Abstract
Synthetic foldamers consisting of β-amino acids offer excellent model systems for examining the effect of backbone flexibility on the dynamics of protein folding. Herein, we study the folding-unfolding kinetics of a β-peptide that folds into a 14-helical structure in water. We find that the T-jump induced relaxation kinetics of this peptide occur on the nanosecond timescales and are noticeably slower than those of alanine based α-helical peptides, and additionally, the relaxation rates show a weaker dependence on temperature. These differences appear to indicate that the folding energy landscapes of these peptides are different. In addition, we find that the amide I′ band of this β-peptide exhibits a sharp feature at about 1612 cm−1, which we believe is a distinct infrared reporter of 14-helix.
Synthetic foldamers1 designed to mimic proteins are not only attractive candidates for novel biological applications, but they also provide an additional framework for studying the protein folding problem.2 For example, foldamers constructed from β-amino acids (i.e., β-peptides) provide an excellent platform for examining the effect of backbone flexibility on the dynamics and the mechanism of protein folding. This is because β-amino acids contain an additional backbone methylene unit compared to their natural α-amino acid counterparts, thus creating an additional backbone torsion angle. Herein, we measure the relaxation kinetics of a 15-residue β-peptide3 in response to a laser-induced temperature-jump (T-jump), aiming to understand how this additional degree of freedom and increased backbone flexibility affect the folding dynamics of monomeric helices and to benchmark the folding rate of β-peptides.
The β-peptide studied here has the following sequence, Y(AKKAEE)2Ad-Asp (all residues are β-amino acids, except the d-Asp), and has been shown to form a monomeric 14-helix in aqueous solution that is stabilized by a series of electrostatic interactions between the β3-hGlu and β3-hLys side chains.3 While a previous study indicated that about 12 residues of this β-peptide are helical at 2 °C,3 its unfolding transition, as measured by circular dichroism (CD) spectroscopy, occurs over a broad range of temperature (Figure S1, Supporting