SUMMARY
Signal termination in two-component systems occurs by loss of the phosphoryl group from the response regulator protein. This review explores our current understanding of the structures, catalytic mechanisms and means of regulation of the known families of phosphatases that catalyze response regulator dephosphorylation. The CheZ and CheC/CheX/FliY families, despite different overall structures, employ identical catalytic strategies using an amide side chain to orient a water molecule for in-line attack of the aspartyl phosphate. Spo0E phosphatases contain sequence and structural features that suggest a strategy similar to the chemotaxis phosphatases but the mechanism used by the Rap phosphatases is not yet elucidated. Identification of features shared by phosphatase families may aid in identification of currently unrecognized classes of response regulator phosphatases.
Introduction
An essential component of any signal transduction system is control of the longevity of the cellular response to a stimulus. The lifetime of the output signaling molecule must be long enough for an effective response but timely signal termination is necessary for the cell to adjust its behavior as conditions change. In two-component systems, detection of a stimulus controls the autophosphorylation of a histidyl residue on a sensor kinase protein. The phosphoryl group is subsequently transferred to an aspartyl residue on the receiver domain of a partner response regulator, which activates the response regulator to execute the cellular response [1,2]. Signal termination involves hydrolysis of the phosphoryl group and return of the response regulator to its unactivated state.
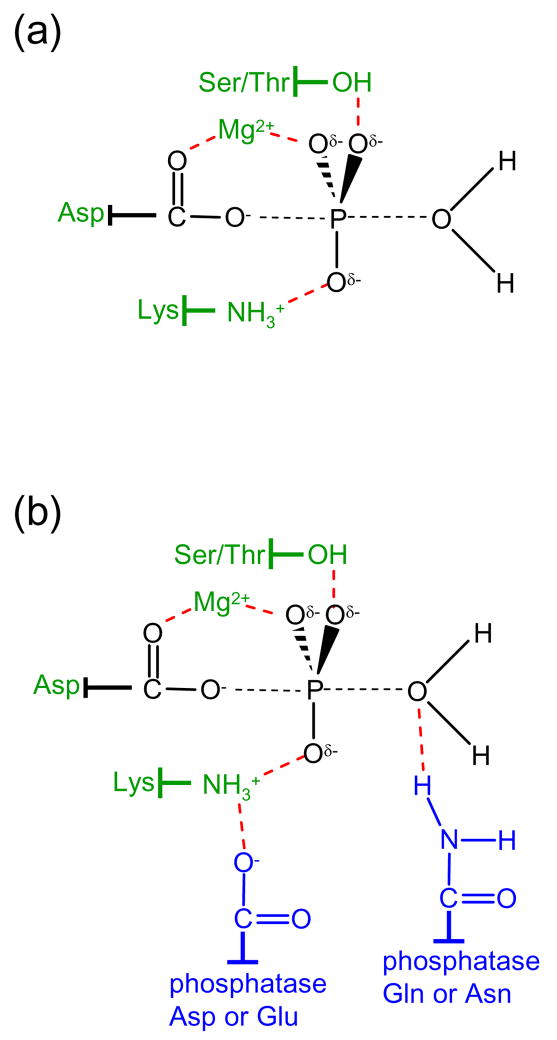
Response regulators contain highly conserved active site geometries centered around the the aspartate residue that undergoes phosphorylation. Hydrolysis of the aspartyl phosphate involves attack of a nucleophilic water molecule on the phosphorous atom with the aspartic acid carboxylate as a leaving group [3]. Phosphorous substitution reactions generally proceed through an in-line mechanism so that the nucleophile attacks from a position colinear with the phosphorus atom and leaving group. A model for the proposed bipyramidal transition state [4,5] for response regulator aspartyl phosphate hydrolysis is shown in Figure 1. For response regulator self-catalyzed dephosphorylation, an active site Mg2+ mediates catalysis [6] and is aided by conserved threonine/serine [7] and lysine [4] residues, perhaps by transition state stabilization (Figure 1a).
Figure 1.
