Abstract
Lipases are involved in the generation of jasmonates, which regulate responses to biotic and abiotic stresses. Two sn-1-specific acyl hydrolases, DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1) and DONGLE (DGL), have been reported to be localized in plastids and to be essential and sufficient for jasmonate biosynthesis in Arabidopsis (Arabidopsis thaliana) leaves. Here, we show that levels of 12-oxo-phytodienoic acid (OPDA) and jasmonic acid in three different DGL RNA interference lines and the dad1 mutant were similar to wild-type levels during the early wound response as well as after Pseudomonas infection. Due to the lack of sn-2 substrate specificity, synthesis of dinor OPDA was not expected and also not found to be affected in DGL knockdown and DGL-overexpressing lines. As reported, DAD1 participates in jasmonate formation only in the late wound response. In addition, DGL protein was found to be localized in lipid bodies and not in plastids. Furthermore, jasmonate levels in 16 additional mutants defective in the expression of lipases with predicted chloroplast localization did not show strong differences from wild-type levels after wounding, except for a phospholipase A (PLA) PLA-Iγ1 (At1g06800) mutant line that displayed diminished wound-induced dinor OPDA, OPDA, and jasmonic acid levels. A quadruple mutant defective in four DAD1-like lipases displayed similar jasmonate levels as the mutant line of PLA-Iγ1 after wounding. Hence, we identify PLA-Iγ1 as a novel target gene to manipulate jasmonate biosynthesis. Our results suggest that, in addition to DAD1 and PLA-Iγ1, still unidentified enzymes with sn-1 and sn-2 hydrolase activity are involved in wound- and pathogen-induced jasmonate formation, indicating functional redundancy within the lipase family.
Plants have a complex hormone-based network to regulate their growth and development and to respond to biotic and abiotic stresses. The latter response requires immediate mobilization of defense signals. Those can be either accomplished by stimulus-triggered release of preformed hormones, prohormones from defined compartments, or by rapid de novo synthesis. In the plant kingdom, jasmonic acid (JA) is a widely distributed oxylipin that is biosynthesized de novo in chloroplasts and peroxisomes in acute stress responses. JA is derived from the triunsaturated fatty acids 18:3 and 16:3 esterified in plastidic galactolipids. According to the classical Vick-Zimmerman pathway (Vick and Zimmerman, 1983; Bell et al., 1995), biosynthesis is initiated by release of 18:3 and 16:3 from galactolipids by a lipase. The fatty acid precursors can then be converted by 13-lipoxygenase (13-LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) to 12-oxo-phytodienoic acid (OPDA; a C18 compound) or dinor-12-oxo-phytodienoic acid (dnOPDA; a C16 compound), respectively. Oxo-phytodienoic acids are not only an intermediate in JA biosynthesis, which is accomplished in peroxisomes through reduction and β-oxidation to yield the common product JA, but also electrophilic mediators with signaling properties different from JA (Stintzi et al., 2001; Böttcher and Pollmann, 2009). In an additional step, JA can be converted into biologically active ligands such as jasmonyl-l-Ile and jasmonyl-l-Trp. With respect to JA biosynthesis, a noteworthy special feature of the model plant Arabidopsis (Arabidopsis thaliana) and a few species within the Brassicaceae family is the accumulation of high levels of OPDA and dnOPDA esterified in plastidic galactolipids. In Arabidopsis, an alternative or additional pathway exists that starts with the action of the 13-LOX on 18:3 and 16:3 esterified in galactolipids (Buseman et al., 2006; Kourtchenko et al., 2007). The resulting hydroperoxy galactolipids may then be substrates for AOS and AOC, yielding galactolipid species containing esterified OPDA and dnOPDA in plastidic membranes. These lipid species were termed arabidopsides and may serve as storage lipids that may allow the rapid release of JA biosynthesis intermediates. In any case and in all plants, lipase activity is essential to supply the JA biosynthesis with either fatty acid precursors or OPDA/dnOPDA.
Jasmonates are involved in developmental processes such as reproduction, for which maintenance of a basal threshold level of JA (about 200 pmol g−1 fresh weight) in Arabidopsis flowers is essential (Ishiguro et al., 2001). Furthermore, rapid formation of a high JA level is observed in acute defense responses. For instance, after wounding, JA levels dramatically increase from almost undetectable levels (below 33 pmol g−1 fresh weight) to approximately 250 pmol g−1 fresh weight within 60 s only. Within 3 min after wounding, OPDA and arabidopside levels increase up to 15-fold and 300-fold, respectively. JA levels increase almost simultaneously, reaching levels of more than 10 nmol g−1 fresh weight after 60 to 90 min (Glauser et al., 2009). Apparently, the entire enzymatic machinery involved in the rapid formation of arabidopsides, OPDA, dnOPDA, and JA has to be constitutively present in Arabidopsis leaves. To provide the JA biosynthesis pathway with precursors (fatty acids and/or dnOPDA/OPDA), the involved lipase(s) has to be rapidly activated through a so far unknown activation mechanism.
Two lipases have been reported to be involved in the wound response. The DEFECTIVE IN ANTHER DEHISCENCE1 gene (DAD1) has been shown to encode a chloroplastic glycerolipid lipase. Initially, DAD1 was characterized as a phospholipase (PLA) A1 involved in JA biosynthesis in Arabidopsis flowers (Ishiguro et al., 2001). DAD1-dependent JA formation is essential for Arabidopsis male fertility. Notably, Arabidopsis dad1 T-DNA insertion mutants are still capable of synthesizing JA even in flowers (about 30 pmol g−1 fresh weight), indicating that more than one lipase contributes to JA biosynthesis, which is, however, insufficient to maintain normal reproduction. DAD1 expression is mainly restricted to stamen filaments and is undetectable in unwounded leaves.
Recently, a homolog of DAD1, the DONGLE protein (DGL; At1G05800; PLA-Iα1), which like DAD1 (At2g44810; PLA-Iβ1) is a member of the PLA-I family of lipases, has been proposed as an essential lipase involved in the early wound response in Arabidopsis leaves. A DGL-overexpressing mutant was shown to display a dwarf phenotype with small round leaves and an extremely high basal JA accumulation (about 20 nmol g−1 fresh weight), increased expression of JA-responsive genes, and increased resistance to the necrotrophic fungus Alternaria brassicicola (Hyun et al., 2008). Expression of DGL was not found in flowers, siliques, or roots and was weak or undetectable by reverse transcription (RT)-PCR analysis in 35-d-old unwounded wild-type leaves (Hyun et al., 2008). However, DGL and DAD1 expression increased transiently upon wounding. DGL expression peaked at 1 h after wounding and returned to almost baseline levels after 2 h, while DAD1 expression peaked at 1 h and remained high for at least 4 h.
The dgl-i and dgl-i dad1 knockdown plants were generated and used to elucidate the biological functions of DGL and DAD1 by Hyun et al. (2008). Without wounding, JA was not detectable in both lines, whereas relatively high basal JA levels (50 pmol g−1 fresh weight) were detected in 35-d-old wild-type and dad1 plants. From these experiments, it was concluded that DGL is sufficient to maintain basal JA levels in Arabidopsis leaves. Kinetic analysis of JA accumulation after wounding revealed that DGL is essential for rapid JA biosynthesis within the first hour after wounding, while DAD1 supports JA biosynthesis in the later wounding phase (Hyun et al., 2008).
The PLA superfamily comprises PLA1 and PLA2 enzymes that hydrolyze glycerolipids at their sn-1 and sn-2 positions, respectively. Based on sequence data and biological properties, PLAs can be grouped into four families: PLA1 that preferentially hydrolyze phosphatidylcholine (PC) and glycerolipids at the sn-1 position, PA-PLA1 that prefer phosphatidic acids, secreted sPLA2 with sn-2 specificity, and patatin-like PAT-PLA with little sn-1/sn-2 specificity that are homologous to animal Ca2+-independent PLA2 (Ryu, 2004). Within the Arabidopsis PLA1 family, the class I family comprises seven PLA1 with predicted plastidic transit peptides: DGL (At1g05800; PLA-Iα1), DAD1 (At2g44810; PLA-Iβ1), At2g31690 (PLA-Iα2), At4g16820 (PLA-Iβ2), At1g06800 (PLA-Iγ1), At2g30550 (PLA-Iγ2), and At1g51440 (PLA-Iγ3; Ryu, 2004).
The glycerolipid acyl hydrolase sn-1 specificity of both DGL1 and DAD1 has been confirmed experimentally (Ishiguro et al., 2001; Hyun et al., 2008; Seo et al., 2009). In contrast to the strict sn-1 specificity, the substrate preference toward different glycerolipids is broad. The lipase DAD1 displayed highest lipolytic activity toward PC (100%), while the activity with monogalactosyldiacylglycerol (MGDG) or triacylglycerol (TAG) was only 16% or 6%, respectively (Ishiguro et al., 2001). In contrast, DGL displayed highest activity with digalactosyldiacylglycerol (DGDG; 100%) as substrate and substantially lower activity (50%–25%) with PC, MGDG, and TAG (Seo et al., 2009).