Information). Although the thermal unfolding of a helical peptide is not a simple two-state process, the apparent van’t Hoff enthalpy for the process provides a mathematically convenient measure of the breadth of the transition. An analysis of the CD melting curve of this β-peptide indicates that its unfolding transition is approximately twice as broad as that of a 19-residue α-helical peptide.4 This broadening presumably reflects the smaller cooperative unit for formation of the 14-helix versus the α-helix. For example, nearly complete helix formation in water can be obtained with an appropriately designed 12-residue 14-helix, which is approximately one half the value required to form a fully stable α-helix. The reduced number of main chain hydrogen-bonds required for formation of a stable 14-helix should lead to a smaller enthalpic change of unfolding.
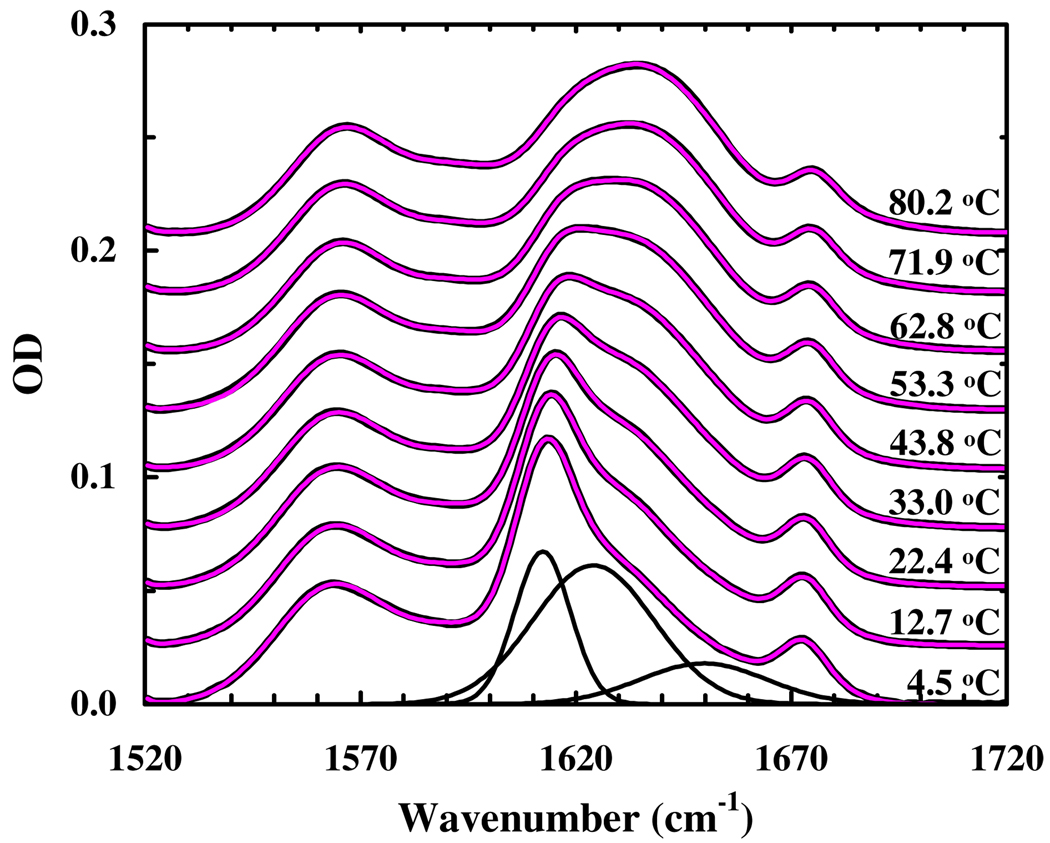
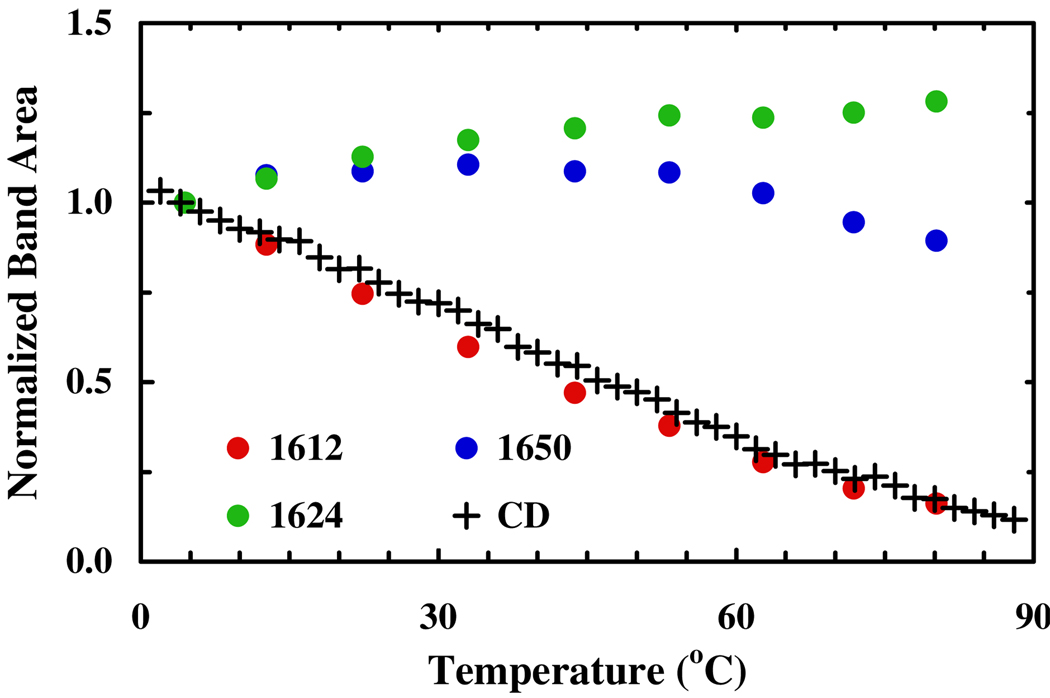
The thermal unfolding transition of this β-peptide is further evaluated by infrared (IR) spectroscopy. As shown (Figure 1), at low temperatures its amide I′ band, which is a well-established IR probe of protein secondary structures, is characterized by a narrow band centered at about 1612 cm−1, whereas at high temperatures the amide I′ band is dominated by a broad feature centered at about 1634 cm−1. Further analysis of these infrared data using a global fitting method5 (see Supporting Information for detail) indicates that the spectra (1520 – 1720 cm−1) can be well fit by six well-separated Gaussian functions centered at 1564 ± 2, 1592 ± 3, 1612 ± 2, 1624 ± 3, 1650 ± 3, 1673 ± 2 cm−1, respectively. Among these bands, the 1564 and 1592 cm−1 components are associated with deprotonated β3-hGlu and d-Asp side chains, respectively, and the 1673 cm−1 band arises from the residual trifluoroacetic acid from peptide synthesis, whereas the remaining three amide I′ bands are related to significantly populated backbone conformations. Since solvated α-helices6,7 show an amide I′ band at around 1632 cm−1, these results indicate that the local electrostatic environment of the backbone carbonyls in the 14-helix is different from that in the α-helix and that IR spectroscopy could be a useful means for distinguishing between helices