Abstract
Classical genetic analysis has revealed that the determinate habit of soybean (Glycine max) is controlled by a recessive allele at the determinate stem (Dt1) locus. To dissect the molecular basis of the determinate habit, we isolated two orthologs of pea (Pisum sativum) TERMINAL FLOWER1a, GmTFL1a and GmTFL1b, from the soybean genome. Mapping analysis indicated that GmTFL1b is a candidate for Dt1. Despite their high amino acid identity, the two genes had different transcriptional profiles. GmTFL1b was expressed in the root and shoot apical meristems (SAMs), whereas GmTFL1a was mainly expressed in immature seed. The GmTFL1b transcript accumulated in the SAMs during early vegetative growth in both the determinate and indeterminate lines but thereafter was abruptly lost in the determinate line. Introduction of the genomic region of GmTFL1b from the indeterminate line complemented the stem growth habit in the determinate line: more nodes were produced, and flowering in the terminal raceme was delayed. The identity between Dt1 and GmTFL1b was also confirmed with a virus-induced gene silencing experiment. Taken together, our data suggest that Dt1 encodes the GmTFL1b protein and that the stem growth habit is determined by the variation of this gene. The dt1 allele may condition the determinate habit via the earlier loss in GmTFL1b expression concomitant with floral induction, although it functions normally under the noninductive phase of flowering. An association test of DNA polymorphisms with the stem growth habit among 16 cultivars suggested that a single amino acid substitution in exon 4 determines the fate of the SAM after floral induction.
The transition from vegetative to reproductive phases at the shoot apical meristem (SAM) is controlled by the interaction of positive and negative regulators, such as LEAFY (LFY), APETALA1 (AP1), and TERMINAL FLOWER1 (TFL1; for review, see Benlloch et al., 2007). LFY and AP1 are the main promoters of floral meristem identity and encode transcription factors; both are expressed throughout the young floral meristem from the earliest stages of development, and after the onset of LFY expression, AP1 is expressed in these meristems (Mandel et al., 1992; Weigel et al., 1992; Maizel et al., 2005). The role played by TFL1 in floral initiation is the opposite of that of LFY and AP1. The Arabidopsis (Arabidopsis thaliana) tfl1 loss-of-function mutants flower early, and the SAM is converted into a terminal flower (Shannon and Meeks-Wagner, 1991; Schultz and Haughn, 1993). Constitutive expression of TFL1 driven by the cauliflower mosaic virus 35S promoter greatly extends the duration of all developmental phases (Ratcliffe et al., 1998). The TFL1 gene in Arabidopsis maintains indeterminate growth of the SAM by delaying the up-regulation of floral meristem identity genes (Ratcliffe et al., 1998).
TFL1 is homologous to phosphatidylethanolamine-binding proteins that play diverse roles related to signaling pathways controlling growth and differentiation (for review, see Benlloch et al., 2007). TFL1 belongs to a small gene family, one of whose members, FLOWERING LOCUS T (FT), is also a regulator of flowering time. However, FT acts in an opposite manner to TFL1; FT promotes flowering and conversion of the SAM to a flower (Kardailsky et al., 1999; Kobayashi et al., 1999). The structures of the TFL1 and FT proteins are very similar; the predicted polypeptides encoded by the Arabidopsis FT and TFL1 genes are 175 and 177 amino acids long, respectively, with differences in only 39 residues from nonconservative changes, including substitutions and insertions/deletions (Ahn et al., 2006). Based on the results of swapping experiments of single amino acids and discrete domains between Arabidopsis TFL1 and FT, the amino acid substitution from His to Tyr at residue 88 in TFL1 (residue 85 in FT) in the ligand-binding pocket and different segments in exon 4 of both genes were found to be responsible for their differential bindings to interactors, resulting in opposite activities for TFL1 and FT (Hanzawa et al., 2005; Ahn et al., 2006).
CENTRORADIALIS (CEN) is an Antirrhinum TFL1 ortholog (Bradley et al., 1996). As with tfl1 mutants, recessive mutations in the CEN gene result in the conversion of the normally indeterminate inflorescence to a determinate condition. However, the time to flowering is not affected in the cen mutants in contrast to the tfl1 mutants (Bradley et al., 1996, 1997). Both TFL1 and CEN are expressed in the subapical region of the shoot meristem; TFL1 is expressed both in vegetative and inflorescence shoot meristems, whereas CEN is only expressed in the inflorescence meristem (Bradley et al., 1996, 1997). The absence of CEN expression in the apex before the floral transition could explain why the cen mutations do not affect flowering time. This additional function of Arabidopsis TFL1 correlates with its expression during the vegetative phase, when it delays the commitment of plants to form an inflorescence; the early flowering in the tfl1 mutants is thus a result of an earlier commitment to form floral meristems (Bradley et al., 1997). On the other hand, the most likely Arabidopsis ortholog of CEN is not TFL1 but the ATC gene (Arabidopsis CEN homolog), another member of the TFL1 clade (Mimida et al., 2001). However, while ATC can functionally substitute for TFL1, as suggested by its ability to complement the tfl1 mutant phenotype when constitutively expressed, ATC loss-of-function mutations do not result in any obvious phenotype, indicating that ATC could be involved in a function other than inflorescence identity (Mimida et al., 2001).
The homologues of Arabidopsis TFL1/Antirrhium CEN have been isolated and their functions have been characterized in many plant species (for review, see Benlloch et al., 2007). Pea (Pisum sativum) contains at least three TFL1/CEN homologues (Foucher et al., 2003). No function has been assigned to PsTFL1b, a likely ortholog of CEN, while PsTFL1a and PsTFL1c are most closely related to Arabidopsis TFL1 and probably have functions identical to those conferred by Arabidopsis TFL1 when the functions of these genes are combined. Mutations in PsTFL1a, also known as DETERMINATE (DET), cause the determination of the main apex without affecting flowering time, in a manner similar to that in cen mutants of Antirrhinum. On the other hand, mutations in PsTFL1c, also known as LATE FLOWERING (LF), cause early flowering without affecting determination. Similar to CEN, DET is expressed in the shoot apex only after the floral transition, while LF expression is also observed in the vegetative apex. Therefore, in pea, the two functions of the Arabidopsis TFL1 gene, flowering time and apex determinacy, seem to be controlled by two different genes (Benlloch et al., 2007).
Soybean (Glycine max) also possesses two types of stem growth habits like pea. One type is the indeterminate stem, in which the terminal bud continues the vegetative activity of SAM during most of the growing season; the inflorescences of this type are axillary racemes (Carlson and Lersten, 1987). The second type is the determinate stem, in which the vegetative activity of SAM ceases when it becomes an inflorescence; this type has both axillary racemes and a terminal raceme.
The stem growth habit influences various agronomical traits. Determinate plant lines, for example, generally reach much shorter heights with increased lodging resistance and have lower lowest-pod heights and more main stem branches per plant than do indeterminate cultivars of similar maturities (Bernard, 1972; Foley et al., 1986; Ablett et al., 1989; Ouattara and Weaver, 1994; Robinson and Wilcox, 1998; Kilgore-Norquest and Sneller, 2000). The determinate lines also have shorter flowering and reproductive periods than do the indeterminate lines of similar maturities, although the difference in time to flowering is trivial (Bernard, 1972; Ouattara and Weaver, 1994; Kilgore-Norquest and Sneller, 2000).
Two genes, Dt1 and Dt2, affect stem termination in soybean (Bernard, 1972). A recessive allele, dt1, and a dominant allele, Dt2, hasten the termination of apical stem growth, which decreases both plant height and number of nodes. Of these, dt1 has a much greater effect. Bernard (1972) also observed an intermediate, semideterminate phenotype distinct from both the indeterminate and determinate phenotypes in hybrid populations between the indeterminate and determinate lines. Those semideterminate plants segregated for the indeterminate and the determinate habits, indicating that Dt1 behaved as a partially dominant gene in the genetic background tested (Bernard, 1972). Another recessive allele, dt1-t, also affects plant phenotype much like the dt1 allele, but it significantly delays stem termination (Thompson et al., 1997). Despite the agronomical importance of stem growth habit for soybean production, our knowledge about this habit is still limited.
In this study, we isolated the TFL1 orthologs from the soybean genome to better understand the mechanisms underlying stem termination and floral initiation in soybean. We here show that a TFL1 ortholog is the Dt1 gene that controls stem growth habit in soybean.
