin signaling to trigger cortical cell division (Tirichine et al., 2007; Frugier et al., 2008).
Nodule formation and functioning are energy-consuming processes, and legumes have evolved several strategies to control the number of nodules. One such strategy, autoregulation of nodulation (AON; Kosslak and Bohlool, 1984), is activated when the first nodules develop and involves systemic signals and shoot-controlled factors (Magori and Kawaguchi, 2009). Insight into this mechanism has been obtained by the identification of supernodulation mutants of soybean (Glycine max; nts-1), M. truncatula (sunn), L. japonicus (har1), and pea (Pisum sativum; sym29), each of which is deficient in an LRR-RLK that is required in shoots for AON (Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005). Auxin might be implicated in this process because sunn1 mutants display a higher level of long-distance shoot-to-root auxin transport than wild-type plants. Importantly, in contrast to the wild type, this transport is not reduced upon nodulation (van Noorden et al., 2006).
The LRR-RLKs responsible for AON belong to the evolutionary clade of group XI RLKs (Shiu and Bleecker, 2001) that includes the Arabidopsis (Arabidopsis thaliana) receptors CLAVATA1 (CLV1), PXY-like1 (PXL1) and PXL2, BARELY ANY MERISTEM1 (BAM1) to BAM3, and the putative tracheary element differentiation inhibitory factor (TDIF) receptor (TDR; Clark et al., 1997; DeYoung et al., 2006; Fisher and Turner, 2007; Hirakawa et al., 2008; Ogawa et al., 2008). These LRR-RLKs bind or putatively recognize CLV3/embryo-surrounding region (CLE) peptides (Hirakawa et al., 2008; Ogawa et al., 2008) that are a group of small (12–13 amino acids) secreted peptides derived from the C-terminal region of preproproteins (Mitchum et al., 2008; Oelkers et al., 2008). The Arabidopsis genome contains 32 gene family members, of which the best studied is CLV3, the peptide ligand for a CLV1-containing cell surface receptor complex. Recognition of the CLV3 peptide is important for stem cell homeostasis within the shoot apical meristem (SAM; Ogawa et al., 2008), whereas TDIF, a phloem-secreted CLE peptide, binds in vitro the PXY/TDR receptor expressed in the procambial cells and inhibits vascular differentiation (Hirakawa et al., 2008).
CLE peptides with related sequences exhibit redundancy, as proven by similarity in gain-of-function phenotypes (Strabala et al., 2006; Jun et al., 2008). At least two groups of CLE peptides can be distinguished. Group I peptides, exemplified by CLV3, result in premature root and shoot meristem growth arrest when exogenously applied or ectopically expressed, indicating that they are promoters of cellular differentiation. Members of group II, exemplified by TDIF, prevent cellular differentiation, as evidenced by the suppression of procambium-to-xylem transdifferentiation in zinnia (Zinnia elegans) cell cultures, and control the rate and orientation of vascular cell division (Ito et al., 2006; Etchells and Turner, 2010). Although these studies would suggest two groups with opposing functions, synergistic actions between these groups of peptides have been demonstrated (Whitford et al., 2008). In Arabidopsis, the proliferation of vascular precursor cells induced by the group II CLE41 peptides is enhanced by the addition of CLE6 peptides that otherwise have an inhibiting effect on root growth when applied at high concentrations and thus belong to group I. Moreover, genetic studies with clv3, clv1, and bam mutants reveal complex spatiotemporally controlled interactions between putative ligand/receptor complexes (DeYoung and Clark, 2008).
Cellular dedifferentiation and differentiation processes act at sequential stages of nodule development. At nodule initiation, pericycle and cortical cells dedifferentiate and divide (van Brussel et al., 1992; Timmers et al., 1999; van Spronsen et al., 2001). When sufficient cells encompassing the nodule primordia have formed, division ceases and the cells differentiate and become infected with rhizobia. Meanwhile, for indeterminate nodules, an apical meristem is established that supplies a constant pool of cells for bacterial infection. Hence, CLE peptides might not only be involved in AON but also regulate the (de)differentiation processes that control nodule development.
We analyzed the role of CLE peptides in M. truncatula nodulation. By specialized BLAST searches, nine peptide genes were identified in the Mt2.0 release in addition to the 16 previously discovered (Cock and McCormick, 2001; Oelkers et al., 2008). Two genes, designated MtCLE12 and MtCLE13, were differentially expressed during nodule development. The expression patterns hint at roles during (de)differentiation throughout nodule development. Moreover, when MtCLE12 and MtCLE13 were ectopically overexpressed, an inhibition of nodulation by long-distance signaling was observed. Furthermore, transgenic plants bearing roots expressing 35S:MtCLE12 or 35S:MtCLE13 had elongated petioles, a systemic effect not observed in sunn-1 mutants, although nodulation was still strongly hampered.
RESULTS
Search for Up-Regulated MtCLE Genes during Nodulation
Besides the 16 putative M. truncatula CLE genes described (Cock and McCormick, 2001; Oelkers et al., 2008), we searched for additional MtCLE genes in the EST databases (www.tigr.org/tdb/tgi/) and in the M. truncatula genomic data (Mt2.0) with a PAM30 tBLASTn homology-based algorithm to identify peptide sequences corresponding to the conserved CLE motif (Supplemental Table S1). Twenty-five MtCLE candidates were identified and designated MtCLE1 to MtCLE25 (Supplemental Fig. S1). The corresponding CLE preproproteins varied in length between 45 and 221 amino acids and had a high level of sequence divergence outside the CLE motif (Supplemental Fig. S1). Except for MtCLE3, all proteins contained an N-terminal signal peptide as predicted by HMM SignalP and neural networks (Bendtsen et al., 2004). Two MtCLE peptide candidates contained multiple CLE domains, namely MtCLE14 and MtCLE22 that had seven and three tandemly arranged CLE domains, respectively.
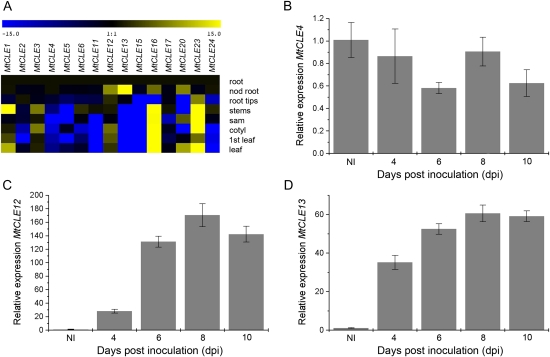
To determine tissue- or organ-specific expression, quantitative reverse transcription (qRT)-PCR was carried out for each MtCLE candidate on cDNAs derived from root elongation zones, nodulated roots (1 month post inoculation), root tips, stems, SAMs, cotyledons, first leaves, and mature leaves. cDNA of root elongation zones was used as the reference tissue. Transcripts were detected for 15 of the 25 identified MtCLE genes, of which 10 required 30 or more PCR cycles, indicating low transcript levels or cell-specific expression. MtCLE2, MtCLE4, MtCLE5, MtCLE11, MtCLE15, and MtCLE24 were mainly expressed in root tissues, MtCLE1, MtCLE16, and MtCLE23 were expressed predominantly in shoot tissues, and MtCLE6, MtCLE12, and MtCLE17 were expressed in several plant tissues (Fig. 1A). For MtCLE3 and MtCLE13, expression was restricted to stems and cotyledons and to nodulated roots, respectively, while for MtCLE20, transcripts were detected in nodulated roots, leaves, and SAMs. Four MtCLE genes (MtCLE12, MtCLE13, MtCLE16, and MtCLE20) were more abundantly transcribed in nodulated than in control roots (Fig. 1A). Biological repeats, however, only consistently confirmed the differential expression of MtCLE12 and MtCLE13.
Figure 1.
Expression analysis of MtCLE genes. A, Heat map of MtCLE expression in different tissues as measured by qRT-PCR. Samples are cDNA from root elongation zones (root), nodulated roots 1 month post inoculation (nod root), root tips, stems, SAMs (sam), cotyledons (cotyl), first leaves (1st leaf), and mature leaves (leaf). B to D, Expression analysis of MtCLE4, MtCLE12, and MtCLE13, respectively, by qRT-PCR on cDNA samples of zone I root tissues of uninoculated plants (NI) and at 4, 6, 8, and 10 dpi.
To study the temporal expression during nodule development, the relative transcript level of each MtCLE gene was analyzed at 4, 6, 8, and 10 d post inoculation (dpi). The elongation zone of uninoculated roots, the nodule initiation site, was used as the reference tissue. MtCLE12 expression was low at 4 dpi (Fig. 1C) but increased until 6 dpi and remained high until 10 dpi. MtCLE13 transcript increased until 10 dpi (Fig. 1D). As we were interested in the function of CLE peptides during nodulation, MtCLE12 and MtCLE13 were selected for further analysis and compared with MtCLE4, given its root-specific expression (Fig. 1, A and B).