formed by α- and β-peptides. In particular, the 1612 cm−1 band appears to be a distinctive IR marker of the 14-helical structure as its intensity (integrated area) shows the same thermal unfolding transition as that obtained from CD spectroscopy (Figure 2). On the other hand, the other two bands, which we tentatively assign to those amide carbonyls that are not hydrogen-bonded to an amide N-H group, are less useful in this regard. Additionally, they are expected to significantly overlap with spectral features largely arising from amide groups in randomly coiled and unfolded peptides. For example, N-methylacetamide, which has been used as a classical model for an exposed amide in unfolded proteins, has an amide I′ band near 1625 cm−1 near 20°C.8 Because of such overlaps, these bands are expected to show a weaker temperature dependence, as observed.
Figure 1.
FTIR spectra of the β-peptide (in the amide I′ region) measured at different temperatures, as indicated. These data have been offset for clarity. The thin red lines represent the best global fits of these data to the model discussed in the text. The black lines represent the spectral components obtained for the 4.5 °C spectrum.
Figure 2.
Integrated area of the three amide I′ components versus temperature, as indicated. Also shown (+) is the normalized CD signal of the β-peptide at 212 nm as a function of temperature (Figure S1, Supporting Information).
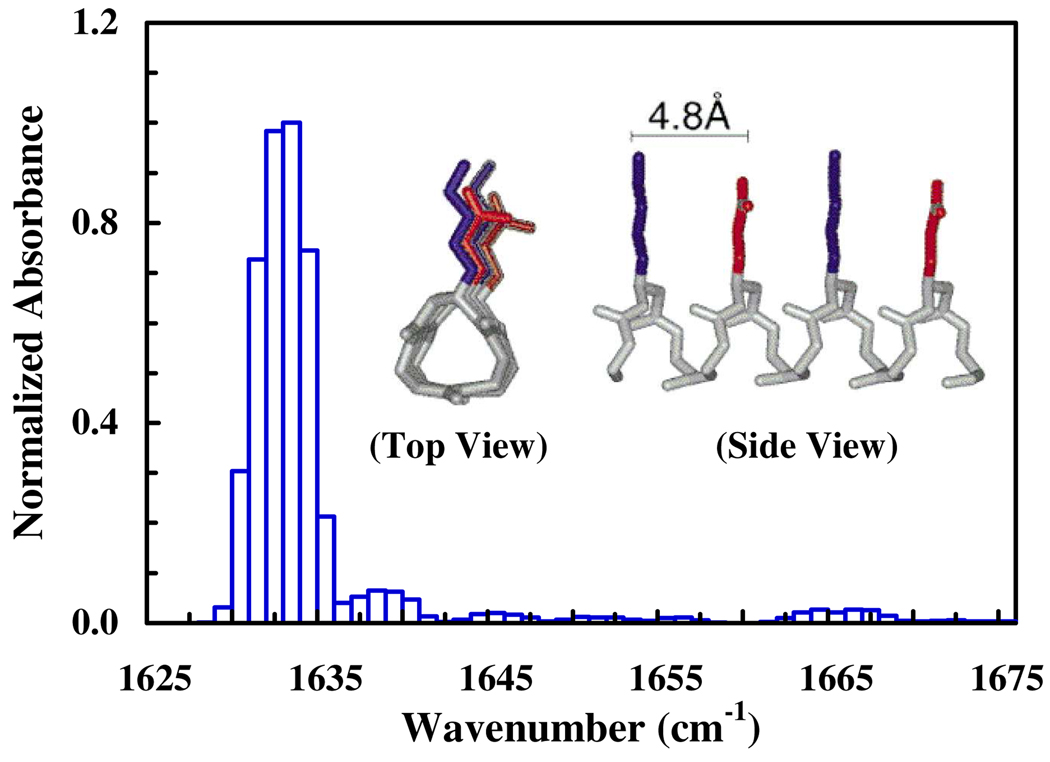
To further verify that the 1612 cm−1 band is a good reporter of the 14-helical conformation, we compute its amide I band based on the local amide I Hamiltonian (LAH).9 The details of the calculations are presented in the Supporting Information. Briefly, an ideal 14-helix, which has the same sequence as the current β-peptide, was generated using a custom computational foldamer design platform (Supporting Information). This helix forms the starting structure for a 1 ns molecular dynamics (MD) simulation. For each of the 10,000 structures obtained from the 1 ns MD trajectory at 300 K, the LAH of the peptide is then constructed, wherein all the diagonal elements are assumed to have a vibrational frequency of 1650 cm−1, whereas the off-diagonal elements are calculated using the transition dipole