Chemistry and catalysis of response regulator dephosphorylation. (a) The proposed bipyramidal transition state for hydrolysis of the aspartyl phosphate group is shown in black with partial bonds represented as dashed lines. Conserved groups proposed to catalyze response regulator autodephosphorylation are green and stabilizing interactions (deduced from structures of response regulators complexed with BeF3−) are represented by red dashed lines. (b) The same coloring scheme applies as in panel (a) but with the addition of conserved residues (blue) provided by phosphatases within the CheZ and CheC/CheX/FliY families. The conserved amide stimulates the reaction by direct interaction with the nucleophilic water molecule thus orienting it for the in-line attack. In CheX-mediated dephosphorylation, a threonine residue on CheY (Thr81 in B. burgdorferi CheY3) also forms a hydrogen bond with the attacking water molecule (not shown).
Although the response regulator active site provides some catalysis for its own dephosphorylation, in many cases, response regulator dephosphorylation largely occurs through catalysis by another protein. Some sensor kinases possess phosphatase activity towards their response regulator [1,8,9]. Alternatively, dephosphorylation of response regulators may be catalyzed by an auxiliary phosphatase, the topic of this review. To date, four different classes of auxiliary phosphatases have been identified– CheZ, CheC/CheX/FliY, Spo0E, and Rap. The classes have been characterized to different extents with detailed structural mechanisms now available for CheZ and CheC/CheX/FliY. Evidence so far suggests that despite different topologies, phosphatases from different classes may use similar strategies to catalyze response regulator dephosphorylation.
CheZ structure and catalytic mechanism
CheZ is probably the best characterized of the response regulator phosphatases, a consequence of its involvement in the well studied E. coli chemotaxis pathway [10,11]. CheZ catalyzes the dephosphorylation of CheY, the response regulator that binds to the flagellar switch to control swimming behavior. The CheZ family is found in the α, β/γ, δ, and ε classes of proteobacteria [12].
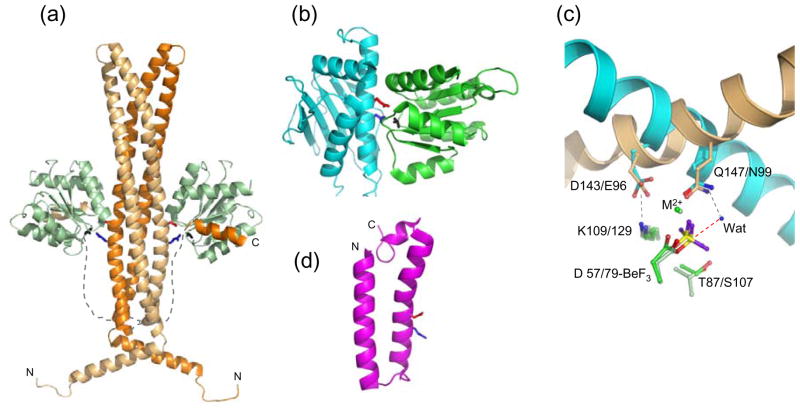
CheZ is a homodimer with a multidomain structure (Figure 2a, [13]). A long four-helix bundle comprised of a helical hairpin from each CheZ chain contains (by symmetry) two catalytic surfaces. Extending from the base of the four-helix bundle are two (one from each monomer) 32-residue linkers followed by a short C-terminal helix (‘C-helix’) [14–16]. The symmetry of the CheZ dimer confers two identical binding sites for phosphorylated CheY (CheYp) and each CheYp interacts with two separate regions of CheZ. The surface of CheYp containing the aspartyl phosphate interacts with the CheZ catalytic surface and the α4β5α5 surface of CheYp binds the CheZ C-helix [13,15,16]. In the absence of CheYp, the linker/C-helix are highly mobile [17]. Because CheYp displays appreciable (~2 μM) affinity for the isolated C-helix peptide [16,18] whereas CheYp interaction with the four-helix bundle is undetectable [13,14], CheYp probably binds first to the CheZ C-helix. This may serve to increase the local concentration of CheYp so that interaction with the CheZ catalytic surface rapidly ensues.
Figure 2.