As shown in Supplemental Table S2, the CLE peptide sequences of MtCLE12 and MtCLE13 differ by four amino acids. We analyzed the homology of MtCLE12, MtCLE13, and MtCLE4 to Arabidopsis CLE peptides. MtCLE12 and MtCLE13 were most similar to AtCLE1 to AtCLE7 and MtCLE4 to AtCLE9/10. Upon L. japonicus nodulation, three CLE genes were described to be up-regulated (Okamoto et al., 2009). Peptides derived from MtCLE12 and MtCLE13 only differed by three and one amino acids (an A/G change at position 4) from the identical LjCLE-RS1 and LjCLE-RS2, respectively.
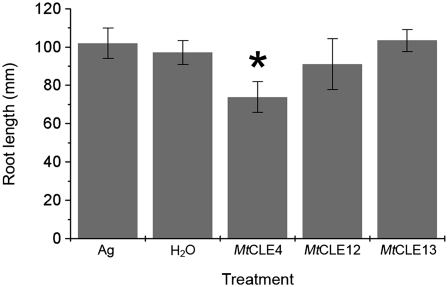
Effects of Exogenous Application of MtCLE4, MtCLE12, and MtCLE13 Peptides on Root Growth and Nodulation
To check the effect on root growth, we supplied 10 μm of chemically synthesized peptides corresponding to the CLE domain of these three proteins (MtCLE4p, MtCLE12p, and MtCLE13p; see “Materials and Methods”) exogenously to the primary roots of 2-d-old seedlings. Root length was measured after 8 d of growth. As controls, seedlings were grown on medium either supplemented or not (no peptide) with a synthetic 16-amino acid peptide corresponding to the C terminus of the Arabidopsis AGAMOUS protein. Primary root length for 16 plants per treatment was measured (Fig. 2). After addition of MtCLE12p or MtCLE13p, an average root length of 91.0 ± 13.2 and 103.4 ± 5.8 mm was observed, respectively, which did not differ significantly from the average root length with (101.9 ± 8.0 mm) and without (97.0 ± 6.2 mm) addition of the control peptide. For the seedlings treated with MtCLE4p, the average primary root length was 73.8 ± 8.0 mm, which is 24% shorter than the controls. The difference in number of lateral roots per primary root was not statistically significant. Exogenous peptide application (10–50 μm) had no effect on the nodule number.
Figure 2.
Effect of in vitro application of MtCLE4, MtCLE12, and MtCLE13 peptides on the root length of M. truncatula. The plants were grown for 6 d on plates containing 10 μm peptides (n = 39 for each treatment). As a control, plants were not treated (H2O) or were treated with the AGAMOUS (Ag) peptide. Data and error bars represent means ± sd. The asterisk marks a statistically different group.
MtCLE12 and MtCLE13 Expression Pattern in Roots and Developing Nodules
Spatial expression patterns of MtCLE12 and MtCLE13 in roots and developing nodules was investigated by promoter:GUS analysis and in situ hybridizations. A 2-kb region upstream of MtCLE12 and MtCLE13 was isolated based on the available genomic data (http://www.ncbi.nlm.nih.gov/) and cloned 5′ to the uidA gene. Transcriptional activation of the uidA gene was visualized by GUS staining.
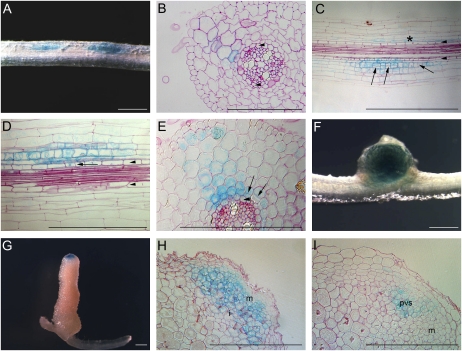
No GUS staining was observed in uninoculated transgenic roots. At 3 dpi, MtCLE13 expression was detected in cell clusters along root zones susceptible to rhizobial infection (Fig. 3A). At that time point, some incipient nodule primordia were present, but not all were at the same stage on a single root. As a result, between the most developed primordia and the root tip, many more incipient nodulation events occurred, and the closer to the root tip, the less developed the primordia. Inoculations with Sm2011-mRFP, carrying the monomeric red fluorescent protein (mRFP), revealed that pMtCLE13:GUS was expressed only in regions of bacterial infection. Careful comparison of the infection events with the GUS staining patterns indicated that the pMtCLE13:GUS expression could be followed down to nodulation events in which only curled colonized root hairs were visible. Sections of these infected regions revealed that these clusters corresponded to early nodulation stages without or with only a few cell divisions. Closest to the root tip, the pMtCLE13:GUS expression was mainly localized in inner cortical cells (Fig. 3, B and C), scattered in outer cortical cells (Fig. 3B), and absent in the pericycle and vascular tissues (Fig. 3, B and C). At positions where cell division was more pronounced in the inner cortex, the GUS staining pattern was more intense in the dividing cells (Fig. 3, C–E). A decreasing gradient of pMtCLE13:GUS expression radiated from the inner to the outer cortical cells and pericycle (Fig. 3, C–E). Expression was highest in the dividing cortical cells, but dividing pericycle cells displayed GUS staining as well. In round, young nodules, GUS staining was seen throughout the central tissue (Fig. 3F). In elongated nodules, it was restricted to the apical region, corresponding to the meristematic and early infection zones (Fig. 3, G and H). Although mostly all the meristematic cells were blue, in some nodules the expression was the highest in cells that corresponded to the provascular system (Fig. 3I). In situ hybridizations revealed a similar expression pattern, with low transcript levels in the meristem and cells of the early infection zone (Supplemental Fig. S2). For some nodules, MtCLE13 transcripts were more clearly detectable in cells of the provascular strands (Supplemental Fig. S2, C and D).
Figure 3.
MtCLE13 promoter activity during nodulation. A, Transgenic pMtCLE13:GUS root segment of the susceptible root zone I at 3 dpi. GUS staining was observed in patches along the root. B, Transverse section through a root segment at an initial stage of nodule formation when still no cell divisions occur. Blue staining is the highest in the inner cortical cells. Arrowheads indicate pericycle. C and D, Longitudinal sections through the root segment shown in A. In the incipient nodulation event (indicated by an asterisk in C), staining is seen in inner cortical cells but not in the pericycle cells. In slightly more developed nodule primordia, where cell divisions are visible in cortex and pericycle (examples marked by arrows in C and D), GUS staining is the strongest in the dividing cells of the cortex but also in the pericycle. E, Transverse section through a young developing nodule primordium at a stage similar to that in D. Cell division is clearly visible in the cortex (arrows). The pericycle cell file is indicated by arrowheads (in C–E). pMtCLE13:GUS is expressed in the cortex and at a low level in the pericycle, with the highest expression in the dividing inner cortical cells. F, A round, young nodule with GUS staining throughout the nodule tissue. G, GUS staining of a mature, elongated nodule. H, Longitudinal section through an elongated nodule. GUS staining is seen in the meristematic tissue and early infection zone. I, Longitudinal section through an elongated nodule, in which the expression is the highest in cells that presumably correspond to the provascular strands. i, Infection zone; m, meristem; pvs, provascular strands. Bars = 1 mm (A, B, D, F, and G) and 0.5 mm (C, E, H, and I).
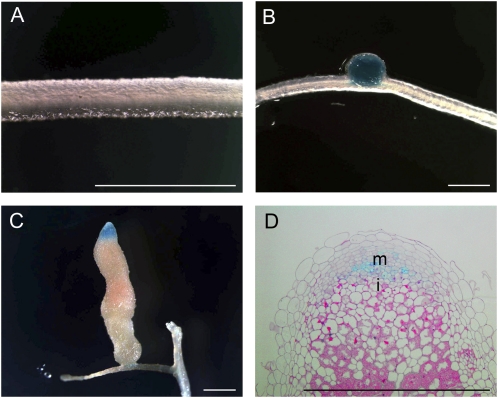
For pMtCLE12:GUS, no expression was detectable at the earliest stages of nodule development (Fig. 4A) but appeared in young round nodules (Fig. 4B). Later in nodule development, pMtCLE12:GUS was restricted to the apical zone of elongated nodules, similar to pMtCLE13:GUS (Fig. 4, C and D).
Figure 4.
MtCLE12 promoter activity during nodulation. A, pMtCLE12:GUS transgenic root segment of the susceptible root zone I at 3 dpi. No GUS staining is observed. B, GUS analysis of a young round nodule. MtCLE12 is expressed throughout. C, GUS analysis of a mature elongated nodule. Blue staining is observed in the apical part. D, Longitudinal section through C. i, Infection zone; m, meristem. Bars = 2 mm.