coupling (TDC) model.9 Upon diagonalization of the LAH, the eigenstates of the exciton system are obtained. As shown (Figure 3), the resultant stick spectrum exhibits a rather narrow peak at around 1633 cm−1, which resembles the sharp feature at around 1612 cm−1 of the experimental spectrum. The difference in the peak position is most likely due to the crude estimate of the diagonal elements of the LAH. In addition, the calculated spectrum clearly underestimates the intensity of other components. This must be due to the simplicity of the current computational treatment, which assumes that all the uncoupled amide I oscillators are degenerate and includes only TDC and the 14-helical conformation. However, in order to treat all the oscillators in a more realistic manner and also to include other peptide conformations in the calculation, more sophisticated computational modelings10 are required, which are beyond the scope of the current study. Nevertheless, the finding that couplings among the amide I oscillators of an ideal 14-helix via the mechanism of TDC produce a sharp amide I peak supports the aforementioned perception that the 1612 cm−1 band is an IR signature of 14-helical β-peptides. The latter notion is further corroborated by the fact that the amide I band of a 12/10/12/10-helix in CD3OD is centered at about 1640 cm−1.11
Figure 3.
Calculated amide I stick spectrum of an ideal 14-helix, whose structure is shown in the inset (adopted form reference 3).
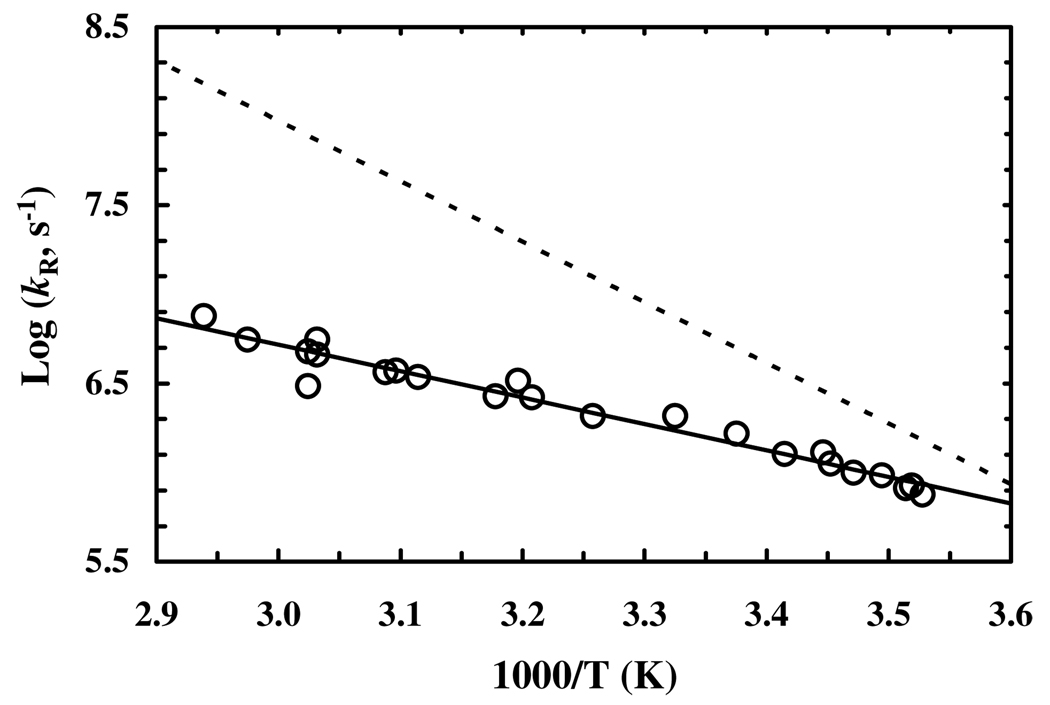
The relaxation kinetics of this β-peptide were measured by a time-resolved IR technique wherein the relaxation process was initiated by a laser-induced T-jump.6 The details of the IR T-jump setup have been described elsewhere.12 The only difference is that in the current study the IR probe is derived form a tunable quantum cascade laser (Daylight Solutions, CA). As shown (Figure S2, Supporting Information), the T-jump induced conformational relaxation kinetics, monitored by the absorbance change of the β-peptide at a single wavelength, 1614 cm−1, can be described by a single-exponential function. Similar to that observed for α-helical peptides,4,13 the relaxation rate of this β-peptide shows an Arrhenius temperature dependence in the temperature