The established high sn-1 substrate specificity of both DGL and DAD1 raises an important question about the lipase involved in dnOPDA formation. The oxylipin dnOPDA occurs endogenously in unwounded Arabidopsis leaves both in free form and in esterified form at the sn-2 position in MGDG and DGDG (Weber et al., 1997; Böttcher and Weiler, 2007). It has been unequivocally shown that dnOPDA is exclusively derived from 7Z,10Z,13Z-hexadecatrienoic acid (16:3), since the Arabidopsis desaturase mutant fad5 is incapable of synthesizing 16:3 and dnOPDA (Weber et al., 1997). In “16:3” plants such as Arabidopsis, 16:3 is synthesized through the “prokaryotic” pathway in plastids and mainly found in the sn-2 position of MGDG and to a lesser extent in the sn-2 position of DGDG (Wallis and Browse, 2002). After wounding, levels of free dnOPDA and OPDA increase simultaneously by more than 15-fold. Levels of dnOPDA and OPDA at 3 min after wounding were found to be 66 and 256 pmol g−1 fresh weight, respectively (Glauser et al., 2009). The sn-1-specific DGL and DAD1 lipases were reported to be sufficient both for maintaining basal levels of JA and wound-induced JA formation, which is compatible with the release of 18:3 or OPDA from the sn-1 position of MGDG and DGDG. However, another lipase remains to be discovered that is capable of releasing 16:3 and/or dnOPDA from the sn-2 position of MGDG and/or DGDG. Since dnOPDA is an alternative precursor for JA synthesis, JA synthesis would not be expected to be completely abolished in mutants deficient in sn-1-specific lipases. Moreover, the relatively weak or absent expression of both DGL and DAD1 suggests that protein levels of both lipases are constitutively low, and this raises the question of how wild-type plants manage to synthesize high levels of jasmonates within a few seconds after wounding (Glauser et al., 2009).
To address these questions, we examined levels of JA, OPDA, and dnOPDA in DGL-overexpressing as well as dgl-i knockdown lines and dad1 mutant plants. However, our data do not support the conclusion that DGL and DAD1 are essential and sufficient for JA synthesis. Therefore, a bioinformatic approach was used to identify unknown putative plastidic lipases. Comparative analyses of oxylipin levels in 16 mutant lines and a quadruple knockout mutant of four members of the PLA-I family in addition to DGL and DAD1 revealed that all were capable of synthesizing JA under basal and wound-induced conditions. Therefore, we propose that lipases in addition to DGL and DAD1 are involved in wound-induced JA formation.
RESULTS
Wound-Induced Jasmonate Accumulation in the dgl-D Mutant Line
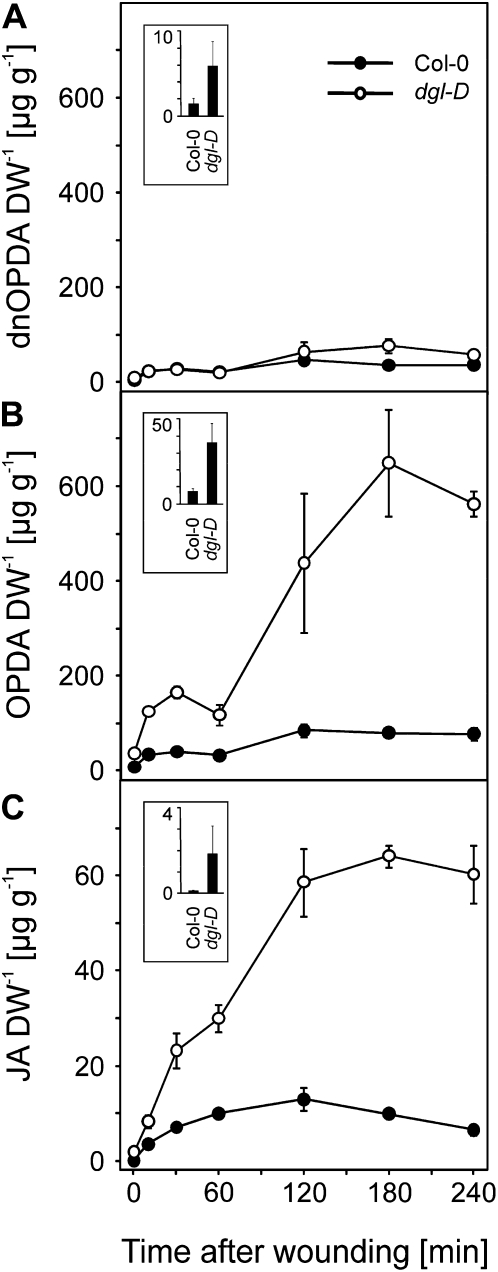
We first investigated jasmonate levels in an activation-tagged mutant Arabidopsis line designated as dgl-D by Hyun et al. (2008) that strongly overexpresses the DGL/PLA-Iα1 (At1g05800) gene. Under basal conditions, levels of JA in the dgl-D line were on average 18 ± 2 times higher and the levels of dnOPDA and OPDA were 4.0 ± 3.1 and 5.1 ± 1.6 times higher, respectively, than in wild-type leaves of 6-week-old plants, which is in agreement with the previously published data. The high basal levels of jasmonates are likely responsible for the morphological and developmental phenotypes of this mutant. After wounding, strong 15- to 60-fold increases of dnOPDA, OPDA, and JA were observed in wild-type plants 60 min after wounding (Fig. 1). However, levels of OPDA and JA were much higher in dgl-D compared with the wild type as early as 15 min after wounding, and levels remained dramatically elevated over the wound-induced wild-type levels throughout the 4-h time course. In contrast, the dgl-D mutation had virtually no impact on the accumulation of dnOPDA throughout the time course, as dnOPDA levels were similar between dgl-D and the wild type. This result is compatible with the known sn-1 substrate specificity of the DGL enzyme, which cannot hydrolyze 16:3 and dnOPDA-containing galactolipids.
Figure 1.
Free oxylipins in dgl-D and wild-type plants after wounding. Comparison of free dnOPDA (A), OPDA (B), and JA (C) in rosette leaves of 6-week-old wild-type plants (black circles) and dgl-D overexpression plants (white circles). Leaves were harvested at time zero and at 10, 30, 60, 120, 180, and 240 min after wounding. Insets display basal jasmonate levels at time zero. Data represent means of three biological replicates ± sd. DW, Dry weight.
Gene Expression and Jasmonate Analyses of Mutant Lines with Reduced PLA-Iα1/DGL Gene Transcript Levels and DAD1 Knockout Plants after Wounding
In parallel to the dgl-D line, we analyzed leaves of 6-week-old plants of the dad1 mutant (Ishiguro et al., 2001) and RNA interference (RNAi)-silenced knockdown lines for the DGL gene in the wound-induced jasmonate synthesis. Since no DGL knockout mutants were available, RNAi lines were generated. A plasmid carrying a gene-specific RNAi construct for silencing of the DGL gene transcript was obtained by the AGRIKOLA consortium (Hilson et al., 2004). The Arabidopsis ecotype Columbia (Col-0) was used for floral vacuum transformation. Two independent transgenic knockdown lines, pla-Iα1 7-1 and 8-1, were selected for further experiments. In addition, the dgl-i line generated by Hyun et al. (2008) was analyzed.
DGL gene transcripts in pla-Iα1 7-1 and 8-1 (Supplemental Fig. S1a) as well as in dgl-i were not detectable by semiquantitative RT-PCR. Analysis of the expression of other PLA-I genes in the pla-Iα1 8-1 line revealed that knockdown of the DGL was specific (i.e. transcripts of other members of the PLA-I family were not diminished; Supplemental Fig. S1b). Since the basal expression of the DGL gene even in wild-type leaves is very low, expression levels were quantified using the more sensitive quantitative RT-PCR technique, which revealed that the DGL transcript levels in untreated leaves of the pla-Iα1 8-1 line and the dgl-i line were on average 24% and 20% of the wild-type level, respectively.