MtCLE12 and MtCLE13 Expression in Nodulation Mutants
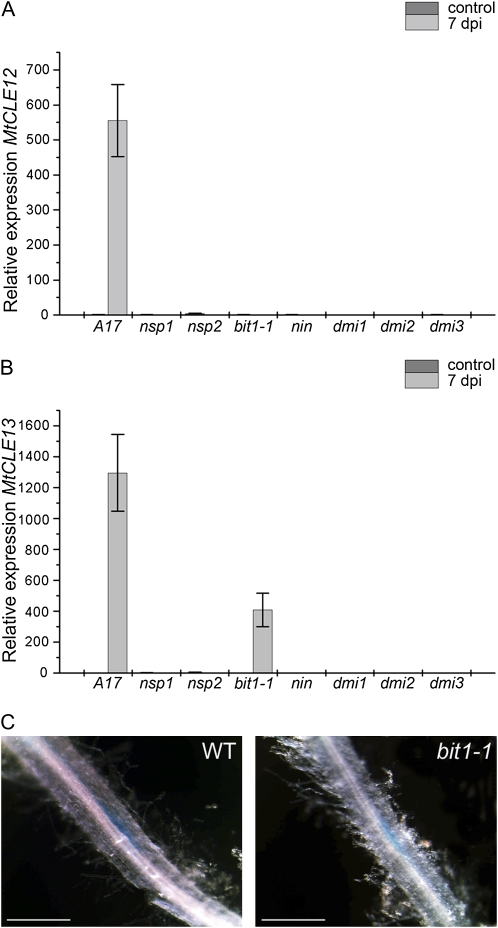
To investigate whether NF signaling is required for the induction of MtCLE12 and MtCLE13 expression, we analyzed the transcript levels by qRT-PCR before and after inoculation of nin, the ERN1 mutant branching infection threads1-1 (bit1-1), nsp1, nsp2, dmi1, dmi2, and dmi3 mutants (Fig. 5). The mutant lines did not develop nodules, except for bit1-1, which formed arrested primordia and infection foci (Andriankaja et al., 2007; Middleton et al., 2007). For MtCLE12, gene expression was induced upon inoculation in wild-type M. truncatula roots (Fig. 5A) but not in roots of the nodulation mutants, and likewise for MtCLE13 transcripts, except in bit1-1, albeit less abundantly than upon wild-type inoculation. The expression levels of MtCLE4 in the different mutants before and after inoculation were the same as those in the inoculated wild-type roots.
Figure 5.
MtCLE12 and MtCLE13 expression in nodulation mutants. A and B, Expression analysis of MtCLE12 and MtCLE13 by qRT-PCR on cDNA samples of zone I root tissues of wild-type plants (A17) and nsp1, nsp2, bit1-1, nin, dmi1, dmi2, and dmi3 mutants before inoculation (control) and at 7 dpi. C, pMtCLE13:GUS activity at 5 dpi in the roots of a wild-type plant (WT) and in a bit1-1 mutant. Bars = 1 mm. [See online article for color version of this figure.]
As a confirmation of the qRT-PCR analysis, pMtCLE13:GUS transgenic roots were generated in each of the mutant backgrounds and analyzed at 5 dpi. Because nodulation events are not synchronized in M. truncatula, consecutive early nodulation stages, ranging from stages with only a few cell divisions to young nodule primordia, could be observed along the root at 5 dpi. GUS staining revealed the typical MtCLE13 cluster pattern in the wild-type roots. The bit1-1 mutant was the only mutant in which blue-stained cortical regions were seen, corresponding to infection (Fig. 5C). The other mutants displayed no GUS staining. These data are in agreement with the qRT-PCR data and reveal that MtCLE13 expression is linked with the NF-induced cortical cell activation.
Induction of MtCLE12 and MtCLE13 Transcription by Auxin and Cytokinin
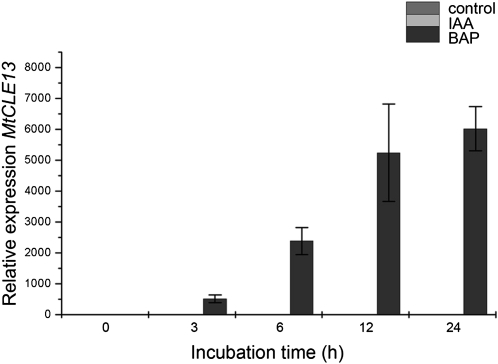
A correct auxin/cytokinin balance is a prerequisite for nodule formation (Oldroyd and Downie, 2008; Ding and Oldroyd, 2009). To determine whether auxins and/or cytokinins affect MtCLE12 and MtCLE13 expression, 10−6 m indole-3-acetic acid (IAA) or 10−7 m 6-benzylaminopurine (BAP) was supplemented to the growth medium of 5-d-old seedlings. The roots of 18 seedlings were harvested under each condition after 0, 3, 6, 12, 24, and 96 h. Roots from plates without hormone addition were used as a negative control.
No significant differences in MtCLE12 expression were detected in treated versus control roots. Auxin addition had no influence on the expression of MtCLE13 (Fig. 6), but in roots treated with 10−7 m BAP, MtCLE13 transcripts were up-regulated after a 3-h treatment and levels further increased with extended treatment times (up to 24 h; Fig. 6).
Figure 6.
Influence of auxin and cytokinin on MtCLE13 expression. qRT-PCR analysis of MtCLE13 expression in cDNA samples of roots grown in the presence of 10−6 m auxin (IAA) or 10−7 m cytokinin (BAP). Growth medium without hormones was used for the control plants. Samples of 5-d-old plants were taken at 0, 3, 6, 12, and 24 h after hormone addition.
Effect of Ectopic Expression of 35S:MtCLE12 and 35S:MtCLE13 on Nodulation
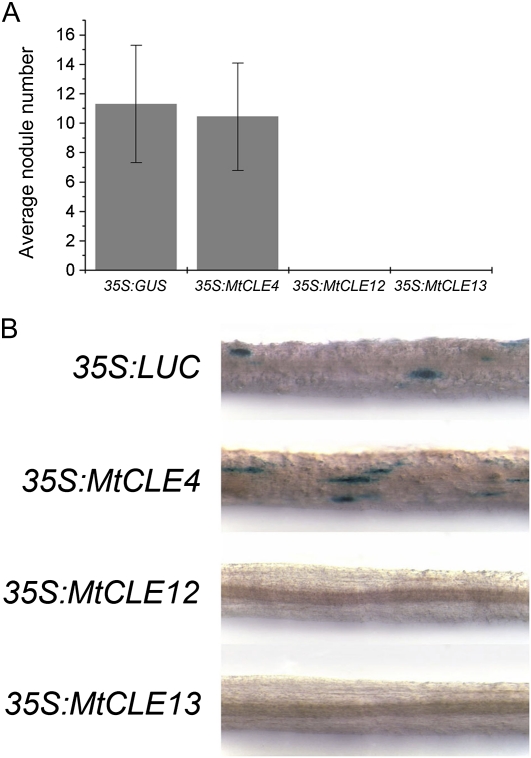
To analyze the functions of MtCLE4, MtCLE12, and MtCLE13, composite plants were made that carried transgenic roots ectopically overexpressing one of the three MtCLE genes. Nodulation of these transgenic roots was assessed at 21 dpi. Nodulation of control transgenic roots (35S:GUS) resulted on average in 11 ± 4 nodules per root (Fig. 7A). On the 35S:MtCLE4 transgenic roots, an average of 10 ± 4 nodules were counted. No nodules were detected on 35S:MtCLE12 and 35S:MtCLE13 transgenic roots (Fig. 7A). qRT-PCR analysis confirmed the ectopic overexpression of the respective constructs.
Figure 7.
Inhibition of nodulation in 35S:MtCLE12 and 35S:MtCLE13 transgenic roots. A, Average nodule number on roots expressing 35S:GUS, 35S:MtCLE4, 35S:MtCLE12, and 35S:MtCLE13 at 21 dpi (n = 28–66). Data and error bars represent means ± sd. B, pMtENOD11:GUS activity in roots expressing 35S:LUC, 35S:GUS, 35S:MtCLE12, and 35S:MtCLE13 at 5 dpi. No staining is visible in the 35S:MtCLE12 and 35S:MtCLE13 samples.
To determine at what stage these two CLE peptides affect nodulation, we investigated whether ectopic overexpression of MtCLE12 and MtCLE13 interferes with S. meliloti NF synthesis. For this purpose, the NF-overproducing strain Gmi6390:2011 (pMH682; Roche et al., 1991) was inoculated on 35S:MtCLE12 and 35S:MtCLE13 transgenic roots (Supplemental Fig. S3). Similar to results obtained with the wild-type strain, nodulation was totally abolished but was unaffected on 35S:MtCLE4 and control 35S:GUS roots.
To assess whether early NF signaling events still take place in roots ectopically overexpressing these CLE genes, we analyzed the transcription of the early marker, ENOD11, using a GUS transcriptional reporter (pENOD11:GUS) in 35S:MtCLE12 and 35S:MtCLE13 transgenic roots (Journet et al., 2001) and compared it with transgenic roots ectopically overexpressing either firefly luciferase (LUC) or MtCLE4. The transgenic roots were inoculated and stained with GUS at 3 dpi. In the 35S:LUC and 35S:MtCLE4 transgenic roots, GUS staining was observed in the epidermal cells at sites of incipient infection (Fig. 7B) but not in 35S:MtCLE12 and 35S:MtCLE13 transgenic roots (Fig. 7B). These results suggest that ectopic expression of MtCLE12 and MtCLE13, but not of MtCLE4, inhibits nodulation at the very early stages of NF signal transduction, before the onset of ENOD11 expression.