Structures of response regulator phosphatases and their active sites. (a) The E. coli CheZ·CheY·BeF3−·Mg2+ complex (pdb 1KMI) with the two chains of the CheZ dimer as light orange and dark orange ribbons and the CheZ linkers sketched as dashed lines. CheY is in light green ribbons. The CheZ catalytic surface is defined by conserved acid (Asp143, red sticks) and amide (Gln147, blue sticks). BeF3− is in black sticks. (b) B. burgdorferi CheX·CheY·BeF3−·Mg2+ (pdb 3HZH) with CheX (cyan ribbon) and CheY3 (green ribbon). CheX conserved acid (Glu96, red) and amide (Asn99, blue) and BeF3− (black) are in stick representation. (c) Overlay of CheZ·CheY·BeF3−·Mg2+ and CheX·CheY3·BeF3−·Mg2+ active sites. The catalytic helices of CheZ (light orange) and CheX (cyan) are shown as ribbons and conserved CheX and CheZ residues are in sticks in the same color as the parent molecule. Select CheY residues are in sticks with E. coli CheY (light green) and B. burgdorferi CheY3 (green). BeF3− is yellow and purple. The ordered water molecule (‘Wat’) in the present CheX·CheY3·BeF3−· Mg2+ structure is a blue sphere and the red dashed line connects the water to the beryllium atom, the predicted path of nucleophilic attack. Interactions revealed in the crystal structures are gray dashed lines. (d) One of the 25 conformers in the NMR structure of B. anthracis BA5174 (pdb 2C0S), a member of the Spo0E family. The Gln (blue) and Asp (red) residues within the conserved (SQELD) motif are shown as sticks.
In the E. coli CheZ·CheY·BeF3−·Mg2+ complex, CheZ Asp143 and Gln147, within a conserved (-DXXXQ-) motif on the catalytic surface, insert into the active site of CheY. Asp143 forms a salt bridge with the conserved CheY active site lysine and Gln147 is oriented towards the BeF3− group. The Gln147 amide nitrogen is in the appropriate position to form a hydrogen bond with a water molecule that is positioned for in-line attack of the phosphorous atom (Figure 2a,c). Orientation of a hydrolytic water molecule by a glutamine residue is the mechanism by which GAP proteins stimulate activity in Ras-like GTPases [19]. Thus, CheZ works by providing an additional functional group to further stimulate the reaction mechanism already in place for CheY autodephosphorylation (Figure 1b). Using this mechanism, CheZ enhances the rate of CheY autodephosphorylation by a factor of about 100 [20].
Regulation of CheZ activity
Plots of CheZ activity versus CheYp concentration are sigmoidal, indicating positive cooperativity [20,21]. Thus CheZ activity is suppressed at low CheYp levels, which could serve as a mechanism to keep intracellular CheYp levels from getting too low. The positive cooperativity is consistent with differential binding affinities of CheYp for the two active sites on the CheZ dimer [20], but the structural basis for the cooperativity is not known. CheZ mutants that exhibit a gain-of-function phenotype cluster on the lower half of the four helix bundle [22] and several representatives of this class eliminate positive cooperativity without significantly changing kcat [20,21]. This suggests that cooperativity is mediated by interactions at the base of the bundle or the linkers that emanate from this region.
There is growing evidence that cellular localization plays a role in regulation of CheZ activity. In E. coli, a subpopulation of CheZ is associated with the transmembrane complex composed of chemoreceptors, the CheA sensor kinase, and the CheW scaffolding protein [23,24]. CheZ localization is mediated by interaction between the hairpin end of the four-helix bundle (Figure 2a) and an exposed hydrophobic helix of CheA-short (CheAs)- a truncated version of CheA [25–27]. Association of CheZ with CheAs increases CheZ phosphatase activity by two to three-fold [28]. Binding between relevant fragments of CheAs and CheZ results in NMR chemical shift perturbations that propagate down the CheZ four-helix bundle to the catalytic site, providing a possible structural explanation for CheZ activation by CheAs [25]. Clustering of CheZ at the cell poles results in a homogenous concentration of CheYp across the length of the cell [29,30] which may be important in synchronization of the rotational direction of flagellar motors.