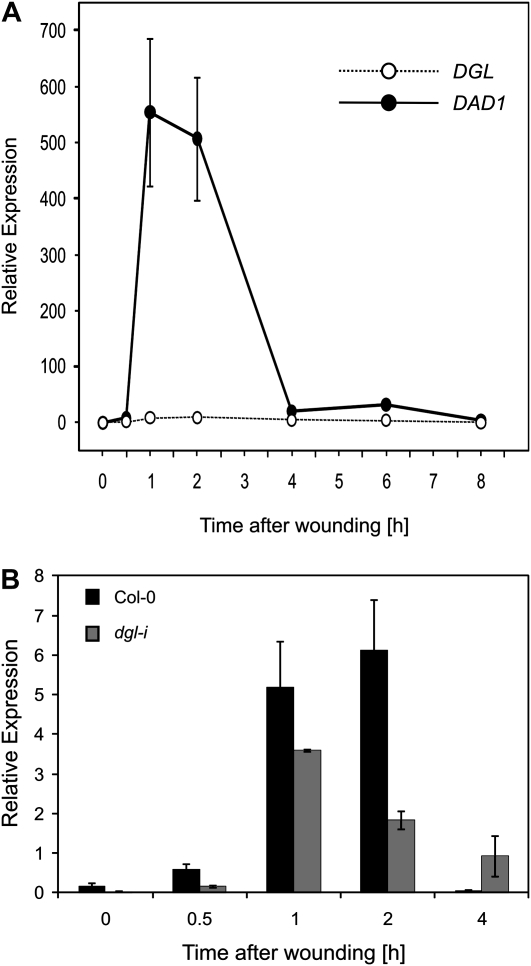
Constitutive and wound-induced expression of DAD1 and DGL/PLA-Iα1 was determined using quantitative RT-PCR in wild-type and mutant plants. In agreement with published data, basal expression of both genes was very low. We found 0.174 ± 0.1 DGL transcripts and 0.423 ± 0.4 DAD1 transcripts per 105 actin transcripts (Fig. 2A). One hour after wounding of wild-type leaves, expression of the DGL and DAD1 genes increased to 5.2 ± 2 and 554 ± 131 transcripts per 105 actin transcripts, respectively. Notably, after wounding, expression of DGL was at all times during the time course substantially lower than DAD1 expression. Constitutive basal expression of DGL in the dgl-i line was found to be 0.033 ± 0.004 transcripts per 105 actin transcripts (Fig. 2B). These findings are in agreement with previously published data by Hyun et al. (2008), albeit we found a somewhat lower accumulation of DGL transcripts after wounding in the wild type, which might be due to different growth conditions or wounding protocols.
Figure 2.
Analyses of transcription levels after wounding. A, Relative gene expression of DGL and DAD1 per 105 actin transcripts in 6-week-old wild-type plant rosette leaves. Leaves were harvested at time zero and at 0.5, 1, 2, 4, 6, and 8 h after wounding. Data represent means of three biological replicates ± sd. B, Relative expression of DGL in rosette leaves of 6-week-old wild-type plants (black bars) and dgl-i RNAi plants (gray bars). Leaves were harvested at time zero and at 0.5, 1, 2, and 4 h after wounding. Data represent means of three biological replicates ± sd.
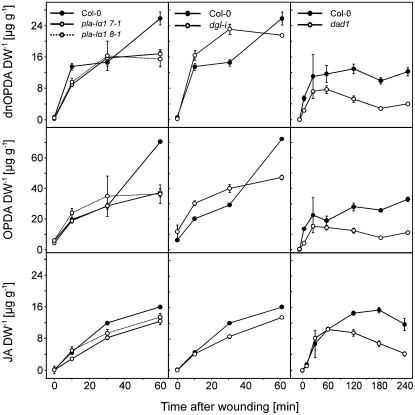
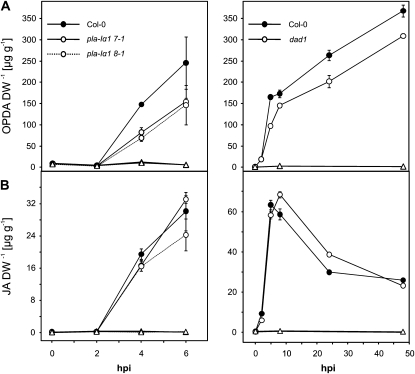
According to the published data on the dgl-i line, complete lack of JA under basal and dramatically diminished JA levels in the early wound response was expected in the three DGL-silenced lines based on Hyun et al. (2008). However, we could not confirm these findings. The dgl-i line had about 65% (0.14 ± 0.025 μg g−1 dry weight) of the wild-type JA level in untreated leaves, about 72% (8.6 ± 0.5 μg g−1 dry weight) of the wild-type JA level 30 min after treatment, and 83% (13.54 ± 0.2 μg g−1 dry weight) of the wild-type JA level 1 h after wounding (Fig. 3). The mutant lines pla-Iα1 7-1 and 8-1 showed similar results. Moreover, OPDA as well as dnOPDA levels within 30 min after wounding did not show significant differences from the wild type either in the mutant line dgl-i or in the two pla-Iα1 lines. One hour after wounding in dgl-i, 65% (46.1 ± 1.4 μg g−1 dry weight) of wild-type levels of OPDA and 83% (21.5 ± 0.4 μg g−1 dry weight) of wild-type levels of dnOPDA were detected. Mutant line pla-Iα1 8-1 had about 51% (36.3 ± 6 μg g−1 dry weight) of wild-type levels of OPDA and 60% (15.5 ± 1.9 μg g−1 dry weight) of wild-type levels of dnOPDA.
Figure 3.
Free oxylipins in DGL knockdown lines and dad1 after wounding. Comparison of free dnOPDA, OPDA, and JA in rosette leaves of 6-week-old wild-type plants pla-Iα1 7-1, pla-Iα1 8-1, dgl-i RNAi plants, and dad1. Leaves were harvested at time zero and at 10, 30, and 60 min after wounding and, for dad1, at 120, 180, and 240 min after wounding. Data represent means of three biological replicates ± sd. DW, Dry weight.
In the knockout mutant dad1, OPDA and JA levels did not significantly differ from the wild type (Fig. 3) in the early phase (0–60 min) of the wound response, as expected (Ishiguro et al., 2001; Hyun et al. 2008). As reported by Hyun et al. (2008), levels of OPDA and JA decreased in the late wound response phase (60–240 min) in dad1, while the levels remained high in the wild type (Fig. 3, right column), indicating that DAD1 is involved in the late phase of JA production. A similar decrease of dnOPDA levels in dad1 but not in wild-type leaves was observed. An alteration in dnOPDA levels by knockout of DAD1 was not expected because, due to its strict sn-1 substrate specificity, DAD1 cannot be directly involved in 16:3 and dnOPDA turnover.
In summary, the measured oxylipin levels in the dad1 mutant support the assumed role of the glycerolipid lipase DAD1 during later stages in wound response (Hyun et al., 2008). However, our data obtained with three independent DGL RNAi lines do not support the concept of an essential role of DGL for maintaining basal levels of JA or for rapid JA formation within a few minutes after wounding. Notably, wound-induced dnOPDA levels were somewhat lower in all DGL and DAD1 transgenic lines at later time points, albeit both enzymes displayed sn-1 specificity and therefore should not be involved directly in the biosynthetic pathway to dnOPDA. Hence, the possibility cannot be excluded that the observed effects of both mutations on jasmonate formation might be indirect.