range of the experiment (Figure 4). However, in comparison with those obtained for α-helical peptides of similar size4,13 and also a nonbiological helical peptide14 two important conclusions can be reached. First, the T-jump induced relaxation process of this β-peptide occurs on a slower timescale. For example, the relaxation time of this β-peptide is 550 ± 75 ns at 300 K, whereas T-jump IR and UV resonance Raman studies have shown that alanine based α-helical peptides typically relax in the range of 50 – 250 ns at this temperature.13 Thus, our results suggest that at room temperature this 14-helical peptide folds slower than alanine based α-helical peptides, although pure α-helix has been suggested to fold on the microsecond timescale.15 Second, the apparent enthalpy of activation, 6.8 ± 1.0 kcal/mol, of the relaxation rate of this β-peptide is significantly smaller than that (~15 kcal/mol) obtained for an α-helical peptide.4 It has been shown that the rate of the helix-coil transition depends on the solvent viscosity;16 and for D2O, this viscosity dependence is expected to contribute up to 4.5 kcal/mol to the total activation energy. Taken together, these results suggest that the folding kinetics of this 14-helical peptide encounters a small, if any, energetic barrier besides the dynamic drag exerted by the solvent molecules on the diffusive motions of the peptide. Thus, it appears that the slower folding time of this β-peptide is due to the additional degree of freedom in β-amino acid backbones, which can effectively increase the entropic penalty for folding. This notion is consistent with the fact that the intercept (~11) of the log(kR) versus 1/T plot of the β-peptide, which contains contributions from the prefactor and entropy of activation, is smaller than that (~18) of the 19-residue alanine-rich peptide.4
Figure 4.
Logarithm of the relaxation rate constant of the β-peptide (open circle) versus 1/T. Linear regression of these data (solid line) yields an activation enthalpy of 6.8 ± 1.0 kcal/mol and an intercept of 11.1 ± 0.3. Also shown (dashed line) is the temperature dependence of the relaxation rate of a 19-residue alanine-rich peptide with a reported apparent activation energy of 15.5 kcal/mol (reproduced from reference 4).