CheC/CheX/FliY structure and catalytic mechanism
The CheC/CheX/FliY family spans diverse phyla within Archaea and Bacteria and contains members that are predicted to operate in two-component systems other than chemotaxis (http://mistdb.com/[31]). CheC/CheX/FliY sequences contain an internal two-fold pseudo-symmetry that is reflected in the overall α/β globular fold (Figure 2b,[32,33]). The folds for Thermotoga maritima CheC and CheX are distinguished from each other by two secondary structural elements that form α-helices in CheC and shorter β-strands in CheX [32]. The additional β-strands in CheX help mediate interactions with a partner chain, so CheC is monomeric and CheX forms a stable homodimer [32,34]. However, the Borrelia burgdorferi CheX dimer dissociates upon binding CheY3 (one of three CheYs encoded in the B. burgdorferi genome) complexed with BeF3− [35].
The conserved domain that defines the CheC/CheX/FliY family is related to a domain within FliM, the flagellar switch protein to which CheYp binds to control cell swimming behavior [32,36,37]. CheX and CheC contain only the conserved domain. In FliY, however, the conserved domain is flanked by two other domains, as in FliM. A C-terminal domain mediates association with the flagellar switch apparatus [38] and a short N-terminal peptide is connected to the central conserved domain via a linker. In FliM, the peptide interacts with the α4β5α5 surface on CheYp [16,39], analogous to the C-helix in CheZ. Removal of the linker and N-helix reduces FliY binding to CheYp [38] so it is probable that FliY uses this additional peptide to enhance substrate binding much like described above for the C-helix of CheZ.
A conserved (-EXXN-) motif is present twice in CheC and FliY, due to the sequence symmetry, but the N-terminal copy is absent, either by truncation or mutation, in CheX. The conserved Glu and Asn within the motif are separated by a single turn on an alpha helix [32]. The crystal structure of the complex between B. burgdorferi CheX and CheY3·BeF3−·Mg2+ [35] showed that the CheX Glu96 carboxylate forms a salt bridge with the CheY3 active site lysine and the CheX Asn99 amide forms a hydrogen bond with an ordered water molecule that is positioned for nucleophilic attack. A CheY3 active site threonine residue- Thr81- (distinct from the conserved threonine/serine) also interacts with the water molecule. Therefore, CheX appears to operate much like CheZ. An acid residue (Glu in CheX, Asp in CheZ) forms a critical salt bridge with the CheY3 conserved active site lysine. An amide residue (Asn in CheX, Gln in CheZ) acts to orient a hydrolytic water for in-line nucleophilic attack of the phosphoryl group (Figure 1b). This mechanism is supported by functional analysis of mutant CheC and CheX proteins from several species [32,40,41].
That CheX and CheZ work by nearly identical mechanisms is evident when overlaying the active sites from the co-crystal structures with CheY substrates (Figure 2c). The functional atoms from the CheX and CheZ side chains (the carboxylate oxygen from the acid residue and the amide nitrogen from the amide residue) are in nearly identical positions with respect to the CheY active site. However, the helices that bear the acid/amide pair on CheX and CheZ are oriented at about a 75° angle with respect to each other. This is an elegant example of “mechanistic” convergent evolution [42,43] as CheX and CheZ execute the same catalytic strategy with distinct structural solutions. Though the catalytic acid and amide are separated by a single helical turn in both proteins, they are separated by two residues in CheX and by three residues in CheZ. By orienting the helix that bears these residues differentially with respect to CheY and by varying the side chain length of the essential residues, the functional atoms end up in virtually identical locations. The mechanism described above should be applicable to the entire CheC/CheX/FliY family. Protein docking analysis predicts that CheYp could interact with both active sites in CheC and FliY without steric clash [35].
Activation of CheC by CheD
CheCs are phylogentically divided into three subclasses [31]. Class I CheCs are often co-translated with CheD, a deamidase that modifies receptors for chemotactic adaptation [11]. Class I CheCs derived from T. maritima or Bacillus subtilis exhibit lower phosphatase activity than CheX [32] or FliY [40] derived from the same species, but the activity of CheC is enhanced by binding CheD [32,33,40]. CheC interacts with CheD via one of the alpha helices that distinguishes CheC from CheX [44] but the mechanism of CheC activation by CheD is not known. CheD may act as an allosteric effector as its binding surface on CheC is removed from both CheYp binding surfaces [35]. Regulatory mechanisms have not yet been identified for CheX or FliY.