Localization Studies
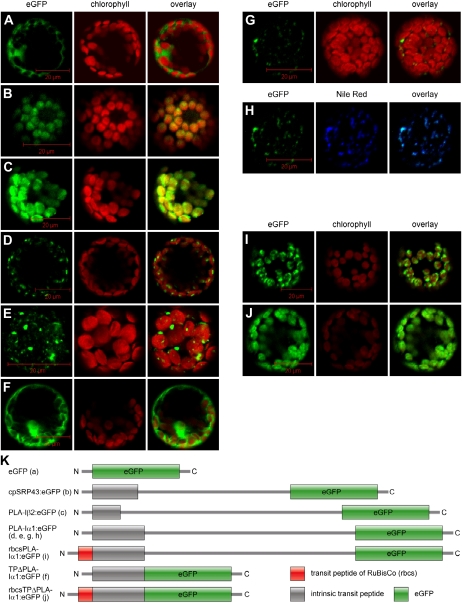
All known enzymes involved in OPDA and dnOPDA biosynthesis in Arabidopsis, such as 13-LOX, AOS, and AOC, are localized in plastids. In addition, the fatty acid precursor of dnOPDA, 16:3, is biosynthesized in plastids and exclusively found esterified in the sn-2 position of MGDG and DGDG. Moreover, the bulk of the cellular OPDA and dnOPDA is found esterified in MGDG and DGDG, termed arabidopsides, which are storage molecules for preformed JA precursors that potentially can be rapidly mobilized when needed (for review, see Böttcher and Pollmann, 2009). Therefore, a plastidic localization of the lipases involved in JA biosynthesis would be expected. Among the seven PLA-I lipases, plastidic localization has unambiguously been shown for PLA-Iβ1 (DAD1) and PLA-Iα2 (a putative TAG lipase; Ishiguro et al., 2001; Padham et al., 2007). Recently, plastidic localization of the other five PLA-I class lipases, including PLA-Iα1 (DGL), has been shown by transient expression of eGFP-tagged fusion proteins (Hyun et al., 2008; Seo et al., 2009). The fluorescence of eGFP fusion proteins appeared as distinct dots colocalized with plastids or localized nearby. The signals of the eGFP-tagged DAD1-like lipases did not completely overlap with the autofluorescence of the chloroplasts, as it was shown for established chloroplastic proteins involved in JA biosynthesis such as DAD1 and AOC1 to AOC4 (Ishiguro et al., 2001; Schaller et al., 2008). Since the reported data (Hyun et al., 2008; Seo et al., 2009) do not unambiguously prove plastidic localization, the subcellular localization of PLA-Iα1 (DGL) was reinvestigated also using eGFP-tagged proteins. As a negative control, the eGFP protein without any fusion partner was employed to visualize cytoplasmatic localization (Fig. 4A). As a positive control, the 43-kD subunit of the chloroplast signal recognition particle (cpSRP43) fused to eGFP was used to show chloroplast localization (Fig. 4B; Schünemann, 2007). Additionally, the full-length PLA-Iβ2 and PLA-Iα1 (DGL) proteins, including their respective 36- and 88-amino acid-long predicted transit peptides (Emanuelsson et al., 1999; Ishiguro et al., 2001), were fused to eGFP. After expression of PLA-Iβ2:eGFP in polyethylene glycol-transformed Arabidopsis protoplasts, we observed a complete overlap of the eGFP fluorescence and chlorophyll autofluorescence (Fig. 4C), thereby verifying the plastidic localization of the PLA-Iβ2 protein (Seo et al., 2009). The PLA-Iα1 (DGL) protein localization was analyzed using the same vectors and conditions as used for PLA-Iβ2. However, the observed signals from the PLA-Iα1:eGFP proteins were significantly smaller in size compared with the autofluorescence signals from plastids and were detected as distinct dots next to the chloroplast (Fig. 4, D and E). To exclude transportation problems due to the size of the eGFP fusion construct, only the predicted transit peptide of PLA-Iα1 was fused to eGFP. The resulting fusion protein TPΔPLA-Iα1:eGFP showed a similar fluorescence distribution as the cytoplasmatic eGFP and did not overlap with the autofluorescence signals from plastids when merged (Fig. 4F). This result indicated that the predicted 88-amino acid plastid transit peptide is not sufficient to facilitate protein import into plastids. Two additional fusion constructs were generated to verify that PLA-Iα1 is not localized within plastids. The cDNA encoding the transit sequence of the small subunit of the chloroplast Rubisco from Pisum sativum was cloned N terminal to full-length PLA-Iα1:eGFP and TPΔPLA-Iα1:eGFP that lacks the endogenous transit sequence of PLA-Iα1 (for schematic overview, see Fig. 4K). These constructs show a clear chloroplast localization (Fig. 4, I and J) and prove that the observed nonchloroplast localization of PLA-Iα1:eGFP is not due to problems associated with the experimental conditions.
Figure 4.
Localization studies of PLA-Iα1:eGFP fusion constructs in Arabidopsis protoplast. In A to G, I, and J, the left column shows the GFP channel and eGFP-tagged proteins (indicated in green), followed by chlorophyll autofluorescence (indicated in red; middle column) and an overlay of both previously described channels (right column). A, The eGFP expression alone is shown as a negative control. B, The plastidic protein cpSPR43 fused to eGFP is shown as a positive control. C, Full-length PLA-Iβ2:eGFP fusion protein. D and E, Full-length PLA-Iα1:eGFP fusion protein in 10× (D) and 40× (E) magnifications. F, Predicted putative transit peptide of PLA-Iα1 tagged with eGFP. G and H, Colocalization experiments with Nile Red-stained lipid bodies. G shows distribution of full-length PLA-Iα1:eGFP fusion protein and chlorophyll autofluorescence, and H shows Nile Red-stained lipid bodies. In H, the left column shows the eGFP channel (indicated in green), followed by Nile Red fluorescence (indicated in blue; middle column) and an overlay of both previously described channels (right column). I, Transit peptide of Rubisco fused to full-length PLA-Iα1 (rbcsPLA-Iα1:eGFP). J, Transit peptide of Rubisco fused to the predicted putative transit peptide of PLA-Iα1 (rbcsTPΔPLA-Iα1:eGFP). K, Schematic overview of the localization constructs.
Instead of a chloroplast localization, we observed colocalization of the full-length PLA-Iα1:eGFP protein with cytoplasmatic lipid bodies (stained with Nile Red) that are often associated with chloroplasts (Fig. 4, G and H; Supplemental Fig. S5). Due to the dynamic nature of the lipid body formation and fusion, not every single lipid body colocalized with PLA-Iα1:eGFP. Similar colocalization patterns of lipid body-associated proteins were recently described for a cytosolic 9-/13-hydroperoxide lyase (De Domenico et al., 2007) and oleosin, which is necessary in lipid body formation (Wahlroos et al., 2003).
Redundancy of Glycerolipid Lipases in Oxylipin Biosynthesis after Wounding
As described above, it is generally assumed that lipases are critically involved in the release of 18:3, 16:3, OPDA, and dnOPDA from galactolipids for biosynthesis of JA. Beside the sn-1-specific DAD1, also lipases with sn-2 or dual sn-1/sn-2 specificity are required to generate free OPDA and dnOPDA. To identify putative galactolipid lipases involved in JA synthesis, we searched for α/β-fold hydrolase, Ser-active lipase, and GDSL lipase sequences in the PFAM (http://pfam.sanger.ac.uk/), Prosite (http://www.expasy.ch/prosite/), and String (http://string.embl.de/; Jensen et al., 2009) databases. Over 219 candidates with predicted hydrolyzing activity were identified and analyzed further for putative plastidic localization using PPDB (http://ppdb.tc.cornell.edu/introduction.aspx) and TargetP (http://www.cbs.dtu.dk/ services/TargetP/; Emanuelsson et al., 1999). In addition to the seven PLA class I (DAD1-like) lipases, 14 putative lipases with predicted plastid transit peptides were identified. For the entire candidate lipases, T-DNA insertion knockout mutants were obtained from the Nottingham Arabidopsis Stock Centre (for details, see “Materials and Methods”). However, we could not generate seeds from the At1g29120 mutants, and two mutants (At5g23530 and At1g20130) did not produce homozygous plants. These three mutants, therefore, were not further analyzed. From the remaining 11 candidate mutants, homozygous plants were generated and used in further experiments (Supplemental Table S1). An antisense knockdown mutant of PLA-Iα2 (At2g31690), which was characterized as plastid triacylglycerol lipase, was obtained from Padham et al. (2007) and shown to display normal basal and wound-induced JA levels (data not shown). The T-DNA mutant line of At1g06800 (pla-Iγ1) was the only one with a Nossen background (Nos). Therefore, Arabidopsis ecotype Nos plants were used as an additional wild-type control. None of the mutant lines showed any phenotype for growth or development under standard growth conditions. All lines described here bolt and set seeds like the corresponding wild types. Seedling sizes were comparable to that of the wild type.