RESULTS
Stem Morphology in Near-Isogenic Lines for the Dt1 Locus
The stem morphology and plant architecture of near-isogenic lines (NILs) for the Dt1 locus, #6-22Dt1 (indeterminate) and #6-22dt1 (determinate), are presented in Figure 1. These NILs were developed from the progeny of the residual heterozygous line (RHL) #6-22, which was derived from a cross between determinate cv Misuzudaizu (MI) and indeterminate cv Moshidou Gong 503 (MO). Determinate line #6-22dt1 produced a terminal raceme with a few flower buds at the tip of the main stem (Fig. 1A), whereas indeterminate line #6-22Dt1 formed a cluster of axillary racemes surrounded by unexpanded leaflets (Fig. 1B). These two lines differ markedly in their plant architecture at maturity when grown outdoors: the determinate NIL was short with fewer nodes because stem growth terminated after flowering, whereas the indeterminate NIL was tall and produced more nodes (Fig. 1C).
Figure 1.
Morphology of the top of stems and plant architecture in determinate and indeterminate soybean NILs. A, Determinate NIL #6-22dt1 (dt1/dt1). A terminal raceme with a few flower buds (arrow) was produced at the tip of the main stem. B, Indeterminate NIL #6-22Dt1 (Dt1/Dt1). A cluster of axillary racemes (arrow) was surrounded by unexpanded leaflets. C, Plant morphology at maturity in the progeny of RHL #6-22. The #6-22dt1 (dt1/dt1) plants were short with a few nodes because of stem termination after stage R1, whereas #6-22Dt1 (Dt1/Dt1) plants were tall and produced more nodes. Plants with intermediate phenotypes were estimated to be heterozygous for the Dt1 locus, based on the FLP in intron 1 of GmTFL1b.
Isolation of Soybean TFL1 Orthologs
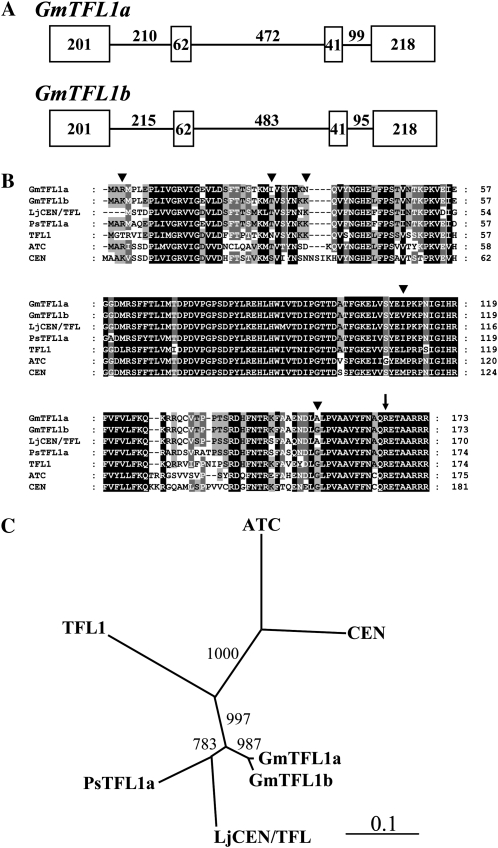
Pea contains at least three TFL1/CEN homologues, of which PsTFL1a controls the determination of the main apex (Foucher et al., 2003). To isolate orthologs of TFL1 from the soybean genome, we screened for soybean ESTs deposited in the GenBank/EMBL/DDBJ databases with the predicted amino acid sequence of PsTFL1a. PCR using primers designed based on the detected soybean EST sequence (AX478027) was performed to survey a bacterial artificial chromosome (BAC) library developed from MI. We detected a positive BAC clone (GMJMiB384F11), which contained a homolog of the EST sequence. By a primer-walking method, we identified a sequence of 4,216 bp containing a soybean TFL1 ortholog. On the other hand, the PCR with the same primer pair and subsequent sequence analysis for genomic DNAs of the determinate breeding line TK780 (TK) and the indeterminate wild soybean (Glycine soja) line Hidaka 4 (H4) identified two TFL1 orthologs, designated GmTFL1a and GmTFL1b. GmTFL1a corresponded to the EST sequence AX478027, and GmTFL1b was identical to the sequence of the TFL1 ortholog embedded in the BAC clone. Flanking sequences for GmTFL1a in TK and H4 were determined using a genome-walking technique composed of ligation of restriction enzyme-digested DNA to adaptors followed by amplification with nested PCR using adaptor-specific and gene-specific primers. The genome-walking approach successfully amplified fragments for the 5′ and 3′ regions of GmTFL1a when applied to the PvuII-digested DNA library and the DraI-digested DNA library, respectively. After sequencing the amplified fragments, we finally obtained contigs of 3,501 bp containing the entire gene region of GmTFL1a. The cDNAs of both GmTFL1a and GmTFL1b were also isolated by reverse transcriptase-mediated (RT)-PCR using RNA extracted from stem tips of indeterminate NIL #6-22Dt1. Comparisons between genomic DNA and cDNA sequences revealed that the GmTFL1a and GmTFL1b genes are both composed of four exons (Fig. 2A), as in the TFL1 genes of other plant species, and are predicted to encode sequences with 173 amino acids (Fig. 2B). The multiple alignment of the predicted amino acid sequences of GmTFL1a, GmTFL1b, Arabidopsis TFL1 (Bradley et al., 1997), Arabidopsis ATC (Mimida et al., 2001), Antirrhinum majus CEN (Bradley et al., 1996), pea PsTFL1a (Foucher et al., 2003), and Lotus japonicus LjCEN/TFL1 (Guo et al., 2006) is presented in Figure 2B. GmTFL1a and GmTFL1b differed in five amino acids and had high similarities to PsTFL1a (85.2% for GmTFL1a and 85.8% for GmTFL1b), LjCEN/TFL1 (86.5% for both GmTFL1a and GmTFL1b), and Arabidopsis TFL1 (75.1% for GmTFL1a and 76.3% for GmTFL1b). A recent origin of GmTFL1a and GmTFL1b is suggested by a phylogenetic tree constructed using a neighbor-joining method (Fig. 2C).
Figure 2.
Structures, predicted amino acid sequences, and phylogenetic tree of soybean TFL1 orthologs GmTFL1a and GmTFL1b. A, Exon/intron structures of GmTFL1a and GmTFL1b. Exon (boxes) and intron sizes are in bp. B, Multiple alignment of the predicted amino acid sequences of GmTFL1a, GmTFL1b, L. japonicus LjCEN/TFL1 (Guo et al., 2006), pea PsTFL1a (Foucher et al., 2003), Arabidopsis TFL1 (Bradley et al.,1997), Arabidopsis ATC (Mimida et al., 2001), and A. majus CEN (Bradley et al., 1996). Highly conserved amino acids are in black, dark gray, or light gray depending on the level of identity (darker = higher level). Numbers on the right refer to amino acid positions in relation to the N-terminal Met of each sequence. Arrowheads above the sequences refer to differences between GmTFL1a and GmTFL1b. The arrow indicates an amino acid substitution from Arg (R) in the Dt1 allele to Trp (W) in the dt1 allele. C, Phylogenetic relationships of TFL1/CEN proteins constructed using the neighbor-joining method with the program ClustalW. Bootstrap values of 1,000 replicates are indicated at the branches of the tree.
Genetic Mapping Suggests That GmTFL1b May Be a Candidate for Dt1
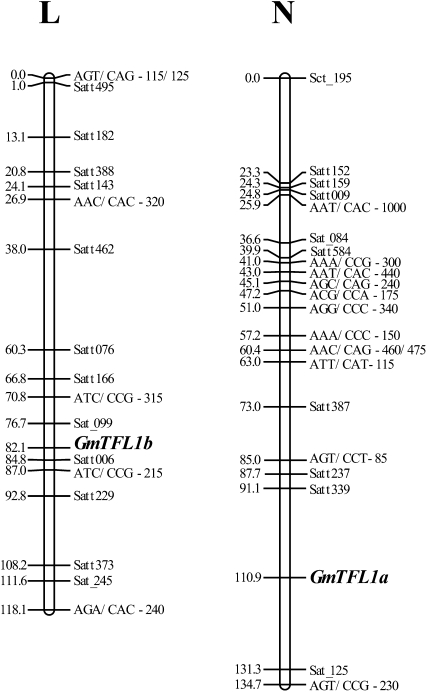
GmTFL1a and GmTFL1b were mapped onto a genetic map that was constructed using recombinant inbred lines (RILs) derived from a cross between TK and H4 (Liu et al., 2007). We developed molecular markers for GmTFL1a (Supplemental Fig. S1A) and GmTFL1b (Supplemental Fig. S1B), which were used to map these genes. GmTFL1a was assigned between simple sequence repeat markers Satt339 and Satt125 in linkage group N (Fig. 3). GmTFL1b was assigned to the vicinity of a simple sequence repeat marker (Satt006) in linkage group L (Fig. 3), the tagging marker for the Dt1 locus (Molnar et al., 2003).