Sporulation
Initiation of endospore formation in B. subtilis and other gram positive bacteria is controlled by a phosphorelay that involves two response regulators, Spo0F and Spo0A [45]. Spo0F is the initial recipient of a phosphoryl group from any of a set of sensor kinases. The phosphoryl group is then transferred from Spo0F to a histidine residue of a histidyl phosphotransferase (Spo0B) and finally to Spo0A, a transcriptional activator of sporulation genes. Two families of response regulator phosphatases were identified by their activities as negative regulators of sporulation [46,47]. Spo0E phosphatases catalyze the dephosphorylation of Spo0A [48] whereas Rap phosphatases catalyze the dephosphorylation of Spo0F [46].
Spo0E phosphatases
Members of the Spo0E family are present primarily in the Bacillales and Clostridia classes within firmicutes [49]. Within the Bacillus family, genomes commonly encode three or four Spo0E family proteins, one of which carries the Spo0E namesake [49,50]. Spo0E proteins are small (50–150 residues) and many contain a conserved -SQELD- sequence motif [50]. NMR structures have been determined for two Spo0E family members from Bacillus anthracis: BA5174, which contains the –SQELD- sequence and BA1655, which instead contains a –SRDLD- motif [50]. Both BA5174 and BA1655 consist of a single alpha helical hairpin with flexible ends (Figure 2d, [50]). BA5174 is a monomer and BA1655 forms a homodimer with the two hairpins arranged in a head to tail orientation to form a four-helix bundle.
There are several indications that Spo0E may use a similar mechanistic strategy as CheZ and CheX. Gln36 and Asp39 within the –SQELD- motif of BA5174 are separated by a single helical turn in the middle of α2 with side chains projecting outwards towards solvent [50] (Figure 2d). This arrangement is reminiscent of the catalytic surfaces of CheX [35] and CheZ [13], but with the acid and amide side chains in the opposite order in the primary sequence. Both the conserved amide (glutamine) and acid (aspartate) residues are essential for optimal catalysis in B. subtilis Spo0E [51] but interestingly, substitution of the Gln results in only partial loss of activity whereas substitution of the Asp results in complete loss of activity, the converse to what is observed with CheZ [13] and CheX [35]. Therefore, the acid (not the amide) residue of Spo0E is proposed to interact with the hydrolytic water molecule, perhaps as a general base [51]. If the acid is the key catalytic residue in Spo0E, that could also explain the phosphatase activity of BA1655, which does not contain the conserved amide.
As described above, the CheX and CheZ phosphatases interact with their response regulator substrates in different orientations depending on how many residues separate the conserved acid/amide pair. In Spo0E, like CheX, the acid and amide are separated by two residues. Therefore, the catalytic helix of Spo0E is predicted to align itself in the same orientation as the CheX helix with respect to its response regulator. The Spo0E binding surface on the Spo0A response regulator, probed by alanine scanning mutagenesis, contains similar secondary structure elements as the CheY binding surfaces CheX [52]. If the Spo0E helix is oriented with the same N→C directionality as CheX and CheZ, the acid residue would be in a position to activate the water, as the biochemistry results suggest [51].
Rap phosphatases
The Rap (response regulator aspartyl phosphatase) family was originally identified by demonstration of phosphatase activity towards the response regulator Spo0F by two homologous B. subtilis sporulation suppressors, RapA and RapB [46]. The B. subtilis genome encodes nine additional homologous proteins (designated RapC-RapK) but recent studies have demonstrated functional variation within the family. Spo0F phosphatase activity has been demonstrated for RapE [53] and RapH [54], which cluster with RapA and RapB in a phylogenetic clade [54]. Other family members- RapC [55], RapF [56], RapG [57], RapH [54], and RapK [58] associate with the DNA-binding effector domain of the ComA or DegU response regulators and inhibit their abilities to regulate transcription. Interestingly, RapH possesses both activities. Because the amino acid sequence motifs that signify phosphatase or effector domain binding activity are not known, we cannot yet predict the function(s) of uncharacterized Rap proteins.