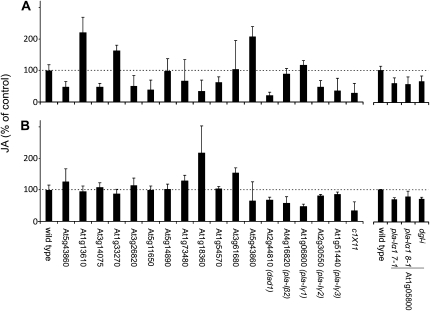
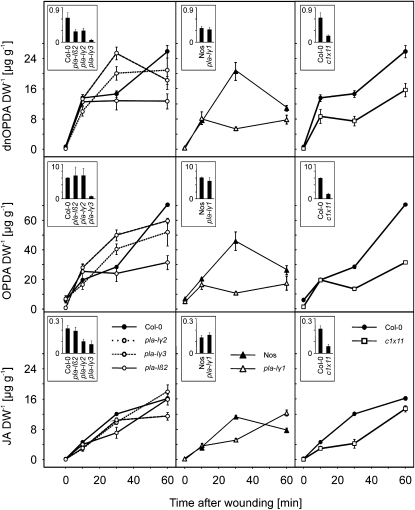
All homozygous lines were analyzed for basal and wound-induced JA levels 30 min after wounding. Data collection for all mutant lines was done in two different laboratories due to availability and workload. To compare the two different data sets, JA levels in untreated and wounded plants were plotted relative to the corresponding wild-type plants, which were grown under the same growth conditions (Fig. 5). Typical JA levels in Arabidopsis Col-0 leaves were 0.21 ± 0.02 μg g−1 dry weight (basal level, mean ± sd, 100%) and 12.08 ± 0.25 μg g−1 dry weight (level 30 min after wounding, mean ± sd, 100%). As shown in Figure 5, basal JA levels and dramatically elevated wound-induced JA levels could be detected in all lipase mutants. Basal levels of JA varied between 22% in the dad1 mutant to 220% of the wild-type level in the mutant line of gene At1g13610. Wound-induced JA levels (30 min) were found in the range of 49% in mutant line pla-Iγ1 to 218% of the wild-type level in the mutant line of gene At1g18360. Less than 65%, with a P value of 0.05 statistically significant reduced wound-induced JA level from the wild type, was monitored in single mutant lines for the genes At1g33270, At4g16820 (PLA-Iβ2), and At1g06800 (PLA-Iγ1). Among the DAD1-like lipases (Fig. 5), pla-Iγ1 displayed the greatest reduction of wound-induced JA levels (49% of wild-type levels) followed by pla-Iβ2 (58%), while pla-Iγ2 and pla-Iγ3 showed no reduced wound-induced JA levels but statistically relevant reduced basal levels, with 48% and 38% of the wild-type basal JA level, respectively. However, none of the tested mutants was completely impaired in JA formation 30 min after wounding. A combination of mutant lines pla-Iβ2, pla-Iγ1, pla-Iγ2, and pla-Iγ3 was chosen to test for functional redundancy of the DAD1-like lipases in JA biosynthesis, because they showed the most severe effects in the early wound response. A kinetic analysis for JA, OPDA, and dnODPA of the four single mutant lines and in addition a quadruple mutant line (pla-Iβ2xpla-Iγ1xpla-Iγ2xpla-Iγ3) termed line c1x11 was performed. The quadruple mutant pla-Iβ2xpla-Iγ1xpla-Iγ2xpla-Iγ3 was gained by serial crosses of the described single mutants. Homozygous T-DNA insertion and loss of detectable transcripts for all four genes were verified in line c1x11, which was used for all experiments (Supplemental Fig. S4). As described above, compared with the wild type, basal levels in the single pla-Iγ2 and pla-Iγ3 mutants had less than 50% of wild-type basal JA and OPDA, and line pla-Iγ2 also had less than 50% of wild-type basal dnOPDA (Fig. 6, insets). Wound-induced jasmonate levels of JA, OPDA, and dnOPDA ranged between 70% and 100% of wild-type levels in both lines. The single mutant line pla-Iβ2 showed significantly reduced JA levels (59% of wild-type level; 7.1 ± 1.4 μg g−1 dry weight) 30 min after wounding and less than 50% of wild-type OPDA and dnOPDA levels 60 min after wounding. Most severe effects were observed in the pla-Iγ1 mutant line 30 min after wounding. Knockout of the PLA-Iγ1 gene resulted in clear reductions of dnOPDA (29% of wild-type level; 5.7 ± 0.4 μg g−1 dry weight), OPDA (25% of wild-type level; 11.2 ± 0.8 μg g−1 dry weight), and JA (49% of wild-type level; 5.4 ± 0.3 μg g−1 dry weight). One hour after wounding, less than 67% of wild-type OPDA and 73% of wild-type dnOPDA levels were detected in mutant line pla-Iγ1. All for pla-Iγ1, given values are statistically significant with a P value of 0.05. As in lines pla-Iγ2 and pla-Iγ3, the reduced basal dnOPDA, OPDA, and JA levels were also seen in the quadruple mutant line c1x11 (less than 29% of wild-type levels). With a P value of 0.05, no differences from wild-type levels 10 min after wounding but less than 50% of wild-type cyclopentanone levels after 30 min and less than 70% of OPDA and dnOPDA wild-type levels were detected 1 h after wounding.
Figure 5.
Comparison of JA contents in lipase mutants after wounding. Relative JA contents in rosette leaves of 6-week-old wild-type plants, mutant lines of putative chloroplast-localized lipases, mutant lines of DAD1-like lipases, and quadruple mutant line c1x11 in untreated leaves (A) and 30 min after wounding (B). Data represent means of at least three biological replicates ± sd.
Figure 6.
Free oxylipins in DAD1-like lipase mutant lines after wounding. Comparison of free dnOPDA, OPDA, and JA in rosette leaves of 6-week-old wild-type plants, in the single mutant lines pla-Iβ2, pla-Iγ2, pla-Iγ3, and pla-Iγ1, and in the quadruple mutant line c1x11. Leaves were harvested at time zero and at 10, 30, and 60 min after wounding. Insets display basal jasmonate levels at time zero. Data represent means of three biological replicates ± sd. DW, Dry weight.
In summary, after wounding, the reduced wound-induced accumulation of JA, OPDA, and dnOPDA in the quadruple line c1x11 was comparable with that seen in the pla-Iγ1 single knockout line, suggesting that PLA-Iγ1 is indeed involved in the early wound response. Based on sequence similarities, PLA-Iγ1 was classified as a class I PLA with predicted sn-1 substrate specificity. Since knockout of PLA-Iγ1 caused a clear reduction in dnOPDA formation also, the protein may display dual sn-1/sn-2 substrate specificity or is not directly involved in galactolipid hydrolysis. Notably, even knockout of four DAD1-like lipases was not sufficient to shut down initial wound-induced JA formation completely. Moreover, JA levels reached nearly wild-type levels in the later stages of the wound response (more than 60 min; Fig. 6), which might in part be due to wound-inducible DAD1 activity. However, it needs to be stressed that PLA-Iγ1 and DAD1 cooperate with additional sn-2- or dual sn-1/sn-2-specific unknown plastidic lipases that are able to release 16:3 and dnOPDA from plastid membranes. Alternatively or additionally, several extraplastidic lipases may become activated after wounding, which may hydrolyze phospholipids and galactolipids originating from disrupted plastids in a rather unspecific way.
DGL and DAD1 Are Not Essential for Jasmonate Biosynthesis in the Interaction of Arabidopsis with Avirulent Pseudomonas syringae Bacteria
Wounding with tweezers causes massive tissue and cell destruction that may lead to unspecific hydrolytic degradation of structural lipids, including plastidic galactolipids. Fatty acids released from damaged organelles may serve as an exogenous or extraplastidic source of substrates for JA biosynthesis. To avoid a situation in which lipases potentially attack membrane lipids that would be inaccessible in an intact cell, elicitors or pathogens can be used to trigger a massive formation of jasmonates. Infection of Arabidopsis Col-0 leaves with the avirulent Pseudomonas syringae pv tomato strain DC3000 expressing the avirulence gene avrRPM1 has been shown to trigger a biphasic jasmonate accumulation that reaches a first maximum 5 to 10 h after infection (Grun et al., 2007). This interaction model was used to study the functions of DGL and DAD1. OPDA and JA levels were determined after infection of wild-type leaves and leaves of the two DGL RNAi lines pla-Iα1 7-1 and 8-1 as well as the dad1 mutant (Fig. 7). We observed a dramatic accumulation of OPDA and JA 4 h after infiltration, which even exceeded the relatively high jasmonate levels after wounding. However, accumulation of OPDA and JA was not affected in the dgl and dad1 mutant lines, suggesting that both enzymes are not involved in P. syringae-triggered jasmonate biosynthesis or that their deficiency can be fully compensated by other redundant lipases.
Figure 7.
Free oxylipins in DGL knockdown lines and the wild type after pathogen treatment. Comparison of free OPDA (A) and JA (B) in rosette leaves of 6-week-old wild-type plants and in pla-Iα1 7-1, pla-Iα1 8-1, and dad1 after Pseudomonas DC3000 (avrRPM1) treatment. Leaves were harvested at 0, 2, 4, 6, 8, 24, and 48 h post infection (hpi). Circles represent the oxylipin amounts in infected plant material, and triangles represent the oxylipin amounts in mock-infiltrated plant material. Data represent means of three biological replicates ± sd. DW, Dry weight.