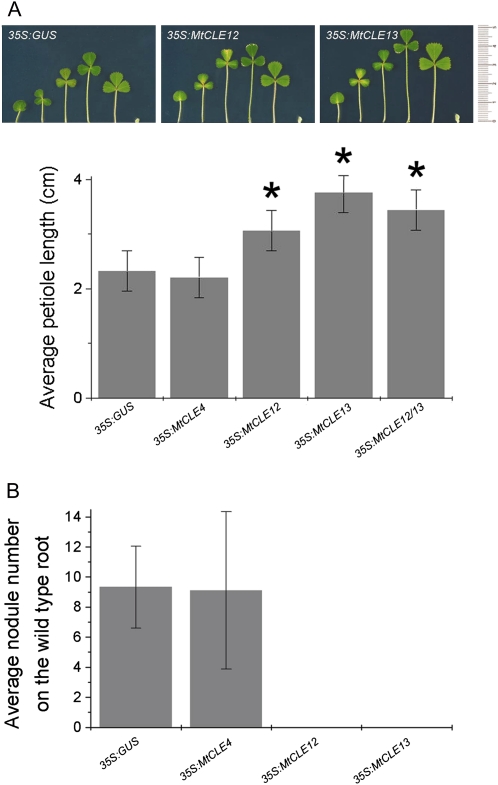
Long-Distance Effects of 35S:MtCLE12 and 35S:MtCLE13 Transgenic Roots on Wild-Type Shoots
While analyzing the effects of 35S:MtCLE12 and 35S:MtCLE13 on the nodule number, we noticed that the petioles in these composite plants were longer than those in controls. To quantify this observation, we compared plants with 35S:MtCLE12, 35S:MtCLE13, and, as controls, 35S:GUS and 35S:MtCLE4 transgenic roots. After growth in nitrogen-rich medium for 40 d after germination, the longest petiole on each plant was measured (Fig. 8A). For each construct, 60 plants were analyzed over three independent experiments. The average petiole length on control and 35S:MtCLE4 composite plants was 2.32 ± 0.36 and 2.21 ± 0.36 cm, respectively, whereas that on 35S:MtCLE12 and 35S:MtCLE13 composite plants was 3.06 ± 0.36 and 3.76 ± 0.36 cm, respectively. To analyze whether ectopic expression of MtCLE12 and MtCLE13 would have a synergistic effect on the petioles of the wild-type shoot, composite plants were made carrying transgenic roots containing both 35S:MtCLE12 and 35S:MtCLE13 constructs (35S:MtCLE12/13). The average petiole length was 3.44 ± 0.36 cm, which is not statistically different from the petiole length measured on composite plants with the 35S:MtCLE13 construct alone (Fig. 8A). qRT-PCR was used to confirm the ectopic overexpression of each construct.
Figure 8.
Long-distance effects of roots expressing 35S:MtCLE12 and 35S:MtCLE13 on petiole length and wild-type root nodulation of composite plants. A, Petioles of one 5-week-old composite plant with roots expressing 35S:GUS, 35S:MtCLE12, or 35S:MtCLE13. The graph represents the average petiole lengths of composite plants carrying 35S:GUS, 35S:MtCLE4, 35S:MtCLE12, 35S:MtCLE13, or 35S:MtCLE12 and 35S:MtCLE13 transgenic roots at 4 weeks post germination (n = 37–51). Data and error bars represent means ± se. Asterisks mark groups statistically different from the control (35S:GUS). B, Average nodule number at 7 dpi on the wild-type main roots of composite plants bearing additional transgenic 35S:GUS (n = 65), 35S:MtCLE4 (n = 10), 35S:MtCLE12 (n = 12), or 35S:MtCLE13 (n = 71) roots. Data and error bars represent means ± sd. [See online article for color version of this figure.]
A longer petiole length could either indicate a specific effect of MtCLE12 and MtCLE13 on petiole growth or an overall faster development. To distinguish between these two hypotheses, developmental stages were assigned to each composite plant according to the method described by Bucciarelli et al. (2006). The results revealed no clear differences in the rate of composite plant development (Supplemental Fig. S4). However, plants with roots expressing 35S:MtCLE13 were out of the range of the control plants and, therefore, deemed to be a little faster in development. Leaf sizes of 35S:MtCLE12 and 35S:MtCLE13 composite plants, calculated with ImageJ (http://rsb.info.nih.gov/ij/), showed no statistically significant differences compared with controls.
Long-Distance Effects of 35S:MtCLE12 and 35S:MtCLE13 Transgenic Roots on Nodulation of Wild-Type Roots
In the Agrobacterium rhizogenes transgenic root assay, cotransformed transgenic roots were identified by screening for GFP roots (see “Materials and Methods”). Often, non-GFP-expressing roots grow on the same composite plant, suggesting that these roots are not cotransformed and contain only endogenous A. rhizogenes T-DNA(s). We repeatedly saw no nodules on these roots when the composite plants carried 35S:MtCLE12 and 35S:MtCLE13 transgenic roots. These observations indicated that the expression of 35S:MtCLE12 or 35S:MtCLE13 in roots might have a negative systemic effect on the nodulation of roots that do not express the constructs but grow on the same plant. Therefore, genomic DNA was prepared from GFP-fluorescent and nonfluorescent roots, and the presence of the GFP gene was analyzed by PCR. In some composite plants (Supplemental Fig. S5B), nonfluorescent roots still contained the GFP gene. Therefore, these observations did not unequivocally demonstrate a negative systemic influence of 35S:MtCLE12 or 35S:MtCLE13 on nodulation.
With a different procedure (see “Materials and Methods”), composite plants were generated with small transgenic roots while the wild-type root was kept intact. At 7 dpi with Sm2011-GFP, a Nod¯ phenotype was observed on the primary wild-type root of 35S:MtCLE12 and 35S:MtCLE13 composite plants (Fig. 8B), and on average nine nodules were counted on the primary wild-type roots of 35S:MtCLE4 and 35S:GUS control plants. Given that plants were grown in an aeroponic system that confined the roots of all the plants in the same compartment, we can rule out that this phenotype is the result of peptide diffusion. These data show that ectopic overexpression of MtCLE12 and MtCLE13 abolishes nodulation not only locally in transgenic roots but also systemically in nontransformed roots of the same plant.
Analysis of the Long-Distance Responses in the sunn-1 Mutant Background
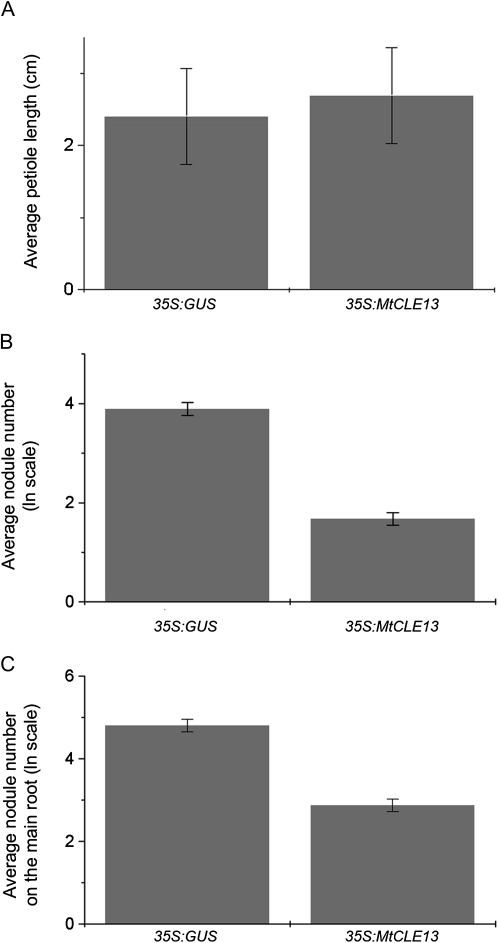
Nodulation is under the control of AON, a systemic response that involves shoot-controlled factors (Magori and Kawaguchi, 2009). Because of the long-distance responses observed when MtCLE12 and MtCLE13 are ectopically overexpressed, we investigated whether MtCLE12 and MtCLE13 are involved in AON and might be perceived by the SUNN receptor. Therefore, we tested whether the long-distance effects provoked by 35S:MtCLE13 expression on petiole length and on nodulation could be observed in the sunn-1 mutant background (Schnabel et al., 2005).
In contrast to the results in the wild-type background, the petiole length did not elongate in composite sunn-1 mutant plants containing roots expressing 35S:MtCLE13 (Fig. 9A). The average petiole length was 2.7 ± 0.7 cm, which is comparable to that of control plants (2.4 ± 0.7 cm; Fig. 9A).