In both α-helical and 14-helical peptides, the unfolding transition can be considered to correspond to the conversion of an ensemble of “folded” states consisting of partially to fully helical conformers into a second ensemble of “unfolded” states consisting of randomly coiled and locally folded states. Interestingly, both CD and IR measurements report a coincident transition for loss of the 14-helix, despite the fact that the amide I transition in the IR would likely have a different dependence on helical length than in CD. In particular, on a per-residue basis, amides that are part of a long helix tend to have significantly stronger chiropotic strength than those found in shorter helices. Thus, the helical content of an ensemble, as probed by CD, tends to be dominated by amides in long helices with smaller contributions from short helices (after normalization for helical length). Thus, the coincidence of similar melting curves obtained using two methods with different sensitivities to helical lengths would suggest that the transition occurs between two states. However, the broadness of the thermal melting transition prevents us from further analyzing the relaxation kinetics according to such a “two-state” scenario. Furthermore, it is also likely that increasing temperature results in a gradual decrease in the mean helical length within the folded conformational ensemble, which would also contribute to the large breadth and low apparent change in enthalpy associated with the transition. Regardless of the mechanism of the process, which could be further investigated using computer simulations,17 the small temperature dependence and slowness of the process relative to the α-helix presumably reflects a more unfavorable entropic loss for 14-helix formation.
One of the hallmarks of natural proteins is the cooperativity of protein folding, which occurs as a result of the generally unfavorable free energy of folding of isolated secondary structural units. Although kinetic intermediates can be observed, thermodynamic stability of individual domains is gained only upon formation of the native tertiary structure. At equilibrium, this cooperativity minimizes the population of partially folded states with isolated elements of secondary structure that otherwise could intermolecularly associate to form domain-swapped oligomers or other intermolecularly misfolded fibril-like structures.18 Thus, it is of interest to understand the folding kinetics and mechanism of stabilization of β-peptides, which form more stable isolated secondary structures.19 The increased stability of β-peptides formed from β3-monosubstituted amino acids is surprising, given the presence of a methylene group inserted into the backbone imparting glycine-like flexibility in the unfolded state. However, the increased flexibility allows fine-tuning of the peptide backbone geometry to provide strain-free stabilization of locally folded and helical secondary structures, and dehydration of the methylene can provide an increased hydrophobic driving force for folding. As a result, for β-peptides the cooperative unit required for helix formation is expected to be shorter than that for α-peptides, which consequently leads to a lower enthalpic requirement for helix formation. On the other hand, the increased backbone flexibility would result in a larger entropic penalty associated with the conformational search and hence a slower conformational relaxation rate, as observed.
In summary, this study presents, to the best of our knowledge, the first experimental results on the folding dynamics of β-peptides. Our results suggest that the folding energy landscape of 14-helix may be distinctly different from that of α-helix, which is in agreement with a recent computer simulation.20 In addition, we find that the amide I′ band of β-peptides could be very useful in distinguishing between various helical conformations. In light of the current strong interest in β-peptide foldamers, we believe that these results should be of general interest, as they provide new insight into the folding kinetics and spectroscopic properties of β-peptides at the secondary structure level.
Supplementary Material
Acknowledgement
We thank the NIH (GM-065978, RR-01348) and the NSF (DMR05-20020) for funding.
Footnotes
Supporting Information Available: Materials, methods, T-jump kinetics, and CD data are available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Feng Gai, Email: gai@sas.upenn.edu.
William F. DeGrado, Email: wdegrado@mail.med.upenn.edu.