Figure 3.
Genetic map positions of soybean TFL1 orthologs GmTFL1a and GmTFL1b. GmTFL1a (in linkage group N) and GmTFL1b (in linkage group L) were mapped using RILs derived from a cross between determinate G. max (TK) and indeterminate G. soja (H4) lines.
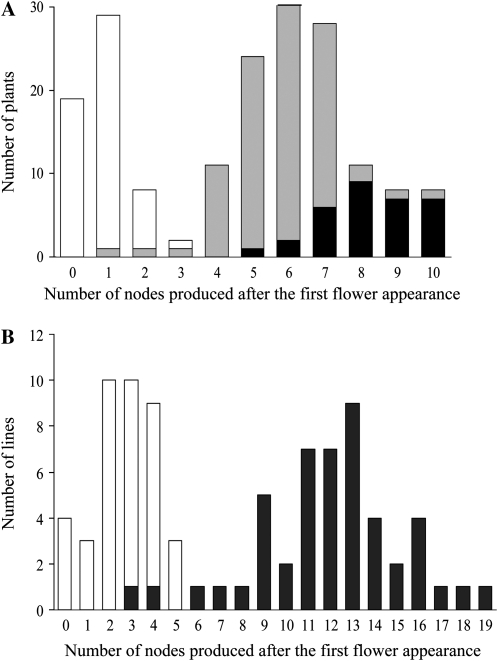
Cosegregation between stem growth habit and GmTFL1b was then tested in two segregating populations: the progeny of RHL #6-22 and the RILs of TK × H4. The stem growth habit was evaluated using the number of nodes produced after the first flower appearance (stage R1; Fehr et al., 1971). The determinate cultivars usually produce only a few nodes after stage R1 because the stem growth terminates shortly after flowering, whereas the indeterminate soybeans produce more nodes (Thseng and Hosokawa, 1972). In the two populations tested, the number of nodes produced after stage R1 had bimodal distributions, suggesting an involvement of a single major gene in the control of stem growth habit (Fig. 4). Genotyping of GmTFL1b revealed that most of the plants/lines that had fewer nodes (less than three for RHL #6-22 and less than five for RILs) were homozygous for the determinate parent-derived GmTFL1b allele, whereas most of the plants/lines that were heterozygous or homozygous for the indeterminate parent-derived GmTFL1b allele had more nodes. The mean values between two homozygotes were highly significant (F = 837.6, P < 8.78E-46 in RHL #6-22 and F = 222.6, P < 1.84E-25 in RILs). Accordingly, the stem growth habit cosegregated with the GmTFL1b genotypes in the two populations. The results obtained from RHL #6-22 further indicated that the heterozygous plants, as a whole, produced fewer nodes than did the plants homozygous for the indeterminate parent-derived GmTFL1b allele (Figs. 1C and 4A), in accordance with the observation of Bernard (1972). The intermediate phenotype in the heterozygous plants suggests that the indeterminate parent-derived GmTFL1b allele exhibits partial dominance, not complete dominance, over the determinate parent-derived GmTFL1b allele.
Figure 4.
Frequency of plants with various numbers of nodes produced after the first flower appearance to show cosegregation between stem growth habit and GmTFL1b. A, Progeny of RHL #6-22. B, RILs derived from a cross between determinate G. max (TK) and indeterminate G. soja (H4) lines. Plants/lines homozygous for the determinate parent-derived GmTFL1b allele (white bars) were almost clearly separated from those having the indeterminate parent-derived allele (gray bars for heterozygote and black bars for homozygote) in the number of nodes produced after the first flower appearance.
Expression Analysis of GmTFL1a and GmTFL1b
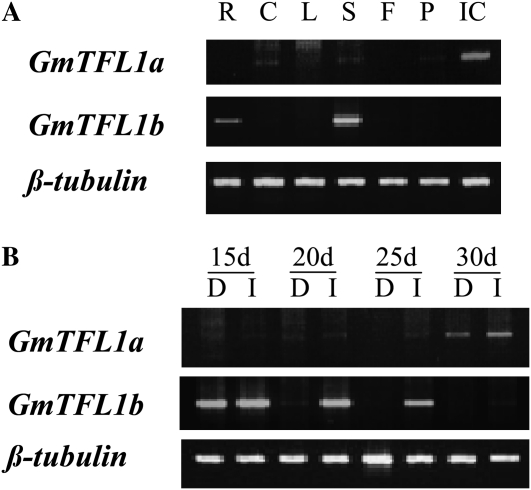
We analyzed the transcription profiles of GmTFL1a and GmTFL1b in different tissues and at different growing stages. Among the tissues tested, both largely differed in their transcription profiles (Fig. 5A). GmTFL1a was expressed highly in immature seed and slightly in the cotyledon and stem tip, whereas GmTFL1b was expressed only in root and stem tip.
Figure 5.
Expression analysis of GmTFL1a and GmTFL1b by RT-PCR. A, Gene expression in root (R), cotyledon (C), leaflet (L), stem tip (S), flower (F), pod (P), and immature cotyledon (IC) of the #6-22Dt1 plants. Transcripts of the β-tubulin gene were amplified as a control. For both paralogs, primers were designed to anneal the 3′ untranslated region and exon 3, which amplify DNA fragments of different sizes from cDNA and genomic DNA, to confirm that no amplification occurred from genomic DNA contaminants in the RNA sample. Note that the expression of GmTFL1a was high in the immature cotyledon but quite low in the stem tip and absent in the root, whereas GmTFL1b was expressed only in the root and the stem tip. B, Expression of GmTFL1a and GmTFL1b in stem tip from 15 to 30 DAE. GmTFL1a in the determinate (D) and indeterminate (I) lines was expressed at a high level only at a later stage of growth after flowering. On the other hand, GmTFL1b was expressed at earlier stages of growth through flowering (25 DAE) in the indeterminate line but was abruptly lost after 15 DAE in the determinate line.
Time course-dependent changes in expression in the stem tip were evaluated in #6-22Dt1 and #6-22dt1 plants grown under a 14.5-h daylength. GmTFL1a was not expressed at earlier stages of growth until 25 d after emergence (DAE), when flowering had not yet started, and was expressed weakly at 30 DAE after flowering started in both indeterminate and determinate lines (Fig. 5B). On the contrary, GmTFL1b was expressed highly at 15 DAE in the two lines. However, the expression in the determinate NIL was abruptly lost and not detected at 20 DAE or later. On the other hand, the GmTFL1b transcripts in the indeterminate NIL were observed until 25 DAE but not at 30 DAE (Fig. 5B). Because both NILs flowered at almost the same time (at 28 DAE), the earlier loss of GmTFL1b expression in the SAM of #6-22dt1 plants may be a factor conditioning the determinate habit. Both mapping and expression analyses strongly suggest that GmTFL1b is a candidate for Dt1.