Figure 9.
Local and systemic responses in the sunn-1 mutant. A, Average length of the longest petiole of composite plants with roots ectopically expressing either 35S:GUS or 35S:MtCLE13 at 4 weeks post germination (n = 12). Data and error bars represent means ± se. B, The ln (logarithmus naturalis) of the nodule number at 7 dpi on 35S:GUS or 35S:MtCLE13 transgenic roots (n = 10–12). C, The ln of the nodule number at 7 dpi on the wild-type main roots of plants bearing 35S:GUS or 35S:MtCLE13 transgenic roots (n = 17–24). Data and error bars represent means ± se.
On average, only 5.3 nodules occurred on sunn1,35S:MtCLE13-transformed roots versus 49.0 nodules on the sunn-1,35S:GUS roots. The long-distance repression of nodulation was investigated in the sunn-1 mutant background with the “hypocotyl-stabbing method” on plants growing under aeroponic conditions. Nodule numbers were counted at 7 dpi with Sm2011-GFP. Nontransformed primary roots of control plants had on average 74.0 nodules versus 17.6 nodules on the main root of sunn-1 plants that expressed 35S:MtCLE13 in transgenic roots (Fig. 9C). Together, these analyses in the sunn-1 mutant background revealed that the petiole elongation was SUNN dependent while the nodulation repression depended only partially on SUNN.
DISCUSSION
CLE Family in M. truncatula
With specialized BLAST searches, 25 MtCLE genes were identified in the Mt2.0 release of the M. truncatula genome that represents approximately 60% of the genome, and more genes are expected upon completion of the genome sequencing. A more accurate picture of the size of the CLE gene family derives from the analysis of L. japonicus and Arabidopsis. The genome of L. japonicus is comparable in size to that of M. truncatula (470 Mb), and currently 91.3% of its genome has been sequenced. Thus far, 39 CLE genes have been identified in L. japonicus (Sato et al., 2008; Okamoto et al., 2009), which is equivalent to the 32 CLE genes in the completely sequenced Arabidopsis genome (157 Mb; Cock and McCormick, 2001; Oelkers et al., 2008).
qRT-PCR detection of 15 MtCLE transcripts revealed six to be expressed specifically in roots, three in shoots, one in nodules, and five across several tissues, hinting at CLE peptide involvement in a multitude of developmental processes throughout the plant (Mitchum et al., 2008). For the remaining 10 MtCLE genes, no transcripts were detected because either they are very lowly expressed and/or respond to specific biotic or abiotic stimuli or they have low abundant cell type-specific expression or are pseudogenes.
As we were interested in CLE peptide function during nodulation, MtCLE12 and MtCLE13 were selected for further analysis because they were up-regulated in nodulated roots. The CLE domain sequences of MtCLE12 and MtCLE13 are very similar, which is indicative of redundant functions, but the gene expression patterns, although partially overlapping, are not identical. For instance, the MtCLE13 expression is specific for nodulation, while MtCLE12 transcripts occur also at low levels in root tips, cotyledons, and first leaves. Moreover, upon inoculation, the MtCLE13 expression is up-regulated much earlier than that of MtCLE12. The promoter of MtCLE13 functions in the NF-activated cortical cells; its activity is later restricted to the nodule primordium and is maintained through nodule maturity in the apical meristematic zone. In contrast, the MtCLE12 promoter activity occurs first in young round nodules and, similar to that of MtCLE13, is later restricted to the apical zone. Finally, the expression of MtCLE13 is induced rapidly by cytokinin, while that of MtCLE12 is unaffected.
In the genome of L. japonicus, three CLE genes have been identified to be up-regulated by rhizobial inoculation (Okamoto et al., 2009). LjCLE-RS1 and LjCLE-RS2 have a high degree of similarity with the CLE domain of MtCLE12 and MtCLE13, but no nodulation-related MtCLE gene was found with a CLE domain similar to that of LjCLE3. Interestingly, LjCLE-RS1 and LjCLE-RS2 are also up-regulated at early stages of nodulation. Based on sequence similarity and expression profiles, MtCLE13 and LjCLE-RS1/LjCLE-RS2 might exert a comparable function during indeterminate and determinate nodule development, respectively.
Studies on the putative CLE peptides of Arabidopsis (Ito et al., 2006; Ni and Clark, 2006; Strabala et al., 2006; Whitford et al., 2008) have shown a direct relationship between CLE domain sequence and induced phenotypes. MtCLE12 and MtCLE13 are most similar to a group of less characterized Arabidopsis CLE peptides (AtCLE1–AtCLE7) that are broadly produced with higher activity levels in the root (Sharma et al., 2003; Ito et al., 2006). Exogenous peptide addition did not suppress procambial-to-xylem cell transdifferentiation in a zinnia cell culture, while application of high, but not low, concentrations of peptides resulted in primary root meristem arrest (Ito et al., 2006; Strabala et al., 2006; Kinoshita et al., 2007; Whitford et al., 2008). Furthermore, ectopic overexpression of this group of peptides resulted in root elongation, mild wus loss-of-function phenotypes, mild distorted leaves, and dwarfing in later growth stages (Strabala et al., 2006). Upon exogenous peptide addition or ectopic overexpression in transgenic roots of MtCLE12 and MtCLE13, neither root growth arrest nor enhanced root elongation was observed. Because root growth analysis of A. rhizogenes-generated roots is technically difficult, phenotypic analysis of transgenic plants carrying heritable 35S:MtCLE12 or 35S:MtCLE13 constructs might help to decipher whether these CLE peptides do indeed induce root growth defects. The CLE domain sequence of MtCLE4 is most highly homologous to type I Arabidopsis CLE peptides, to which CLV3 belongs. This class of CLE peptides causes root meristem arrest upon exogenous application or ectopic overexpression. As expected, exogenous MtCLE4p application inhibited root growth by 24%.
Nodule-Related MtCLE12 and MtCLE13 Expression Is Linked with Differentiation and Dedifferentiation Processes
If MtCLE13 were to be involved in nodulation, its transcription would depend on early nodulation signaling components. Quantitative detection of MtCLE13 transcripts across different nodulation mutants revealed that functional DMI1, DMI2, DMI3, NSP1, NSP2, and NIN, but not ERN1, proteins are necessary for MtCLE13 expression. Interestingly, the ERN1 mutant, bit1-1, is the only mutant in which cell division is initiated and small arrested primordia are observed, indicative for active NF signaling toward the cortex and pericycle (Andriankaja et al., 2007; Middleton et al., 2007). MtCLE13 transcript expression was quickly induced by cytokinin, but not by auxin, the former being the most important hormone for primordium initiation (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). These data show that MtCLE13 transcript expression is positioned downstream of early NF signaling components and suggest a role downstream of cytokinin perception in the control of organ development.
pMtCLE13:GUS analysis indicated that MtCLE13 transcripts are expressed at very early stages of infection within nodulation-susceptible zones of the root cortex. The expression pattern is reminiscent of the signal gradient hypothesis, in which opposing signal gradients from the vasculature and from NF signaling at the epidermis would delimit the cellular landscape to form a nodule (Smit et al., 1995; Heidstra et al., 1997; van Spronsen et al., 2001). Because MtCLE13 is induced by cytokinin, its expression pattern presumably reflects an internal cytokinin gradient in the cortex. Consequently, MtCLE13 peptides might serve as subsequent intercellular signals to control downstream responses. Once cell division was initiated, MtCLE13 expression was the strongest in the dividing cortical and pericycle cells. CLE peptides have been shown to regulate the balance between cell division and differentiation (Ito et al., 2006; Kondo et al., 2006; Simon and Stahl, 2006; Whitford et al., 2008). Therefore, the MtCLE13 peptide gradient might maintain cell division and/or cell identity during the course of nodule development. Analysis of downstream responses to MtCLE13 peptide overexpression via genome-wide expression analysis might provide further insight into MtCLE13 function.
The expression of MtCLE12 is also correlated with cell division and differentiation. Expression of MtCLE12 during nodule development was first observed throughout the mature primordium, where MtCLE13 expression also was detected. The late expression pattern of MtCLE12 was confirmed by the absence of expression in the inoculated nodulation mutants, because none of the tested mutants developed nodule primordia of a stage corresponding with the MtCLE12 expression. By which trigger MtCLE12 expression is induced is thus far unknown, but application of neither cytokinin nor auxin could activate MtCLE12 expression. The transcription factor MtHAP2-1, located in nodule meristematic tissues, is essential for meristem differentiation (Combier et al., 2006). It would be interesting to determine whether a functional MtHAP2-1 is necessary for MtCLE12 induction or whether MtCLE12 regulates MtHAP2-1 expression.
In mature nodules, both MtCLE12 and MtCLE13 are expressed apically, in a zone comprising meristematic cells and cells of the early infection zone. In some nodules, MtCLE13 expression was higher in the provascular strands of the nodule meristem than in other meristematic cell types. Expression of MtCLE12 and MtCLE13 in the nodule apex again suggests a regulatory role for their encoded peptides in nodule meristem cell proliferation and/or differentiation.