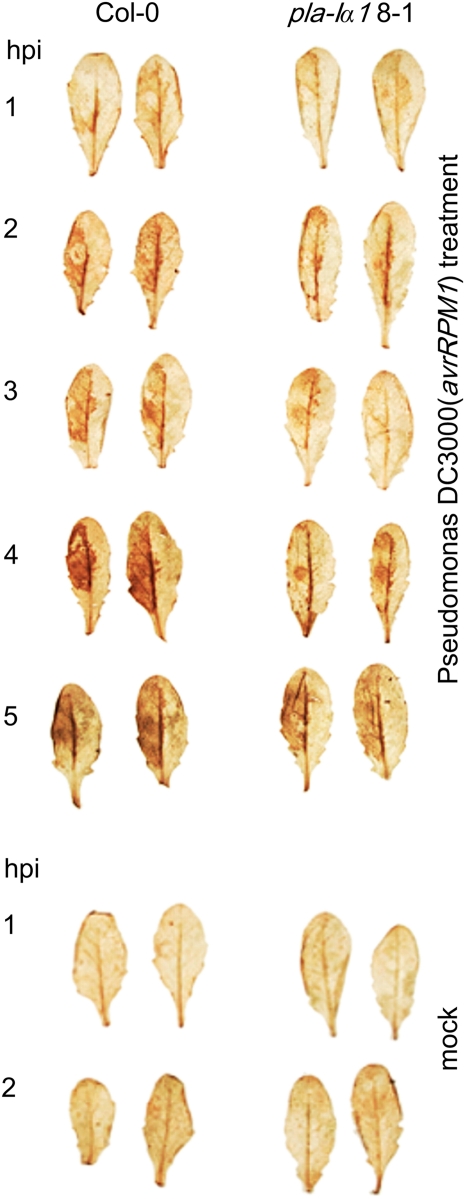
In wild-type plants, the infiltration site appeared yellow to gray and the infiltrated tissue was dead 3 d after infiltration. The knockdown lines pla-Iα1 7-1 and 8-1 showed a slower and reduced development of these symptoms in comparison with the wild-type plants within the first 2 d after infiltration. Although retardation in cell death in both mutant lines was observed several times, no reliable bacterial titer 2 d after infiltration can be given, due to very high variation in the mutant lines. Nevertheless, 3 d after treatment, no phenotypic differences among pla-Iα1 7-1, pla-Iα1 8-1, and Col-0 could be observed (Supplemental Fig. S2). Also, no differences in bacterial growth between analyzed lines were detected at day 3 (Supplemental Fig. S3).
Avirulent pathogens also elicit reactive oxygen species (ROS) accumulation (Lamb and Dixon, 1997), which is dependent on PLA activity (Meijer and Munnik, 2003; Andersson et al., 2006). To investigate whether the pla-Iα1 8-1 line was impaired in the production of ROS due to lacking lipase activity, we stained infiltrated leaves from the wild type and pla-Iα1 8-1 with diaminobenzidine to visualize ROS accumulation (Fig. 8) after P. syringae treatment (Thordal-Christensen et al., 1997). We observed differences in staining intensity between 1 and 4 h after treatment. Staining of infiltrated leaves of the pla-Iα1 8-1 knockdown line was not as intense as in the wild type, which was an indicator of lower levels of ROS in pla-Iα1 8-1. However, 5 to 6 h after infiltration, the staining of pla-Iα1 8-1 leaves was similar to that of wild-type leaves.
Figure 8.
Occurrence of ROS after infiltration with Pseudomonas DC3000 (avrRPM1). Leaves of wild-type and transgenic pla-Iα1 8-1 plants were infiltrated with a 5 × 107 cfu mL−1 suspension of bacteria. To visualize ROS produced, leaves were harvested at the indicated time points (hpi, h post infection) and stained with 3,3′-diaminobenzidine. [See online article for color version of this figure.]
Hence, P. syringae infection of Arabidopsis leaves triggers a massive JA formation, but we did not observe an impact of DGL or DAD1 gene expression deficiency on JA biosynthesis and bacterial growth, while ROS production and symptom development appeared to be delayed in the early phase of the interaction. As for the wound response, our data do not suggest an essential role for PLA-Iα1/DGL and DAD1 in JA biosynthesis upon pathogen infection.
DISCUSSION
In the acute wound and stress response, jasmonate synthesis has to rely on constitutively present biosynthetic enzymes and is controlled only at the level of substrate availability (Laudert and Weiler, 1998; Park et al., 2002). Immediately after wounding, lipid acyl hydrolases provide membrane-bound fatty acids and oxo-phytodienoic acids as substrates for JA synthesis. After initiation of JA biosynthesis, most involved enzymes have been shown to be up-regulated at the transcriptional level (Park et al., 2002). This may increase the pathway capacity (i.e. the ability to synthesize high amounts of jasmonates) at later time points during prolonged or repetitive stress episodes (Wasternack, 2007).
Galactolipids are exclusively found in the outer and inner membranes as well as thylakoid membranes of plastids. Undoubtedly, they are substrates for acyl hydrolases, since levels of free dnOPDA generated from 16:3 (exclusively found in galactolipids at the sn-2 position) increase after wounding. In addition, galactolipids containing OPDA at the sn-1/sn-2 positions and dnOPDA at the sn-2 position (arabidopsides) are rapidly turned over during stresses. Therefore, they are thought to represent a store of preformed prohormones that can be rapidly mobilized by acyl hydrolases. Release of 16:3 and dnOPDA from galactolipids requires an acyl hydrolase capable of cleaving sn-2 ester bonds.
There are several indications that the acyl-hydrolyzing enzymes involved in signal generation are localized within the chloroplast and are associated with thylakoid or inner envelope membranes. The enzymes 13-LOX, AOS, and AOC are all localized in chloroplasts, and there is strong evidence that they act not only on free 16:3 and 18:3 fatty acids in Arabidopsis but rather initially generate arabidopsides. Arabidopsides dramatically accumulate after wounding, which suggests that they are initially formed in thylakoid and inner envelope membranes from which dnOPDA and OPDA can be released by plastidic lipases. Furthermore, for JA synthesis during flower development, it has been shown that the responsible lipase DAD1 is localized in plastids (Ishiguro et al., 2001). Therefore, we focused our study on putative plastidic lipases.
First, the lipases DGL and DAD1 were investigated because these enzymes were reported to be essential for JA synthesis in Arabidopsis leaves after wounding (Ishiguro et al., 2001; Hyun et al., 2008). Also, in tobacco (Nicotiana tabacum), a plastidic member of the PLA1 family, designated GLA1-I (FJ821553), involved directly or indirectly in early wound- and elicitor-induced JA formation was recently identified (Kallenbach et al., 2010). GLA1 shares 46% and 37% identity with Arabidopsis DAD1 and DGL, respectively, and also displays sn-1 substrate specificity toward PC, MGDG, and TAG. In gla1 knockdown plants, JA levels showed reduction of wound-induced JA levels by more than 75% 20 min after wounding or 1 h after elicitation compared with wild-type tobacco plants. In agreement with Hyun et al. (2008), the Arabidopsis dad1 mutant showed lower JA levels at late time points and the DGL-overexpressing line showed elevated levels of JA. However, knockdown of DGL did not have the reported strong effects on basal and early wound-induced JA levels. These discrepancies might be due to different reasons. First, already the basal levels show high variability. Basal JA levels reported in the literature range from undetectable levels (Glauser et al., 2009) up to 500 pmol g−1 fresh weight (Hyun et al., 2008). This is probably due to differences in growth conditions and the physiological as well as the developmental status of the analyzed plants or leaves. Second, variations of the wounding protocol might result in different effects on gene expression and oxylipin levels. Nevertheless, our data do not support an essential function of DGL in the early wound response. The investigation of oxylipin levels after infection with P. syringae also showed no strong differences between the wild type, dad1, and dgl mutant lines (Fig. 7). This indicates that DAD1 and DGL are not involved in pathogen-induced JA formation.
Moreover, based on the sn-1 specificity of DAD1 and DGL, it would not be expected that down-regulation or knockout results in a complete lack of wound-triggered JA increase. The dnOPDA levels increase after wounding or pathogen infection in the wild type. Due to the limited substrate specificity of DGL and DAD1, additional sn-2 or unspecific sn-1/sn-2 lipases must exist and provide free dnOPDA, which in turn would allow JA synthesis in the absence of DGL and DAD1. Since levels of all jasmonates including dnOPDA are reduced in the late phase of the wound response in DGL and DAD1 mutants, these lipases may have an indirect effect on JA formation.
There are more examples for mutants that have supposedly indirect effects on JA levels. The JA-overaccumulating mutant cev1 is defective in cellulose synthase CeSA3 (Ellis and Turner, 2001; Ellis et al., 2002), and joe1/cpl1 is defective in an RNA polymerase II C-terminal domain phosphatase (Jensen et al., 2002; Matsuda et al., 2009). However, neither of these two proteins is involved in JA biosynthesis directly.
On the search for lipases involved in jasmonate biosynthesis in Arabidopsis, we identified by in silico analysis 14 lipases in addition to the seven DAD1-like lipases that are predicted to be localized in chloroplasts. From all these candidate lipases, we analyzed in total 18 different mutant lines in which a single lipase was knocked down or knocked out. However, albeit some of these lines displayed reduced wound-induced JA levels, all of these lines still accumulated JA after wounding (Fig. 5). On the transcript level, none of the DAD1-like lipases and only three of the additional lipases display induced expression (more than 2-fold) at 30 min after wounding, according to Genevestigator (Zimmermann et al., 2004). Among the Arabidopsis lipase knockdown and knockout lines analyzed (Fig. 5) and the triacylglycerol lipase PLA-Iα2 (data not shown), only the knockdown line pla-Iγ1 was identified as a lipase, which significantly contributes to dnOPDA and OPDA formation within the first hour in wound response, while DAD1 contributes to jasmonate formation in the late wound response. Potentially, PLA-Iγ1 could be involved in the release of preformed cyclopentenone jasmonates from arabidopsides directly; however, this issue and the sn-1/sn-2 substrate specificity remain to be clarified
These experiments suggest that several redundant plastidic lipases are involved directly or indirectly in jasmonate biosynthesis after wounding.