Transformation of a Determinate Cultivar with the 4,216-bp Genomic Region Covering the Functional GmTFL1b Allele
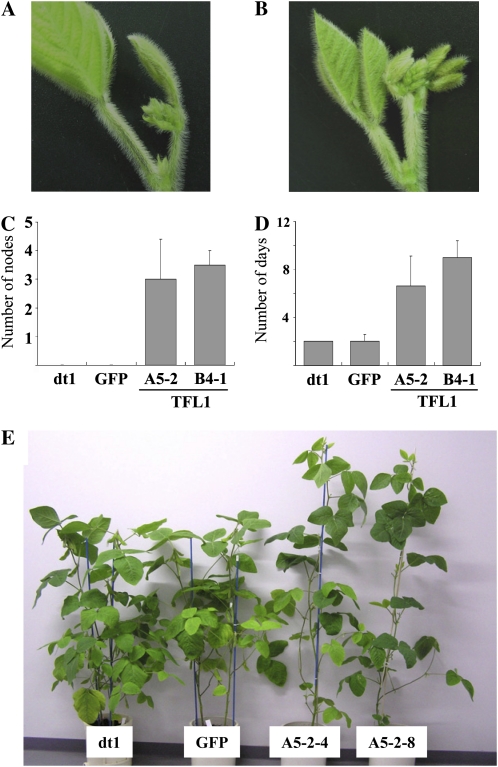
To confirm the identity between Dt1 and GmTFL1b, we transformed the determinate cv Kariyutaka (KA; genotype dt1/dt1) with the 4,216-bp genomic region containing the GmTFL1b allele from the indeterminate cv MO. Expression of the GFP gene, located adjacent to the GmTFL1b transgene in the T-DNA region (Supplemental Fig. S2), was used as a marker for successful transformation. We analyzed GFP-positive T2 plants in the progeny of two independent transgenic T1 plants, A5-2 and B4-1. A T2 line transformed with the pMDC123-GFP construct (GFP plant) was used for the negative control for morphological evaluation. This line contains the GFP gene but no GmTFL1b transgene. KA (dt1) and GFP plants had already formed a terminal raceme at the stem tip at stage R1 (Fig. 6A). They did not produce any additional nodes, and a flower in the terminal raceme opened 2 d later (Fig. 6, C and D). In the T2 plants transformed with the pMDC123-GFP-Dt1 construct, the terminal raceme was still surrounded by unexpanded leaflets crowded together by short internodes at stage R1 (Fig. 6B). The T2 plants produced an average of 3.0 (A5-2) and 3.5 (B4-1) additional nodes at maturity (Fig. 6C), took longer for a flower in the terminal raceme to open (Fig. 6D), and were taller (Fig. 6E). These observations indicated that the GmTFL1b allele from the indeterminate cv MO could complement the determinate habit of KA.
Figure 6.
Complementation of the dt1 allele by transformation. A, Morphology at the stem tip of determinate cv KA, B, The T2 plants carrying the GmTFL1b genomic region. KA plants formed a terminal raceme at the tip of the main stem, whereas T2 plants formed terminal and axillary racemes with unexpanded leaflets. C, Number of nodes produced after the first flower appearance (stage R1). KA plants (dt1) and GFP-transformed T3 plants (GFP) produced no additional nodes after stage R1, whereas the T2 plants (A5-2 and B4-1), on average, produced 3.2 nodes (A5-2) and 3.5 nodes (B4-1). D, Number of days from stage R1 until a flower in the terminal raceme opened. In KA and GFP plants, flowering started at node 5 or 6, and a flower in the terminal raceme opened 2 d later, whereas T2 plants took longer for the flower in the terminal raceme to open. E, Plant morphology of transgenic plants. T2 plants (A5-2-4 and A5-2-8) were taller and produced more nodes than did KA and GFP plants.
Virus-Induced Gene Silencing-Induced Suppression of Vegetative Activity of Terminal Buds
Evidence supporting the identity between Dt1 and GmTFL1b was also obtained from a virus-induced gene silencing (VIGS) experiment. The #6-22Dt1 plants were inoculated with a recombinant virus that contained a 139-nucleotide portion of exon 4 of GmTFL1b (Supplemental Fig. S3). For a negative control, the #6-22Dt1 plants were inoculated with a recombinant virus that contained a portion of the GFP gene, because viral replication is sometimes affected by the presence of an insert. Weak mosaic symptoms appeared on leaflets as a consequence of viral infection. However, the infection itself did not influence stem termination of infected plants.
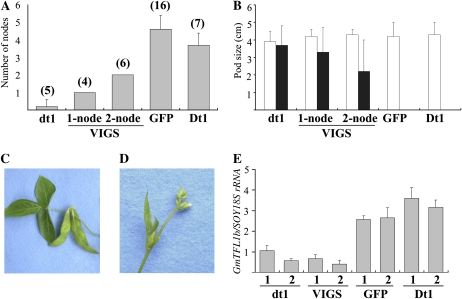
Mock-inoculated #6-22Dt1 plants produced an average of 3.6 ± 0.8 nodes after the first flower opened (stage R1), whereas the mock-inoculated #6-22dt1 plants produced an average of 0.2 nodes (Fig. 7A). All 16 plants of #6-22Dt1 that were infected with the virus carrying the partial GFP sequence had phenotypes similar to the mock-inoculated #6-22Dt1 plants; they produced an average of 4.6 ± 0.8 nodes after stage R1 (Fig. 7A). On the other hand, 21 plants that were infected with the virus carrying a 139-nucleotide portion of exon 4 of GmTFL1b had various phenotypic responses: four plants produced only one additional node (one-node plants), six produced two nodes (two-node plants), and the remaining plants produced three to five nodes after stage R1 (Fig. 7A; Supplemental Fig. S4). The GmTFL1b VIGS also influenced the transition from the vegetative to the reproductive phase in the stem tip. There was no visible difference in flowering time; all tested plants had produced pods of a similar size (approximately 4.0 cm long) at nodes 5 to 6 at 42 DAE (Fig. 7B). The mock-inoculated #6-22dt1 plants produced pods almost the same size at the terminal node (Fig. 7C). On the other hand, the mock-inoculated #6-22Dt1 plants and the #6-22Dt1 plants infected by the virus carrying the GFP sequence still produced flower buds (Fig. 7D). GmTFL1b VIGS-induced, one-node, and two-node #6-22Dt1 plants produced pods 1.0 to 4.8 cm long at the terminal node, as did #6-22dt1 (Fig. 7B). Thus, the GmTFL1b VIGS induced an earlier transition from vegetative to reproductive phases; stems terminated earlier and pods formed earlier at stem tips, supporting the identity between Dt1 and GmTFL1b.
Figure 7.
VIGS-induced suppression of vegetative activity of terminal buds. A, Reduction by VIGS of numbers of nodes produced after stage R1. The #6-22Dt1 plants mock inoculated (Dt1) and infected by the virus carrying the GFP sequence (GFP) produced an average of 3.6 and 4.6 nodes, respectively. On the other hand, of the 21 #6-22Dt1 plants infected by the virus carrying the GmTFL1b constructs (VIGS), four produced only one node after floral initiation (1-node) and six formed two nodes (2-node). The number of plants tested is given in parentheses above the bars. B, Pod size at lower nodes (white bars) and terminal node (black bars). All plants produced pods of approximately 4 cm at nodes 5 and 6, independent of the Dt1 genotypes and treatments. C, Pods of #6-22dt1 plants (dt1) were almost the same size as those at lower nodes. D, Stem tip morphology for the #6-22Dt1 plants mock inoculated (Dt1) and those infected by the virus carrying the GFP sequence (GFP). Those plants maintained vegetative activity and did not form any pods at the stem tip. VIGS-induced 1-node and 2-node plants produced pods of 2.0 cm or more at the terminal node. E, Suppression of the GmTFL1b expression in root by VIGS. Expression as evaluated with real-time PCR was low in #6-22dt1 plants (dt1) compared with the #6-22Dt1 plants mock inoculated (Dt1) and infected by the virus carrying the GFP sequence (GFP). GmTFL1b expression in VIGS plants (VIGS) was similar to levels in the determinate plants. Two plants were analyzed for each treatment. Data represent means for three replications with se.
Down-regulation of GmTFL1b expression by VIGS was confirmed in roots, a tissue that was expected to be suitable for evaluating the effect of VIGS at the mRNA level of GmTFL1b because TFL1 is known to be widely expressed in the Arabidopsis root (Arabidopsis eFP Browser [http://www.bar.utronto.ca/efp/cgi-bin/efpWeb.cgi]; Winter et al., 2007), in contrast to the SAM, where it is expressed only in the central regions (Conti and Bradley, 2007). In real-time PCR to evaluate the expression level of GmTFL1b in root at 20 DAE, expression was lower in the determinate #6-22dt1 plants than in the indeterminate #6-22Dt1 plants (Fig. 7E). VIGS down-regulated the mRNA level of the GmTFL1b gene to the level observed in the #6-22dt1 plants. Infection of the #6-22Dt1 plants with the virus carrying the partial GFP sequence did not influence GmTFL1b expression, excluding the possibility of a nonspecific effect of viral infection.