MtCLE12, MtCLE13, or a Peptide with a Related Sequence Might Control Nodule Number
The lack of nodule development in transgenic roots ectopically overexpressing either MtCLE12 or MtCLE13 hints at a role for CLE signaling in controlling nodule number. Nodulation on 35S:MtCLE12 and 35S:MtCLE13, but not on 35S:MtCLE4, roots was totally abolished at the level of NF perception: no MtENOD11 expression and no developing nodules were seen upon inoculation of transgenic roots. This suppressive effect on nodulation was not phenocopied by exogenous application of synthetic peptides. One possible reason could be related to a role for posttranslational hydroxyprolination and subsequent arabinosylation on CLE peptide bioactivity, as suggested for Arabidopsis CLE peptides AtCLE1 to AtCLE7 (Strabala et al., 2006; Ohyama et al., 2009). Moreover, CLV3 arabinosylation has been found to be critical for high-affinity binding to the CLV1 ectodomain (Ohyama et al., 2009).
Nodule number is controlled by different processes. Our results suggest that CLE peptides are specifically involved in AON because nodule development was inhibited systemically in wild-type roots of composite plants containing roots ectopically overexpressing either MtCLE12 or MtCLE13. Moreover, ectopic expression of MtCLE12 and MtCLE13 in roots promoted petiole elongation in wild-type shoots of these composite plants. Taken together, these results demonstrate that MtCLE12 and MtCLE13 peptides can activate physiological responses at significant distances from the site of transgene expression. In the sunn-1 mutant background, no petiole elongation was observed, but the nodule number was strongly suppressed instead of a complete nodule development inhibition, like that observed in wild-type primary roots. The sunn-1 mutant might possess residual SUNN activity that could potentially allow MtCLE12 or MtCLE13 perception and downstream signaling at a level sufficient for only a mild suppression of nodulation but insufficient for promotion of petiole elongation. The ectopic expression of LjCLE-RS1 or LjCLE-RS2 of L. japonicus strongly reduces nodulation locally and systemically (Okamoto et al., 2009). This inhibitive effect on nodulation was abolished in the hypernodulating1-4 (har1-4) mutant roots, HAR1 being an orthologous gene to SUNN (Okamoto et al., 2009). The phenotypic differences between mutants could potentially be attributed to differences in allele functionality, because har1-4 bears a missense mutation in the LRR ectodomain and sunn-1 is mutated in the kinase domain (Kawaguchi et al., 2002; Krusell et al., 2002; Nishimura et al., 2002; Schnabel et al., 2005). Accordingly, a missense mutation in the LRR ectodomain of CLV1 has a stronger phenotype than a mutation in the kinase domain (Diévart et al., 2003). Moreover, the har1-4 mutant allele causes a more severe nodulation phenotype than the har1-5 allele, which is mutated in the intracellular kinase domain (Kawaguchi et al., 2002; Krusell et al., 2002; Nishimura et al., 2002; Schnabel et al., 2005). Thus, the differences in phenotype might simply be due to residual activities of the mutant protein or to a potentially dominant negative effect on interacting receptors, as for mutant alleles of CLV1 (Diévart et al., 2003).
Because SUNN belongs to class XI of LRR-RLKs, to which a few CLE peptide receptors belong (PXY/TDR and CLV1), it is tempting to propose that SUNN might be part of the receptor complex for MtCLE12 or MtCLE13. However, grafting studies have shown that SUNN and its orthologs are active in the shoot to invoke AON via a shoot-localized mechanism (Nutman, 1952; Delves et al., 1986; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005). Consequently, if the MtCLE12 and MtCLE13 peptides, which are produced upon nodulation in the root, could be perceived by SUNN, long-distance root-to-shoot translocation of the peptides would have to occur, which is in contradiction to the short-distance signaling activities proposed for many CLE peptides (Fukuda et al., 2007; Hirakawa et al., 2008; Whitford et al., 2008; Miwa et al., 2009; Stahl et al., 2009).
Alternatively, the various CLE peptide bioactivities observed upon ectopic overexpression of MtCLE12 or MtCLE13 in roots might mimic the signaling of structurally related CLE peptides that are produced in the shoot, the place of action of SUNN and its orthologs. Because the strong 35S promoter is expressed in the root vasculature, the ectopically produced CLE peptides might be systemically spread throughout the plant and be perceived by the SUNN protein in the shoot. As the SUNN proteins might also be produced in the root vasculature (Schnabel et al., 2005; Nontachaiyapoom et al., 2007), we cannot rule out that the misexpressed CLE proteins in the root vasculature are locally perceived by SUNN to provoke the observed long-distance effects. The shoot-expressed CLE gene might be MtCLE12 itself, because expression analysis has revealed that MtCLE12 is also expressed in first leaves and cotyledons, the site of SUNN activity. In this scenario, MtCLE12 and MtCLE13 activity within the nodule would not be linked with AON but rather with balancing proliferation and differentiation, as supported by the observed expression patterns.
It is equally possible that MtCLE12 and MtCLE13 perception occurs locally in the root, resulting in secondary signals that travel to the shoot, where they are recognized to provoke AON and petiole elongation. Interestingly, the expression of MtCLE12 and MtCLE13 coincides with the activation and progression of AON that is first initiated when the first nodule primordia are formed and is strengthened as more nodules develop (Nutman, 1952; Delves et al., 1986; Takats, 1990; Wopereis et al., 2000; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005; Li et al., 2009). Besides the long-distance effect on nodulation, 35S:MtCLE12 and 35S:MtCLE13 transgenic roots induced elongation of the petioles of the composite plants. Insights into petiole growth might hint at downstream responses of MtCLE12 and MtCLE13 signaling. Petiole length elongation is influenced by auxin, ethylene, abscisic acid, and gibberellins (Cox et al., 2004; Millenaar et al., 2009; Pierik et al., 2009).
In conclusion, CLE peptides most probably play different roles in nodulation. Expression patterns hint at roles during cellular differentiation processes, both at the onset of nodulation and later during nodule meristem development and subsequent homeostasis. Moreover, intertwined or not, the functional analyses imply a role for MtCLE peptides in AON. In Arabidopsis, the CLE peptide signaling is intricate and mediated by different receptor complexes (Miwa et al., 2009; Stahl et al., 2009). In silico analysis of the M. truncatula genome has revealed additional genes that belong to class XI of LRR-RLKs, and expression profiling indicates that they are up-regulated in the nodule. Future studies will investigate the individual role of each of these peptides and their corresponding receptors during the nodulation process and will provide valuable insight into nodule development, nodule cell type determination, and regulation of nodule number.
MATERIALS AND METHODS
Biological Material
Medicago truncatula ‘Jemalong A17’ and ‘J5’ as well as nin, bit1-1, nsp1, nsp2, dmi1, dmi2, dmi3, and sunn-1 mutants (Catoira et al., 2000; Oldroyd and Long, 2003; Marsh et al., 2007; Middleton et al., 2007) and pENOD11:GUS transgenic seeds (Journet et al., 2001) were grown and inoculated as described (Mergaert et al., 2003). Sinorhizobium meliloti 1021, Sm1021 pHC60-GFP (Cheng and Walker, 1998), Sm1021 pQE81-dsRedT3 (Bevis and Glick, 2002), Sm2011 pBHR-mRFP (Smit et al., 2005), Sm2011 pHC60-GFP (Cheng and Walker, 1998), and Sm2011 pMH682-Gmi6390 (Roche et al., 1991) were grown at 28°C in yeast extract broth medium (Vervliet et al., 1975) supplemented with 10 mg L−1 tetracycline for the Sm1021 pHC60-GFP, Sm1021 pQE81-dsRedT3, Sm2011 pBHR-mRFP, Sm2011 pHC60-GFP, and Sm1021 pMH682-Gmi6390 strains.
PCR fragments corresponding to the full-length open reading frames of MtCLE4, MtCLE12, and MtCLE13 were amplified from M. truncatula cDNA and cloned in pB7WG2D driven by the cauliflower mosaic virus 35S promoter (De Loose et al., 1995; Karimi et al., 2002). The vector pK7m34GW2-8m21GW3D was used for the simultaneous ectopic expression of MtCLE12 and MtCLE13 (Karimi et al., 2007). For promoter:GUS analysis, a 2-kb region upstream of MtCLE12 and MtCLE13 was isolated from genomic DNA based on the available genomic data (http://www.ncbi.nlm.nih.gov/). The promoters were fused to the uidA gene in pKm43GWRolDC1 (Karimi et al., 2002). Primers used for amplification are presented in Supplemental Table S3.