It is not known whether the Rap proteins that possess Spo0F phosphatase activity operate with a similar mechanism as CheC/CheX/FliY and CheZ. A conserved sequence motif that could potentially interact with Spo0F similarly to the acid/amide pair used in catalysis by CheZ and CheC/CheX/FliY has not been identified in Rap proteins. Although structures of Rap proteins have not been determined, Rap sequences contain a tandem series of six tetratrico peptide repeats (TPR) that are predicted to be highly helical [55,59,60]. The interaction surface on Spo0F for RapB determined by alanine scanning mutagenesis [61] contains residues within the β1α1, β3α3, β4α4 loops and the first few turns of α1, suggesting that RapB binds directly to the surface of Spo0F that contains the phosphoryl group. Like CheX and CheZ, RapA and B are homodimers in solution and each dimer binds two molecules of Spo0F [60].
The genes that encode many of the Rap proteins are transcriptionally coupled to an adjacent gene that encodes a Phr pre-peptide that, in its mature pentapeptide form, has specific inhibitory activity towards its partner Rap [62–64]. Interestingly, Phr peptides are able to inhibit Rap proteins that possess phosphatase [53,64] or effector domain binding activity [55,56]. Competitive binding experiments indicate that PhrA and Spo0F compete for the same binding surface on the RapA phosphatase [60]. Likewise, the PhrC [55]and PhrF [56] pentapeptides bind directly to RapC and RapF, respectively, which inhibits their interaction with the effector binding domain of ComA. However, the modes of interaction of the Phr pentapeptides with their cognate Rap proteins and the mechanisms of inhibition of both phosphatase and effector domain activities have not yet been elucidated.
Conclusion
Co-crystal structures supported by biochemical functional analysis has been tremendously informative in revealing catalytic mechanisms for the CheZ and CheC/CheX/FliY families of response regulator phosphatases. Strikingly, the two classes, despite distinct overall structures, use a virtually identical mechanism that is mediated by a conserved acid/amide pair. Analogous characterization of representatives of the Spo0E and Rap families promise comparable levels of detail and will inform us as to whether the acid/amide-mediated mechanism is broadly conserved. Identification of cellular mechanisms for the regulation of response regulator phosphatases and the structural basis for the regulation are areas of current investigation (Box 1). With several levels of regulation already established for the CheZ phosphatase, it is likely that the activities of the other response regulator phosphatase families are also carefully regulated within the cell.
Box 1. Current questions regarding response regulator phosphatases.
Is the catalytic mechanism used by CheZ and CheC/CheX/FliY families of phosphatases-mediated by an acid/amide pair- also used by the Spo0E and Rap families?
What are other classes of response regulator phosphatases?
What is the structural basis for the positive cooperativity displayed by CheZ?
Are the activities of FliY, CheX or Spo0E phosphatases regulated? If so, how?
How do Rap phosphatases bind Spo0F for dephosphorylation? How do Rap phosphatases bind ComA or DegU for inhibition of DNA-binding? Are there amino acid sequence motifs to predict which activities a Rap protein will possess?
How do Phr pentapeptides inhibit the activity of Rap proteins?