Sequence Polymorphism of GmTFL1b between Determinate and Indeterminate Lines
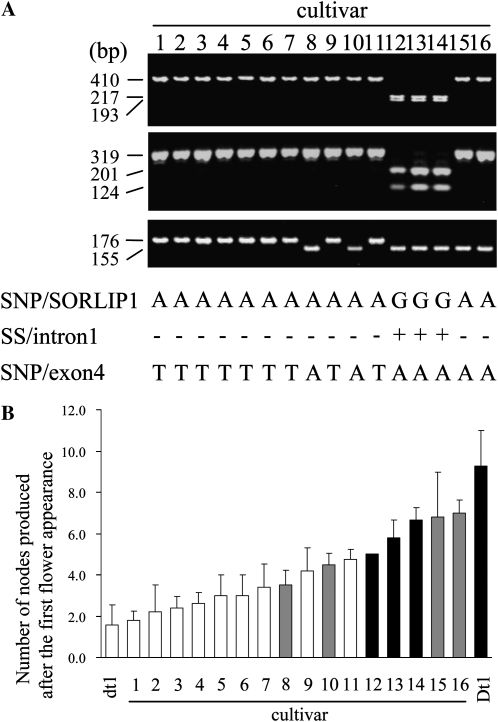
Because the results obtained from mapping, expression analysis, transformation, and VIGS experiments all indicated that the gene responsible for Dt1 is GmTFL1b, we tried to determine which DNA polymorphism(s) conditioned the determinate growth habit. In addition to the three lines (MI, TK, and H4) used for gene isolation, we sequenced the 4,216-bp GmTFL1b region of two indeterminate cultivars, MO and Harosoy (HA), and a determinate NIL of HA, Hdt1. We detected 15 linked sequence polymorphisms among the six lines tested in the 4,216-bp GmTFL1b sequence (Supplemental Fig. S5). Of these, two single-nucleotide polymorphisms (SNPs; one at a cis-element referred to as a sequence overrepresented in light-induced promoters 1 [SORLIP1; Hudson and Quail, 2003] in the promoter [position −1,487] and the other in exon 4 [position 1,283]) and a sequence substitution (SS) in intron 1 (position 279) were detected consistently between determinate and indeterminate lines. The indeterminate lines possessed two SORLIP1 cis-elements: one at 1,487 nucleotides (Supplemental Fig. S5) and the other at 1,306 nucleotides (data not shown) upstream of the start ATG codon. The first SORLIP1 was absent in the three determinate lines as a result of the SNP in the motif. The SNP in exon 4 caused an amino acid substitution at residue 166, from Arg in the Dt1 allele to Trp in the dt1 allele (Fig. 2).
Association of Stem Growth Habit with DNA Polymorphisms of GmTFL1b
We performed an association test between stem growth habit and the three linked DNA polymorphisms using 16 soybean cultivars (Fig. 8A). Genotyping of the three DNA polymorphisms placed nine cultivars into a haplotype, which shared an identical pattern with the dt1 allele (dt1 haplotype; nos. 1–7, 9, and 11 in Fig. 8A), and three into another haplotype, which shared the same pattern with the Dt1 allele (Dt1 haplotype; nos. 12–14 in Fig. 8A). The remaining four had a new combination of polymorphisms; they had the same SNP (A) at exon 4 as the Dt1 allele did, but the SNP at the SORLIP cis-element and the SS in intron 1 were identical to those in the dt1 allele (nos. 8, 10, 15, and 16 in Fig. 8A). The cultivars with the dt1 haplotype had fewer nodes produced after stage R1, whereas those with the Dt1 haplotype produced more nodes after stage R1 (Fig. 8B). The cultivars with the new haplotype varied in their tendency for stem termination; two (nos. 8 and 10) had fewer nodes, and two (nos. 15 and 16) had more nodes. Of the three polymorphisms, the SNP in exon 4 was the most closely associated with stem growth habit; the five accessions with the most nodes (nos. 12–16) all had the A nucleotide at the SNP like HA (Dtl/Dt1), whereas the seven with the fewest nodes (nos. 1–7) all had the T nucleotide like Hdt1 (dt1/dt1). Cultivar Peking, which is reported to have the third allele at the Dt1 locus dt1-t (Thompson et al., 1997), was genotyped for the three diagnostic polymorphic markers and found to have the same haplotype as the Dt1 allele (data not shown).
Figure 8.
Association of stem growth habit with DNA polymorphisms of GmTFL1b. A, Genotypes for 16 early-maturing cultivars at three linked DNA polymorphisms detected consistently between determinate and indeterminate lines. B, The number of nodes produced after stage R1. Based on the deduced genotypes for three DNA markers, the cultivars were classified into three haplotypes: dt1 haplotype (white bars), Dt1 haplotype (black bars), and a new haplotype (gray bars). Cultivars are as follows: 1, Rokujunichimame; 2, Ishikarishiro 1 go; 3, Aochi (Natsu); 4, Wasekeburi; 5, Iwateyagi 1 go; 6, Wasekin; 7, Baihualudadou; 8, Keshuang; 9, Chashouryu; 10, Chamoshidou; 11, Kairyokimusume; 12, Baihua; 13, Shauilihong; 14, Lindiansuoyiling; 15, Chuandou; 16, Zihua 1. Harosoy (Dt1) and its NIL for dt1 (dt1) are included for comparison.
DISCUSSION
The Soybean Stem Growth Habit Gene, Dt1, Is a TFL1 Ortholog
We isolated two orthologs of PsTFL1a, which controls the determinate habit in pea, from the soybean genome and designated them as GmTFL1a and GmTFL1b. These orthologs had a high identity for their amino acid sequences but quite different transcriptional profiles. GmTFL1b was expressed in root and stem tip, like Arabidopsis TFL1, whereas GmTFL1a was mainly expressed in immature seed. Genetic mapping further allocated GmTFL1b near the Dt1 locus, which had been mapped previously (Molnar et al., 2003). Actually, we found that GmTFL1b cosegregated with stem growth habits in two segregating populations. These findings thus led to the hypothesis that Dt1 encodes GmTFL1b.
In this study, we confirmed the identity between Dt1 and GmTFL1b using transformation and VIGS approaches. Transgenic T2 plants of determinate cv KA with the GmTFL1b allele from indeterminate cv MO had a delayed transition from vegetative to reproductive phases at the stem tip, indicating that the GmTFL1b allele from the indeterminate cultivar could complement the determinate growth habit of cv KA. On the other hand, the VIGS of GmTFL1b induced earlier stem termination and pod formation at the stem tip of the indeterminate plants, although its effect varied with individual plants. While no phenotypic change was detected in the indeterminate plants that were infected with the virus containing a partial GFP sequence, VIGS of GmTFL1b successfully induced the phenotypic change from the indeterminate to the determinate habit. Both results, therefore, support that Dt1 encodes GmTFL1b. As with other plant species, the function of TFL1 in meristem identity may be conserved in soybean.
DNA Polymorphism That Causes the Allelic Difference at the Dt1 Locus
The finding that the 4,216-bp genomic region from the indeterminate cv MO was sufficient to complement the function deficient in the determinate cv KA further suggests that a causal DNA polymorphism(s) that conditions the determinate growth habit is involved in this region. Three of the 15 linked DNA polymorphisms in the region were observed consistently between determinate and indeterminate lines. The association analysis of the three polymorphisms with stem growth habit further suggests that a nonsynonymous mutation in exon 4 was more consistent with the difference in the number of nodes produced after stage R1. The SNP causes the substitution from Arg in the Dt1 allele to Trp in the dt1 allele at residue 166. Arg at this residue is conserved not only across the TFL1/CEN genes of various plant species but also in other members of the phosphatidylethanolamine-binding protein family such as FT and BFT, suggesting that Arg plays a crucial role in their functions. FT and TFL1 have been considered to compete for a common interacting partner(s), which has some intermediate level of activity in the absence of FT or TFL1 (Pnueli et al., 2001; Abe et al., 2005; Wigge et al., 2005; Ahn et al., 2006). A current model predicts that FT and TFL1 bind to a bZIP transcription factor, FD, to activate or repress the expression of AP1, respectively (Ahn et al., 2006). The substitution from Arg to Trp at residue 166 of GmTFL1b, therefore, might cause a functional deficiency in the binding affinity with FD, possibly under competitive interaction with FT. The same amino acid substitution from Arg to Trp in the CCT (for CO, CO-like, and TOC1) domain, which controls protein-protein interactions, is also known to be involved in the mutants of an Arabidopsis CONSTANS gene (co-7; Robson et al., 2001) and a wheat VRN2 gene (vrn2; Yan et al., 2004).