For the qRT-PCR analysis, M. truncatula J5 plants were grown in vitro in square petri dishes (12 × 12 cm) on nitrogen-poor SOLi agar (Blondon, 1964). Root tips, SAMs, cotyledons, and first leaves were harvested after 7d; mature leaves, stems, and roots from plants grown in perlite and watered with nitrogen-poor SOLi medium after 1 month; and nodulated roots from plants inoculated with Sm1021 pHC60-GFP after 1 month. For the analysis of temporal expression during nodulation, nodules were obtained 4 to 10 dpi from plants grown in pouches, watered with nitrogen-poor SOLi medium, and inoculated with Sm1021 pHC60-GFP. Infection threads were visible from 4 dpi on, nodule primordia at 6 dpi, and small nodules at 8 dpi. Two days later, at 10 dpi, slightly bigger nodules were observed. Tissue was collected by visualizing the green fluorescent bacteria with a MZFLII stereomicroscope (Leica Microsystems) equipped with a blue light source and a Leica GFP Plus filter set (λex = 480/40, λem = 510 nm LP barrier filter). Zone I of uninoculated roots was isolated at the same developmental stage as the 4-dpi stage.
In Silico Identification of M. truncatula CLE Genes
BLAST searches were done at The Institute for Genomic Research (www.tigr.org/tdb/tgi/) or at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/). The CLE family was identified by repetitive searches similar to those conducted by Cock and McCormick (2001). Repetitive searches were done with the Medicago Gene Index (MGI) at the Dana-Farber Cancer Institute (release 9.0) first with the tBLASTn PAM30 algorithm for the Arabidopsis CLE box consensus (RXXPXXPXPXH). The first identified sequence, MtCLE1 (GenBank EST AW586793), which was confirmed to encode a MtCLE-like peptide, was based on the predicted peptide length (less than 150 amino acids), the presence of a C-terminally localized CLE box, and an N-terminal signal peptide as predicted by HMM SignalP and neural networks (Bendtsen et al., 2004). This first sequence was used to repeat the same search, each time with an additional homologous CLE box sequence, until no unknown family members were found in the EST data. These CLE box sequences were used in the same iterative BLAST searches to identify additional putative CLE peptides from the partially completed genomic sequence (Mt2.0). Sequences were aligned with AlignX within the VectorNTI Advance version 10 suite of programs (http://www.invitrogen.com).
RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. After a DNase treatment, the samples were purified through NH4Ac (5 m) precipitation, quality controlled, and quantified with a Nanodrop spectrophotometer (Isogen). RNA (2 μg) was used for cDNA synthesis with the SuperScript Reverse Transcriptase Kit (Invitrogen). The samples were diluted 50 times and stored at −20°C until further use. The qRT-PCR experiments were done on a LightCycler 480 (Roche Diagnostics), and SYBR Green was used for detection. All reactions were done in triplicate and averaged. The total reaction volume was 5 μL (2.5 μL of master mix, 0.25 μL [5 μm] of each primer, and 2 μL of cDNA). Cycle threshold values were obtained with the accompanying software, and data were analyzed with the 2−ΔΔCt method (Livak and Schmittgen, 2001). The relative expression was normalized against the constitutively expressed 40S ribosomal S8 protein (TC100533; MGI). Primers used (Supplemental Table S3) were unique in the MGI version 9.0 and the Medicago EST Navigation System databases (Journet et al., 2002). Each experiment was repeated at least three times with independent biological tissue.
Statistical Analysis
To estimate the genotype effects on developmental stage, petiole length, and root length, the linear mixed model (random terms underlined) y = μ + genotype + experiment + ϵ was fitted to the data, where y represents the variable, μ is the overall mean, genotype is the fixed genotype effect, experiment represents random experimental effects, and ϵ is the random error. Statistical significance of genotype effects was assessed by a Wald test. In the case of the nodule number in sunn-1 plants, a generalized linear mixed model, of the form y = μ + genotype + experiment + ϵ with a Poisson distribution and a logarithmic link, was fitted to the data. Again, the statistical significance of genotype effects was assessed by a Wald test. All analyses were done with Genstat (http://www.vsni.co.uk/software/genstat/).
In Vitro Application of MtCLE Synthetic Peptides, Auxins, and Cytokinins
Peptides (AGAMOUS, APNNHHYSSAGRQDQT; MtCLE4, KRGVpSGANPLHNR; MtCLE12, DRLSpGGpNHIHN; and MtCLE13, DRLSpAGpDPQHNG; lowercase p indicates hydroxylated Pro), with a purity greater than 89% (ServiceXS), were dissolved in a filter-sterilized sodium phosphate buffer (pH 6, 50 mm [43.5 mm NaH2PO4 and 6.2 mm Na2HPO4]). Two-day-old Jemalong J5 seedlings were grown in vitro in square petri dishes (12 × 12 cm) on agar HP 696-7470 (Kalys) containing the SOLi medium supplemented with 1 mm NH4NO3 and 10 μm peptides (Fiers et al., 2005). The plants were cultured at 25°C with a 16-h photoperiod and 70 μE m−2 m−1 light intensity per day. The roots were covered with aluminum foil for light protection. After 8 d of growth, the root length of 16 plants under each condition was measured from the root tip of the primary root to the base of the hypocotyls with the ImageJ 1.40b program (http://rsb.info.nih.gov/ij/). The experiment was repeated three times with comparable results.
Auxins (10−6 IAA) and cytokinins (10−7 BAP) were diluted in dimethyl sulfoxide and supplemented to the medium of 5-d-old, in vitro-grown plants. As a control, plants were grown without supplemented hormones. The growth conditions of the seedlings were the same as above. After 0, 3, 6, 12, and 24 h of incubation, the roots of 18 plants under each condition were harvested and analyzed by qRT-PCR. The experiment was repeated twice with comparable results.
Agrobacterium rhizogenes-Mediated Transgenic Root Transformation
The protocol was adapted from Boisson-Dernier et al. (2001). Approximately 48 h after germination, the radicle was sectioned at 5 mm from the root tip with a sterile scalpel. Sectioned seedlings were infected by coating the freshly cut surface with the binary vector-containing A. rhizogenes Arqua1 strains. The A. rhizogenes strain was grown at 28°C for 2 d on solid yeast extract broth medium with the appropriate antibiotics (Quandt et al., 1993). The infected seedlings were placed on agar (Kalys) containing the SOLi medium supplemented with 1 mm NH4NO3, in square petri dishes (12 × 12 cm) placed vertically for 5 d at 20°C with a 16-h photoperiod and light at 70 μE m−2 s−1. Subsequently, plants were placed on the same medium between brown paper at 25°C and under identical light conditions. One and 2 weeks later, plants were screened for transgenic roots and characterized by GFP fluorescence with a MZFLII stereomicroscope (Leica Microsystems) equipped with a blue light source and a Leica GFP Plus filter set. One main transgenic root was retained per composite plant. Four weeks after infection, plants were transferred to an aeroponic system, pouches, or perlite-containing pots and incubated with SOLi medium. Three to 7 d after planting, composite plants were inoculated. The petiole lengths were measured on plants grown under the same conditions but incubated with nitrogen-containing ISV medium (E.-P. Journet, D. Barker, M. Harrison, and E. Kondorosi, unpublished data). Forty days after germination, the longest petiole of each plant was scored by ImageJ (http://rsb.info.nih.gov/ij/).
In some experiments, the main root was kept on the juvenile plant and infected by stabbing the hypocotyls with a fine needle containing an A. rhizogenes culture and cotransformed as described above, after which the plants were grown for 2 weeks at 25°C with a 16-h photoperiod and light at 70 μE m−2 s−1. After the plants had been transferred to an aeroponic system for 7 d, nodulation was analyzed on the main, untransformed root of plants bearing GFP-positive hairy roots.
Histochemical Localization of GUS Activity
GUS activity in cotransformed roots and nodules was analyzed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate (Van den Eede et al., 1992). Roots and nodules were vacuum infiltrated during 20 min and subsequently incubated in GUS buffer at 37°C. Incubation lasted 3.5 and 7 h for pMtCLE13-GUS and pMtCLE12-GUS, respectively. After staining, root nodules were fixed, dehydrated, embedded with the Technovit 7100 kit (Heraeus Kulzer) according to the manufacturer's instructions, and sectioned with a microtome (Reichert-Jung). The 3-μm-thick sections were mounted on coated slides (Sigma-Aldrich). For tissue-specific staining, sections were submerged in a 0.05% (w/v) ruthenium red solution (Sigma-Aldrich), washed in distilled water, and dried. Finally, sections were mounted with Depex (BDH Chemicals). Photographs were taken with a Diaplan microscope equipped with bright- and dark-field optics (Leitz). GUS activity of pENOD11:GUS roots was visualized after 7 h of incubation.
In Situ Hybridization
Ten-micrometer sections of paraffin-embedded nodules were hybridized as described (Goormachtig et al., 1997). Nodules were harvested, incubated in fixation buffer, and maintained twice for 15 min under vacuum. A 35S-labeled antisense probe against the complete open reading frame of MtCLE13 was produced according to standard procedures (Sambrook et al., 1989). The probe was cloned into pBluescript KS+ (Stratagene) and further digested with HindIII restriction enzyme to yield templates for radioactive antisense probe production with T3 RNA polymerase (Invitrogen).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of the MtCLE genes.