To this end, however, the possibility that extraplastidic lipases are involved in wound- and pathogen-induced JA formation cannot be excluded. Both MGDG and DGDG are present in the inner and the outer chloroplast envelope membranes and therefore could also be substrates for cytosolic lipases that associate with the outer chloroplast membrane. Notably, levels of galactolipid species containing sn-1 OPDA and sn-2 dnOPDA (MGDG, arabidopsides A and E; DGDG, arabidopside C) are high under basal conditions and increase dramatically within a few seconds after wounding (Glauser et al., 2009). Therefore, these species are expected to be synthesized in situ at the inner chloroplast envelope or thylakoid membrane due to the localization of 13-LOX, AOS, and AOC (Buseman et al., 2006; Kourtchenko et al., 2007). Similar to MGDG, arabidopsides may be translocated to the outer envelope membrane, where they might be also accessible for cytosolic lipases.
Notably, a cytosolic glycerolipid lipase, designated AtPLA1 (At1g61850), has been reported to provide precursors for basal JA production, suggesting the involvement of more than one lipase in basal JA formation. Disruption of AtPLA1 by T-DNA insertion resulted in about 50% reduced levels of free 18:3 and JA (compared with the wild type) but did not reduce wound- or pathogen-induced JA accumulation (Yang et al., 2007). AtPLA1 encodes a protein with a patatin catalytic domain but an overall sequence more similar to mammalian Ca2+-independent iPLA2 than other Arabidopsis patatin-like and iPLA2-like proteins. AtPLA1 and patatin-like acyl hydrolases possess nonspecific sn-1/sn-2 acyl hydrolase activity toward several types of lipids, including phospholipids and galactolipids. However, there is no direct evidence for an involvement of patatin-like enzymes in the production of JA in response to wounding or pathogens.
Jasmonate synthesis is known to be controlled at the level of substrate availability; therefore, lipases have a key regulatory function. Mutant lines of 18 different acyl hydrolases including DGL and DAD1 were analyzed for wound-induced jasmonate levels. However, none of the lipase mutants defective in a single lipase and even a quadruple mutant line was completely compromised in JA formation under basal and wound-induced conditions. Instead, our results indicate that several lipases with both sn-1 and sn-2 (or dual sn-1/sn-2) galactolipid substrate specificity participate in JA formation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
In all experiments, plants were grown on soil under standard short-day conditions (8 h of light/16 h of dark, 20°C–25°C, 200 μE m−2 s−1). The Arabidopsis (Arabidopsis thaliana) Col-0 and Nos ecotypes were used as control lines. Seeds from T-DNA mutant lines impaired in genes PLA-Iβ2 (SM_3_20786), PLA-Iγ1 (RATM12_1251_1), PLA-Iγ2 (SALK_003105), PLA-Iγ3 (SALK_004710), At5g43860 (SAIL_646_E09), At1g13610 (SAIL_897_D11), At3g14075 (SALK_014323), At1g33270 (SALK_139011C), At3g26820 (SALK_139280), At5g11650 (SAIL_716_F08), At5g14890 (SALK_079693), At1g73480 (SALK-095195c), At1g18360 (SALK-072002c), At1g54570 (SALK_076354c), and At3g61680 (SALK_147687c) were obtained from the Nottingham Arabidopsis Stock Centre (www.arabidopsis.org). Seeds from dgl-i and dgl-D were obtained from I. Lee (National Research Laboratory of Plant Developmental Genetics, Department of Biological Sciences, Seoul National University). The knockdown mutant lines with reduced transcript levels of PLA-Iα1/DGL (At1g05800) were generated using a binary vector from the AGRIKOLA project (www.agrikola.org; Hilson et al., 2004) purchased from the Nottingham Arabidopsis Stock Centre (stock no. N246587). The Agrobacterium tumefaciens strain GV3101::pMP90 carrying the RNAi construct was used to transform Col-0 plants under vacuum according to Bechtold and Pelletier (1998). Seeds of transformed lines were grown on Murashige and Skoog plates, transferred to soil after 2 weeks, and selected by spraying with 50 mg L−1 BASTA solution (Bayer CropScience). Neither pla-Iα1 7-1 nor pla-Iα1 8-1 showed any phenotype for growth or development under standard growth conditions. They bolt and set up seeds as wild-type Col-0. Also, seedling sizes were comparable. The dgl-i line was reported to display larger seedling size (Hyun et al., 2008), which, however, was not observed under our growth conditions. To gain the quadruple mutant pla-Iβ2xpla-Iγ1xpla-Iγ2xpla-Iγ3, first pla-Iβ2 was crossed with pla-Iγ1 and with pla-Iγ3 and homozygous pla-Iβ2xpla-Iγ1andpla-Iβ2xpla-Iγ3 double mutants were selected by growing on antibiotics-containing media and verified by PCR. Second, pla-Iβ2xpla-Iγ1andpla-Iβ2xpla-Iγ3 were crossed, resulting in the pla-Iβ2xpla-Iγ1xpla-Iγ3 triple mutant. In a third crossing step, the verified homozygous triple mutant was crossed with pla-Iγ2. Additional information about the primers used for verification of the mutants is given in Supplemental Table S1.
Gene Expression Studies
Total RNA was extracted from leaves using the RNA-T Easy Kit (Omega) following the manufacturer's protocol. Quantitative RT-PCR was performed with 1 μg of RNA, and Moloney murine leukemia virus reverse transcriptase RNase H Minus (point mutant; Fermentas) was used. Analyses were run on a Realplex Mastercyclerepgradient S Lightcycler system (Eppendorf) according to the manufacturer's recommendations and using actin2 (At5g09810) and actin8 (At1g49240) for standardization. For full information about the primers, see Supplemental Table S2.
Wounding and Pathogen Treatment
The leaves of 6-week-old plants were wounded by squeezing two to three times per leaf with tweezers. The avirulent strain Pseudomonas syringae pv tomato strain DC3000 (avrRPM1) was used for all of the pathogen experiments. For analysis of free jasmonates and ROS, pressure infiltration with a 5 × 107 colony-forming units (cfu) mL−1 suspension of bacteria was used for lines pla-Iα1 7-1 and 8-1. Pathogen infiltrations were performed as described by Katagiri et al. (2002). For line dad1, a 108 cfu mL−1 suspension of bacteria was used. Pathogen infiltrations were performed as described by Katagiri et al. (2002), except that in the bacteria resuspension medium, 10 mm MgSO4 was used instead of 10 mm MgCl2. To visualize the accumulation of produced ROS, leaves were stained with 3,3′-diaminobenzidine according to Thordal-Christensen et al. (1997).
Quantitation of Free Jasmonates
For jasmonate analysis, 200 to 300 mg of fresh plant material was extracted with 1.5 mL of 2-isopropanol at 80°C for 15 min. Samples were centrifuged for 10 min at 14,000 rpm, and the supernatant was collected. The remaining plant material was reextracted twice with 1.5 mL of chloroform:2-isopropanol (2:1, v/v) and 1.5 mL of chloroform:methanol (1:2, v/v) until the plant material appeared white. Samples were centrifuged after each extraction step, and the supernatants were combined. The extract was dried under a stream of nitrogen at 60°C. For quantification of jasmonates, 80 ng of [18O2]JA and 100 ng of [18O2]OPDA were added as internal standards for JA and dnOPDA/OPDA, respectively. Samples were dissolved in 60 μL of 1 mm ammonium acetate in methanol, and 8 μL was analyzed by liquid chromatography-tandem mass spectrometry.
Ultra HPLC (UPLC)/tandem mass spectrometry analyses were performed on a Waters Micromass Quattro Premier triple quadrupole mass spectrometer with an electrospray interface coupled to an Acquity UPLC system (Waters). The separation was carried out using a Waters UPLC BEH C18 column (2.1 × 30 mm, 1.7-μm particle size with a 2.1- × 5-mm guard column) with the following solvent system: A = 1 mm aqueous ammonium acetate, B = acetonitrile. A gradient elution was performed at a flow rate of 0.3 mL min−1 at 27°C: 5% to 60% B in 3 min followed by 100% B for 2 min and reconditioning at 5% B for 3 min. The electrospray source was operated in the negative ionization mode (ESI−) at a capillary voltage of 3.0 kV and 120°C. Nitrogen and argon were used as desolvation and collision gas, respectively. The optimized conditions were as follows: temperature, 400°C; desolvation gas flow, 800 mL min−1; collision gas flow, 0.27 mL min−1. Collision energies (CE) and dwell times (DT) were specific for each compound/internal standard pair; the parameters used were [18O2]JA CE-17, DT 25 ms, JA CE-17, DT 25 ms, [18O2]OPDA CE-26, DT 25 ms, OPDA CE-26, DT 25 ms, and dnOPDA CE-16, DT 25 ms. Data were acquired and analyzed using Mass Lynx version 4.1 software. Analysis of the compounds was based on appropriate multiple reaction monitoring of ion pairs for labeled and endogenous JA, OPDA, and dnOPDA using the following mass transitions: [18O2]JA 213 > 63, JA 209 > 59, [18O2]OPDA 295 > 165, OPDA 291 > 165, dnOPDA 263 > 165.