Acknowledgments
My research on response regulator phosphatases is funded by NIH grant GM050860 (to Robert B. Bourret).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of the review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1 ●●.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009:133–154. doi: 10.1146/annurev.micro.091208.073214. A clear and thorough review that would serve as a superb introduction to the field of two-component signal transduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolanin PM, Webre DJ, Stock JB. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry. 2003;42:14075–14082. doi: 10.1021/bi034883t. [DOI] [PubMed] [Google Scholar]
- 4.Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 5.Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 6.Lukat GS, Stock AM, Stock JB. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 7.Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenney L. Phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010:xxx–xxx. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourjik V. Receptor clustering and signal processing in E coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Kirby JR. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol. 2009;63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 12.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 ●.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nature Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. The co-crystal structure of CheZ·CheY·BeF3−·Mg2+ revealed the CheZ catalytic mechanism mediated by a conserved acid/amide pair. [DOI] [PubMed] [Google Scholar]
- 14.Blat Y, Eisenbach M. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- 15.Guhaniyogi J, Robinson VL, Stock AM. Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J Mol Biol. 2006;359:624–645. doi: 10.1016/j.jmb.2006.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J Mol Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 17.Silversmith RE. High mobility of carboxyl-terminal region of bacterial chemotaxis phosphatase CheZ is diminished upon binding divalent cation or CheY-P substrate. Biochemistry. 2005;44:7768–7776. doi: 10.1021/bi0501636. [DOI] [PubMed] [Google Scholar]
- 18.Guhaniyogi J, Wu T, Patel SS, Stock AM. Interaction of CheY with the C-terminal peptide of CheZ. J Bacteriol. 2008;190:1419–1428. doi: 10.1128/JB.01414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Zhang XC. GTP hydrolysis mechanism of Ras-like GTPases. J Mol Biol. 2004;340:921–932. doi: 10.1016/j.jmb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Silversmith RE, Levin MD, Schilling E, Bourret RB. Kinetic characterization of catalysis by the chemotaxis phosphatase CheZ. Modulation of activity by the phosphorylated CheY substrate. J Biol Chem. 2008;283:756–765. doi: 10.1074/jbc.M704400200. [DOI] [PubMed] [Google Scholar]
- 21.Blat Y, Gillespie B, Bren A, Dahlquist FW, Eisenbach M. Regulation of phosphatase activity in bacterial chemotaxis. J Mol Biol. 1998;284:1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]
- 22.Sanna MG, Simon MI. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. J Bacteriol. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 25 ●.Hao S, Hamel D, Zhou H, Dahlquist FW. Structural basis for the localization of the chemotaxis phosphatase CheZ by CheAS. J Bacteriol. 2009;191:5842–5844. doi: 10.1128/JB.00323-09. NMR study that determined the mode of interaction between CheZ and CheAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantwell BJ, Manson MD. Protein domains and residues involved in the CheZ/CheAS interaction. J Bacteriol. 2009;191:5838–5841. doi: 10.1128/JB.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor C, Matsumura P, Campos A. The CheZ binding interface of CheAS is located in alpha-helix E. J Bacteriol. 2009;191:5845–5848. doi: 10.1128/JB.00294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connor C, Matsumura P. The accessibility of Cys-120 in CheAs is important for the binding of CheZ and enhancement of CheZ phosphatase activity. Biochemistry. 2004;43:6909–6916. doi: 10.1021/bi035592n. [DOI] [PubMed] [Google Scholar]
- 29.Vaknin A, Berg HC. Single-cell FRET imaging of phosphatase activity in the Escherichia coli chemotaxis system. Proc Natl Acad Sci U S A. 2004;101:17072–17077. doi: 10.1073/pnas.0407812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipkow K, Andrews SS, Bray D. Simulated diffusion of phosphorylated CheY through the cytoplasm of Escherichia coli. J Bacteriol. 2005;187:45–53. doi: 10.1128/JB.187.1.45-53.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 ●.Muff TJ, Ordal GW. The diverse CheC-type phosphatases: chemotaxis and beyond. Mol Microbiol. 2008;70:1054–1061. doi: 10.1111/j.1365-2958.2008.06482.x. A review of the CheC/CheX/FliY family with additional bioinformatics analysis that characterizes the diversity and breadth of the family. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32 ●●.Park SY, Chao X, Gonzalez-Bonet G, Beel BD, Bilwes AM, Crane BR. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell. 2004;16:563–574. doi: 10.1016/j.molcel.2004.10.018. The high resolution crystal structures of CheC and CheX revealed the novel fold of the CheC/CheX/FliY protein family. Location of conserved acid/amide residues on exposed helices and establishment of functional roles for both residues hinted at a CheZ-like mechanism, later confirmed by the CheX/CheY co-crystal structure. [DOI] [PubMed] [Google Scholar]