Supplemental Figure S2. MtCLE13 transcript accumulation in mature nodules by in situ hybridization.
Supplemental Figure S3. Average nodule number on roots ectopically expressing GUS, MtCLE4, MtCLE12, or MtCLE13 at 11 dpi with a NF-overproducing S. meliloti strain.
Supplemental Figure S4. Log10 of the developmental stage of composite plants carrying roots ectopically expressing GUS, MtCLE4, MtCLE12, MtCLE13, or MtCLE12 and MtCLE13.
Supplemental Figure S5. Detailed genotypic analysis of transgenic roots obtained by A. rhizogenes transformation.
Supplemental Table S1. MtCLE identification.
Supplemental Table S2. Comparison of the CLE peptide sequences of MtCLE4, MtCLE12, and MtCLE13 with the CLE peptide sequences from Arabidopsis CLE genes and from LjCLE-RS1, LjCLE-RS2, and LjCLE3.
Supplemental Table S3. Primers used in the analysis.
Supplementary Material
Acknowledgments
We thank René Geurts (Wageningen University), Pascal Gamas and Clare Gough (Institut de la Recherche Agronomique-Toulouse), Doug Cook (University of California, Davis), and Giles Oldroyd (John Innes Institute) for S. meliloti strains and M. truncatula mutants, and our colleagues Annick De Keyser and Christa Verplancke for skillful technical assistance, Lorin Spruyt, Katja Katzer, Assia Saltykova, and Jorik Verbiest for their input during their master projects and theses, Wilson Ardiles for sequence analysis, Marnik Vuylsteke for help with the statistical analysis, Giel Van Noorden for critical reading of the manuscript, and Martine De Cock and Karel Spruyt for help in preparing the paper and figures, respectively.
References
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de CarvalhoNiebel F. (2007) AP2ERF transcription factors mediate Nod factor-dependent Mt ENOD11 activation in root hairs via a novel cisregulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS. (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20: 83–87 [DOI] [PubMed] [Google Scholar]
- Blondon F. (1964) Contribution à l'étude du développement de graminées fourragères: ray-grass et dactyle. Rev Gen Bot 71: 293–381 [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boot KJM, van Brussel AAN, Tak T, Spaink HP, Kijne JW. (1999) Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Mol Plant Microbe Interact 12: 839–844 [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciarelli B, Hanan J, Palmquist D, Vance CP. (2006) A standardized method for analysis of Medicago truncatula phenotypic development. Plant Physiol 142: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet E-P, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-P, Walker GC. (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180: 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S. (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J-P, Frugier F, de Billy F, Boualem A, ElYahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP21 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MCH, Benschop JJ, Vreeburg RAM, Wagemaker CAM, Moritz T, Peeters AJM, Voesenek LACJ. (2004) The roles of ethylene, auxin, abscisic acid, and gibberellin in the hyponastic growth of submerged Rumex palustris petioles. Plant Physiol 136: 2948–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Loose M, Danthinne X, Van Bockstaele E, Van Montagu M, Depicker A. (1995) Different 5′ leader sequences modulate β-glucuronidase accumulation levels in transgenic Nicotiana tabacum plants. Euphytica 85: 209–216 [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. (1986) Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors. Plant Physiol 82: 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. (2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45: 1–16 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Clark SE. (2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE. (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Oldroyd GED. (2009) Positioning the nodule, the hormone dictum. Plant Signal Behav 4: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Turner SR. (2010) The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C-M. (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Turner S. (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K. (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci 13: 115–120 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Hirakawa Y, Sawa S. (2007) Peptide signaling in vascular development. Curr Opin Plant Biol 10: 477–482 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GED. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormachtig S, AlvesFerreira M, Van Montagu M, Engler G, Holsters M. (1997) Expression of cell cycle genes during Sesbania rostrata stem nodule development. Mol Plant Microbe Interact 10: 316–325 [DOI] [PubMed] [Google Scholar]
- Heidstra R, Yang WC, Yalcin Y, Peck S, Emons A, van Kammen A, Bisseling T. (1997) Ethylene provides positional information on cortical cell division but is not involved in Nod factorinduced root hair tip growth in Rhizobium-legume interaction. Development 124: 1781–1787 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E-P, ElGachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, GianinazziPearson V. (2001) Medicago truncatula ENOD11: a novel RPRPencoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Journet E-P, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer M-J, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al. (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Fiume E, Fletcher JC. (2008) The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci 65: 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY™ vectors for Agrobacteriummediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, ImaizumiAnraku H, Koiwa H, Niwa S, Ikuta A, Syono K, Akao S. (2002) Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 15: 17–26 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. (2007) Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48: 1821–1825 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kosslak RM, Bohlool BB. (1984) Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiol 75: 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Li D, Kinkema M, Gresshoff PM. (2009) Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. J Plant Physiol 166: 955–967 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception in rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Magori S, Kawaguchi M. (2009) Longdistance control of nodulation: molecules and models. Mol Cells 27: 129–134 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED. (2007) Medicago truncatula NIN is essential for rhizobialindependent nodule organogenesis induced by autoactive calcium/calmodulindependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Sautter C, Rolfe BG, Djordjevic MA. (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. (2003) A novel family in Medicago truncatula consisting of more than 300 nodulespecific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, van Zanten M, Cox MCH, Pierik R, Voesenek LACJ, Peeters AJM. (2009) Differential petiole growth in Arabidopsis thaliana: photocontrol and hormonal regulation. New Phytol 184: 141–152 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Davis EL. (2008) Diverse and conserved roles of CLE peptides. Curr Opin Plant Biol 11: 75–81 [DOI] [PubMed] [Google Scholar]
- Miwa H, Kinoshita A, Fukuda H, Sawa S. (2009) Plant meristems: CLAVATA3/ESRrelated signaling in the shoot apical meristem and the root apical meristem. J Plant Res 122: 31–39 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE. (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu G-J, Kouchi H, ImaizumiAnraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al. (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429 [DOI] [PubMed] [Google Scholar]
- Nontachaiyapoom S, Scott PT, Men AE, Kinkema M, Schenk PM, Gresshoff PM. (2007) Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloemspecific expression in transgenic plants. Mol Plant Microbe Interact 20: 769–780 [DOI] [PubMed] [Google Scholar]
- Nutman PS. (1952) Studies on the physiology of nodule formation. III. Experiments on the excision of roottips and nodules. Ann Bot (Lond) 16: 79–101 [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294; erratum Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Science 319: 901 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. (2009) Nod factor/nitrateinduced CLE genes that drive HAR1mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Long SR. (2003) Identification and characterization of nodulationsignaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HRM, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP. (2003) Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Pierik R, Keuskamp DH, Sasidharan R, Djakovic-Petrovic T, de Wit M, Voesenek LACJ. (2009) Light quality controls shoot elongation through regulation of multiple hormones. Plant Signal Behav 4: 755–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt H-J, Pühler A, Broer I. (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminatetype nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptorlike kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EMH, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extends the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé J-C. (1991) Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipooligosaccharide signals. Cell 67: 1131–1143 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet E-P, de CarvalhoNiebel F, Duc G, Frugoli J. (2005) The Medicago truncatula SUNN gene encodes a CLV1like leucinerich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, IturbeOrmaetxe I, Carroll BJ, Gresshoff PM. (2003) Longdistance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC. (2003) The Arabidopsis CLV3like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Stahl Y. (2006) Plant cells CLEave their way to differentiation. Science 313: 773–774 [DOI] [PubMed] [Google Scholar]
- Smit G, de Koster CC, Schripsema J, Spaink HP, van Brussel AA, Kijne JW. (1995) Uridine, a cell division factor in pea roots. Plant Mol Biol 29: 869–873 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factorinduced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Strabala TJ, O'Donnell PJ, Smit A-M, AmpomahDwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HCC, Hudson KR. (2006) Gainoffunction phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats ST. (1990) Early autoregulation of symbiotic root nodulation in soybeans. Plant Physiol 94: 865–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers ACJ, Auriac M-C, Truchet G. (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW. (1992) Induction of preinfection thread structures in the leguminous host plant by mitogenic lipooligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- Van den Eede G, Deblaere R, Goethals K, Van Montagu M, Holsters M. (1992) Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant Microbe Interact 5: 228–234 [DOI] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen PC, Grønlund M, Pacios Bras C, Spaink HP, Kijne JW. (2001) Cell biological changes of outer cortical root cells in early determinate nodulation. Mol Plant Microbe Interact 14: 839–847 [DOI] [PubMed] [Google Scholar]
- Vervliet G, Holsters M, Teuchy H, Van Montagu M, Schell J. (1975) Characterization of different plaqueforming and defective temperate phages in Agrobacterium strains. J Gen Virol 26: 33–48 [DOI] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, de Bruijn FJ, Stougaard J, Szczyglowski K. (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yang W-C, de Blank C, Meskiene I, Hirt H, Bakker J, van Kammen A, Franssen H, Bisseling T. (1994) Rhizobium Nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6: 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.