References
- 1.(a) Gellman SH. Acc. Chem. Res. 1998;31:173. [Google Scholar]; (b) Seebach D, Beck AK, Bierbaum DJ. Chem. & Biodivers. 2004;1:1111. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]; (c) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bautista AD, Craig CJ, Harker EA, Schepartz A. Curr. Opin. Chem. Biol. 2007;11:685. doi: 10.1016/j.cbpa.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Daura X, Jaun B, Seebach D, van Gunsteren WF, Mark AE. J. Mol. Biol. 1998;280:925. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]; (b) Raguse TL, Lai JR, LePlae PR, Gellman SH. Org. Lett. 2001;3:3963. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]; (c) Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2002;124:11564. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]; (d) Daniels DS, Petersson EJ, Qui JX, Schepartz A. J. Am. Chem. Soc. 2007;129:1532. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2001;123:5162. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 4.Huang CY, Klemke JW, Getahun Z, DeGrado WF, Gai F. J. Am. Chem. Soc. 2001;123:9235. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]
- 5.Zhu YJ, Fu XR, Wang T, Tamura A, Takada S, Saven JG, Gai F. Chem. Phys. 2004;307:99. [Google Scholar]
- 6.Gilmanshin R, Williams S, Callender RH, Woodruff WH, Dyer RB. Biochemistry. 1997;36:15006. doi: 10.1021/bi970634r. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee S, Chowdhury P, Gai F. J. Phys. Chem. B. 2007;111:4596. doi: 10.1021/jp0689060. [DOI] [PubMed] [Google Scholar]
- 8.Manas ES, Getahun Z, Wright WW, DeGrado WF, Vanderkooi JM. J. Am. Chem. Soc. 2000;122:9883. [Google Scholar]
- 9.(a) Krimm S, Bandekar J. Adv. Protein Chem. 1986;38:181. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]; (b) Torii H, Tasumi MJ. J. Chem. Phys. 1992;96:3379. [Google Scholar]; (c) Hamm P, Lim M, Hochstrasser RM. J. Phys. Chem. B. 1998;102:6123. [Google Scholar]; (d) Woutersen S, Hamm P. J. Phys.: Condens. Matter. 2002;14:R1035. [Google Scholar]; (e) Chung HS, Tokmakoff A. J. Phys. Chem. B. 2006;110:2888. doi: 10.1021/jp053956a. [DOI] [PubMed] [Google Scholar]
- 10.(a) Gnanakaran S, Hochstrasser RM, Garcia AE. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9229. doi: 10.1073/pnas.0402933101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Scheurer C, Piryatinski A, Mukamel S. J. Am. Chem. Soc. 2001;123:3114. doi: 10.1021/ja003412g. [DOI] [PubMed] [Google Scholar]
- 11.Hamm P, Woutersen S, Rueping M. Helv. Chim. Acta. 2002;85:3883. [Google Scholar]
- 12.Huang CY, Getahun Z, Zhu YJ, Klemke JW, DeGrado WF, Gai F. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2788. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Lednev IK, Karnoup AS, Sparrow MC, Asher SA. J. Am. Chem. Soc. 1999;121:8074. doi: 10.1021/ja003381p. [DOI] [PubMed] [Google Scholar]; (b) Wang T, Du D, Gai F. Chem. Phys. Lett. 2003;370:842. [Google Scholar]; (c) Wang T, Zhu YJ, Getahun Z, Du DG, Huang CY, DeGrado WF, Gai F. J. Phys. Chem. B. 2004;108:15301. doi: 10.1021/jp037272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang WY, Prince RB, Sabelko J, Moore JS, Gruebele M. J. Am. Chem. Soc. 2000;122:3248. [Google Scholar]
- 15.Mikhonin AV, Asher SA, Bykov SV, Murza A. J. Phys. Chem. B. 2007;111:3280. doi: 10.1021/jp0654009. [DOI] [PubMed] [Google Scholar]
- 16.Jas GS, William EA, Hofrichter J. J. Phys. Chem. B. 2001;105:261. [Google Scholar]
- 17.Daura X, Gademann K, Schafer H, Jaun B, Seebach D, van Gunsteren WF. J. Am. Chem. Soc. 2001;123:2393. doi: 10.1021/ja003689g. [DOI] [PubMed] [Google Scholar]
- 18.(a) Ogihara NL, Ghirlanda G, Bryson JW, Gingery M, DeGrado WF, Eisenberg D. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1404. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ghirlanda G, Lear JD, Ogihara NL, Eisenberg D, DeGrado WF. J. Mol. Biol. 2002;319:243. doi: 10.1016/S0022-2836(02)00233-4. [DOI] [PubMed] [Google Scholar]
- 19.(a) Seebach D, Overhand M, Kuhnle FNM, Martinoni B. Helv. Chim. Acta. 1996;79:913. [Google Scholar]; (b) Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; (c) Seebach D, Hook DF, Glattli A. Biopolymers. 2006;84:23. doi: 10.1002/bip.20391. [DOI] [PubMed] [Google Scholar]
- 20.Keller B, Gattin Z, van Gunsteren WF. Proteins: Struct. Funct. Bioinform. 2010 doi: 10.1002/prot.22685. DOI 10.1002/prot.22685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.