The other two polymorphisms may also be involved in different controls of stem growth habit between the Dt1 and dt1 alleles. We found two SORLIP1 cis-elements in the promoter region of the GmTFL1b allele of the indeterminate lines, one of which was absent in the determinate lines due to the SNP in the motif. SORLIP1 is the most common cis-element in SORLIPs in the promoter sequences of Arabidopsis genes that are induced or repressed by far-red light (Hudson and Quail, 2003). The loss of a SORLIP1 element might thus result in the weak or incomplete functioning in the phytochrome A-regulated signal transduction of GmTFL1b expression. It may also be worth addressing the role of the SS in intron 1 in controlling GmTFL1b expression. The Arabidopsis FT gene is regulated by FLOWERING LOCUS C, a MADS box transcription factor, through binding to the first intron in the FT gene (Searle et al., 2006). By developing NILs for different haplotypes that have different combinations of DNA polymorphisms at the SORLIP1 cis-element and intron 1, we will be able to find clues for understanding the possible roles of these mutations in the control of stem growth habit in soybean.
Predicted Function of GmTFL1b in the Control of Stem Growth Habit and Flowering
The TFL1 gene in Arabidopsis maintains the indeterminate growth of the SAM by inhibiting the expression of the floral meristem identity genes LFY and AP1 (for review, see Benlloch et al., 2007). In the tfl1 loss-of-function mutant, however, LFY and AP1 are expressed in the inflorescence meristems and conversely suppress TFL1 expression (Bradley et al., 1997; Liljegren et al., 1999; Ratcliffe et al., 1999; Ferrándiz et al., 2000). Consequently, the tfl1 mutants flower early, and the SAM converts to a terminal flower (Shannon and Meeks-Wagner, 1991; Schultz and Haughn, 1993). Unlike in the Arabidopsis tfl1 mutants, the dt1 allele may not be a loss-of-function allele because the determinate and indeterminate lines did not differ visibly in the stem growth habit, particularly in the earlier vegetative growth stages and in plants grown under a short photoperiod that causes very early maturing (Bernard, 1972) or under the noninductive condition of flowering such as a long photoperiod. Rather, our expression analysis led us to hypothesize that the dt1 protein can function similarly to the Dt1 protein, at least in noninductive phases of flowering, and that the dt1 allele may condition the determinate habit via an earlier loss in GmTFL1b expression, which may be caused by a decrease in binding affinity with interactors such as FD, possibly under competitive interaction with FT. This hypothesis is consistent with the suggestion of Carlson and Lersten (1987) that the difference between the two types is not due to the way that stem growth is terminated but rather is due to the timing of the termination of stem growth.
In contrast to its role in stem termination, GmTFL1b did not visibly affect time to flowering, either in the transformation or in the VIGS experiments, suggesting that Dt1 is not involved in time to flowering. In fact, the NILs for the Dt1 locus developed in this study flowered almost at the same time, agreeing well with the observation of Bernard (1972) that the dt1 isoline tested flowered only 1 to 2 d earlier than the Dt1 isoline. This lack of a marked difference is in a sharp contrast to Arabidopsis TFL1, which affects the time to flowering by controlling the timing of formation of the inflorescence meristem (Bradley et al., 1997), although GmTFL1b was expressed in a vegetative stage like Arabidopsis TFL1. Soybean usually forms axillary racemes from nodes 5 or 6 and sometimes higher toward the stem tip, and flowering starts in the lowest raceme. The formation of axillary racemes might not be directly related to the phase transition in the SAM, which is under the control of GmTFL1b. The role of TFL1 orthologs in time to flowering may thus vary with the expression pattern (Bradley et al., 1996, 1997) and inflorescence growth patterns.
In summary, our results demonstrate that Dt1 encodes the GmTFL1b protein and that the stem growth habit is determined by the variation of this gene. The dt1 allele may condition the determinate habit of the SAM possibly via the earlier loss in GmTFL1b expression concomitant with floral induction, although the dt1 allele functions normally under the noninductive phase of flowering. Further studies are needed to determine the role of amino acid substitution at exon 4 and how the GmTFL1b transcript level is down-regulated by floral induction in the determinate plants to terminate the stem growth. Furthermore, the following findings are suggestive in understanding the control of stem growth habit in the indeterminate plants. First, down-regulation of the GmTFL1 transcript by VIGS suppressed the indeterminate growth at SAMs in indeterminate plants. Second, the GmTFL1b transcript was reduced after flowering even in the indeterminate plants, resulting in the termination of stem growth. The fate of SAM in the indeterminate plants might also be determined by quantitative control of the GmTFL1b transcripts, as in the determinate plants.
MATERIALS AND METHODS
Plant Materials
Two sets of NILs for the Dt1 locus were used to isolate and characterize the soybean (Glycine max) orthologs of TFL1. These lines were #6-22Dt1 (Dt1/Dt1) and #6-22dt1 (dt1/dt1) as well as HA (L58-266; Dt1/Dt1) and its NIL for dt1, Hdt1 (L62-973; dt1/dt1). The former set of NILs was developed from RHL #6-22 for the Dt1 locus in F8 RILs derived from a cross between the determinate cv MI and the indeterminate forage soybean MO (Watanabe et al., 2004). Another set of RILs, which was derived from a cross between the determinate breeding line TK and the indeterminate wild soybean (Glycine soja) line H4 (Liu et al., 2007), was used for mapping the TFL1 orthologs into soybean linkage groups. Cosegregation between stem growth habit and the candidate gene for Dt1 was tested in two segregating populations: the progeny of RHL #6-22, consisting of 181 plants, and the 96 RILs from TK × H4. Sixteen early-maturing cultivars were used for an association test of stem growth habit with three DNA polymorphisms of the candidate gene, which were detected consistently in three sets of comparisons between indeterminate and determinate lines #6-22Dt1 and #6-22dt1, HA and Hdt1, and H4 and TK. Determinate cv KA was used for transformation experiments.
Phenotypic Evaluation of Stem Growth Habits
Stem growth habit was evaluated according to the method of Thseng and Hosokawa (1972), which measured the number of nodes produced after the first flower appearance (stage R1; Fehr et al., 1971). Generally, determinate soybeans produce only a few nodes after floral initiation because stem growth terminates shortly after flowering, whereas the indeterminate soybeans produce more nodes.
Isolation and Sequence Analysis of the Soybean Orthologs of TFL1
To isolate the genomic regions covering the TFL1 orthologs, we first surveyed soybean ESTs deposited in the GenBank/EMBL/DDBJ databases with the predicted amino acid sequence of a pea (Pisum sativum) PsTFL1a (AY340579). Then, a BAC library developed from MI (Xia et al., 2005) was screened using primers TFL1-F and TFL1-R designed based on the detected soybean EST sequence (AX478027). We detected a positive BAC clone (MiB348F11) and sequenced the 4.2-kb region covering a TFL1 ortholog with a primer-walking method. The same primer pair was also used for amplifying the TFL1 orthologs by PCR from the genomic DNA of TK and H4. The fragments amplified were cloned into a pGEM T-Easy vector (Promega). By sequence analysis of the clones from TK and H4, we identified two TFL1 paralogs designated GmTFL1a and GmTFL1b; GmTFL1b was identical to the ortholog embedded in the BAC clone.
Because no positive BAC clone was detected for GmTFL1a in the MI BAC library, a genome-walking technique using the BD GenomeWalker Universal Kit (Clontech) was applied to obtain 5′ and 3′ sequences of GmTFL1a as described previously (Liu et al., 2008). Briefly, genomic DNA from TK and H4 was digested in separate reactions with blunt-end endonucleases: DraI, EcoRV, PvuII, and SspI. The ends of the DNA in each digested pool were ligated to an adaptor sequence. A long PCR was then performed using adaptor primers and paralog-specific primers according to the manufacturer's instructions. The products from the PCR were cloned and sequenced.