- 33.Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 34.Motaleb MA, Miller MR, Li C, Bakker RG, Goldstein SF, Silversmith RE, Bourret RB, Charon NW. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J Bacteriol. 2005;187:7963–7969. doi: 10.1128/JB.187.23.7963-7969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35 ●●.Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0911185107. in press. Co-crystal structure of CheX·CheY·BeF3−·Mg2+ demonstrated a conserved mechanism for the CheC/CheX/FliY and CheZ families of phosphatases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby JR, Kristich CJ, Saulmon MM, Zimmer MA, Garrity LF, Zhulin IB, Ordal GW. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol. 2001;42:573–585. doi: 10.1046/j.1365-2958.2001.02581.x. [DOI] [PubMed] [Google Scholar]
- 37.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci USA. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J Biol Chem. 2003;278:48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- 39.Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nature Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 40.Muff TJ, Ordal GW. The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem. 2007;282:34120–34128. doi: 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- 41.Muff TJ, Foster RM, Liu PJ, Ordal GW. CheX in the three-phosphatase system of bacterial chemotaxis. J Bacteriol. 2007;189:7007–7013. doi: 10.1128/JB.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doolittle RF. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 43.Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg MJ. Convergent evolution of enzyme active sites is not a rare phenomenon. J Mol Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 45.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 46.Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 47.Perego M, Hoch JA. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J Bacteriol. 1991;173:2514–2520. doi: 10.1128/jb.173.8.2514-2520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlsen KL, Grimsley JK, Hoch JA. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci U S A. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubey GP, Narayan A, Mattoo AR, Singh GP, Kurupati RK, Zaman MS, Aggarwal A, Baweja RB, Basu-Modak S, Singh Y. Comparative genomic study of spo0E family genes and elucidation of the role of Spo0E in Bacillus anthracis. Arch Microbiol. 2009;191:241–253. doi: 10.1007/s00203-008-0446-7. [DOI] [PubMed] [Google Scholar]
- 50 ●●.Grenha R, Rzechorzek NJ, Brannigan JA, de Jong RN, Ab E, Diercks T, Truffault V, Ladds JC, Fogg MJ, Bongiorni C, et al. Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli. J Biol Chem. 2006;281:37993–38003. doi: 10.1074/jbc.M607617200. NMR structures of two Spo0E proteins revealed the helical hairpin fold and the possibility of dimerization to form a four-helix bundle. [DOI] [PubMed] [Google Scholar]
- 51.Diaz AR, Stephenson S, Green JM, Levdikov VM, Wilkinson AJ, Perego M. Functional role for a conserved aspartate in the Spo0E signature motif involved in the dephosphorylation of the Bacillus subtilis sporulation regulator Spo0A. J Biol Chem. 2008;283:2962–2972. doi: 10.1074/jbc.M709032200. [DOI] [PubMed] [Google Scholar]
- 52.Stephenson SJ, Perego M. Interaction surface of the Spo0A response regulator with the Spo0E phosphatase. Mol Microbiol. 2002;44:1455–1467. doi: 10.1046/j.1365-2958.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 53.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 55.Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol. 2003;49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 56 ●.Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol. 2005;187:4353–4361. doi: 10.1128/JB.187.13.4353-4361.2005. A model for the three dimensional structure of RapC based on its tetratrico peptide repeat sequence coupled with RapC biochemical analysis sets the stage for future structural and functional analyses of the Rap phosphatases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol. 2003;49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- 58.Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Ishikawa S, Core L, Perego M. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J Biol Chem. 2002;277:20483–20489. doi: 10.1074/jbc.M201086200. [DOI] [PubMed] [Google Scholar]
- 61.Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry. 1998;37:16538–16545. doi: 10.1021/bi981340o. [DOI] [PubMed] [Google Scholar]
- 62.Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 63 ●.Solomon JM, Lazazzera BA, Grossman AD. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. An elegant study using a classical biochemical approach to identify what is now known as the PhrC pentapeptide as a molecule that plays a key role in control of competence and sporulation in Bacillus subtilis. [DOI] [PubMed] [Google Scholar]
- 64.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]