Localization Studies
The various eGFP fusion constructs were generated using the pSP-EGFP vector (Pollmann et al., 2006) by cloning the open reading frame or truncated versions, which were then amplified by PCR from a plasmid containing the corresponding template using gene-specific primers with additional restriction sites. The cDNA encoding the transit sequence of the small subunit of chloroplast Rubisco from Pisum sativum (residues 1–58; full-length Rubisco) were amplified by PCR from pNH7 (T. Bals and D. Schünemann, unpublished data) using primers containing unique restriction sites for KpnI and BamHI. The PCR product was restricted and cloned into the KpnI/BamHI sites of PLA-Iα1:eGFP and TPΔPLA-Iα1:eGFP. The sequences of all the fusion constructs were verified by commercial DNA sequencing. The primers used are listed in Supplemental Table S3. All molecular techniques were performed using standard protocols according to Sambrook et al. (1989) or Ausubel et al. (2000). All localization studies were done with Arabidopsis mesophyll protoplasts that were isolated from leaves of 4-week-old plants and transiently transfected according to Gerdes et al. (2006). The transfected protoplasts were analyzed with a confocal laser scanning system (Zeiss LSM 510 Meta). A krypton/argon laser that produced an excitation light of 488 nm together with a meta-detector system with emission filters set to 500 to 530 nm for GFP and 650 to 798 nm for chlorophyll allowed for the simultaneous detection of GFP- and chlorophyll-mediated fluorescence. To detect Nile Red fluorescence, an excitation wavelength of 488 nm was used, and the emission was recorded by a filter set to 560 to 615 nm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PLA-Iα1 (At1g05800), PLA-Iα2 (At2g31690), PLA-Iβ1 (At2g44810), PLA-Iβ2 (At4g16820), PLA-Iγ1 (At1g06800), PLA-Iγ2 (At2g30550), PLA-Iγ3 (At1g51440), At5g43860, At1g13610, At3g14075, At3g26820, At5g11650, At5g14890, At1g33270, At1g73480, At1g18360, At1g54570, and At3g61680.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Verification of pla-Iα1 knockdown lines.
Supplemental Figure S2. Gene-silencing phenotype of pla-Iα1 lines after treatment with Pseudomonas DC3000 (avrRPM1).
Supplemental Figure S3. Bacterial growth of Pseudomonas DC3000 (avrRPM1).
Supplemental Figure S4. Verification of quadruple mutant pla-Iβ2xpla-Iγ1xpla-Iγ2xpla-Iγ3, line c1x11.
Supplemental Figure S5. Colocalization studies of eGFP fusion proteins in Arabidopsis protoplast.
Supplemental Table S1. Primers used for verification of T-DNA insertion mutants and the pla-Iα1 knockdown line.
Supplemental Table S2. Primers used for gene expression analyses.
Supplemental Table S3. Primers used for cloning and verification of eGFP fusion constructs.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Christine Boettcher (Ruhr-Universität Bochum) for providing standards, lipids, and technical support as well as Petra Duechting and Silke Funke (Ruhr-Universität Bochum) for helpful technical assistance. We also thank Thomas Dandekar (Bioinformatic Division, Universität Wuerzburg) for help with the in silico analyses. We are very grateful to Ilha Lee (Seoul National University) for providing the dgl-D and dgl-i lines and for discussion of results. The Pseudomonas strains were kindly provided by M. Grant from the University of Exeter.
References
- Andersson MX, Hamberg M, Kourtchenko O, Brunnstrom A, McPhail KL, Gerwick WH, Gobel C, Feussner I, Ellerstrom M. (2006) Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside. J Biol Chem 281: 31528–31537 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmann JD, Smith JA, Struhl K, (1995) Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley Interscience, New York, pp 9.7.12–9.7.18 [Google Scholar]
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration . Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE. (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Pollmann S. (2009) Plant oxylipins: plant responses to 12-oxo-phytodienoic acid governed by its specific structural and functional properties. FEBS J 276: 4693–4704 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Weiler EW. (2007) Cyclo-oxylipin-galactolipids in plants: occurrence and dynamics. Planta 226: 629–637 [DOI] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, Roth MR, Esch SW, Shah J, Williams TD, et al. (2006) Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol 142: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico S, Tsesmetzis N, Di Sansebastiano GP, Hughes RK, Casey R, Santino A. (2007) Subcellular localisation of Medicago truncatula 9/13-hydroperoxide lyase reveals a new localisation pattern and activation mechanism for CYP74C enzymes. BMC Plant Biol 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes L, Bals T, Klostermann E, Karl M, Philippar K, Huenken M, Soll J, Schünemann D. (2006) A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J Biol Chem 281: 16632–16642 [DOI] [PubMed] [Google Scholar]
- Glauser G, Dubugnon L, Mousavi SA, Rudaz S, Wolfender JL, Farmer EE. (2009) Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem 284: 34506–34513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun C, Berger S, Matthes D, Mueller MJ. (2007) Early accumulation of non-enzymatically synthesized oxylipins in Arabidopsis after infection with P. syringae. Funct Plant Biol 34: 1–7 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Choi S, Hwang HJ, Yu J, Nam SJ, Ko J, Park JY, Seo YS, Kim EY, Ryu SB, et al. (2008) Cooperation and functional diversification of two closely related galactolipase genes for jasmonic acid (JA) biosynthesis. Dev Cell 14: 183–192 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. (2001) The defective in anther dehiscence1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AB, Raventos D, Mundy J. (2002) Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J 29: 595–606 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. (2009) STRING 8: a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. (2010) Nicotiana attenuata SIPK, WIPK, NPR1 and fatty acid-amino acid conjugates participate in the induction of JA biosynthesis by affecting early enzymatic steps in the pathway . Plant Physiol 152: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. Meyerowitz EM, Somerville CR, , The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0039, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko O, Andersson MX, Hamberg M, Brunnstrom A, Gobel C, McPhail KL, Gerwick WH, Feussner I, Ellerstrom M. (2007) Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonic acid (JA) signaling dependence. Plant Physiol 145: 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Laudert D, Weiler EW. (1998) Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signaling. Plant J 15: 675–684 [DOI] [PubMed] [Google Scholar]
- Matsuda O, Sakamoto H, Nakao Y, Oda K, Iba K. (2009) CTD phosphatases in the attenuation of wound-induced transcription of jasmonic acid biosynthetic genes in Arabidopsis. Plant J 57: 96–108 [DOI] [PubMed] [Google Scholar]
- Meijer HT, Munnik T. (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Padham AK, Hopkins MT, Wang TW, McNamara LM, Lo M, Richardson LGL, Smith MD, Taylor CA, Thompson JE. (2007) Characterization of a plastid triacylglycerol lipase from Arabidopsis. Plant Physiol 143: 1372–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW. (2006) Subcellular localisation and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Ryu SB. (2004) Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci 9: 229–235 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [Google Scholar]
- Schaller F, Zerbe P, Reinbothe S, Reinbothe C, Hofmann E, Pollmann S. (2008) The allene oxide cyclase family of Arabidopsis thaliana: localization and cyclization. FEBS J 275: 2428–2441 [DOI] [PubMed] [Google Scholar]
- Schünemann D. (2007) Mechanisms of protein import into thylakoids of chloroplasts. Biol Chem 388: 907–915 [DOI] [PubMed] [Google Scholar]
- Seo YS, Kim EY, Kim JH, Kim WT. (2009) Enzymatic characterization of class 1 DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Lett 583: 2301–2307 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB. (1997) Subcellular localisation of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Vick BA, Zimmerman DC. (1983) The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem Biophys Res Commun 111: 470–477 [DOI] [PubMed] [Google Scholar]
- Wahlroos T, Soukka J, Denesyuk A, Wahlroos R, Korpela T, Kilby NJ. (2003) Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis 35: 125–132 [DOI] [PubMed] [Google Scholar]
- Wallis JG, Browse J. (2002) Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res 41: 254–278 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonic acid (JA)s: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE. (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94: 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY, Devaiah SP, Pan XQ, Isaac G, Welti R, Wang XM. (2007) AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J Biol Chem 282: 18116–18128 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.