The transcripts covering the entire coding regions of the two paralogs were also amplified from the cDNA synthesized from RNAs of stem tip, including SAM of the #6-22Dt1 line by RT-PCR. Total RNA was isolated according to the method of Napoli et al. (1990), except that we removed genomic DNA from the RNA fraction using DNase I (Takara Bio). The cDNA was synthesized from total RNA essentially as described previously (Koseki et al., 2005). The cDNA synthesis reaction mixture was prepared by mixing 4 μL of 5× reaction buffer (250 mm Tris-HCl [pH 8.3], 375 mm KCl, and 15 mm MgCl2), 2 μL of 0.1 m dithiothreitol, 0.5 μL of RNaseOUT inhibitor (Invitrogen), 1 μL of 100 μm B26 primer (Supplemental Table S1), 4 μL of 2.5 mm deoxyribonucleotide triphosphates, the RNA solution, and water to a final volume of 19 μL. After the addition of 1 μL of reverse transcriptase (Moloney murine leukemia virus; Invitrogen), the cDNA synthesis was performed at 42°C for 1 h. The reverse transcriptase was inactivated by heating the sample at 99°C for 1 min. Amplification reactions were performed using the cDNA as a template with primers TFL1a-RT-F and TFL1a-RT-R for GmTFL1a and TFL1b-RT-F and TFL1b-RT-R for GmTFL1b. Amplified products were cloned and sequenced. The primers used for isolation of the TFL1 genes are listed in Supplemental Table S1. The predicted amino acid sequences were aligned using the ClustalW Multiple Sequence Alignment program version 1.8 (http://clustalw.genome.jp; Thompson et al., 1994). A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) based on protein sequences deduced from the nucleotide sequences of TFL1 orthologs.
Genetic Mapping of GmTFL1a and GmTFL1b
GmTFL1a and GmTFL1b were mapped onto a genetic map constructed using the RILs derived from the cross between TK and H4, which covered 2,383 centimorgan in length with 282 markers (Liu et al., 2007) for the 20 linkage groups in the soybean consensus map (Cregan et al., 1999). The linkage map is available via the National BioResource Project database, Legume Base-Glycine max/soja (http://www.shigen.nig.ac.jp/bean/glycinesoja/top/top.jsp).
A SNP in intron 3 was scored with a derived cleaved amplified polymorphic sequence (dCAPS) marker for mapping the GmTFL1a gene. PCR using primers TFL1a-in3-F and TFL1a-R amplified 240-bp fragments from the genomic DNA of TK and H4. Only the fragments amplified from H4 were digested into 221- and 19-bp fragments with MaeII. Ten microliters of the PCR products was treated with the enzyme overnight, separated by electrophoresis on a 3% NuSieve 3:1 agarose gel (Takara Bio), and visualized with ethidium bromide under UV light. For mapping the GmTFL1b gene, a fragment length polymorphism (FLP) in intron 1, which detected a six-nucleotide difference in length due to a SS in intron 1, was used to map GmTFL1b and was analyzed by PCR using primers TFL1b-in1-F and TFL1b-in1-R; the forward primer was labeled with a fluorescent dye. The FLP was detected by the ABI 377 sequencer with GeneScan software (Applied Biosystems). Primers used for mapping of the TFL1 genes are listed in Supplemental Table S1.
The data were incorporated into the map using the Map Manager program QTXb17. Marker order and distance were determined using the Kosambi function and a criterion of 0.001 probability.
Cosegregation of Stem Growth Habit with the Dt1 Locus
Cosegregation of the stem growth habit with GmTFL1b was assayed for the progeny of RHL #6-22 in an experimental field at Chiba University (Matsudo, Japan) in 2003 and for the RILs of TK × H4 in a greenhouse at Hokkaido University (Sapporo, Japan) in 2004. The FLP or CAPS marker at intron 1 was used to score the GmTFL1b genotype of each plant/line. In the CAPS marker, the amplified fragments by PCR with the same primer set as the FLP marker were digested with AflII, separated on a 2% agarose gel, and visualized with ethidium bromide under UV light. Only the amplified fragment from the Dt1 allele was digested into 201- and 124-bp fragments with AflII. We used a one-way analysis of variance to test for significant differences in the mean number of nodes produced after stage R1 between the GmTFL1b genotypes.
Analysis of Gene Expression by RT-PCR
We analyzed tissue-specific expression and time course-dependent expression at the stem tip for the two GmTFL1 paralogs. Total RNA was isolated from root, cotyledon, leaflet, stem tip, flower, pod, and immature cotyledon of the #6-22Dt1 plants for tissue-specific expression analysis and from the stem tip of different growing stages of both #6-22Dt1 and #6-22dt1 plants for the time course-dependent expression analysis. In the latter experiment, plants were grown in a growth chamber with a constant air temperature of 25°C and average photon flux of 300 μmol photons m−2 s−1 with a daylength of 14.5 h. Bulk stem tips from four plants were sampled every 5 d starting at 15 DAE.
The cDNA was synthesized from total RNA as described. Transcripts of the β-tubulin gene were amplified using PCR with primers β-tub-F and β-tub-R from synthesized cDNA as a control for the RT-PCR of GmTFL1a and GmTFL1b transcripts. The PCR consisted of 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 4 min. RT-PCR was performed using a common forward primer in exon 3 of both paralogs (TFL1a/b-RT-F) and a paralog-specific reverse primer in the 3′ untranslated region (TFL1a-RT-R for GmTFL1a and TFL1b-RT-R for GmTFL1b). PCR with these primers amplifies a 274-bp fragment and a 269-bp fragment for GmTFL1a and GmTFL1b, respectively, which are distinct from those amplified from genomic DNAs (373-bp fragment for GmTFL1a and 364-bp fragment for GmTFL1b). The PCR consisted of 32 cycles of 94°C for 30 s, 58°C for 20 s, and 72°C for 30 s, with a final extension at 72°C for 4 min. The RT-PCR products were separated by electrophoresis on a 1% agarose gel and visualized with ethidium bromide under UV light. The primers used for RT-PCR are listed in Supplemental Table S1.
Transformation of a 4,216-bp Genomic GmTFL1b Sequence of the Dt1 Line
Soybeans were transformed according to the method of Sato et al. (2007). Determinate cv KA was transformed with the binary vector including the genomic region of GmTFL1b from MO using Agrobacterium tumefaciens strain EHA105. A 4,216-bp genomic region that contained the putative promoter and coding region of GmTFL1b was amplified from the genomic DNA of MO using primers TFL1b-transform-F and TFL1b-transform-R. The PCR was performed with KOD plus (Toyobo) using the manufacturer's instructions. The A-tailed amplified fragment was cloned into the pGEM T-Easy vector, and the sequence was confirmed. After excision by EcoRI digestion, the inserted fragment containing the genomic region of GmTFL1b was cloned into the site between the GFP and bar (phosphinothricin resistance gene) cassettes in binary vector pMDC123-GFP (Supplemental Fig. S2). The construct having the 4,216-bp genomic region from MO was designated as pMDC123-GFP-Dt1. A T2 line homozygous for the GFP transgene, which possessed the pMDC123-GFP vector, was used as a negative control. Transformed and nontransformed KA plants were grown in a growth room with a constant air temperature of 23°C and average photon flux of 270 μmol photons m−2 s−1 with a daylength of 16 h.
Virus-Induced Silencing of GmTFL1
Virus-induced silencing of GmTFL1b was done essentially as described previously (Nagamatsu et al., 2007, 2009). The targeted sequences in VIGS were 139-bp sense and antisense sequences of exon 4 of the GmTFL1b gene, which overlapped in a 100-bp sequence (Supplemental Fig. S3). The sense and antisense sequences were amplified from the genomic DNA of MO as template by PCR using primer sets TFL1b-sen-F/TFL1b-sen-R and TFL1b-ant-F/TFL1b-ant-R, respectively. In both cases, a MluI site (ACGCGT) was attached at the 5′ part of the reverse primers for the subsequent plasmid construction. After digestion of the PCR products with MluI, each fragment was cloned into the StuI site and the MluI site of the CMV2-A1 vector (Otagaki et al., 2006). Two plasmids each containing the sense and antisense GmTFL1b sequences were used in the VIGS experiment. In vitro transcription of viral RNA was done as described previously (Nagamatsu et al., 2007, 2009). Leaves of 4-week-old plants of Nicotiana benthamiana that had been dusted with Carborundum were rub inoculated with the in vitro-generated transcripts. The unifoliolate leaves of the 7-d-old 6-22Dt1 plants were then inoculated with sap from the mixed-infected leaves of N. benthamiana plants, which contained both the sense and antisense plasmids. Successful infection of the N. benthamiana and soybean plants without deletion of the inserted sequences was confirmed by RT-PCR of the viral RNA.
After infection, plants were grown in a growth chamber with a constant air temperature of 25°C, average photon flux of 300 μmol photons m−2 s−1, and daylength of 14.5 h. Quantitative RT-PCR was also carried out to analyze the down-regulation of GmTFL1b expression by VIGS in roots of plants at 20 d after emergence. The cDNA was synthesized from total RNA as described except that random 9-mer oligonucleotides (TaKaRa) were used as primers. A forward primer in exon 4 (TFL1b-realtime-F) and a reverse primer in the 3′ untranslated region (TFL1b-RT-R) were used. Primers SOY18SrRNA-861F and SOY18SrRNA-1013R were used for amplifying soybean 18S ribosomal RNA transcripts as an internal control. Quantitative RT-PCR was done as described previously (Nagamatsu et al., 2007). The primers used for RT-PCR are listed in Supplemental Table S1.
DNA Polymorphism Analysis and Association Test
In addition to the three lines (MI, TK, and H4) used for gene isolation, we sequenced the 4,216-bp GmTFL1b region for two indeterminate cultivars, MO and HA, and a determinate NIL of HA, Hdt1. Then, an association test was performed between stem growth habit and DNA polymorphisms detected consistently between determinate and indeterminate lines using 16 soybean cultivars. The DNA polymorphisms analyzed were two SNPs, one located in a SORLIP1 cis-element in the putative promoter region and the other located in exon 4, which caused an amino acid substitution, and a SS in intron 1. The cis-elements in the promoter region of GmTFL1b were analyzed using the program PLACE (http://www.dna.affrc.go.jp/PLACE), and the SNP at the SORLIP1 cis-element was analyzed with a CAPS marker. PCR using primers TFL1b-pro-F and TFL1b-pro-R amplified 410-bp fragments from the genomic DNA of determinate and indeterminate lines. Only the amplified fragments from the Dt1 allele were digested into 217- and 193-bp fragments with NdeI. The SNP in exon 4 was scored with a dCAPS marker. The 176-bp products amplified by PCR using primers TFL1b-ex4-F and TFL1b-ex4-R were digested with XbaI into 155- and 21-bp fragments in the Dt1 allele, but they were not digested in the dt1 allele. In both cases, 10 μL of the PCR products was treated with the enzyme overnight, separated by electrophoresis on a 1% agarose gel or a 3% NuSieve 3:1 agarose gel, and visualized with ethidium bromide under UV light. The SS in intron 1 was analyzed with the CAPS marker as described. Stem growth habit for six plants of each of 16 soybean accessions was evaluated in an experimental field at Hokkaido University in 2006.
Sequence data from this article can be found at the GenBank/EMBL/DDBJ data libraries under accession numbers AB511820 and AB511821 for the genomic sequences of the MI allele (dt1) of GmTFL1b and the TK allele of GmTFL1a, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DNA polymorphisms used for mapping.
Supplemental Figure S2. Structures of the plasmids pMDC123-GFP and pMDC123-GFP-Dt1.
Supplemental Figure S3. The 139-bp sense and antisense sequences in exon 4 of GmTFL1b used for VIGS.
Supplemental Figure S4. Stem growth habit of 21 indeterminate #6-22Dt1 plants that were infected with the recombinant viruses that contained 139-nucleotide sense or antisense sequences of exon 4 of the GmTFL1b gene.
Supplemental Figure S5. Positions of polymorphic sites and their flanking sequences in the GmTFL1a (A) and GmTFL1b (B) genomic sequences.
Supplemental Table S1. List of PCR primers used in this study.
Supplementary Material
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ablett GR, Beversdorf WD, Dirks VA. (1989) Performance and stability of indeterminate and determinate soybean in short-season environments. Crop Sci 29: 1428–1433 [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Settano-Mislata A, Madueño F. (2007) Floral initiation and inflorescence architecture: a comparative view. Ann Bot (Lond) 100: 659–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard RL. (1972) Two genes affecting stem termination in soybeans. Crop Sci 12: 235–239 [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. (1996) Control of inflorescence architecture in Antirrhinum. Nature 379: 791–797 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Carlson JB, Lersten NR. (1987) Reproductive morphology. Boerma HR, Specht JE, , Soybeans: Improvement, Production, and Uses, Ed 2. Agronomy Monograph No. 16. American Society of Agronomy/Crop Science Society of America/Soil Science Society of America, Madison, WI, pp 95–134 [Google Scholar]
- Conti L, Bradley D. (2007) Terminal Flower 1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, et al. (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39: 1464–1490 [Google Scholar]
- Fehr WR, Caviness CE, Burmood DT, Pennington JS. (1971) Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci 11: 929–931 [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. (2000) Redundant regulation of meristem identity and plant architecture by FRUITFUL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Foley TC, Orf JH, Lambert JW. (1986) Performance of related determinate and indeterminate soybean isolines. Crop Sci 26: 5–8 [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Pameau C. (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhao Z, Chen J, Hu X, Luo D. (2006) A putative CENTRORADIALIS/TERMINAL FLOWER 1-like gene, Ljcen1, plays a role in phase transition in Lotus japonicus. J Plant Physiol 163: 436–444 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH. (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequences and microarray data. Plant Physiol 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kilgore-Norquest L, Sneller CH. (2000) Effect of stem termination on soybean traits in southern U.S production systems. Crop Sci 40: 83–90 [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A. (2005) The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by the sequence-specific degradation of the chalcone synthase RNA. Plant Cell Physiol 46: 1879–1883 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinypich A, Ditta GS, Yanofsky MF. (1999) Interaction among APETALA1, LEAFY, and TERMINAL FLOWER 1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fujita T, Yan ZH, Sakamoto S, Xu DH, Abe J. (2007) QTL mapping of domestication related traits in soybean (Glycine max). Ann Bot (Lond) 100: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J. (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260–263 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W. (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336 [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Rai S, Charette M, Cober ER. (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4 and E7 maturity genes in soybean. Genome 46: 1024–1036 [DOI] [PubMed] [Google Scholar]
- Nagamatsu A, Masuta C, Matsuura H, Kitamura K, Abe J, Kanazawa A. (2009) Down-regulation of flavonoid 3′-hydroxylase gene expression by virus-induced silencing in soybean reveals the presence of a threshold mRNA level associated with pigmentation in pubescence. J Plant Physiol 166: 32–39 [DOI] [PubMed] [Google Scholar]
- Nagamatsu A, Masuta C, Senda M, Matsuura H, Kasai A, Hong JS, Kitamura K, Abe J, Kanazawa A. (2007) Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol J 5: 778–790 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otagaki S, Arai M, Takahashi A, Goto K, Hong JS, Masuta C, Kanazawa A. (2006) Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol 23: 259–265 [Google Scholar]
- Ouattara S, Weaver DB. (1994) Effect of growth habit on yield and agronomic characteristics of late-planted soybean. Crop Sci 34: 870–873 [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. (2001) Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Amaya I, Vincert CA, Rothetein S, Carpenter R, Coen ES, Bradley DJ. (1998) A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES. (1999) Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Robinson SL, Wilcox JR. (1998) Comparison of determinate and indeterminate soybean near-isolines and their response to row spacing and planting date. Crop Sci 38: 1554–1557 [Google Scholar]
- Robson F, Costa MMR, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, Putterill J, Coupland G. (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato H, Yamada T, Kita Y, Ishimoto M, Kitamura K. (2007) Production of transgenic plants and their early seed set in Japanese soybean variety, Kariyutaka. Plant Biotechnol 24: 533–536 [Google Scholar]
- Schultz EA, Haughn GW. (1993) Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119: 745–765 [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Bernard RL, Nelson RL. (1997) A third allele at the soybean dt1 locus. Crop Sci 37: 757–762 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thseng FS, Hosokawa S. (1972) Significance of growth habit in soybean breeding. I. Varietal differences in characteristics of growth habit. Jpn J Breed 22: 261–268 [Google Scholar]
- Watanabe S, Tajuddin T, Yamanaka N, Hayashi M, Harada K. (2004) Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed Sci 54: 399–407 [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyetowitz EM. (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pittograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Sato H, Watanabe S, Kawasaki S, Harada K. (2005) Construction and characterization of BAC library of soybean. Euphytica 141: 129–137 [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.